Abstract
Malignant liver tumors, including primary malignant liver tumors and liver metastases, are among the most frequent malignancies worldwide. The disease carries a poor prognosis and poor overall survival, particularly in cases involving liver metastases. Consequently, the early detection and precise differentiation of malignant liver tumors are of paramount importance for making informed decisions regarding patient treatment. Significant research efforts are currently directed towards the development of diagnostic tools for different types of cancer using minimally invasive techniques. A prominent area of focus within this research is the evaluation of circulating microRNA, for which dysregulated expression is well documented in different cancers. Combining microRNAs in panels using serum or plasma samples derived from blood holds great promise for better sensitivity and specificity for detection of certain types of cancer.
Keywords: malignant liver tumors, primary malignant liver tumors, liver metastases, serum, plasma, circulating microRNA, tissue of origin determination
1. Introduction
Malignant liver tumors are among the most common malignancies worldwide. They include both primary liver malignancies and liver metastases and are often characterized by poor prognosis and low overall survival rate. Hepatocellular carcinoma (HCC) is the predominant primary liver malignancy, followed by intrahepatic cholangiocarcinoma (CCA) [1,2,3,4]. The incidence of liver metastases exceeds that of primary liver tumors [5]. Liver metastases can arise from a variety of malignancies, including carcinomas, melanomas, lymphomas, sarcomas, and germ cell tumors [5,6]. Among these, carcinomas are the most common (92%), with adenocarcinoma being the most common subtype (75%) [6]. The main sources of liver metastases are colorectal carcinomas, followed by pancreatic, breast, lung, and gastric carcinomas [4,6].
Differentiation between HCC and CCA as well as liver metastasis is usually straightforward. However, occasionally pathologists are challenged by difficult cases, such as small biopsies, different histological subtypes or inconclusive immunophenotype [5]. The interplay of factors such as the etiology of liver disease, the rate of cancer progression, and molecular diversity both between metastatic specimens and within the same tumor mass complicates the differentiation of malignant liver tumors. In some cases, the primary tumor site or tissue of origin (TOO) of a metastatic tumor remains unclear and is therefore classified as a carcinoma of unknown primary (CUP) [6,7]. It should be noted that most CUPs are detected in the liver [7]. The data show that up to 24% to 50% of CUP patients have liver metastases and that these patients are associated with an increased mortality rate [7,8,9]. In addition, patients with liver metastases often have a worse overall survival prognosis compared to patients with metastases to other anatomic sites [3].
Because prognosis and treatment are often based on the type of primary cancer, the differentiation between primary liver malignancies and liver metastases and the determination of TOO in CUP in the liver are of vital importance [1,3,6,10].
2. MicroRNA in Cancer
MicroRNAs (miRNAs) represent a class of small non-coding RNAs, typically between 20 and 22 nucleotides in length. These intriguing molecules are derived from various genomic sources, including intergenic regions and exon sequences within non-coding transcription units. It is noteworthy that a substantial proportion, up to 60%, of known miRNAs arise from intronic sequences nestled within either protein-coding genes or non-coding transcription units. MiRNAs can be found encoded as individual genes or clustered together in genomic regions. In some instances, miRNA clusters are co-regulated and co-transcribed, hinting at intricate regulatory mechanisms [11]. In the human genome, miRNAs are estimated to constitute more than 3% of the entire set of genes. Functionally, miRNAs exhibit remarkable diversity, impacting various facets of gene regulation. However, their principal mode of action in mammals primarily involves the inhibition of mRNA translation through base-pairing interactions with the 3’-UTR (untranslated region) of target mRNAs. In the complex milieu of animal cells, individual miRNAs can exert their influence over numerous mRNA targets, sometimes numbering as many as 200 predicted targets per miRNA. Additionally, a single mRNA can be subject to regulation by multiple miRNAs [12].
In the context of cancer research, miRNAs have emerged as pivotal gene-specific regulators, bearing similarities in their activities to a multitude of protein transcription factors known to be crucial players in the transformation of normal cells into malignant ones. MiRNAs wield their influence over various stages of gene expression, including transcription, mRNA stability, and mRNA translation. Notably, cancer cells exhibit genetic and epigenetic alterations compared to their non-malignant counterparts, and miRNAs are increasingly recognized as central players in mediating these distinctions. Genome-wide profiling endeavors have unveiled distinct miRNA signatures unique to specific cancer types, underscoring the diagnostic potential of these molecules. Combining miRNA markers with other biomarkers holds promise for enhancing cancer risk assessment, detection, and prognosis. Moreover, specific genetic polymorphisms have been linked to the susceptibility of developing various types of cancer. Hence, there is a growing imperative to integrate genomic mutations with miRNA markers to formulate comprehensive marker panels that offer more accurate risk assessment and early diagnosis in the realm of cancer research and clinical practice [13,14,15].
3. Circulating Tumor miRNA
A significant breakthrough has been the identification of miRNAs as potential biomarkers in serum or plasma, offering a minimally invasive approach to cancer screening. Therefore, understanding the characteristics of secretory miRNAs and their utility in cancer detection is of paramount importance [16,17].
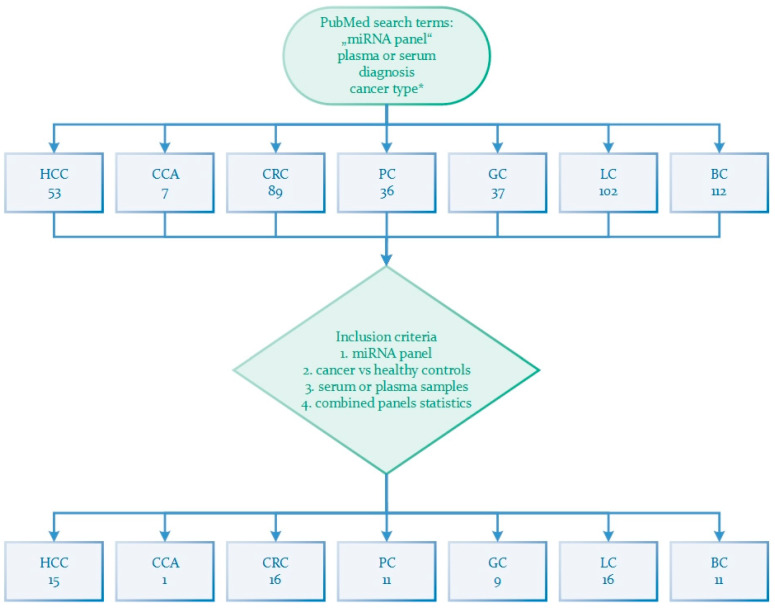
In recent years, miRNAs have gained significant attention among researchers exploring their potential as biomarkers for cancer diagnosis and prognosis. To systematically prepare this review paper, we conducted a comprehensive search of the literature on PubMed. We combined the terms “miRNA panel” with “plasma” or “serum” and “diagnosis” for each type of cancer selected in this review; these are the primary liver cancers HCC and CCA and liver-metastasizing cancers colorectal cancer (CRC), pancreatic cancer (PC), gastric cancer (GC), lung cancer (LC), and breast cancer (BC) (Figure 1). In total, seven separate searches were performed.
Figure 1.
Schematic presentation of workflow. The literature search was performed in PubMed with terms “miRNA panel”, plasma or serum, diagnosis and term for each cancer included in this review. Seven separate searches resulted in different numbers of studies from which we selected those that met the inclusion criteria. HCC: hepatocellular carcinoma; CCA: cholangiocarcinoma; CRC: colorectal cancer; PC: pancreatic cancer; GC: gastric cancer; LC: lung cancer; BC: breast cancer; *: HCC, CCA, CRC, PC, GC, LC, or BC.
Our search yielded the following results: HCC 53, CCA 7, CRC 89, PC 36, GC 37, LC 102, and BC 112. We meticulously reviewed these studies and selected those that employed a panel of miRNAs for the detection of each cancer type, distinguishing them from healthy controls. We further selected original articles that tested their selected miRNA panels on human serum or plasma samples and evaluated a combined panels of selected miRNA markers. Furthermore, by combining different miRNAs from each cancer panel, we explore the potential to create a more specific blood-based panel capable of detecting multiple cancers simultaneously, offering a promising avenue for comprehensive cancer screening.
4. Circular miRNA in Primary Liver Cancers
4.1. Hepatocellular Carcinoma
We categorized miRNA panels into three groups. The first group consisted of eight panels exclusively featuring miRNAs. In the second group, there were three panels where miRNAs were combined with lncRNA and mRNA. The third group involved four panels where miRNAs were combined with α-fetoprotein (AFP).
It is worth noting that several miRNAs were consistently utilized across multiple miRNA panels. These shared miRNAs are miR-126, miR-21, miR-122, miR-125b, miR-375, miR-206, miR-192, miR-223, miR-26a, and miR-27a. Additionally, AFP was a recurring component in different panels (Table 1).
Table 1.
An overview of circulating miRNA panels for detection of HCC.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA Panels | |||||
| miR-122 | Plasma | 457 HCC, 167 HC | ↑ | AUC = 0.941 | [18] |
| miR-192 | ↑ | ||||
| miR-21 | ↑ | ||||
| miR-223 | ↓ | ||||
| miR-26a | ↓ | ||||
| miR-27a | ↓ | ||||
| miR-801 | ↓ | ||||
| miR-206 | Serum | 261 HCC, 173 HC | ↑ | AUC = 0.887 (95% CI = 0.850–0.918) Sensitivity = 85.55% Specificity = 73.3% |
[19] |
| miR-141-3p | ↑ | ||||
| miR-433-3p | ↑ | ||||
| miR-1228-5p | ↑ | ||||
| miR-199a-5p | ↓ | ||||
| miR-122-5p | ↓ | ||||
| miR-192-5p | ↓ | ||||
| miR-26a-5p | ↓ | ||||
| miR-214-5p | Serum | 224 HCC, 84 HC | ↓ | AUC = 0.95 with 95% CI Sensitivity = 83.2% Specificity = 96.9% Accuracy = 86.8% |
[20] |
| miR-125b | ↓ | ||||
| miR-1269 | ↑ | ||||
| miR-375 | ↓ | ||||
| miR-27b-3p | Serum | 212 HCC, 110 HC | ↑ | AUC = 0.823 (p < 0.0001) |
[21] |
| miR-192-5p | ↑ | ||||
| miR-375 | Serum | 149 HCC, 149 HC | ↑ | AUC = 0.995 (95% CI: 0.985–1) |
[22] |
| miR-10a | ↑ | ||||
| miR-122 | ↑ | ||||
| miR-423 | ↑ | ||||
| miR-4661-5p | Serum exosomes | 84 HCC, 26 HC | ↑ | AUC = 0.942 (95% CI = 0.895–0.972) Sensitivity = 84.5% Specificity = 89.3% PPV = 88.8% NPV = 85.2% |
[23] |
| miR-4746-5p | ↑ | ||||
| miR-126 | Serum | 34 HCC, 25 HC | ↑ | AUC = 1.00 SE = 0 p-value < 0.001 |
[24] |
| miR-21 | ↑ | ||||
| miR-30c | ↑ | ||||
| miR-193b | ↑ | ||||
| miR-122 | ↑ | ||||
| miR-222 | ↑ | ||||
| miR-125b | ↑ | ||||
| miR-10b | Serum | 27 HCC, 50 HC | ↑ | AUC = 0.94 (95% CI: 0.89–0.99) |
[25] |
| miR-181a | ↓ | ||||
| miR-106b | ↑ | ||||
| miRNA + lncRNA + mRNA panels | |||||
| miR-16-2 | Serum | 78 HCC, 42 HC | ↑ | Sensitivity = 79.5% Specificity = 100% |
[26] |
| miR-21-5p | ↑ | ||||
| lncRNA-CTBP | ↑ | ||||
| mRNA LAMP2 | ↑ | ||||
| miR-1262 | Serum exosomes | 60 HCC, 18 HC | ↓ | Sensitivity = 100% Specificity = 76.7% PPV = 81.1% NPV = 100% Accuracy = 88.3% |
[27] |
| lncRNA-RP11-513I15.6 | ↓ | ||||
| mRNA RAB11A | ↓ | ||||
| miR-4764-5p | Serum | 49 HCC, 36 HC | ↓ | Sensitivity = 100% Specificity = 76.7% PPV = 81.1% NPV = 100% Accuracy = 88.3% |
[28] |
| lncRNA-RP11-156p1.3 | ↓ | ||||
| mRNA RFTN1 | ↓ | ||||
| miRNA + AFP panels | |||||
| miR-122 | Serum | 192 HCC, 95 HC | ↑ | AUC = 1 | [29] |
| miR-885-5p | ↑ | ||||
| miR-29b | ↓ | ||||
| AFP | |||||
| miR-92-3p | Serum | 115 HCC, 40 HC | ↑ | AUC = 0.988 | [30] |
| miR-107 | ↑ | ||||
| miR-3126-5p | ↓ | ||||
| AFP | |||||
| miR-125b | Serum | 90 HCC, 30 HC | ↓ | AUC = 0.936 (CI = 0.878ߝ0.995) Sensitivity = 0.907 Specificity = 0.933 |
[31] |
| miR-223 | ↓ | ||||
| miR-27a | ↓ | ||||
| miR-26a | ↓ | ||||
| AFP | |||||
| miR-206 | Plasma | 38 HCC, 20 HC | ↑ | AUC = 0.989 (CI = 0.919-1.000) |
[32] |
| miR-126 | ↓ | ||||
| AFP | |||||
HCC: hepatocellular carcinoma; HC: healthy controls; AFP: α-fetoprotein; AUC: area under receiver operating characteristic curve; SE: standard deviation; PPV: positive predictive value; NPV: negative predictive value; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression.
4.1.1. miRNA-Only Panels
In our review of miRNA-only panels for HCC detection, all the panels exhibited high diagnostic accuracy, with area under receiver operating characteristic curve (AUC) values ranging from 0.887 to 1.00 (Table 1). These panels consisted of two to eight miRNAs per panel [18,19,20,21,22,23,24,25]. The development of these miRNA panels followed diverse methodologies. Some researchers utilized microarrays [18] or gene expression arrays [22] for the initial screening process, while others leveraged datasets from the Gene Expression Omnibus (GEO) or The Cancer Genome Atlas (TCGA) [23] or employed sequencing techniques [19]. In the process of panel development, some studies took a more straightforward approach [20,24,25], while others adopted a phased strategy. One study employed a two-phase approach [22], and others extended it to a three-phase approach [18,19,23].
Although some panels exhibited high AUC, like the studies of Ali et al. [24] and Jiang et al. [25], their sample cohorts were quite small, 34 and 27 HCC cases, respectively. To obtain more objective results, these two panels should undergo additional testing. More reliable results are those from studies that used multi-phase testing on larger cohorts, such as the studies performed by Zhou et al. [18], Tan et al. [19] and Zhu et al. [21].
The panel with the most included samples was one from the study of Zhou et al. that used microarrays to screen 723 microRNAs in 137 plasma samples for diagnosing HCC. The panel was tested on a training cohort and then validated using an independent cohort, providing a high diagnostic accuracy for HCC [18].
4.1.2. miRNA Panels Combined with lncRNA and mRNA
Compared to miRNA-only panels, the panels where miRNAs were combined to lncRNA and mRNA had sensitivities from 79.5% to 100% and specificities from 76.7% to 100% (Table 1) [26,27,28]. However, it is worth noting that the statistical data, while promising, were derived from cohorts with relatively smaller sample sizes compared to the miRNA-only panels, with sample numbers ranging from 49 to 78 HCC cases. These studies also followed a straightforward approach without validation cohorts; therefore, they should be tested on independent cohort to confirm their statistical value.
4.1.3. miRNA Panels Combined with AFP
AFP is one of the most widely used biomarkers since it was first introduced in the 1960s; nevertheless, its sensitivity value to diagnose HCC is around 60% and the specificity is still inadequate [33]. In up to one-third of HCC cases, serum levels of AFP remain normal; furthermore, elevation in AFP can also occur in some benign liver diseases as well as other tumors (germinal cell tumor) [34].
AFP was included to improve the statistical value of the panels, which is confirmed by the high value of the AUCs, ranging from 0.936 to 1 (Table 1) [29,30,31,32]. Although none of the panels combined with AFP had a multi-phase approach, the study performed by Zekri et al. had a big cohort of 192 HCC cases, and their panels also exhibited the best statistic potential [29].
4.2. Cholangiocarcinoma
For CCA, we could only include one panel, as the other search results did not meet our inclusion criteria, which required a combined statistical score (Table 2).
Table 2.
An overview of circulating miRNA panels for detection of CCA.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miR-10b-3p | plasma | 48 CCA, 20 HC | ↑ | AUC = 0.781 (95% CI: 0.585–0.914) Sensitivity = 83.3% Specificity = 75.0% PPV = 71.4% NPV = 85.7% |
[35] |
| miR-26b-3p | ↑ | ||||
| miR-27a-3p | ↑ | ||||
| miR-106b-3p | ↑ | ||||
| miR-219a-3p | ↑ | ||||
| miR-338-5p | ↑ | ||||
| miR-421 | ↑ |
CCA: cholangiocarcinoma; HC: healthy controls; AUC: area under receiver operating characteristic curve; PPV: positive predictive value; NPV: negative predictive value; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression.
Wada et al. identified a seven-miRNA panel from publicly available datasets. This panel underwent testing on 241 tissue samples from two clinical cohorts, comprising a training set (n = 177), a validation set (n = 64), and matched plasma samples (n = 68). The panel successfully discriminated CCA from healthy individuals with an AUC of 0.781 [35]. It is worth noting that only three out of seven miRNAs in this panel are unique to CCA, namely miR-219a, miR-338, and miR-421. The other four miRNAs are also found in some of the panels used for discriminating HCC from healthy individuals.
5. Circular miRNA in Liver-Metastasizing Primary Cancers
The liver is a common site for metastasis, in part due to its unique and diverse cellular and architectural composition that renders the liver hospitable to tumor cells. According to epidemiology studies, far more common secondary tumor deposits originate from colon and lung cancers. However, pancreatic, gastric, breast and prostate cancers are also known to spread to the liver. Since the presence of liver metastases is associated with worse survival, accurate and prompt diagnosis is crucial [36,37,38]. Not rarely does a pathologist face difficulties in differentiating metastases of unknown origin from primary liver tumors.
5.1. Colorectal Cancer
The miRNA panels detecting CRC were divided into two groups. The first group consisted of 13 panels exclusively featuring miRNAs. The second group had three panels, which included miRNAs, lncRNA, and mRNA. In all panels detecting CRC, we found some miRNAs included in more panels; these miRNAs are miR-21, miR-27a, miR-143, and miR-145 (Table 3).
Table 3.
An overview of circulating miRNA panels for detection of CRC.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-23a-3p | Serum | 427 CRC, 276 HC | ↑ | AUC = 0.877 Sensitivity = 81.4% Specificity = 81% PPV = 80.6% NPV = 81.8% Accuracy = 81.2% |
[39] |
| miR-27a-3p | ↑ | ||||
| miR-142-5p | ↑ | ||||
| miR-376c-3p | ↑ | ||||
| miR-19a-3p | Serum | 196 CRC, 138 HC | ↑ | AUC = 0.87 | [40] |
| miR-21-5p | ↑ | ||||
| miR-425-5p | ↑ | ||||
| miR-145 | Serum | 175 CRC, 130 HC | ↓ | AUC = 0.886 (95% CI = 0.850–0.921) |
[41] |
| miR-106a | ↑ | ||||
| miR-17-3p | ↑ | ||||
| miR-27a | Exosomes | 170 CRC, 130 HC | ↑ | AUC = 0.801 | [42] |
| miR-130a | ↑ | ||||
| miR-193a-5p | Plasma | 149 CRC, 110 HC | ↑ | AUC = 0.88 (95% CI = 0.82–0.93) |
[43] |
| miR-210 | ↑ | ||||
| miR-513a-5p | ↑ | ||||
| miR-628-3p | ↑ | ||||
| miR-103a-3p | Plasma | 139 CRC, 132 HC | ↑ | AUC = 0.895 | [44] |
| miR-127-3p | ↑ | ||||
| miR-151a-5p | ↑ | ||||
| miR-17-5p | ↑ | ||||
| miR-181a-5p | ↑ | ||||
| miR-18b-5p | ↑ | ||||
| miR-30e-3p | Serum | 137 CRC, 145 HC | ↑ | AUC = 0.883 Sensitivity = 0.800 Specificity = 0.787 |
[45] |
| miR-146a-5p | ↑ | ||||
| miR-148a-3p | ↓ | ||||
| miR-203a-3p | Serum | 135 CRC, 135 HC | ↑ | AUC = 0.893 Sensitivity = 81.25% Specificity = 73.33% |
[46] |
| miR-145-5p | ↓ | ||||
| miR-375-3p | ↓ | ||||
| miR-200c-3p | ↓ | ||||
| miR-18a | Plasma | 130 CRC, 244 HC | ↑ | AUC = 0.745 (95% CI = 0.708–0.846) |
[47] |
| miR-20a | ↑ | ||||
| miR-21 | ↑ | ||||
| miR-29a | ↑ | ||||
| miR-92a | ↑ | ||||
| miR-106b | ↑ | ||||
| miR-133a | ↑ | ||||
| miR-143 | ↑ | ||||
| miR-145 | ↑ | ||||
| miR-409-3p | Plasma | 124 CRC, 117 HC | ↑ | AUC = 0.897 | [48] |
| miR-7 | ↓ | ||||
| miR-93 | ↓ | ||||
| miR-144-3p | Plasma | 101 CRC, 134 HC | ↓ | Sensitivity = 93.8% Specificity = 91.3% |
[49] |
| miR-425-5p | ↓ | ||||
| miR-1260b | ↓ | ||||
| miR-601 | Plasma | 90 CRC, 58 HC | ↓ | AUC = 0.792 Sensitivity = 83.3% Specificity = 69.1% |
[50] |
| miR-760 | ↓ | ||||
| miR-126 | Plasma | 50 CRC, 150 HC | ↓ | AUC = 0.906 | [51] |
| miR-139 | ↓ | ||||
| miR-143 | ↓ | ||||
| miR-595 | ↑ | ||||
| miRNA + lncRNA + mRNA panels | |||||
| miR-20b-5p | Plasma | 597 CRC, 585 HC | ↑ | AUC = 0.954 (95% CI = 0.913–0.994) |
[52] |
| miR-329-3p | ↑ | ||||
| miR-374b-5p | ↑ | ||||
| miR-503-5p | ↑ | ||||
| lncRNA-XLOC_001120 | ↑ | ||||
| lncRNA-ENSG00000243766.2 | ↑ | ||||
| miR-3940-5p | Plasma | 70 CRC * | ↓ | Sensitivity = 100% Specificity = 88.6% PPV = 100% NPV = 85% Accuracy = 93.07% |
[53] |
| lncRNA-SNHG14 | ↑ | ||||
| mRNA-NAP1L2 | ↑ | ||||
| miR-59 | Serum | 70 CRC, 20 HC | ↓ | Sensitivity = 100% Specificity = 61.7% PPV = 75.3% NPV = 100% Accuracy = 83.1% |
[54] |
| lncRNA-RP11-909B2.1 | ↑ | ||||
| mRNA L3MBTL1 | ↑ | ||||
CRC: colorectal carcinoma; HC: healthy controls; AUC: area under receiver operating characteristic curve; PPV: positive predictive value; NPV: negative predictive value; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression; *: it was impossible to deduce the number of healthy controls included in the study.
5.1.1. miRNA-Only Panels
Panels demonstrating diagnostic potential for distinguishing between CRC and healthy controls exhibited AUC values ranging from 0.745 to 0.906 (Table 3). In the discovery phase, various methodologies were employed, including bioinformatics utilizing data from GEO and TCGA [42,43,51], gene expression arrays [40,41,47], and sequencing [39]. The studies used two-phase [47], three-phase [40,41,43,45,48,49], and four-phase testing [39,40,42,44,46].
The study by Li et al. stood out with the highest AUC, employing a four-miRNA panel selected using GEO and TCGA data. This panel was validated on both tissue and plasma samples, showing great promise. However, it is important to note that their cohort was relatively modest, consisting of 50 CRC cases. Thus, it is advisable that the panel undergoes independent cohort testing to validate the observed statistics [51].
In contrast, the study by Vychytilova-Faltejskova et al. featured the largest cohort, making it a highly reliable study based on the number of samples. It was conducted in three phases and utilized sequencing for testing. The panel was assessed in a total of 427 CRC patients and 276 healthy donors. The discovery phase, conducted through Illumina small RNA sequencing, identified fifty-four significantly dysregulated microRNAs in the sera of CRC patients compared to healthy individuals (p-value < 0.01). This study established a diagnostic four-microRNA signature with an AUC of 0.877, effectively distinguishing early-stage CRC patients from healthy individuals [39].
5.1.2. miRNA Panels Combined with lncRNA
In our exploration, we uncovered three distinct panels that incorporated both miRNA and other RNAs. Among these panels, one particularly stood out, demonstrating an impressive AUC of 0.954. This panel featured a combination of four miRNAs and two lncRNAs, tested on a substantial cohort of 597 CRC patients and 585 healthy controls (HC). These RNAs exhibited notable upregulation in the plasma of CRC patients compared to healthy individuals. This specific panel exhibited exceptional performance with an AUC of 0.996 in the training set and 0.954 in the validation set [52]. However, it is important to note that two additional panels were conducted that integrated miRNA, lncRNA, and mRNA with high sensitivities, albeit they were tested on considerably smaller cohorts [53,54] when compared to the comprehensive panel designed by Li et al. [52].
5.2. Pancreatic Cancer
The miRNA panels designed for PC detection can be categorized into three groups: miRNA-only panels, panels combining miRNAs with the carbohydrate antigen 19-9 (CA19-9) tumor marker, and panels combining miRNAs with proteins (Table 4).
Table 4.
An overview of circulating miRNA panels for detection of PC.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-122 | Plasma | 409 PC, 312 HC | ↑ | AUC = 0.93 (95% CI = 0.90–0.96) |
[55] |
| miR-34a | ↑ | ||||
| miR-145 | ↑ | ||||
| miR-636 | ↑ | ||||
| miR-223 | ↑ | ||||
| miR-26b | ↑ | ||||
| miR-885-5p | ↑ | ||||
| miR-150 | ↑ | ||||
| miR-126 | ↑ | ||||
| miR-505 | ↑ | ||||
| miR-122-5p | Plasma | 216 PC, 220 HC | ↑ | AUC = 0.937 | [56] |
| miR-125b-5p | ↑ | ||||
| miR-192-5p | ↑ | ||||
| miR-193b-3p | ↑ | ||||
| miR-221-3p | ↑ | ||||
| miR-27b-3p | ↑ | ||||
| miR-30c-5p | Plasma | 168 PC, 124 HC | ↑ | AUC = 0.93 | [57] |
| miR-340-5p | ↑ | ||||
| miR-335-5p | ↑ | ||||
| miR-23b-3p | ↑ | ||||
| miR-142-3p | ↑ | ||||
| miR-145-5p | ↑ | ||||
| miR-200b-3p | ↑ | ||||
| miR-429 | ↑ | ||||
| miR-1260b | ↑ | ||||
| miR-145-3p | ↑ | ||||
| miR-216b-5p | ↑ | ||||
| miR-200a-3p | ↑ | ||||
| miR-217-5p | ↑ | ||||
| let-7b-5p | Plasma | 129 PC, 107 HC | ↑ | AUC = 0.910 | [58] |
| miR-192-5p | ↑ | ||||
| miR-19a-3p | ↑ | ||||
| miR-19b-3p | ↑ | ||||
| miR-223-3p | ↑ | ||||
| miR-25-3p | ↑ | ||||
| miR-574-3p | Plasma | 90 PC, 154 HC | ↑ | AUC = 0.96 (95% CI = 0.92–1.00) |
[59] |
| miR-885-5p | ↑ | ||||
| miR-144-3p | ↓ | ||||
| miR-130b-3p | ↑ | ||||
| miR-334a-5p | ↑ | ||||
| miR-24-3p | ↑ | ||||
| miR-106b-5p | ↓ | ||||
| miR-22-5p | ↑ | ||||
| miR-451a | ↓ | ||||
| let-7d-3p | ↑ | ||||
| miR-101-3p | ↓ | ||||
| miR-26a-5p | ↓ | ||||
| miR-197-3p | ↑ | ||||
| miR-423-3p | ↑ | ||||
| miR-122-5p | ↑ | ||||
| miR-125a-3p | Plasma | 77 PC, 65 HC | ↑ | AUC = 0.862 | [60] |
| miR-4530 | ↑ | ||||
| miR-92a-2-5p | ↑ | ||||
| miRNA + CA19-9 panels | |||||
| miR-16 | Serum | 471 PC, 248 HC | * | AUC = 0.94 (95% CI = 0.90–0.97) Sensitivity = 85 %Specificity = 98% Accuracy = 89% |
[61] |
| miR-18a | |||||
| miR-20a | |||||
| miR-24 | |||||
| miR-25 | |||||
| miR-27a | |||||
| miR-29c | |||||
| miR-30a-5p | |||||
| miR-191 | |||||
| miR-323-3p | |||||
| miR-345 | |||||
| miR-483-5p | |||||
| CA19-9 | |||||
| miR-16 | Plasma | 138 PC, 68 HC | ↑ | AUC = 0.979 (95% CI = 0.962–0.996) |
[62] |
| miR-196a | ↑ | ||||
| CA19-9 | ↑ | ||||
| miR-34a-5p | Plasma | 136 PC, 73 HC | ↑ | AUC = 0.94 (95% CI = 0.89–0.98) |
[63] |
| miR-130a-3p | ↑ | ||||
| miR-222-3p | ↑ | ||||
| CA19-9 | ↑ | ||||
| miR-130a-3p | Plasma | 68 PC, 61 HC | ↑ | AUC = 0.986 (95% CI = 0.972–1.000) |
[64] |
| miR-21-5p | ↑ | ||||
| miR-223-3p | ↑ | ||||
| miR-7975 | ↑ | ||||
| miR-8069 | ↑ | ||||
| CA19-9 | ↑ | ||||
| miRNA + protein panels | |||||
| miR-1246 | Serum | 131 PC, 20 HC | ↑ | Sensitivity = 100% (95% CI = 95%–100%) Specificity = 80% (95% CI: 67%–90%) |
[65] |
| miR-4644 | ↑ | ||||
| miR-3976 | ↑ | ||||
| miR-4306 | ↑ | ||||
| CD44v6 | ↑ | ||||
| Tspan8 | ↑ | ||||
| MET | ↑ | ||||
| CD104 | ↑ | ||||
PC: pancreatic cancer; HC: healthy controls; CA19-9: carbohydrate antigen 19-9; AUC: area under receiver operating characteristic curve; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression; *: the data on down- or upregulation of miRNA was not presented in publication.
Interestingly, several miRNAs consistently appear in multiple panels. These shared miRNAs include miR-16, miR-24, miR-34a, miR-122, miR-130a, miR-145, miR-223, and miR-885. The tumor marker CA19-9 is also included in more panels.
5.2.1. miRNA-Only Panels
A total of six panels exclusively comprised miRNAs for PC detection. The AUCs observed in these panels were between 0.862 and 0.96 (Table 4). The approaches used in the discovery phase of these panels consisted of machine learning methods [57], bioinformatics [60], gene expression arrays [55,56,58,59] and sequencing [57]. The panels were tested in two phases [57], three phases [55,59,60], or four-phases [56,58].
The panel with the best statistic for discriminating between PC and healthy individuals was proposed by Franklin et al. with an AUC of 0.96. This study also had the most miRNAs in the panel—15 miRNAs. The study was designed as a multi-phase study, although it had somewhat smaller sample size and should be further independently tested on larger patient cohorts [59]. The panel with the largest cohort of samples used to discriminate among PC and healthy controls included 409 PC cases and 312 healthy individuals. The panel was composed of 10 miRNAs, and the samples were divided into discovery, training, and validation cohorts. The training cohort exhibited an AUC of 0.93 (95% CI: 0.90–0.96); these results were comparably robust in the validation cohort, with an AUC of 0.93 (95% CI: 0.89–0.97) [55].
5.2.2. miRNA Panels Combined with CA19-9
Although it is one of the most widely used tumor markers, CA19-9 is not exclusive to PC, since its levels may be significantly increased in cases of benign biliary conditions, especially those with obstructive jaundice, as well as some other malignancies such as hepatocellular, gastric, colonic, esophageal, and other non-gastrointestinal cancers, and its interpretation should correlate with other markers [66].
Nevertheless, incorporating CA19-9 into miRNA panels has shown promising results in enhancing the detection of PC. Four notable studies have explored this combined approach. Three of the studies used gene expression arrays in the discovery phase of the study [61,63,64]. Although not having the highest AUC, the most reliable is the panel proposed by Johansen et al., who employed a comprehensive three-phase discovery process. Initially, they utilized the TaqManVR Human MicroRNA assay to identify 34 differentially expressed miRNAs between PC patients and healthy controls. These miRNAs were subsequently tested in a training cohort, leading to the construction of a diagnostic panel comprising 12 miRNAs. Among these, Index III, when combined with CA19-9, exhibited remarkable diagnostic performance with an AUC of 0.94 (0.90–0.97), sensitivity of 85%, specificity of 98%, and overall accuracy of 89% for distinguishing PC patients from healthy individuals [61].
5.2.3. miRNA Panels Combined with Proteins
We included one more panel for detection of PC where four miRNAs were combined with four proteins. The selection process was performed on exosomes of PC cell lines and evaluated on PC exosomes of patients. This panel shows great promise, with a sensitivity of 100% for distinguishing PC from healthy controls [65].
5.3. Gastric Cancer
The miRNA panels designed for detection of GC compared to healthy controls can be divided into two groups: those composed solely of miRNAs and those combining miRNAs with the lncRNA (Table 5).
Table 5.
An overview of circulating miRNA panels for detection of GC.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-140 | Serum | 424 GC, 468 HC | * | AUC = 0.92 (95% CI = 0.88–0.96) Sensitivity = 87.0% (95% CI = 0.794–0.925) Specificity = 68.5% (95% CI = 0.670–0.698) |
[67] |
| miR-183 | |||||
| miR-30e | |||||
| miR-103a | |||||
| miR-126 | |||||
| miR-93 | |||||
| miR-142 | |||||
| miR-21 | |||||
| miR-29c | |||||
| miR-424 | |||||
| miR-181a | |||||
| miR-340 | |||||
| miR-10b-5p | Serum/exosomes | 205 GC, 167 HC/30 GC, 28 HC | ↑ | AUC = 0.702 | [68] |
| miR-132-3p | ↑ | ||||
| miR-185-5p | ↑ | ||||
| miR-195-5p | ↑ | ||||
| miR-20a-3p | ↑ | ||||
| miR-296-5p | ↑ | ||||
| miR-19b-3p | Serum exosomes | 130 GC, 130 HC | ↑ | AUC = 0.814 | [69] |
| miR-106a-5p | ↑ | ||||
| miR-16 | Plasma | 124 GC, 160 HC | ↑ | AUC = 0.812 | [70] |
| miR-25 | ↑ | ||||
| miR-92a | ↑ | ||||
| miR-451 | ↑ | ||||
| miR-486-5p | ↑ | ||||
| miR-21 | Plasma | 115 GC, 60 HC | ↑ | AUC = 0.887 (95% CI = 0.83–0.943) Sensitivity = 84.8% Specificity = 79.2% |
[71] |
| miR-93 | ↑ | ||||
| miR-106a | ↑ | ||||
| miR-106b | ↑ | ||||
| miR-21 | Serum | 92 GC, 89 HC | ↑ | AUC = 0.919 (95% CI = 0.863-0.975) |
[72] |
| miR-31 | ↓ | ||||
| miR-92a | ↓ | ||||
| miR-181b | ↓ | ||||
| miR-203 | ↓ | ||||
| miR-221 | Serum | 82 GC, 82 HC | ↑ | Sensitivity = 82.4% Specificity = 58.8% |
[73] |
| miR-744 | ↑ | ||||
| miR-376c | ↑ | ||||
| miR-7641 | Plasma | 62 GC, 90 HC | ↓ | AUC = 0.799 (95% CI = 0.691–0.908) p < 0.001 |
[74] |
| miR-425-5p | ↓ | ||||
| miR-1180-3p | ↓ | ||||
| miR-122-5p | ↓ | ||||
| miRNA + lncRNA panels | |||||
| miR-675-5p | Plasma | 62 GC, 40 HC | ↓ | AUC = 0.927 (95% CI = 0.85–0.96) p < 0.0001 Sensitivity = 88.78% Specificity = 85% |
[75] |
| H19 | ↑ | ||||
| MEG3 | ↓ | ||||
GC: gastric cancer; HC: healthy controls; AUC: area under receiver operating characteristic curve; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression; *: the data on down- or upregulation of miRNA was not presented in publication.
Some miRNAs are included in more panels; these are miR-21, miR-92a, miR-93 and miR-106a.
5.3.1. miRNA-Only Panels
We included eight miRNA-only panels for distinguishing GC from healthy individuals (Table 5). Interestingly, the discovery phase included only two methods; one was gene expression arrays [67,68,70,73] and the other microarrays [74]. The panels were composed of 2 to 12 miRNAs, with a statistical importance of AUC from 0.702 to 0.92 (Table 5). The statistical rigor is better when multi-phase studies are adopted, which was performed in cases with two-phase studies [71,72,73,74], three-phase studies [67,68,69], and four-phase studies [70].
The study with the highest AUC, the largest cohort and the most extensive study we uncovered for detection of GC using miRNA panels was a comprehensive three-phase, multicenter investigation involving a total of 5248 subjects from Singapore and Korea. The biomarker discovery and verification phases were accomplished through comprehensive serum miRNA profiling and multivariate analysis of 578 miRNA candidates in retrospective cohorts of 682 subjects. Subsequently, a clinical assay was developed and rigorously validated in a prospective cohort of 4566 subjects. The culmination of this research effort resulted in the creation of a clinical assay for the detection of GC based on a robust 12-miRNA biomarker panel. This panel demonstrated exceptional performance with an AUC of 0.93 (95% CI: 0.90–0.95) in the discovery cohort and an AUC of 0.92 (95% CI: 0.88–0.96) in the verification cohort. In the prospective study, the assay exhibited an overall sensitivity of 87.0% (95% CI: 79.4–92.5%) at a specificity level of 68.4% (95% CI: 67.0–69.8%), ultimately yielding an AUC of 0.848 (95% CI: 0.81–0.88) [67]. This study has already completed the clinical trial phase and is now available as a test named The GASTROClear. The test is labeled as an in vitro diagnostic medical device (IVD) for the detection of gastric-neoplasia-associated miRNA biomarkers in human serum and has been shown to detect 87% of all GC, including up to 89% of early-stage GC [76].
5.3.2. miRNA Panels Combined with lncRNA
In miRNA panels for GC detection, there was only one panel with a combination of miRNAs and lncRNAs, comprising miR-675-5p, H19, and MEG3. Although Ghaedi et al. tested several miRNAs, this was the final selected panel that could discriminate among GC and healthy subjects with an AUC of 0.927; the sample size used for testing was quite small, therefore further testing should be performed to validate the statistical performance of the test [75].
5.4. Lung Cancer
The miRNA panels for the detection of LC were divided into panels with only miRNAs and panels with combination of miRNAs and lncRNAs. Several miRNAs were included in many panels, such as miR-205, miR-215, miR-200b, miR-375, miR-486, and miR-1299 (Table 6).
Table 6.
An overview of circulating miRNA panels for detection of LC.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| let-7a-5p | Serum | 744 NSCLC, 944 HC | ↓ | AUC = 0.973 (95% CI = 0.947–0.987) |
[77] |
| miR-375 | ↓ | ||||
| miR-1-3p | ↑ | ||||
| miR-1291 | ↑ | ||||
| miR-214-3p | ↑ | ||||
| miR-17 | Plasma | 676 NSCLC, 456 HC | ↓ | AUC = 0.873 (95% CI = 0.843–0.899) Sensitivity = 81% Specificity = 80% |
[78] |
| miR-190b | ↓ | ||||
| miR-19a | ↓ | ||||
| miR-19b | ↓ | ||||
| miR-26b | ↓ | ||||
| miR-375 | ↑ | ||||
| miR-451a | Serum exosomes | 434 LUAD, 149 HC | ↑ | AUC = 0.965 | [79] |
| miR-194-5p | ↑ | ||||
| miR-486-5p | ↑ | ||||
| miR-193b | Serum | 154 NSCLC, 45 HC | ↑ | AUC = 0.993 (95% CI 0.979–1.000) p < 0.001 |
[80] |
| miR-301 | ↑ | ||||
| miR-14 | ↑ | ||||
| miR-200b | ↑ | ||||
| miR-9-3p | Serum exosomes | 147 NSCLC, 149 HC | ↑ | AUC = 0.878 | [81] |
| miR-205-5p | ↑ | ||||
| miR-210-5p | ↑ | ||||
| miR-1269a | ↑ | ||||
| miR-146b | Serum | 128 NSCLC, 30 HC | ↑ | AUC = 0.96 Accuracy = 92.005 |
[82] |
| miR-205 | ↑ | ||||
| miR-29c | ↑ | ||||
| miR-30b | ↑ | ||||
| miR-340 | Plasma | 120 NSCLC, 120 HC | ↓ | AUC = 0.862 Sensitivity = 78.33% Specificity = 77.5% |
[83] |
| miR-450b-5p | ↑ | ||||
| miR-125a-5p | Serum | 118 NSCLC, 135 HC | ↓ | AUC = 0.936 Sensitivity = 87.5% Specificity = 87.5% |
[84] |
| miR-25 | ↓ | ||||
| miR-126 | ↓ | ||||
| miR-142-5p | Serum | 112 LUAD, 120 HC | ↑ | AUC = 0.933 (95% CI = 0.884–0.965) Sensitivity = 82.93% Specificity = 96.67% |
[85] |
| miR-409-3p | ↓ | ||||
| miR-223-3p | ↑ | ||||
| miR-146a-5p | ↓ | ||||
| let-7b-5p | Plasma | 46 NSCLC, 41 HC | ↓ | AUC = 0.868 Sensitivity = 80% Specificity = 80% |
[86] |
| let-7e-5p | ↑ | ||||
| miR-23a-3p | ↓ | ||||
| miR-486-5p | ↓ | ||||
| miR-215-5p | Serum | 39 NSCLC, 32 HC | ↓ | AUC = 0.8013 Sensitivity = 67% Specificity = 68% |
[87] |
| miR-1299 | ↓ | ||||
| miR-205-5p | ↑ | ||||
| miR-1246 | ↑ | ||||
| miR-520c-3p | Serum exosomes | 36 NSCLC, 36 HC | ↑ | AUC = 0.857 (95% CI, 0813–0.901) p < 0.0001 |
[88] |
| miR-1274b | ↑ | ||||
| miR-145 | Serum | 30 NSCLC, 20 HC | ↑ | AUC = 1 Sensitivity = 100% Specificity 100% |
[89] |
| miR-382 | ↑ | ||||
| miR-21 | Serum | 28 NSCLC, 17 HC | ↑ | AUC = 0.91 (95% CI = 0.80–1.0) |
[90] |
| miR-223 | ↓ | ||||
| miR-205-5p | Serum | 20 SCLC, 32 HC | ↓ | AUC = 0.948 Sensitivity = 90.00% Specificity = 93.75% |
[87] |
| miR-1299 | ↓ | ||||
| miR-215-5p | ↓ | ||||
| miR-141-3p | ↓ | ||||
| miR-200b-5p | ↓ | ||||
| miRNA + lncRNA panels | |||||
| miR-1254 | Serum | 156 NSCLC, 107 HC | ↓ | AUC = 0.844 (95% CI = 0.778–0.91) Sensitivity = 93.3% Specificity = 73.2% |
[91] |
| miR-485-5p | ↓ | ||||
| miR-574-5p | ↓ | ||||
| MALAT1 | ↓ | ||||
| miR-150 | Serum | 30 NSCLC, 15 HC | ↓ | AUC = 0.784 Sensitivity = 80% Specificity = 80% |
[92] |
| linc00673 | ↑ | ||||
LC: lung cancer; NSCLC: non-small cell lung cancer cancer; SCLC: small cell lung cancer; LUAD: lung adenocarcinoma; HC: healthy controls; AUC: area under receiver operating characteristic curve; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression.
5.4.1. miRNA-Only Panels
We included 14 studies with miRNA-only panels for detection of LC from healthy individuals (Table 6). Some studies described discovery phases with methods of gene expression array [78,80,88], sequencing [79,86,87], or bioinformatics [81,83,85]. The studies were two-phased [80,82,83,86,87,88,89,90] and three-phased [77,78,79,81,85].
For the detection of LC, the panels comprised from two to six miRNAs per panel, with the statistical significance of AUC from 0.8013 to 1 (Table 6). Although the AUC was 1, we have to acknowledge that the sample size was quite small, with only 30 NSCLC and 20 healthy control samples [89]. In LC, we have three panels with cohort sizes over 400 cases. These studies exhibit AUC from 0.873 to 0.973 [77,78,79].
The biggest cohort study included 1132 participants and was divided into a training cohort (n = 565) and independent validation cohort (n = 461). For the screening phase, a microarray was used that helped identify six microRNAs (miR-17, miR-190b, miR-19a, miR-19b, miR-26b, and miR-375), providing high diagnostic accuracy in discriminating LC patients from healthy individuals with AUCs of 0.873 and 0.868 for the training and validation cohort, respectively [78].
The second biggest cohort included 744 NSCLC cases and 944 matched controls. A miRNA panel for NSCLC detection was developed with validation on three cohorts. Ying et al. discovered 35 candidate miRNAs, of which 22 were verified, and subsequently developed a 5-miRNA panel that detected NSCLC with AUCs between 0.936 and 0.984 in the discovery and verification cohorts. The panel was validated in three independent cohorts with AUCs of 0.973, 0.916, and 0.917. The sensitivity of the miRNA panel was 81.3% [77].
Finally, Yao et al. attempted to construct a miRNA panel for lung adenocarcinoma (LUAD) detection. The starting material was plasma-derived extracellular vesicles (EVs). They observed upregulation of miR-451a, miR-194-5p, and miR-486-5p in EVs from LUAD patients, compared to healthy controls. The AUC of the combined panel was 0.9650 [79].
5.4.2. miRNA Panels Combined with lncRNA
In detection of LC in blood samples, there were two studies that besides miRNA included lncRNA [91,92]. The panel with better statistical value had an AUC of 0.861; although its cohort was larger, it was still modest compared to studies with bigger cohorts described in miRNA-only panels. Therefore, further validation would be needed to confirm the statistical value of the panel [91].
5.5. Breast Cancer
We found 12 miRNA panels for detection of BC patients compared to normal, of which only one was composed of miRNAs and lncRNA and the others were composed of only miRNAs. We also identified miRNAs included in more than one panel, which are miR-9, miR-19b, miR-20b, miR-92a, miR-106a, and miR-133a (Table 7).
Table 7.
An overview of circulating miRNA panels for detection of BC.
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-133a-3p | Serum | 540 BC, 502 HC | ↑ | AUC = 0.915 Accuracy = 82.3% Sensitivity = 72.2% Specificity = 91.5% |
[93] |
| miR-497-5p | ↑ | ||||
| miR-24-3p | ↑ | ||||
| miR-125b-5p | ↑ | ||||
| miR-377-3p | ↓ | ||||
| miR-374c-5p | ↓ | ||||
| miR-324-5p | ↓ | ||||
| miR-19b-3p | ↓ | ||||
| let-7b-5p | Plasma | 257 BC, 257 HC | ↑ | AUC = 0.978 | [94] |
| miR-122-5p | ↑ | ||||
| miR-146b-5p | ↑ | ||||
| miR-210-3p | ↑ | ||||
| miR-215-5p | ↑ | ||||
| miR-127-3p | Plasma | 247 BC, 140 HC | ↑ | AUC = 0.81 (95% CI = 0.75–0.88) |
[95] |
| miR-376a | ↑ | ||||
| miR-652 | ↑ | ||||
| miR-148b | ↑ | ||||
| miR-376c | ↑ | ||||
| miR-409-3p | ↑ | ||||
| miR-801 | ↑ | ||||
| miR-106a-5p | Serum | 204 BC, 202 HC | ↑ | AUC = 0.93 (95% CI = 0.911–0.964) Sensitivity = 87% Specificity = 89% |
[96] |
| miR-19b-3p | ↑ | ||||
| miR-20b-5p | ↑ | ||||
| miR-92a-3p | ↑ | ||||
| miR-106a-3p | Plasma | 200 BC, 200 HC | ↑ | AUC = 0.88 (95% CI = 0.855–0.923) Sensitivity = 82% Specificity = 79% |
[96] |
| miR-106a-5p | ↑ | ||||
| miR-20b-5p | ↑ | ||||
| miR-92a-2-5p | ↑ | ||||
| miR-92a | Serum | 164 BC, 132 HC | ↑ | AUC = 0.91 | [97] |
| miR-133a | ↑ | ||||
| miR-9-5p | Serum | 135 BC, 125 HC | ↑ | AUC = 0.880 Sensitivity = 86.25% Specificity = 81.25% |
[98] |
| miR-34b-3p | ↓ | ||||
| miR-146a-5p | ↓ | ||||
| miR-9 | Plasma | 62 BC, 20 HC | ↑ | AUC = 0.88 (95% CI = 0.78–0.99) Sensitivity = 96.8% Specificity = 80% |
[99] |
| miR-16 | ↑ | ||||
| miR-21 | ↑ | ||||
| miR-429 | ↑ | ||||
| miR-451 | Serum | 60 BC, 29 HC | ↓ | AUC = 0.953 Sensitivity = 94.7% Specificity = 82.8% |
[100] |
| miR-148a | ↓ | ||||
| miR-27a | ↓ | ||||
| miR-30b | ↓ | ||||
| miR-145 | Plasma | 41 BC, 32 HC | ↑ | AUC = 0.97 (95% CI = 0.929–1.000) Sensitivity 97% Specificity 91% |
[101] |
| miR-425-5p | ↑ | ||||
| miR-139-5p | ↑ | ||||
| miR-130a | ↑ | ||||
| miR-142-5p | Serum | 31 BC, 16 HC | ↑ | AUC = 0.8387 Sensitivity = 93.33% Specificity = 68.75% |
[102] |
| miR-320a | ↑ | ||||
| miR-4433b-5p | ↑ | ||||
| miRNA + lncRNA panels | |||||
| let-7a | Serum | 158 BC, 107 HC | ↓ | AUC = 0.968 PPV = 0.97 NPV = 0.85 |
[103] |
| miR-155 | ↑ | ||||
| miR-574-5p | ↑ | ||||
| MALAT1 | ↑ | ||||
BC: breast cancer; HC: healthy controls; AUC: area under receiver operating characteristic curve; CI: confidence interval; ↑: upregulated expression; ↓: down-regulated expression.
5.5.1. miRNA-Only Panels
We included 11 studies with miRNA-only panels for the detection of BC from healthy individuals (Table 6). The methods used in the discovery phase were gene expression array [94,95], sequencing [102], bioinformatics [98,99], or microarrays [97,101]. Some studies used two-phase [95,102], three-phase [93,96,97,98], or four-phase [94] approaches. The studies selected for this review had panels for BC detection composed of two to eight miRNAs with statistical values of AUC between 0.8387 and 0.978 (Table 7).
Interestingly, the best statistical value belonging to the panel with the second-largest cohort. Using an Exiqon panel, Li et al. selected candidate miRNAs in a screening phase, followed by analysis in training, testing, and external validation phases. They identified five plasma miRNAs with significantly different expression levels between BC patients and healthy individuals. This panel achieved AUCs of 0.683, 0.966, and 0.978 for the training, testing, and external validation sets, respectively [94].
Among the studies of miRNA panels for BC detection, several had large cohorts. The larges was divided into cohorts of discovery phase (n = 289) and two validation phases (n = 374 and n = 379). The researchers identified and validated 30 miRNAs with dysregulated expression in BC. An optimized eight-miRNA panel consistently performed well across all cohorts, achieving an AUC of 0.915, accuracy of 82.3%, sensitivity of 72.2%, and specificity of 91.5% [93].
5.5.2. miRNA Panels Combined with lncRNA
Only one panel in BC was a combination of miRNAs and lncRNAs. The study was conducted in two phases, consisting of training and validation sets. The selected panel consisted of three miRNAs and one lncRNA. The AUCs were 0.960 and 0.968 for the training and validation sets, respectively [103].
6. miRNA Specificity in Cancers
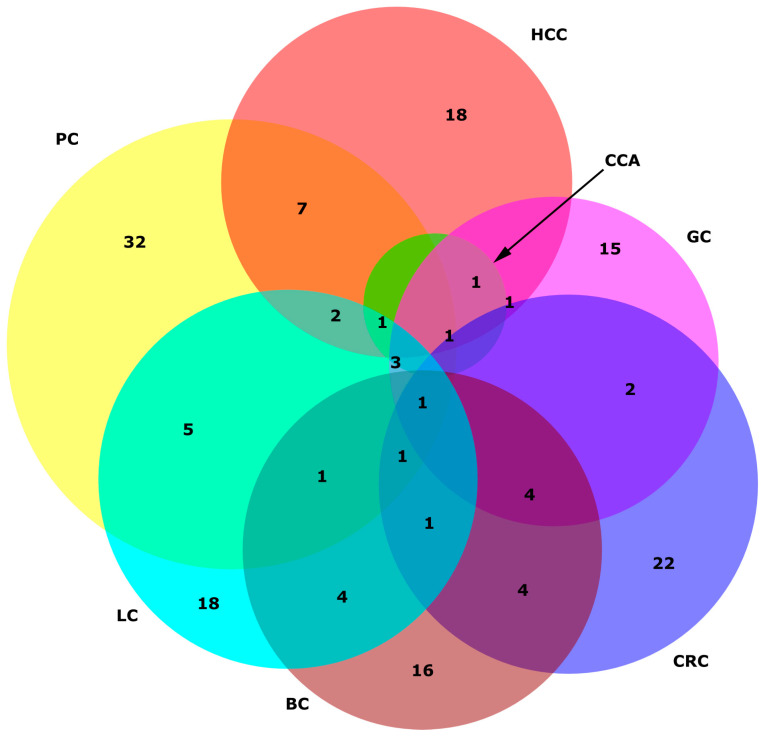
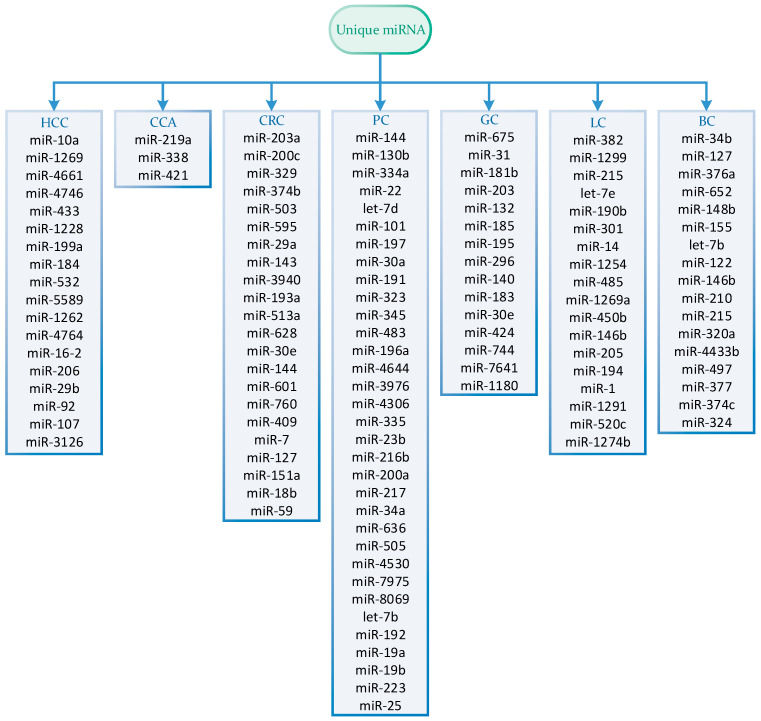
From the numerous panels featured in this review, our objective was to identify miRNAs that exhibit specificity for each distinct cancer type, as well as those that are recurrently included in panels across different cancer types. Figure 2 illustrates the intersections among all miRNAs found within panels of various cancer types. It is evident from the figure that certain miRNAs appear to be specific to each cancer type. However, we must acknowledge that the data utilized to generate this figure are sourced from the panels presented within this study. As a result, there remains a possibility that miRNAs designated as unique here may still exhibit differential expression in other cancer types. The cancer-specific miRNAs are presented in Figure 3.
Figure 2.
Venn diagram of miRNAs included in miRNA panels presented in this review. Red circle HCC: hepatocellular carcinoma: lime circle CCA: cholangiocarcinoma: blue circle CRC: colorectal cancer: yellow circle PC: pancreatic cancer: fuchsia circle GC: gastric cancer: aqua circle LC: lung cancer: maroon circle BC: breast cancer; ∩: intersection. The intersects among these seven sets are: HCC ∩ CCA ∩ CRC ∩ PC ∩ GC: miR-106b; HCC ∩ CCA ∩ CRC ∩ PC ∩ BC: miR-27a; HCC ∩ CCA ∩ PC ∩ LC: miR-26b; HCC ∩ CCA ∩ GC: miR-10b; HCC ∩ CRC ∩ PC ∩ GC ∩ LC: miR-126; HCC ∩ CRC ∩ PC ∩ GC ∩ LC ∩ BC: miR-21; HCC ∩ CRC ∩ GC: miR-181a; HCC ∩ CRC ∩ LC: miR-375; HCC ∩ PC: miR-30c, miR-222, miR-423, miR-27b, miR-192, miR-885, miR-26a; HCC ∩ PC ∩ GC: miR-221, miR-122; HCC ∩ PC ∩ LC: miR-193b, miR-223; HCC ∩ PC ∩ BC: miR-125b; HCC ∩ LC: miR-214, miR-141, let-7b; HCC ∩ BC: miR-801; CRC ∩ PC: miR-18a, miR-1260b; CRC ∩ PC ∩ GC: miR-20a; CRC ∩ PC ∩ GC ∩ LC ∩ BC: miR-142; CRC ∩ PC ∩ LC ∩ BC: miR-145; CRC ∩ PC ∩ BC: miR-130a; CRC ∩ GC: miR-93, miR-103a; CRC ∩ GC ∩ BC: miR-106a, miR-92a, miR-376c, miR-425; CRC ∩ LC: miR-17, miR-210, miR-23a, miR-19a; CRC ∩ LC ∩ BC: miR-146a; CRC ∩ BC: miR-20b, miR-139, miR-133a, miR-148a; PC ∩ GC ∩ LC: miR-25, miR-29c, miR-340; PC ∩ GC ∩ BC: miR-16; PC ∩ LC: miR-451a, miR-1246, miR-200b, miR-150, miR-125a; PC ∩ LC ∩ BC: miR-574; PC ∩ BC: miR-24, miR-429, miR-92a-2, let-7b; GC ∩ LC: miR-486; GC ∩ LC ∩ BC: miR-19b; GC ∩ BC: miR-451; LC ∩ BC: miR-409, miR-9, miR-30b, let-7a. Created using DeepVenn [104].
Figure 3.
Unique miRNAs presented in each cancer included in miRNA panels from studies selected in this review. HCC: hepatocellular carcinoma; CCA: cholangiocarcinoma; CRC: colorectal cancer; PC: pancreatic cancer; GC: gastric cancer; LC: lung cancer; BC: breast cancer.
This intriguing revelation underscores the versatility and potential cross-application of certain miRNAs as valuable diagnostic biomarkers across diverse cancer types. The miRNAs highlighted in Figure 2 appear to transcend tissue-specific boundaries, suggesting broader implications in the field of cancer detection and diagnostics.
In our assessment of panels with the largest cohorts, we found that for GC detection, the commercially available panel includes four cancer-specific miRNAs (miR-140, miR-183, miR-30e, and miR-424), while six other miRNAs are common to multiple cancer types [67]. This strategy of selecting miRNAs for panels appears effective, as it takes into account both shared miRNAs across cancer types and those specific to certain cancers. In panels for BC detection, we also identified cancer-specific miRNAs. In the study with the largest cohort, the eight-miRNA panel included four cancer-specific miRNAs (miR-497, miR-377, miR-374c, and miR-324) [93]. In the study with the second largest cohort, four out of five miRNAs were cancer-specific (miR-122, miR-146b, miR-210, and miR-215) [94].
The PC panel included three cancer-specific miRNAs (miR-34a, miR-636, and miR-505) out of ten miRNAs in the panel [55]. Similarly, the CCA panel contained three out of seven miRNAs that were cancer-specific (miR-219a, miR-338, and miR-421) [35]. In LC, the first three largest cohort studies each included at least one cancer-specific miRNA: miR-1 in a five-miRNA panel in the largest cohort study [77], miR-190b in a six-miRNA panel in the second-largest cohort study [78], and miR-194 in a three-miRNA panel in the third study [79]. Interestingly, we did not observe cancer-specific miRNAs in HCC [18] and CRC [39], as the most statistically reliable panels did not include the miRNAs we identified as cancer-specific.
This observation opens up exciting avenues for further research and exploration into commonalities and shared molecular signatures that may underlie various cancer types, ultimately paving the way for more universal and robust cancer detection strategies.
7. Conclusions and Future Perspectives
We performed a systematic review of the literature of miRNA panels able to detect primary liver cancers, HCC and CCA, and liver-metastasizing cancer, which includes CRC, GC, PC, LC and BC. The ability to distinguish between primary liver cancers and metastatic liver cancers presents an intricate diagnostic challenge of paramount importance. Our research approach was centered on providing a comprehensive overview of existing studies and their findings. However, we recognize that further in-depth research is essential to unravel the intricacies of miRNA deregulation in specific cancer types as compared to others. Specifically, the identification of miRNAs that exhibit distinct deregulation patterns in specific cancer types compared to their counterparts holds immense potential. These identified miRNAs will be fundamental for the development of miRNA panels tailored for discriminating between different cancer types. This approach assumes a critical role in cancer diagnostics, particularly in the context of distinguishing between primary liver cancers and liver-metastasizing cancers. The ability to make this distinction is pivotal, as it has direct implications for clinical management and prognosis, particularly in cases where the origin of the cancer is initially uncertain.
In essence, our systematic review of the literature serves as a foundational step, highlighting the need for further research endeavors that focus on pinpointing specific miRNAs linked to distinct cancer types. With these insights, we can develop miRNA panels that hold the promise of significantly enhancing our ability to differentiate between primary and metastatic liver cancers, ultimately leading to more accurate diagnoses and tailored treatment strategies. Although most of studies have been made on primary tumors and healthy tissue, possible miRNA panels that hold the promise of significantly enhancing our ability to differentiate between primary and metastatic liver cancers could be proposed, ultimately leading to more accurate diagnoses and personalized treatment strategies.
Author Contributions
Conceptualization, B.R. and N.H.; resources, B.R. and N.H.; data curation, B.R. and N.H.; writing—original draft preparation, B.R. and N.H.; writing—review and editing, B.R. and N.H.; supervision, N.H.; funding acquisition, N.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Slovenian Research and Innovation Agency under research core funding No. P3-0054 and project J3-3070.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Ananthakrishnan A., Gogineni V., Saeian K. Epidemiology of primary and secondary liver cancers. Semin. Interv. Radiol. 2006;23:47–63. doi: 10.1055/s-2006-939841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Draškovič T., Zidar N., Hauptman N. Circulating Tumor DNA Methylation Biomarkers for Characterization and Determination of the Cancer Origin in Malignant Liver Tumors. Cancers. 2023;15:839. doi: 10.3390/cancers15030859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsilimigras D.I., Brodt P., Clavien P.-A., Muschel R.J., D’Angelica M.I., Endo I., Parks R.W., Doyle M., de Santibañes E., Pawlik T.M. Liver metastases. Nat. Rev. Dis. Primers. 2021;7:27. doi: 10.1038/s41572-021-00261-6. [DOI] [PubMed] [Google Scholar]
- 5.Centeno B.A. Pathology of liver metastases. Cancer Control. 2006;13:13–26. doi: 10.1177/107327480601300103. [DOI] [PubMed] [Google Scholar]
- 6.de Ridder J., de Wilt J.H.W., Simmer F., Overbeek L., Lemmens V., Nagtegaal I. Incidence and origin of histologically confirmed liver metastases: An explorative case-study of 23,154 patients. Oncotarget. 2016;7:55368–55376. doi: 10.18632/oncotarget.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Wouw A.J., Janssen-Heijnen M.L., Coebergh J.W., Hillen H.F. Epidemiology of unknown primary tumours; incidence and population-based survival of 1285 patients in Southeast Netherlands, 1984–1992. Eur. J. Cancer. 2002;38:409–413. doi: 10.1016/S0959-8049(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 8.Ayoub J.P., Hess K.R., Abbruzzese M.C., Lenzi R., Raber M.N., Abbruzzese J.L. Unknown primary tumors metastatic to liver. J. Clin. Oncol. 1998;16:2105–2112. doi: 10.1200/JCO.1998.16.6.2105. [DOI] [PubMed] [Google Scholar]
- 9.Pavlidis N., Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379:1428–1435. doi: 10.1016/S0140-6736(11)61178-1. [DOI] [PubMed] [Google Scholar]
- 10.Varadhachary G.R., Talantov D., Raber M.N., Meng C., Hess K.R., Jatkoe T., Lenzi R., Spigel D.R., Wang Y., Greco F.A., et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J. Clin. Oncol. 2008;26:4442–4448. doi: 10.1200/JCO.2007.14.4378. [DOI] [PubMed] [Google Scholar]
- 11.Pillai R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pino M.S., Chung D.C. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 14.Kaikkonen M.U., Lam M.T., Glass C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Kosaka N., Iguchi H., Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zen K., Zhang C.-Y. Circulating MicroRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J., Yu L., Gao X., Hu J., Wang J., Dai Z., Wang J.F., Zhang Z., Lu S., Huang X., et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 19.Tan Y., Ge G., Pan T., Wen D., Chen L., Yu X., Zhou X., Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS ONE. 2014;9:e107986. doi: 10.1371/journal.pone.0107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elemeery M.N., Badr A.N., Mohamed M.A., Ghareeb D.A. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J. Gastroenterol. 2017;23:3864–3875. doi: 10.3748/wjg.v23.i21.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H.T., Liu R.B., Liang Y.Y., Hasan A.M.E., Wang H.Y., Shao Q., Zhang Z.C., Wang J., He C.Y., Wang F., et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37:888–896. doi: 10.1111/liv.13356. [DOI] [PubMed] [Google Scholar]
- 22.An Y., Gao S., Zhao W.C., Qiu B.A., Xia N.X., Zhang P.J., Fan Z.P. Novel serum microRNAs panel on the diagnostic and prognostic implications of hepatocellular carcinoma. World J. Gastroenterol. 2018;24:2596–2604. doi: 10.3748/wjg.v24.i24.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho H.J., Baek G.O., Seo C.W., Ahn H.R., Sung S., Son J.A., Kim S.S., Cho S.W., Jang J.W., Nam S.W., et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020;9:5459–5472. doi: 10.1002/cam4.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali H.E.A., Abdel Hameed R., Effat H., Ahmed E.K., Atef A.A., Sharawi S.K., Ali M., Abd Elmageed Z.Y., Abdel Wahab A.H. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2017;41:e51–e62. doi: 10.1016/j.clinre.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L., Cheng Q., Zhang B.H., Zhang M.Z. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening: A validation set from China. Medicine. 2015;94:e603. doi: 10.1097/MD.0000000000000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Tawdi A.H., Matboli M., Shehata H.H., Tash F., El-Khazragy N., Azazy Ael S., Abdel-Rahman O. Evaluation of Circulatory RNA-Based Biomarker Panel in Hepatocellular Carcinoma. Mol. Diagn. Ther. 2016;20:265–277. doi: 10.1007/s40291-016-0200-9. [DOI] [PubMed] [Google Scholar]
- 27.Abd El Gwad A., Matboli M., El-Tawdi A., Habib E.K., Shehata H., Ibrahim D., Tash F. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J. Cell. Biochem. 2018;119:8600–8610. doi: 10.1002/jcb.27109. [DOI] [PubMed] [Google Scholar]
- 28.Ali H.S., Boshra M.S., El Meteini M.S., Shafei A.E., Matboli M. lncRNA- RP11-156p1.3, novel diagnostic and therapeutic targeting via CRISPR/Cas9 editing in hepatocellular carcinoma. Genomics. 2020;112:3306–3314. doi: 10.1016/j.ygeno.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Zekri A.N., Youssef A.S., El-Desouky E.D., Ahmed O.S., Lotfy M.M., Nassar A.A., Bahnassey A.A. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016;37:12273–12286. doi: 10.1007/s13277-016-5097-8. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Li T., Qiu Y., Zhang T., Guo P., Ma X., Wei Q., Han L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine. 2017;96:e5642. doi: 10.1097/MD.0000000000005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo D., Chen L., Liu X., Wang X., Xi Q., Luo Y., Zhang N., Guo H. Combination of miR-125b and miR-27a enhances sensitivity and specificity of AFP-based diagnosis of hepatocellular carcinoma. Tumour Biol. 2016;37:6539–6549. doi: 10.1007/s13277-015-4545-1. [DOI] [PubMed] [Google Scholar]
- 32.Wu X., Wan R., Ren L., Yang Y., Ding Y., Wang W. Circulating MicroRNA Panel as a Diagnostic Marker for Hepatocellular Carcinoma. Turk. J. Gastroenterol. 2022;33:844–851. doi: 10.5152/tjg.2022.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trevisani F., D’Intino P.E., Morselli-Labate A.M., Mazzella G., Accogli E., Caraceni P., Domenicali M., De Notariis S., Roda E., Bernardi M. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: Influence of HBsAg and anti-HCV status. J. Hepatol. 2001;34:570–575. doi: 10.1016/S0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 34.Han L.L., Lv Y., Guo H., Ruan Z.P., Nan K.J. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J. Gastroenterol. 2014;20:10249–10261. doi: 10.3748/wjg.v20.i30.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada Y., Shimada M., Morine Y., Ikemoto T., Saito Y., Baba H., Mori M., Goel A. A blood-based noninvasive miRNA signature for predicting survival outcomes in patients with intrahepatic cholangiocarcinoma. Br. J. Cancer. 2022;126:1196–1204. doi: 10.1038/s41416-022-01710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horn S.R., Stoltzfus K.C., Lehrer E.J., Dawson L.A., Tchelebi L., Gusani N.J., Sharma N.K., Chen H., Trifiletti D.M., Zaorsky N.G. Epidemiology of liver metastases. Cancer Epidemiol. 2020;67:101760. doi: 10.1016/j.canep.2020.101760. [DOI] [PubMed] [Google Scholar]
- 37.Clark A.M., Ma B., Taylor D.L., Griffith L., Wells A. Liver metastases: Microenvironments and ex-vivo models. Exp. Biol. Med. 2016;241:1639–1652. doi: 10.1177/1535370216658144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 39.Vychytilova-Faltejskova P., Radova L., Sachlova M., Kosarova Z., Slaba K., Fabian P., Grolich T., Prochazka V., Kala Z., Svoboda M., et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941–950. doi: 10.1093/carcin/bgw078. [DOI] [PubMed] [Google Scholar]
- 40.Zhu M., Huang Z., Zhu D., Zhou X., Shan X., Qi L.W., Wu L., Cheng W., Zhu J., Zhang L., et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget. 2017;8:17081–17091. doi: 10.18632/oncotarget.15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Liu Y., Wang C., Deng T., Liang H., Wang Y., Huang D., Fan Q., Wang X., Ning T., et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci. Rep. 2015;5:12921. doi: 10.1038/srep12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Pan B., Sun L., Chen X., Zeng K., Hu X., Xu T., Xu M., Wang S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2018;27:746–754. doi: 10.1158/1055-9965.EPI-18-0067. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura K., Hernández G., Sharma G.G., Wada Y., Banwait J.K., González N., Perea J., Balaguer F., Takamaru H., Saito Y., et al. A Liquid Biopsy Signature for the Detection of Patients with Early-Onset Colorectal Cancer. Gastroenterology. 2022;163:1242–1251.e2. doi: 10.1053/j.gastro.2022.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Zhu M., Shan X., Zhou X., Wang T., Zhang J., Tao J., Cheng W., Chen G., Li J., et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene. 2019;687:246–254. doi: 10.1016/j.gene.2018.11.055. [DOI] [PubMed] [Google Scholar]
- 45.Peng X., Wang J., Zhang C., Liu K., Zhao L., Chen X., Huang G., Lai Y. A three-miRNA panel in serum as a noninvasive biomarker for colorectal cancer detection. Int. J. Biol. Markers. 2020;35:74–82. doi: 10.1177/1724600820950740. [DOI] [PubMed] [Google Scholar]
- 46.Huang G., Wei B., Chen Z., Wang J., Zhao L., Peng X., Liu K., Lai Y., Ni L. Identification of a four-microRNA panel in serum as promising biomarker for colorectal carcinoma detection. Biomark. Med. 2020;14:749–760. doi: 10.2217/bmm-2019-0605. [DOI] [PubMed] [Google Scholar]
- 47.Luo X., Stock C., Burwinkel B., Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS ONE. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S., Xiang J., Li Z., Lu S., Hu J., Gao X., Yu L., Wang L., Wang J., Wu Y., et al. A plasma microRNA panel for early detection of colorectal cancer. Int. J. Cancer. 2015;136:152–161. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 49.Tan Y., Lin J.J., Yang X., Gou D.M., Fu L., Li F.R., Yu X.F. A panel of three plasma microRNAs for colorectal cancer diagnosis. Cancer Epidemiol. 2019;60:67–76. doi: 10.1016/j.canep.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q., Huang Z., Ni S., Xiao X., Xu Q., Wang L., Huang D., Tan C., Sheng W., Du X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS ONE. 2012;7:e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S.Q., Xie L.Y., Cai Z.M., Wei H.T., Xie M.Z., Hu B.L., Ning S.F. Systematic analyzing a five- miRNA panel and its diagnostic value of plasma expression in colorectal cancer. Mol. Biol. Rep. 2023;50:7253–7261. doi: 10.1007/s11033-023-08642-8. [DOI] [PubMed] [Google Scholar]
- 52.Li J., Feng Y., Heng D., Chen R., Wang Y., Xu Z., Zhang D., Zhang C., Zhang Y., Ji D., et al. Circulating non-coding RNA cluster predicted the tumorigenesis and development of colorectal carcinoma. Aging. 2020;12:23047–23066. doi: 10.18632/aging.104055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matboli M., Shafei A.E., Ali M.A., El-Din Ahmed T.S., Naser M., Abdel-Rahman T., Anber N., Ali M. Role of extracellular LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal cancer. Arch. Physiol. Biochem. 2021;127:479–485. doi: 10.1080/13813455.2019.1650070. [DOI] [PubMed] [Google Scholar]
- 54.Samir N., Matboli M., El-Tayeb H., El-Tawdi A., Hassan M.K., Waly A., El-Akkad H.A.E., Ramadan M.G., Al-Belkini T.N., El-Khamisy S., et al. Competing endogenous RNA network crosstalk reveals novel molecular markers in colorectal cancer. J. Cell. Biochem. 2018;119:6869–6881. doi: 10.1002/jcb.26884. [DOI] [PubMed] [Google Scholar]
- 55.Schultz N.A., Dehlendorff C., Jensen B.V., Bjerregaard J.K., Nielsen K.R., Bojesen S.E., Calatayud D., Nielsen S.E., Yilmaz M., Holländer N.H., et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 56.Zhou X., Lu Z., Wang T., Huang Z., Zhu W., Miao Y. Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: A miRNA expression analysis. Gene. 2018;673:181–193. doi: 10.1016/j.gene.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura K., Zhu Z., Roy S., Jun E., Han H., Munoz R.M., Nishiwada S., Sharma G., Cridebring D., Zenhausern F., et al. An Exosome-based Transcriptomic Signature for Noninvasive, Early Detection of Patients with Pancreatic Ductal Adenocarcinoma: A Multicenter Cohort Study. Gastroenterology. 2022;163:1252–1266.e2. doi: 10.1053/j.gastro.2022.06.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou X., Wei J., Huang Z., Zhou X., Lu Z., Zhu W., Miao Y. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019;8:2810–2822. doi: 10.1002/cam4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franklin O., Jonsson P., Billing O., Lundberg E., Öhlund D., Nyström H., Lundin C., Antti H., Sund M. Plasma Micro-RNA Alterations Appear Late in Pancreatic Cancer. Ann. Surg. 2018;267:775–781. doi: 10.1097/SLA.0000000000002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seyed Salehi A., Parsa-Nikoo N., Roshan-Farzad F., Shams R., Fathi M., Asaszadeh Aghdaei H., Behmanesh A. MicroRNA-125a-3p, -4530, and -92a as a Potential Circulating MicroRNA Panel for Noninvasive Pancreatic Cancer Diagnosis. Dis. Markers. 2022;2022:8040419. doi: 10.1155/2022/8040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johansen J.S., Calatayud D., Albieri V., Schultz N.A., Dehlendorff C., Werner J., Jensen B.V., Pfeiffer P., Bojesen S.E., Giese N., et al. The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int. J. Cancer. 2016;139:2312–2324. doi: 10.1002/ijc.30291. [DOI] [PubMed] [Google Scholar]
- 62.Liu J., Gao J., Du Y., Li Z., Ren Y., Gu J., Wang X., Gong Y., Wang W., Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer. 2012;131:683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 63.Dittmar R.L., Liu S., Tai M.C., Rajapakshe K., Huang Y., Longton G., DeCapite C., Hurd M.W., Paris P.L., Kirkwood K.S., et al. Plasma miRNA Biomarkers in Limited Volume Samples for Detection of Early-stage Pancreatic Cancer. Cancer Prev. Res. (Phila.) 2021;14:729–740. doi: 10.1158/1940-6207.CAPR-20-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang G., Qiu J., Xu J., Xiong G., Zhao F., Cao Z., Chen G., Liu Y., Tao J., Zheng L., et al. Using a microRNA panel of circulating exosomes for diagnosis of pancreatic cancer: Multicentre case-control study. Br. J. Surg. 2023;110:908–912. doi: 10.1093/bjs/znac375. [DOI] [PubMed] [Google Scholar]
- 65.Madhavan B., Yue S., Galli U., Rana S., Gross W., Müller M., Giese N.A., Kalthoff H., Becker T., Büchler M.W., et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 66.Tsen A., Barbara M., Rosenkranz L. Dilemma of elevated CA 19-9 in biliary pathology. Pancreatology. 2018;18:862–867. doi: 10.1016/j.pan.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 67.So J.B.Y., Kapoor R., Zhu F., Koh C., Zhou L., Zou R., Tang Y.C., Goo P.C.K., Rha S.Y., Chung H.C., et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut. 2021;70:829–837. doi: 10.1136/gutjnl-2020-322065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Z., Zhu D., Wu L., He M., Zhou X., Zhang L., Zhang H., Wang W., Zhu J., Cheng W., et al. Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer Epidemiol. Biomark. Prev. 2017;26:188–196. doi: 10.1158/1055-9965.EPI-16-0607. [DOI] [PubMed] [Google Scholar]
- 69.Wang N., Wang L., Yang Y., Gong L., Xiao B., Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 2017;493:1322–1328. doi: 10.1016/j.bbrc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 70.Zhu C., Ren C., Han J., Ding Y., Du J., Dai N., Dai J., Ma H., Hu Z., Shen H., et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer. 2014;110:2291–2299. doi: 10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao G., Jiang T., Liu Y., Huai G., Lan C., Li G., Jia G., Wang K., Yang M. Droplet digital PCR-based circulating microRNA detection serve as a promising diagnostic method for gastric cancer. BMC Cancer. 2018;18:676. doi: 10.1186/s12885-018-4601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang S., Wang J., Li J., Luo Q., Zhao M., Zheng L., Dong X., Chen C., Che Y., Liu P., et al. Serum microRNA expression profile as a diagnostic panel for gastric cancer. Jpn. J. Clin. Oncol. 2016;46:811–818. doi: 10.1093/jjco/hyw085. [DOI] [PubMed] [Google Scholar]
- 73.Song M.Y., Pan K.F., Su H.J., Zhang L., Ma J.L., Li J.Y., Yuasa Y., Kang D., Kim Y.S., You W.C. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS ONE. 2012;7:e33608. doi: 10.1371/journal.pone.0033608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu X.L., Ren L.F., Wang H.P., Bai Z.T., Zhang L., Meng W.B., Zhu K.X., Ding F.H., Miao L., Yan J., et al. Plasma microRNAs as potential new biomarkers for early detection of early gastric cancer. World J. Gastroenterol. 2019;25:1580–1591. doi: 10.3748/wjg.v25.i13.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghaedi H., Mozaffari M.A.N., Salehi Z., Ghasemi H., Zadian S.S., Alipoor S., Hadianpour S., Alipoor B. Co-expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene. 2019;687:135–142. doi: 10.1016/j.gene.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 76.GASTROClear Blood Test for Early Detection. [(accessed on 10 October 2023)]. Available online: https://gastroclear.mirxes.com.
- 77.Ying L., Du L., Zou R., Shi L., Zhang N., Jin J., Xu C., Zhang F., Zhu C., Wu J., et al. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc. Natl. Acad. Sci. USA. 2020;117:25036–25042. doi: 10.1073/pnas.2006212117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu S., Kong H., Hou Y., Ge D., Huang W., Ou J., Yang D., Zhang L., Wu G., Song Y., et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer. 2018;123:44–51. doi: 10.1016/j.lungcan.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 79.Yao B., Qu S., Hu R., Gao W., Jin S., Liu M., Zhao Q. A panel of miRNAs derived from plasma extracellular vesicles as novel diagnostic biomarkers of lung adenocarcinoma. FEBS Open Bio. 2019;9:2149–2158. doi: 10.1002/2211-5463.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nadal E., Truini A., Nakata A., Lin J., Reddy R.M., Chang A.C., Ramnath N., Gotoh N., Beer D.G., Chen G. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci. Rep. 2015;5:12464. doi: 10.1038/srep12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X., Jiang X., Li J., Wang J., Binang H., Shi S., Duan W., Zhao Y., Zhang Y. Serum exosomal miR-1269a serves as a diagnostic marker and plays an oncogenic role in non-small cell lung cancer. Thorac. Cancer. 2020;11:3436–3447. doi: 10.1111/1759-7714.13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X., Zhang Q., Zhang M., Su W., Wang Z., Li Y., Zhang J., Beer D.G., Yang S., Chen G. Serum microRNA Signature Is Capable of Early Diagnosis for Non-Small Cell Lung Cancer. Int. J. Biol. Sci. 2019;15:1712–1722. doi: 10.7150/ijbs.33986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Y., Jing H., Zhang J. MicroRNA-340 and MicroRNA-450b-5p: Plasma Biomarkers for Detection of Non-Small-Cell Lung Cancer. J. Environ. Public. Health. 2022;2022:8024700. doi: 10.1155/2022/8024700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Wang P., Yang D., Zhang H., Wei X., Ma T., Cheng Z., Hong Q., Hu J., Zhuo H., Song Y., et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin. Lung Cancer. 2015;16:313–319.e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 85.Wang J., Zhang C., Peng X., Liu K., Zhao L., Chen X., Yu H., Lai Y. A combination of four serum miRNAs for screening of lung adenocarcinoma. Hum. Cell. 2020;33:830–838. doi: 10.1007/s13577-020-00346-6. [DOI] [PubMed] [Google Scholar]
- 86.Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., Liu L., Lin B., Su H., Zhao L., et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 87.Geng X., Tsou J.H., Stass S.A., Jiang F. Utilizing MiSeq Sequencing to Detect Circulating microRNAs in Plasma for Improved Lung Cancer Diagnosis. Int. J. Mol. Sci. 2023;24:10277. doi: 10.3390/ijms241210277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong Y., Ding X., Bian Y., Wang J., Zhou W., Wang X., Li P., Shen Y., Wang J.J., Li J., et al. Discovery and validation of extracellular vesicle-associated miRNAs as noninvasive detection biomarkers for early-stage non-small-cell lung cancer. Mol. Oncol. 2021;15:2439–2452. doi: 10.1002/1878-0261.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Toukhy S.E., El-Daly S.M., Kamel M.M., Nabih H.K. The diagnostic significance of circulating miRNAs and metabolite profiling in early prediction of breast cancer in Egyptian women. J. Cancer Res. Clin. Oncol. 2023;149:5437–5451. doi: 10.1007/s00432-022-04492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bedard E.L.R., Abraham A.G., Joy A.A., Ghosh S., Wang X., Lim A., Shao D., Loebenberg R., Roa W.H. A Novel Composite Biomarker Panel For Detection Of Early Stage Non-small Cell Lung Cancer. Clin. Investig. Med. 2021;44:E15–E24. doi: 10.25011/cim.v44i1.36016. [DOI] [PubMed] [Google Scholar]
- 91.Peng H., Wang J., Li J., Zhao M., Huang S.K., Gu Y.Y., Li Y., Sun X.J., Yang L., Luo Q., et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–242. doi: 10.1016/j.lfs.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 92.Baran K., Kordiak J., Jabłoński S., Brzeziańska-Lasota E. Panel of miR-150 and linc00673, regulators of CCR6/CCL20 may serve as non-invasive diagnostic marker of non-small cell lung cancer. Sci. Rep. 2023;13:9642. doi: 10.1038/s41598-023-36485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zou R., Loke S.Y., Tang Y.C., Too H.P., Zhou L., Lee A.S.G., Hartman M. Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br. J. Cancer. 2022;126:472–481. doi: 10.1038/s41416-021-01593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li M., Zou X., Xia T., Wang T., Liu P., Zhou X., Wang S., Zhu W. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019;8:7006–7017. doi: 10.1002/cam4.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cuk K., Zucknick M., Madhavan D., Schott S., Golatta M., Heil J., Marmé F., Turchinovich A., Sinn P., Sohn C., et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS ONE. 2013;8:e76729. doi: 10.1371/journal.pone.0076729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li M., Zhou Y., Xia T., Zhou X., Huang Z., Zhang H., Zhu W., Ding Q., Wang S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018;170:257–270. doi: 10.1007/s10549-018-4757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chan M., Liaw C.S., Ji S.M., Tan H.H., Wong C.Y., Thike A.A., Tan P.H., Ho G.H., Lee A.S. Identification of circulating microRNA signatures for breast cancer detection. Clin. Cancer Res. 2013;19:4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 98.Chen X., Li X., Wang J., Zhao L., Peng X., Zhang C., Liu K., Huang G., Lai Y. Breast invasive ductal carcinoma diagnosis with a three-miRNA panel in serum. Biomark. Med. 2021;15:951–963. doi: 10.2217/bmm-2020-0785. [DOI] [PubMed] [Google Scholar]
- 99.Kim M.W., Park S., Lee H., Gwak H., Hyun K.A., Kim J.Y., Jung H.I., Il Kim S. Multi-miRNA panel of tumor-derived extracellular vesicles as promising diagnostic biomarkers of early-stage breast cancer. Cancer Sci. 2021;112:5078–5087. doi: 10.1111/cas.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luo J., Zhao Q., Zhang W., Zhang Z., Gao J., Zhang C., Li Y., Tian Y. A novel panel of microRNAs provides a sensitive and specific tool for the diagnosis of breast cancer. Mol. Med. Rep. 2014;10:785–791. doi: 10.3892/mmr.2014.2274. [DOI] [PubMed] [Google Scholar]
- 101.Itani M.M., Nassar F.J., Tfayli A.H., Talhouk R.S., Chamandi G.K., Itani A.R.S., Makoukji J., Boustany R.N., Hou L., Zgheib N.K., et al. A Signature of Four Circulating microRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int. J. Mol. Sci. 2021;22:6121. doi: 10.3390/ijms22116121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ozawa P.M.M., Vieira E., Lemos D.S., Souza I.L.M., Zanata S.M., Pankievicz V.C., Tuleski T.R., Souza E.M., Wowk P.F., Urban C.A., et al. Identification of miRNAs Enriched in Extracellular Vesicles Derived from Serum Samples of Breast Cancer Patients. Biomolecules. 2020;10:150. doi: 10.3390/biom10010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang S.K., Luo Q., Peng H., Li J., Zhao M., Wang J., Gu Y.Y., Li Y., Yuan P., Zhao G.H., et al. A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients with Breast Cancer. Med. Sci. Monit. 2018;24:2476–2488. doi: 10.12659/MSM.909453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hulsen T. DeepVenn—a web application for the creation of area-proportional Venn diagrams using the deep learning framework Tensorflow.js. arXiv. 2022 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.