Abstract
To understand the ecological and genetic role of viruses in the marine environment, it is critical to know the infectivity of viruses and the types of interactions that occur between marine viruses and their hosts. We isolated four marine phages from turbid plaques by using four indigenous bacterial hosts obtained from concentrated water samples from Mamala Bay, Oahu, Hawaii. Two of the rod-shaped bacterial hosts were identified as Sphingomonas paucimobilis and Flavobacterium sp. All of the phage isolates were tailed phages and contained double-stranded DNA. Two of the phage isolates had morphologies typical of the family Siphoviridae, while the other two belonged to the families Myoviridae and Podoviridae. The head diameters of these viruses ranged from 47 to 70.7 nm, and the tail lengths ranged from 12 to 146 nm. The burst sizes ranged from 7.8 to 240 phage/bacterial cell, and the genome sizes, as determined by restriction digestion, ranged from 36 to 112 kb. The members of the Siphoviridae, T-φHSIC, and T-φD0, and the member of the Myoviridae, T-φD1B, were found to form lysogenic associations with their bacterial hosts, which were isolated from the same water samples. Hybridization of phage T-φHSIC probe with lysogenic host genomic DNA was observed in dot blot hybridization experiments, indicating that prophage T-φHSIC was integrated within the host genome. These phage-host systems are available for use in studies of marine lysogeny and transduction.
Direct viral enumeration techniques have revealed high levels of viral particles in many regions and many habitats of the marine environment (3, 10, 28–30). Moreover, viruses have been found to contribute significantly to the mortality of bacterioplankton and phytoplankton, indicating that they are important in microbial ecosystems (10). However, direct observations of viruses yield little information concerning the infectivity of the viruses, their genetic diversity, or their ability to enter into lysogenic interactions with their hosts. It is the last issue that we have become interested in over the past several years.
There is recent evidence that lysogeny is a common occurrence in the marine environment. When a series of diverse marine environments were examined for prophage induction by using mitomycin C and UV radiation, up to 38% of the bacterial population contained inducible prophage (17). Examination of 88 random marine bacterial isolates indicated that more than 40% of these isolates contained inducible prophage or defective phagelike particles (16). Of nearly 300 marine phage isolates tested, 29 were identified as temperate phage isolates by Moebus (22). Hidaka and Shirahama (13) have also described isolation of a temperate phage from marine mud in Kagoshima Bay, Japan.
We were interested in isolating marine temperate phage-host systems to study lysogeny in the marine environment, as well as the potential for phage-mediated gene transfer (transduction) in the marine environment. The interactions between marine viruses and their hosts may significantly influence the genetic diversity and composition of marine microbial communities. We attempted to isolate several temperate phage-host systems from water samples from Mamala Bay, Oahu, Hawaii; our goal was to establish transduction systems with indigenous marine bacterial hosts and bacteriophages. The characteristics of these phage-host systems are described here.
MATERIALS AND METHODS
Isolation of bacterial hosts and phages.
Twenty-liter water samples were collected from surface and subsurface waters of Ke’ehi Lagoon and Sand Island sewage outfall offshore in Mamala Bay, Hawaii, on 14 to 17 February 1994 by pumping water into acid-washed carboys on a small boat. The water samples were concentrated by vortex flow filtration (15) from 20 liters to approximately 50 ml within 3 h of collection. Bacteria were isolated from the concentrated samples on artificial seawater agar plates containing (per liter) 5 g of peptone and 1 g of yeast extract (ASWJP+PY) (23). Each of the colonies was reisolated three times to ensure the purity of the bacterial isolates. Each bacterial isolate was then used as a host for isolation of temperate phages from the same concentrated sample or samples from nearby locations.
For temperate phage isolation, 1 or 0.1 ml of a concentrated microbial community from seawater was mixed with 1 ml of a potential host culture in a tube containing 3 ml of soft agar at 47°C. The mixture was poured over an ASWJP+PY plate to form a thin layer. After incubation overnight at 28°C, turbid plaques were picked from the plates, and each individual plaque was reisolated three times to ensure the purity of the phage isolate. Phage lysates were produced by eluting the top agar overlay plates. Stock samples were stored at 4°C after filtration through 0.2-μm-pore-size filters.
Characterization of marine bacterial isolates.
Gram staining was performed with each bacterial host by using a Fisher Diagnostics Gram Stain Set (Fisher Scientific, Pittsburgh, Pa.) and the manufacturer’s recommended procedures. Bacterial morphologies were examined with a transmission electron microscope (TEM) at magnifications of ×9,000 to ×15,000. Gram-negative rod-shaped bacteria were further tested by using an API-NFT bacterial identification kit (BioMerieux Viteck, Inc., Hazelwood, Mo.) and the manufacturer’s recommended procedures.
TEM examination of phage morphology.
One drop of a freshly prepared phage lysate was spotted onto a Formvar–carbon-coated 200-mesh TEM grid (Electron Microscopy Sciences, Fort Washington, Pa.). The edge of the grid was touched with a piece of Whatman filter paper to drain away the excess fluid. The grid was then stained with a 2% uranyl sulfate solution for 30 s, washed with 1 drop of deionized water for 10 s, and dried in air before examination with a Hitachi model 500 TEM. The microscope magnification was calibrated by using 50-nm polystyrene beads (nanosphere; Electron Microscope Sciences) as size standards. Photomicrographs of phages were taken at magnifications of ×48,000 to ×100,000. Morphological characteristics of phages were compiled from multiple photomicrographs of phage particles in order to minimize size or shape anomalies.
Determination of phage genome size by restriction enzyme digestion.
Phage DNAs were extracted from 10- to 30-ml portions of phage lysates by using a Wizard Lambda Preps DNA purification system (Promega, Madison, Wis.) and the manufacturer’s recommended procedures. Briefly, freshly prepared phage lysates were digested with RNase A and DNase I at 37°C for 15 min and precipitated with polyethylene glycol 8000 on ice for 30 min. The precipitates were resuspended in 500 μl of phage buffer containing 150 mM NaCl, 40 mM Tris-HCl (pH 7.4), and 10 mM MgSO4. Phage DNA was purified by using a mini Purification Resin column (Promega) and was eluted from the column with 80°C deionized water. The purified phage DNA was stored at 4°C until restriction enzyme digestion.
For restriction enzyme digestion, approximately 1 μg of each phage DNA was digested in a total volume of 40 μl. Both uncut DNA and restriction enzyme-cut DNA were visualized on agarose gel electrophoresis gels (0.4, 1, and 1.5% agarose) stained with ethidium bromide (26). The molecular weight of each phage DNA was then estimated by using the sizes of large fragments determined from 0.4% gels, the sizes of medium fragments determined from 1% gels, and the sizes of small fragments determined from 1.5% gels. A 1-kb DNA ladder (12.2 to 0.5 kb; Gibco BRL, Gaithersburg, Md.), HindIII-digested λ DNA fragments (23 to 0.56 kb; Promega), and a high-molecular-weight DNA marker (48.5 to 8.3 kb; Gibco BRL) were used as molecular weight markers to calculate the sizes of the phage DNA fragments by using linear regression and inverse prediction.
One-step growth experiments.
The burst sizes and one-step growth curves were determined as described by Weiss et al. (31), with minor modifications. One milliliter of each overnight culture was transferred to 20 ml of fresh ASWJP+PY and incubated with shaking for about 2 h, until the A600 was ∼0.6 and the viable cell counts were around 108 cells/ml. One-milliliter portions of each bacterial culture were then transferred to fresh tubes and mixed with phage at a multiplicity of infection of 1. Each mixture was incubated at room temperature for 20 min to allow phage adsorption. After this adsorption period, the cells were pelleted to wash away the unattached phage. Infected cells were diluted to 10−5 in 10 ml of artificial seawater and incubated at 28°C with shaking. Samples were withdrawn over time for use in an infectious center assay in which the soft agar overlay method was used. Bacterial viable counts were determined before the bacteria were mixed with phage and at the end of the experiment. Burst size was estimated from triplicate experiments by using the following equation:
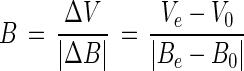 |
where B is the burst size, ΔV represents changes in the viral number, ΔB represents changes in the bacterial number, Ve is the viral number at the end of the experiment, V0 is the viral number at the beginning of the experiment, Be is the bacterial number at the end of the experiment, and B0 is the bacterial number at the beginning of the experiment.
Purification of plasmid DNA and chromosomal DNA.
Plasmid DNA and chromosomal DNA were purified from bacterial cells by using a Wizard Plasmid DNA Preps kit and a Wizard Genomic DNA Preps kit (Promega), respectively, and the manufacturer’s recommended procedures. Special precautions were taken to extract DNA from potential lysogens; these precautions included washing potential lysogenic cells three times before DNA extraction to avoid contaminating DNA preparations with free phage DNA. The purity of the chromosomal DNA in these preparations was similar to the purity of chromosomal DNA purified with a cesium chloride gradient. When chromosomal DNA purified with the Prep kits was added to cesium gradients, only a single sharp band was observed in each sample after centrifugation. Concentrations of DNA were measured fluorometrically by using Hoechst 33258 dye as described previously (24).
Probe labeling and dot blot hybridization.
A 100-bp AccI digestion fragment of temperate phage T-φHSIC DNA was randomly cloned into Riboprobe vector pGEM4Z (Promega) by using the cloning protocol recommended by the manufacturer. A 35S-RNA probe was prepared by transcription of the fragment with T7 RNA polymerase by using 35S-UTP (9).
Various concentrations of phage DNA, plasmid DNA, and chromosomal DNA were dotted onto charged nylon membrane filters (Zetaprobe; Bio-Rad, Richmond, Calif.). The filters were baked in a vacuum oven for 2 h at 80°C, rewet with 0.4 M Tris-HCl (pH 8), and then hybridized overnight with the phage probe at 42°C as previously described (9). Each filter was washed once for 5 min at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 1 mM dithiothreitol and then three times for 1 h at 65°C in PSE (0.25 M sodium phosphate, 2% sodium dodecyl sulfate, 1 mM EDTA; pH 7.4) and three times for 30 min at 65°C in PES (40 mM sodium phosphate, 1% sodium dodecyl sulfate, 1 mM EDTA; pH 7.4). The hybridization signal was detected by autoradiography.
RESULTS
Isolation of bacterial hosts and phages from marine environments.
Table 1 shows the results of isolation of bacterial hosts and phages from marine environments. Four bacterial isolates were found to be able to serve as hosts for isolation of turbid plaques. Each bacterium was named after the sampling station. Bacteriophage isolates were also named by using a similar system, except that the prefix T-φ (representing temperate phage) was used. Although one of the phage isolates was later shown to be lytic, the name of the phage was not changed. In most cases the phage was isolated from the same sample as that from which the host bacterium was isolated. One bacterium, designated D1B, was also a host for a phage isolate (T-φD0-3) from a sample obtained from a nearby environment (Ke’ehi Lagoon). T-φD0-3 had the same plaque morphology as T-φD1B and was later shown to produce the same restriction enzyme band patterns. The phage isolates were host specific, and none of the viruses cross-infected other hosts tested.
TABLE 1.
Isolation of bacterial hosts and phages from Mamala Bay, Oahu, Hawaii
| Isolation site for bacteria | Bacterial isolate (host) | Turbid plaque isolation site | Corresponding temperate phage |
|---|---|---|---|
| Ke’ehi Lagoon | HSIC | Ke’ehi Lagoon | T-φHSIC |
| D0 | Ke’ehi Lagoon | T-φD0 | |
| Sand Island outfall (15 m subsurface) | D1B | Sand Island outfall (15 m subsurface) | T-φD1B |
| Ke’ehi Lagoon | T-φD0-3 | ||
| Sand Island outfall | D2S | Sand Island outfall | T-φD2S |
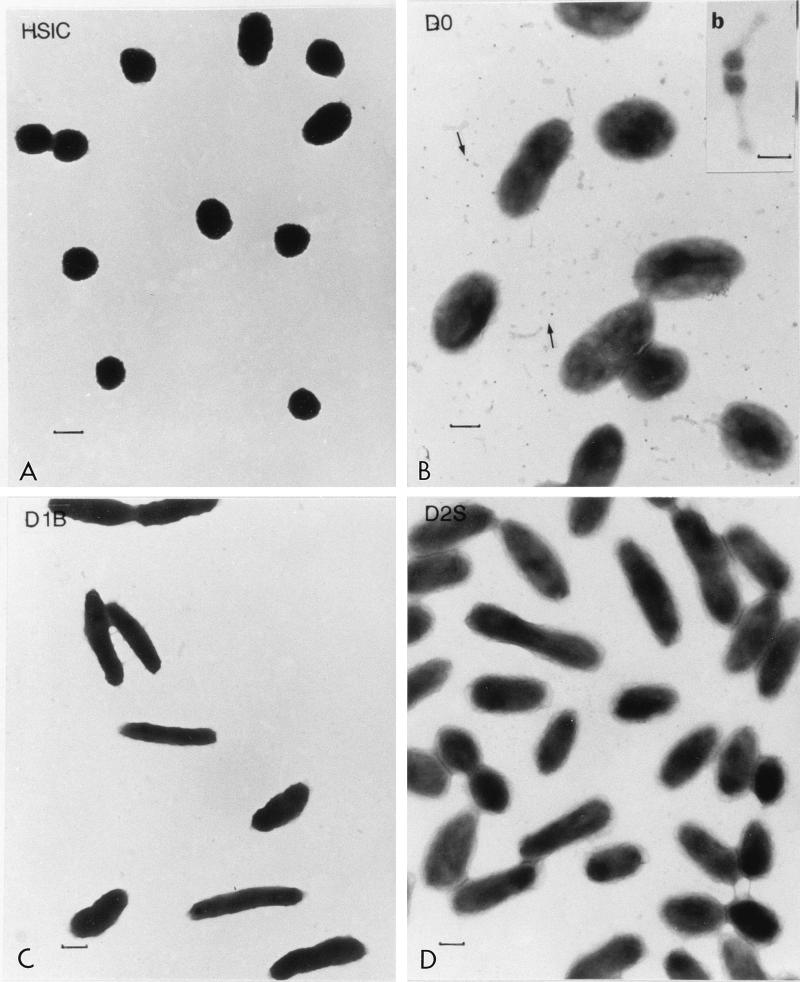
The bacterial isolates were identified by using the API-NFT kit, and Fig. 1 shows the morphology of these isolates. Bacterial isolate HSIC was spherical (Fig. 1A), while the other three isolates were either short fat rods or slender rods (Fig. 1B through D), and all of the bacteria were gram negative. Only rod-shaped bacteria were characterized further by the API-NFT test, because of the limitations of this test. Host bacterium D0 showed an excellent match with Sphingomonas paucimobilis. D1B belonged to the genus Flavobacterium and was tentatively identified as Flavobacterium breve or Flavobacterium weeksella. D2S possessed a capsular layer (Fig. 1D) and may belong to the genus Pseudomonas, Aeromonas, or Shewanella. Clonal cultures of isolate D0 inoculated from a single colony always contained phage particles (Fig. 1B), suggesting that this bacterial isolate was a lysogen upon isolation. Phage particles found in the cell culture were slightly different from isolate T-φD0 particles in that the heads were smaller (average size, 39 ± 1.15 nm [n = 4], compared to 54.3 ± 3.7 nm [n = 4]); their tails were about the same length as T-φD0 tails (average length, 86.5 ± 6.24 nm) but appeared to be wider than T-φD0 tails (Fig. 2D).
FIG. 1.
Morphologies of bacterial isolates from Mamala Bay. (A) HSIC. (B) D0 and phage particles in the cell culture (indicated by arrows). (b) Phage particles found in the D0 culture. Bar = 50 nm. (C) D1B. (D) D2S. (A, B, C, and D) Bars = 0.5 μm.
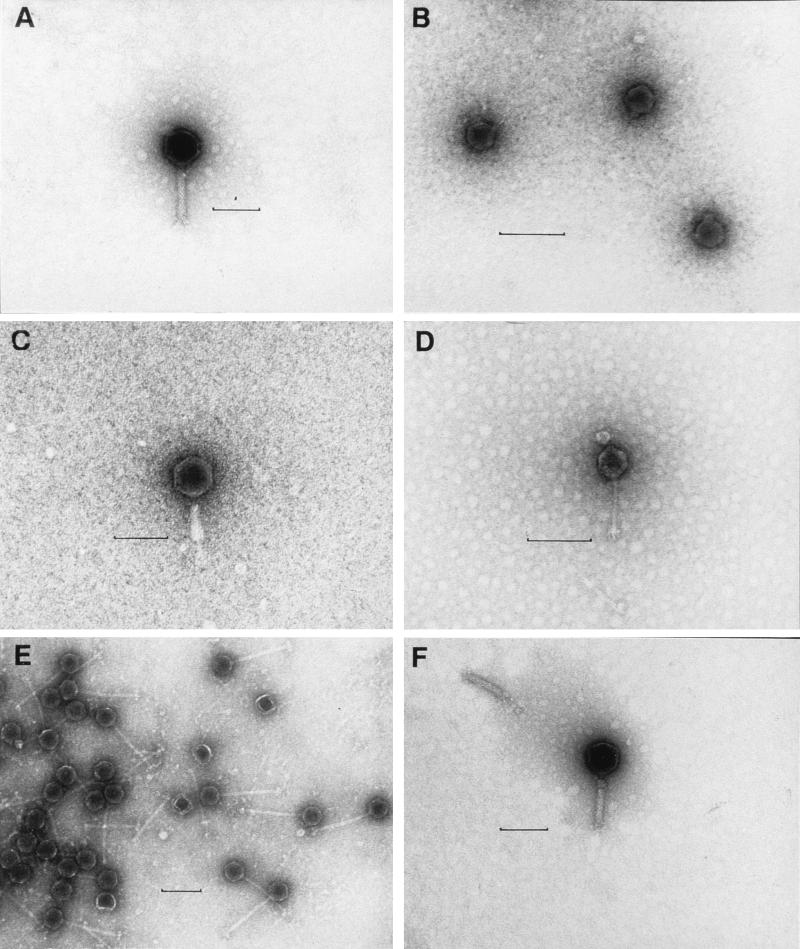
FIG. 2.
Electron photomicrographs of phages isolated from Mamala Bay. (A) T-φD0-3. (B) T-φD2S. (C) T-φD1B. (D) T-φD0. (E) T-φHSIC. (F) T-φD0-3. Bars = 100 nm.
All of the phage isolates formed turbid plaques on the bacterial host lawns, and turbid plaques are the typical plaque morphology of temperate phages. Phage T-φD0 formed plaques with completely turbid centers and poorly defined edges. Phage T-φHSIC plaques had small clear centers and wide halos of bacterial growth, and the edge of each plaque was defined by a sharp, narrow ring. Small-plaque mutants and large clear-plaque mutants were often seen in the phage preparations, and the number of plaque mutants increased with prolonged storage of the phage lysates in a refrigerator. Phage T-φD0-3 formed very turbid plaques which occasionally had tiny clear centers. Phage T-φD2S plaques had relatively large clear centers and halos wider than that of T-φD0-3; unlike the T-φHSIC plaques, there was not a clear ring around each halo. The temperate nature of T-φD2S is questionable (see Discussion).
Figure 2 shows electron photomicrographs of the temperate phages, and phage sizes are given in Table 2. The phage head sizes ranged from 47 to 70.7 nm, while the tail sizes ranged from 12 to 146 nm. Each T-φHSIC phage had a long flexible tail, which appeared to be noncontractile (Fig. 2E). Each T-φD0-3 phage apparently had a contractile tail (Fig. 2A and F) and was similar in morphology to T-φD1B (Fig. 2C); both of these phages belong to the family Myoviridae. Each T-φD0 phage had a very thin flexible tail with no observable collar structure (Fig. 2D). Both T-φD0 and T-φHSIC belong to the family Siphoviridae. Each T-φD2S phage (Fig. 2B) had a very short tail, which is typical of members of the family Podoviridae.
TABLE 2.
Morphological characteristics of phages isolated from Mamala Bay, Oahu, Hawaii
| Temperate phage | Family | Head size (nm) | Tail size (nm) |
|---|---|---|---|
| T-φHSIC | Siphoviridae | 47 ± 3.7 (n = 7) | 146 ± 3.7 (n = 7) |
| T-φD0 | Siphoviridae | 54.3 ± 3.7 (n = 4) | 87.9 ± 6.4 (n = 3) |
| T-φD1B | Myoviridae | 70.7 ± 4.8 (n = 5) | 108.5 ± 7.8 (n = 5) |
| T-φD2S | Podoviridae | 49.1 ± 3.9 (n = 7) | 12 ± 1.2 (n = 3) |
Genome sizes and types of phage isolates.
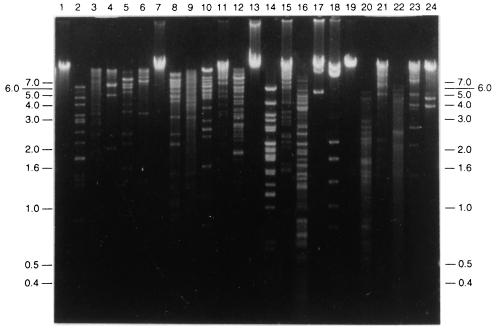
All of the phage isolates were double-stranded DNA viruses, as indicated by restriction enzyme digestion results (Fig. 3). At least seven restriction enzymes were used for each purified phage DNA (Table 3). Interestingly, none of the phage DNAs could be digested by restriction endonuclease BamHI, but the reason for this resistance was not determined. T-φD0 and T-φD2S DNAs were resistant to EcoRI and BglII, respectively. T-φHSIC DNA was found to be resistant to restriction digestion by XbaI, KpnI, SalI, PstI, SacI, and SmaI (data not shown). Some restriction enzymes digested phage DNA into more than 30 fragments which appeared as a DNA smear on the agarose gel, while other enzymes digested phage DNA into many fragments of similar size, resulting in smearing in a region of the gel. Only clear restriction patterns were used to determine molecular weight. Phage genome sizes were determined by adding the sizes of the restriction fragments from each enzyme digestion. Table 3 shows the estimated phage genome sizes as determined by restriction digestion. The genome size of T-φHSIC was about 36 kb, as determined with BglII- and EcoRI-digested fragments; HpaI digestion of T-φHSIC DNA gave a slightly lower value (33 kb). Some of the HpaI-digested fragments were more intense than the other fragments on the gel (Fig. 3), possibly because of multiple bands at the same molecular weight. Such similar bands may have also occurred in HpaI-digested T-φD0 and T-φD2S DNAs. The size of the T-φD0 genome was about 71 kb, as determined by BglII, EcoRV, and AccI digestion. T-φD1B DNA had the same restriction pattern as phage T-φD0-3 DNA (data not shown), suggesting these phages were the same phage or closely related phages isolated from different locations. The T-φD1B genome size averaged 112 kb, as determined by restriction digestion with HpaI, BglII, and EcoRV, while the genome size of T-φD2S was 65 kb (Table 3).
FIG. 3.
Phage DNAs digested with restriction endonucleases to determine molecular weights. Lane 1, uncut T-φD0 DNA; lanes 2 through 6, T-φD0 DNA cut with HpaI, HindIII, BglII, AccI, and EcoRV, respectively; lane 7, uncut T-φD1B DNA; lanes 8 through 12, T-φD1B DNA cut with HapI, AccI, BglII, EcoRI, and EcoRV, respectively; lane 13, uncut T-φD2S DNA; lanes 14 through 18, T-φD2S DNA cut with HpaI, HindIII, AccI, EcoRI, and EcoRV, respectively; lane 19, uncut T-φHSIC DNA; lanes 20 through 24, T-φHSIC cut with HpaI, HindIII, AccI, EcoRI and EcoRV, respectively. The positions of molecular weight standards (in kilobases) are indicated on both sides of the gel.
TABLE 3.
Estimated phage genome sizes as determined by restriction enzymes digestion
| Phage | Estimated molecular sizes (kb) as determined with:
|
||||||
|---|---|---|---|---|---|---|---|
| HpaI | BglII | EcoRV | AccI | EcoRI | HindIII | BamHI | |
| T-φHSIC | 33 (17)a,b | 37 (3) | Smearc | Smearc | 36 (9) | Smearc | No sitesd |
| T-φD0 | 43 (16)b | 71 (7) | 72 (8) | 69 (11) | No sitesd | Smearc | No sitesd |
| T-φD1B | 110 (20) | 113 (19) | 114 (30) | Smearc | Smearc | Smearc | No sitesd |
| T-φD2S | 52 (21)b | No sitesd | 65 (8) | 64.4 (26) | Smearc | Smearc | No sitesd |
The numbers in parentheses are numbers of fragments.
Doublets or triplets may have occurred in the digested phage DNA.
DNA was digested into too many fragments, or fragments having similar sizes were concentrated in a region of the gel.
DNA could not be digested by the restriction enzyme used.
Phage one-step growth curves.
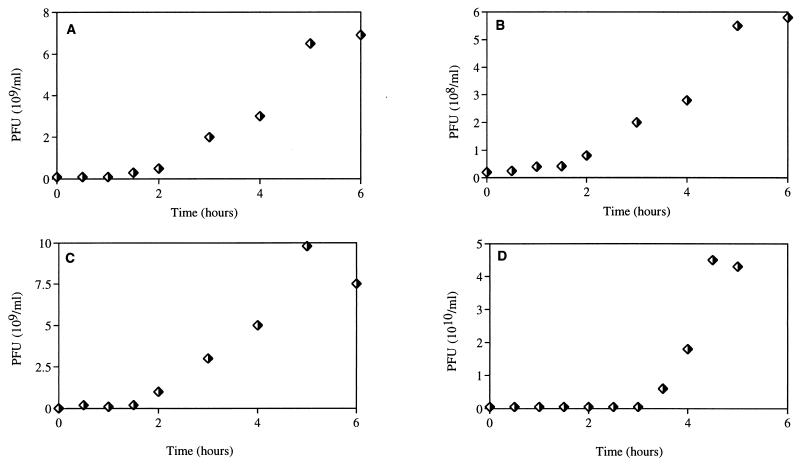
Figure 4 shows the one-step growth curves for the phage isolates. T-φD0 had the smallest burst size of the four phages examined (ca. 7.8), while T-φD2S had the largest burst size (240). The burst sizes for T-φHSIC and T-φD1B were ca. 47 and 176, respectively. The latent period of these phages ranged from 90 to 180 min (Fig. 4).
FIG. 4.
One-step growth curves for phage isolates from Mamala Bay. (A) T-φHSIC. (B) T-φD0. (C) T-φD1B. (D) T-φD2S.
Lysogenic characteristics.
Bacteria isolated from the turbid plaque centers or halo areas were resistant to phage infection, suggesting that the bacterial hosts might be lysogenized with the phages. Among the four phage-host systems, only the relationship between temperate phage T-φHSIC plaques, and its host, HSIC, was characterized in detail. Bacteria picked from the halo areas of T-φHSIC plaques, designated L-HSIC, were isolated by preparing at least three consecutive streaks on fresh agar plates to ensure the purity of the isolate. L-HSIC had a different colony morphology than wild-type HSIC. HSIC colonies had smooth surfaces and were rounded and opaque. L-HSIC colonies were more transparent, had uneven edges, and appeared wrinkly after incubation for 48 h. L-HSIC cell cultures inoculated from a single colony shed viruses into the medium which were infectious to wild-type HSIC.
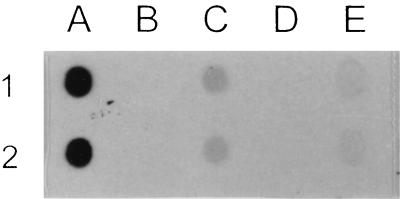
To confirm that phage T-φHSIC DNA was inside the L-HSIC cells, a dot blot of DNA from T-φHSIC, plasmid preparations of HSIC and L-HSIC, and chromosomal preparations of HSIC and L-HSIC were probed with 35S-labeled phage T-φHSIC probe (Fig. 5). Strong hybridization of the probe with phage DNA was found, as expected. Hybridization of the probe with L-HSIC plasmid DNA and weak hybridization with L-HSIC chromosomal DNA were also observed. The weak hybridization of L-HSIC chromosomal DNA was not a result of contamination of plasmid DNA in the preparation because chromosomal DNA preparations produced only a single band on a cesium chloride gradient when an attempt was made to repurify the DNA (data not shown). The hybridization of the probe with L-HSIC plasmid DNA was also not a result of contamination of chromosomal DNA, because the concentration of plasmid DNA dotted was one-fourth the concentration used in the chromosomal dot, yet resulted in a higher degree of hybridization. No hybridization was detected with HSIC plasmid and chromosomal DNA.
FIG. 5.
Dot blot hybridization preparations probed with the phage T-φHSIC DNA probe. Lane A, 0.4 μg of T-φHSIC DNA; lane B, 2.1 μg of plasmid DNA from HSIC; lane C, 0.5 μg of plasmid DNA from L-HSIC; lane D, 2.1 μg of chromosomal DNA from HSIC; lane E, 1.8 μg of chromosomal DNA from L-HSIC. Rows 1 and 2 are replicates.
DISCUSSION
Incorporation of viruses into the budget of microbial C transfer by using the lytic phage-host interaction model resulted in an imbalance in the carbon budget (5). Therefore, the dynamics of temperate phage-host relationships in the marine environment should be considered. Previously, we have shown that lysogenic bacteria were abundant among marine bacterial isolates (16), suggesting that many marine bacteriophages may be temperate in nature. However, isolation of temperate phages from marine environments was rare. Moebus tested nearly 300 marine phage isolates and found that only 29 were temperate (22). However, his phage collections were obtained by using a liquid nutrient enrichment isolation method (21) which favors isolation of lytic phages (8, 14). Kokjohn et al. (18) suggested that high numbers of lytic phages found in the aquatic environment were an artifact of nutrient enrichment isolation methods.
We isolated four different types of turbid plaques on native marine bacterial hosts from concentrated seawater samples obtained from Mamala Bay. Two of the turbid plaques resembled the classical turbid plaques of temperate phages (8). T-φHSIC plaque morphology is similar to the plaque morphology of a Bacillus subtilis phage described by Romig and Brodesky (25). However, these authors did not determine if phage DNA was integrated into the host genome. In this study, using dot blot hybridization and probing, we detected T-φHSIC DNA inside the host cell, suggesting that this phage isolate is a temperate phage. Plaques of T-φD2S are similar to the plaques of Rhizobium leguminosarum phage 3H (27), the plaques of the Klebsiella pneumoniae capsule bacteriophage XIV (20), and the plaques of the M-1 phage of Rhizobium japonicum (7). Lindberg (20) described bacteriophages isolated from capsulated species of bacteria with plaque morphology consisting of a large clear center surrounded by a crater-like depression or halo in the bacterial lawn. This halo comprised an area in which the bacterial capsular material was destroyed by the phage-induced polysaccharide depolymerase but bacterial viability was not affected (2, 7). A similar phenomenon was observed with the D2S phage-host system, since bacterial strain D2S was encapsulated. The turbid halos around T-φD2S plaques may have been the result of bacteria with damaged capsular material rather than lysogeny. Thus, T-φD2S may be lytic.
As determined by the API-NFT test, bacterial host D0 was identified as S. paucimobilis (previously known as Pseudomonas paucimobilis). Pseudomonas phages have been routinely isolated from the marine environment (12). Bacterium D1B was identified as a Flavobacterium sp. strain. There are two prior reports of isolation of Flavobacterium marine phages, both from Kagoshima Bay, Japan (11, 12).
It is not surprising that all of our phage isolates are tailed phages. According to a literature review by Børsheim (4), most marine phages in culture have tails. However, the head sizes of our phage isolates were smaller than the head sizes of most of the known marine bacteriophages. Børsheim’s review indicated that the sizes of the majority of cultured marine phages range from 60 to 100 nm. Børsheim suggested that most of the marine phages in seawater belong to groups that are not cultivable, because the viruses from seawater samples were dominated by 30- to 60-nm particles. Most of our phage isolates fell into the 30- to 60-nm range and appeared to be representatives of Børsheim’s suggested noncultivable group.
A survey of 52 cultured marine phages by Børsheim suggested that there is great variation in burst size; the average marine phage burst size is 185, and burst sizes range from 5 to 610. The burst sizes of our phage isolates fell within this range.
The latent periods of phages nt-1 and nt-6, which were isolated from a salt marsh, are 50 and 60 min, respectively, under optimal conditions (32). However, the latent periods increased to 170 and 120 min, respectively, when the temperate was 10°C below the optimal temperature (33). A phage infecting Vibrio fischeri MJ-1 had a latent period of 25 min (19), while the latent periods for two bacteriophages isolated from the North Sea were 150 and 180 min (6). The latent periods of our phage isolates are comparable to these previously determined latent periods.
It is difficult to compare the genome sizes of our phage isolates with the genome sizes of previously isolated marine phages, because most reports of marine viruses have not characterized the phage nucleic acids. Ackermann and DuBow (1) suggested that nucleic acid type and gross morphology are the most important properties for phage description and classification and that less emphasis should be placed on molecular weight and restriction endonuclease patterns.
The results of dot blot hybridization suggest that the viral DNA was maintained as a prophage inside lysogenized cells. We interpret the hybridization signal of the phage gene probe with the lysogenized host chromosomal DNA as an indication that phage DNA is integrated within the host genome. The hybridization signal was weak compared with the hybridization signal of phage DNA, suggesting that only one copy or a few copies of phage DNA were integrated in the chromosome. Because the size of our probe is only 100 bp, it can hybridize with only a small region of the phage DNA.
Lysogen L-HSIC was spontaneously induced to lytic replication at a high frequency, and more than 106 free phage particles per ml were found in the supernatant of an overnight culture. The hybridization observed in the plasmid DNA fraction may have been present because the phage DNA was maintained as an autonomous plasmid or may have been the result of phage DNA that was injected into the cell but was unable to replicate because of the presence of a vegetative replication repressor produced by the prophage. Lysogens are immune to lytic infection by viruses that are closely related to the prophage but are not resistant to phage DNA injection. The presence of free phage in the culture medium may have created a positive pressure for the maintenance of the lysogens. Wild-type strain HSIC contained a plasmid upon isolation (detected by plasmid prep and gel electrophoresis) (data not shown). The lysogen L-HSIC plasmid DNA preparation may have contained both original plasmid DNA and phage DNA that was in the cell but not in the chromosome. The copy number of phage DNA in the plasmid preparation was apparently higher than the copy number of chromosomal DNA but lower than the copy number of pure phage DNA on a per-weight basis. This explains the greater hybridization observed in the plasmid preparation than in the chromosomal DNA preparation.
The results of this study suggest that temperate phages can be easily found as members of the microbial community. We are currently collecting samples from the Gulf of Mexico for isolation of temperate phage-host systems. Phage isolates from Mamala Bay share many similar properties with other marine phage isolates, while also remaining unique. The interaction of temperate phages and the microbial population in the marine environment may contribute significantly to microbial genetic diversity and composition by conversion and transduction. The indigenous phage-host systems isolated in this study have been used to study marine phage gene transduction (17a).
ACKNOWLEDGMENTS
This research was supported by grants OCE 9502775, OCE 95P00774, and OCE 9115942 from the National Science Foundation. Funds were also provided by the Mamala Bay Study Commission and by a Knight Fellowship and a Gulf Charitable Trust Fellowship to S.C.J.
REFERENCES
- 1.Ackermann H W, DuBow M S. Viruses of prokaryotes. 1. General properties of bacteriophages. Boca Raton, Fla: CRC Press; 1987. [Google Scholar]
- 2.Barnet Y M, Humphrey B. Exopolysaccharide depolymerase induced by Rhizobium bacteriophage. Can J Microbiol. 1975;21:1647–1650. doi: 10.1139/m75-239. [DOI] [PubMed] [Google Scholar]
- 3.Bergh Ø, Børsheim K Y, Bratbak G, Helda M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 4.Børsheim K Y. Native marine bacteriophages. FEMS Microbiol Ecol. 1993;102:141–159. [Google Scholar]
- 5.Bratbak G, Heldal M, Thingstad T F, Riemann B, Haslund O H. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar Ecol Prog Ser. 1992;83:273–280. [Google Scholar]
- 6.Chen P K, Citarella R V, Salazar O, Colwell R R. Properties of two marine bacteriophages. J Bacteriol. 1966;91:1136–1139. doi: 10.1128/jb.91.3.1136-1139.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandekar A M, Modi V V. Interaction between Rhizobium japonicum phage M-1 and its receptor. Can J Microbiol. 1987;24:685–688. doi: 10.1139/m78-115. [DOI] [PubMed] [Google Scholar]
- 8.Freifelder D. Molecular biology. Boston, Mass: Jones and Bartlett, Inc.; 1987. [Google Scholar]
- 9.Frischer M E, Thrumond J M, Paul J H. Natural plasmid transformation in a high-frequency-of-transformation marine Vibrio strain. Appl Environ Microbiol. 1990;56:3439–3444. doi: 10.1128/aem.56.11.3439-3444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrman J A, Suttle C A. Viruses in marine planktonic systems. Oceanography. 1993;6:57–63. [Google Scholar]
- 11.Hidaka T. Isolation of marine bacteriophages from sea water. Bull Jpn Soc Sci Fish. 1971;37:1199–1206. [Google Scholar]
- 12.Hidaka T. Characterization of marine bacteriophages newly isolated. Mem Fac Fish Kagoshima Univ. 1993;22:47–61. [Google Scholar]
- 13.Hidaka T, Shirahama T. Preliminary characterization of a temperate phage system isolated from marine mud. Mem Fac Fish Kagoshima Univ. 1974;23:137–148. [Google Scholar]
- 14.Jacob F, Wollman E L. Induction of phage development in lysogenic bacteria. Cold Spring Harbor Symp Quant Biol. 1953;18:101–121. doi: 10.1101/sqb.1953.018.01.019. [DOI] [PubMed] [Google Scholar]
- 15.Jiang S C, Thurmond J M, Pichard S L, Paul J H. Concentration of microbial populations from aquatic environments by vortex flow filtration. Mar Ecol Prog Ser. 1992;80:101–107. [Google Scholar]
- 16.Jiang S C, Paul J H. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar Ecol Prog Ser. 1994;104:163–172. [Google Scholar]
- 17.Jiang S C, Paul J H. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar Ecol Prog Ser. 1996;142:27–38. [Google Scholar]
- 17a.Jiang, S., and J. H. Paul. Unpublished data.
- 18.Kokjohn T A, Sayler G S, Miller R V. Attachment and replication of Pseudomonas aeruginosa bacteriophages under conditions simulating aquatic environments. J Gen Microbiol. 1991;137:661–666. [Google Scholar]
- 19.Levisohn R, Moreland J, Nelson L H. Isolation and characterization of a generalized transducing phage for the marine luminous bacterium Vibrio fischeri MJ-1. J Gen Microbiol. 1987;133:1577–1582. [Google Scholar]
- 20.Lindberg A A. Bacterial surface carbohydrates and bacteriophage adsorption. In: Sutherland I E, editor. Surface carbohydrates of the prokaryotic cells. London, United Kingdom: Academic Press; 1977. pp. 289–356. [Google Scholar]
- 21.Moebus K. A method for the detection of bacteriophages from ocean water. Helgol Meeresunters. 1980;34:375–391. [Google Scholar]
- 22.Moebus K. Lytic and inhibition responses to bacteriophages among marine bacteria, with special reference to the origin of phage-host systems. Helgol Meeresunters. 1983;36:375–391. [Google Scholar]
- 23.Paul J H. The use of Hoechst dyes 33258 and 33342 for enumeration of attached and pelagic bacteria. Appl Environ Microbiol. 1982;43:939–949. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul J H, Meyers B. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romig W R, Brodesky A M. Isolation and preliminary characterization of bacteriophages for Bacillus subtilis. J Bacteriol. 1961;82:135–141. doi: 10.1128/jb.82.1.135-141.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Staniewski R. Morphology and general characteristics of phage active against Rhizobium. J Basic Microbiol. 1987;27:155–165. [Google Scholar]
- 28.Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi seas. Mar Ecol Prog Ser. 1996;131:287–300. [Google Scholar]
- 29.Torrella F, Morita R Y. Evidence by electron micrographs for a high incidence of bacteriophage particles in the waters of Yaquina Bay, Oregon: ecological and taxonomical implications. Appl Environ Microbiol. 1979;37:774–778. doi: 10.1128/aem.37.4.774-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinbauer M G, Fuks D, Puskaric S, Peduzzi P. Diel, seasonal and depth-related variability of viruses and dissolved DNA in the northern Adriatic Sea. Microb Ecol. 1995;30:25–41. doi: 10.1007/BF00184511. [DOI] [PubMed] [Google Scholar]
- 31.Weiss B D, Capage M A, Kessel M, Benson S A. Isolation and characterization of a generalized transducing phage for Xanthomonas campestris pv. campestris. J Bacteriol. 1994;176:3354–3359. doi: 10.1128/jb.176.11.3354-3359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zachary A. Physiology and ecology of bacteriophages of the marine bacterium Beneckea nariegens: salinity. Appl Environ Microbiol. 1976;31:415–422. doi: 10.1128/aem.31.3.415-422.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zachary A. An ecological study of bacteriophages of Vibrio natriegens. Can J Microbiol. 1978;24:321–324. doi: 10.1139/m78-053. [DOI] [PubMed] [Google Scholar]