Abstract
This article describes the first successful detection of airborne Mycoplasma hyopneumoniae under experimental and field conditions with a new nested PCR assay. Air was sampled with polyethersulfone membranes (pore size, 0.2 μm) mounted in filter holders. Filters were processed by dissolution and direct extraction of DNA for PCR analysis. For the PCR, two nested pairs of oligonucleotide primers were designed by using an M. hyopneumoniae-specific DNA sequence of a repeated gene segment. A nested PCR assay was developed and used to analyze samples collected in eight pig houses where respiratory problems had been common. Air was also sampled from a mycoplasma-free herd. The nested PCR was highly specific and 104 times as sensitive as a one-step PCR. Under field conditions, the sampling system was able to detect airborne M. hyopneumoniae on 80% of farms where acute respiratory disease was present. No airborne M. hyopneumoniae was detected on infected farms without acute cases. The chance of successful detection was increased if air was sampled at several locations within a room and at a lower air humidity.
Mycoplasma hyopneumoniae, the etiologic agent of enzootic pneumonia (EP) in pigs, causes major economic losses in the pig industry worldwide (21). Several countries have established EP-free herds as parts of national pig health schemes (12, 13). However, despite rigid biosecurity measures, many of these herds become reinfected with EP each year. As a consequence of these often-unexplained reinfections, the hypothesis of airborne agent transmission between farms has been tested in a number of field studies. It was demonstrated that the risk of reinfection of EP-free herds was associated with the distance to conventional pig farms in the neighborhood and with the sizes of these farms as well as with the concentration of pigs in the area (11, 24, 26). These risk factors indicate an airborne transmission process. However, the pathogen has never been isolated from the air. Early attempts by Tamàsi (25) were only partially successful. An isolate that looked morphologically similar to M. hyopneumoniae colonies was described, but the identity of the organism remained undetermined.
Accurate diagnosis of EP is essential to prevent infection from spreading. The most commonly used diagnostic methods are serological analyses such as direct or blocking enzyme-linked immunosorbent assay (6, 9, 16, 18, 23) and direct detection of the organism in clinical samples by immunofluorescence (10). In recent years, molecular genetic techniques which have improved sensitivity and specificity have become available (3, 16). The molecular genetic methods benefit from the fact that the microorganisms do not have to be alive at the time of analysis. This makes the tools attractive for use in detection by air sampling techniques, for example, air filtration, which may adversely affect survival.
Air filtration is one of the simplest and cheapest air sampling techniques available for the investigation of bioaerosols (4). It has been used successfully in combination with PCR assays (19). The objective of this project was to establish a highly sensitive and specific nested PCR method and to develop a filtration-based air sampling technique for the detection of M. hyopneumoniae in the air.
MATERIALS AND METHODS
Strain growth conditions and DNA extraction.
The porcine Mycoplasma and Acholeplasma strains used in this study are listed in Table 1. M. hyopneumoniae and Mycoplasma flocculare were grown in Friis medium (8). The other strains were grown in B medium (7). The cells were cultivated until the end of the exponential phase of growth, harvested by centrifugation at 20,000 × g for 20 min, washed three times in TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.5), and resuspended in 1/100 of the original volume of TE buffer. Titration of the viable cells (estimated as CFU per milliliter) was performed by spreading samples of sequential 10-fold dilutions on solid Friis medium (8) and counting the colonies after 10 days of incubation.
TABLE 1.
Porcine Mycoplasma and Acholeplasma strains used in this study and their reaction in the nested PCR assay
| Species | Strain | Sourcea | PCR result
|
||
|---|---|---|---|---|---|
| Outer | Inner | Nested | |||
| M. hyopneumoniae | ATCC 25934 | ATCC (type strain) | + | + | + |
| NCTC10110 | NCTC (type strain) | + | + | + | |
| J (JF 184) | UK | + | + | + | |
| BQ 14 | France | + | +b | + | |
| EP-S 924 | Switzerland | + | +b | + | |
| EP-S 938 | Switzerland | + | + | + | |
| EP-S 939 | Switzerland | + | + | + | |
| EP-S 946 | Switzerland | + | + | + | |
| EP-S 223 | Switzerland | + | + | + | |
| 232 | Camden, NSW, Australia | + | + | + | |
| YZ | Unknown | + | +b | + | |
| Beaufort | Camden, NSW, Australia | + | +b | + | |
| Sue | Camden, NSW, Australia | + | + | + | |
| C173512 | Camden, NSW, Australia | + | + | + | |
| OMZ407 | Camden, NSW, Australia | + | + | + | |
| Hungary 1 | Budapest, Hungary | + | +b | + | |
| M. flocculare | Ms 42 | MRC (type strain) | − | − | − |
| ZH1 | Zürich, Switzerland | − | − | − | |
| ZH2 | Zürich, Switzerland | − | − | − | |
| JF1628 | Cambridge, UK | − | − | − | |
| JF7337 | Cambridge, UK | − | − | − | |
| M. hyorhinis | BTS-7 | MRC (type strain) | − | − | − |
| SEP200 | Stockholm, Sweden | − | − | − | |
| F44 | Stockholm, Sweden | − | − | − | |
| 31-53 | Ottawa, Canada | − | − | − | |
| M. hyosynoviae | S16 | MRC (type strain) | − | − | − |
| M60 | Stockholm, Sweden | − | − | − | |
| A. axanthum | S-743 | MRC (type strain) | − | − | − |
| A. granularum | BTS-39 | MRC (type strain) | − | − | − |
| A. laidlawii | PG8 | MRC (type strain) | − | − | − |
ATCC, American Type Culture Collection (Rockville, Md.); NCTC, National Collection of Type Cultures (London, Great Britain); UK, United Kingdom; NSW, New South Wales; MRC, Mycoplasma Reference Center (Aarhus, Denmark).
The DNA sequence of the 808-bp fragment was determined with the Taq DyeDeoxy Terminator Cycle sequencing kit (Applied Biosystems/Perkin-Elmer Cetus) and oligonucleotides MHP950-2L and MHP950-2R.
In order to obtain pure genomic DNA of mycoplasmal cultures, cells were harvested by centrifugation, washed in TE buffer, and resuspended in 1/10 of the original volume of TE buffer. A volume of 100 μl of resuspended cells (1010 cells/ml in TE buffer) was lysed by addition of 500 μl of GES buffer (5 M guanidium thiocyanate, 100 mM EDTA, 0.5% N-lauroylsarcosine [nastelle sarcosyl]) for 10 min at room temperature, cooled on ice, and then mixed with 250 μl of 7.5 M ammonium acetate, pH 7.7. The lysate was then extracted three times with 500 μl of phenol-chloroform-isoamyl alcohol (49.5:49.5:1) (Fluka Chemicals, Buchs, Switzerland). The DNA was then precipitated by the addition of 0.7 volume of isopropanol and collected by centrifugation at 10,000 × g for 15 min at 4°C in an Eppendorf centrifuge. The DNA pellet was washed three times with 80% ethanol, dried, and resuspended in 100 μl of TE buffer. The DNA concentration was determined spectrophotometrically with a model 2105 GeneQuantII (Pharmacia Biotech, Uppsala, Sweden).
Air sampling system.
Air was sampled with polyethersulfone membranes (47-mm diameter) with a pore size of 0.2 μm (Supor200; Gelman Sciences, Ann Arbor, Mich.) and mounted in filter holders (Schleicher & Schuell GmbH, Dassel, Germany). The air was pumped at a rate of 18.3 to 20.0 liters/min with a vacuum pressure pump (Millipore, Bedford, Mass.). The airflow in the filter system was controlled with an in-line rotameter (Messerli Messtechnik, Riehen, Switzerland).
In order to determine the sensitivity of detection of mycoplasmas on the filters, we filtered 1-ml samples of a consecutively 10-fold-diluted culture of M. hyopneumoniae NCTC10110 to obtain samples with concentrations ranging from 106 to 0 viable cells/ml.
An experimental aerosol of M. hyopneumoniae was generated by nebulizing a formaldehyde-inactivated culture in a closed 0.54-m3 chamber with a commercial nebulizer (DP10; DPMedical, Medela, Baar, Switzerland) with a vaporization rate of approximately 2 ml/min. The plume was sampled for 10 s and 1 and 6 min with the sampling system described above. The experimental setup captured approximately 1/10 of the volume of the evaporated material per time unit.
Air sample processing for PCR assay.
The filters from the air samplings and the artificially contaminated filters were thoroughly dried, folded, and dissolved in 5 ml of chloroform by vortexing in a 15-ml Falcon tube (catalog no. 2059; Becton Dickinson, Lincoln Park, N.J.). Drying was necessary to ensure complete dissolution of the polyethersulfone membranes. The DNA was then extracted by the addition of 3.3 ml of TE buffer and shaking for 10 min at room temperature. Phase separation was achieved by centrifugation for 10 min at 10,000 × g. After recovery of the aqueous phase, the chloroform phase was extracted a second time with 2 ml of TE buffer. Subsequently the aqueous phases were combined and extracted with 5 ml of phenol-chloroform-isoamyl alcohol (49.5:49.5:1). After phase separation by centrifugation for 10 min at 10,000 × g, the aqueous phase was recovered, mixed with 8 ml of chilled ethanol and 400 μl of 3M sodium (pH 5.5), and cooled at −20°C for 10 min to precipitate the DNA. The precipitated DNA was recovered by centrifugation for 20 min at 10,000 × g, dried, and resuspended in 50 μl of TE buffer.
Design of specific oligonucleotide primers and PCRs.
The oligonucleotide primers used in the PCRs are based on the sequence data for the 1,023-bp repeated element MHYP1-03-950 (GenBank/EMBL accession no. AF004388) (7a). This sequence has been shown to be specific for the species M. hyopneumoniae and is present at one to seven copies per chromosome. Seven copies of MHYP1-03-950 were shown to be present in the M. hyopneumoniae type strain, NCTC10110. The repeated element MHYP1-03-950 does not contain sequences typical for insertion sequences or known multicopy gene families. Southern blot analysis of genomic DNA of M. flocculare, Mycoplasma hyorhinis, Mycoplasma hyosynoviae, and Acholeplasma laidlawii with a labelled probe of MHYP1-03-950 did not show hybridization signals under low-stringency conditions (7a). Two nested pairs of oligonucleotide primers (Table 2) were designed with the primer analysis software OLIGO 4 (National Biosciences, Plymouth, Minn.). The outer primer pair (MHP950-1L and MHP950-1R) in the first reaction gave an amplification product of 913 bp, and the inner primer pair (MHP950-2L and MHP950-2R) in the second reaction gave a product of 808 bp. Both primer pairs were designed to be used at an annealing temperature of 52°C for practical reasons.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer | Sequence | Location (bp)b |
|---|---|---|
| MHP950-1L | AGGAACACCATCGCGATTTTTA | 46–67 |
| MHP950-1R | ATAAAAATGGCATTCCTTTTCA | 958–937 |
| MHP950-2L | CCCTTTGTCTTAATTTTTGAA | 102–123 |
| MHP950-2R | GCCGATTCTAGTACCCTAATCC | 909–888 |
All primers were annealed at 52°C.
Based on nucleotide sequence AF004388.
The PCRs were carried out in a DNA thermal cycler (GeneAmp 9600; Perkin-Elmer Cetus) in 50 μl of a reaction mixture (10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl, 0.005% Tween 20, 0.005% Nonidet P-40 detergent, a 170 μM concentration of each deoxynucleoside triphosphate, 0.25 μM [each] forward and reverse primers, 1.25 U of Taq polymerase) with 5 μl of extracted DNA from the filters or 2 ng of purified DNA (17) from cultured mycoplasmas as a template. The amplification consisted of 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min. In the nested PCR assay, the first reaction was done with primers MHP950-1L and MHP950-1R. The second (nested) PCR was performed with primers MHP950-2L and MHP950-2R, with 1 μl of amplification product from the first reaction as a template. The universal primers 16SUNI-L and 16SUNI-R, which amplify the 16S rRNA gene rrs (15), were used in control PCRs under the same amplification conditions. The PCR amplification products were analyzed by gel electrophoresis on 0.7% agarose gels and visualized after staining with ethidium bromide on a UV transilluminator according to standard protocols (2). Bacteriophage λ DNA digested with HindIII was used as molecular mass standard. The sensitivity of the method was determined by the analysis of artificially contaminated filters with 0.1-ml samples of a culture of M. hyopneumoniae NCTC10110 at 108 viable cells/ml which was sequentially diluted from 10 to 1010 times in culture medium. The same extraction protocols as for filters from the air sampling were used.
DNA sequence analysis of the 808-bp PCR product of the inner reaction was done with the PRISM Ready Reaction DyeDeoxy Terminator Cycle sequencing kit (Applied Biosystems/Perkin-Elmer Cetus, Norwalk, Conn.) with oligonucleotides MHP950-2L and MHP950-2R in an ABI Prism 310 genetic analyzer (Applied Biosystems/Perkin-Elmer Cetus).
Field sampling.
Air was sampled in pig rooms on seven commercial farms, in one pig room in an animal clinic, and in one room in a research facility (specific-pathogen-free [SPF] facility, negative control). Farms where EP was present were either SPF farms with a laboratory-confirmed reinfection with M. hyopneumoniae but whose pigs showed no acute clinical signs (three farms) or conventional farms whose pigs had acute respiratory problems diagnosed by the local veterinarian (four farms). A detailed description of the farms and rooms is given in Table 3.
TABLE 3.
Description of Swiss pig farms and rooms sampled
| Farm no. | Room no. | Air vol (m3/pig) | Air temp (°C) | % Relative humidity | No. of coughs/100 pigs/10 min | Age of pigs (wk) | Description of facility |
|---|---|---|---|---|---|---|---|
| 1 | 1a | 16.25 | 21.0 | 60 | 25.0 | (Mixed) | Pig facility at animal hospital |
| 2 | 1 | 2.31 | 18.6 | 84 | 0 | 11 | Integrated, SPF farm reinfected with EP |
| 3 | 1 | 6.00 | 20.0 | 65 | 0 | (Mixed) | SPF research unit (negative control) |
| 4 | 1a | 4.88 | 20.2 | 91 | 4.0 | 19–22 | Growing farm that purchased pigs from several sources; acute respiratory problems in pigs |
| 2 | 2.95 | NMb | NM | 3.0 | 15 | ||
| 3a | 2.92 | 18.8 | 75 | 17.0 | 17 | ||
| 4a | 2.83 | NM | NM | 10.0 | 14–16 | ||
| 5 | 1 | 2.00 | 23.2 | 85 | 7.0 | 16 | Integrated, SPF farm reinfected with EP |
| 6 | 1 | 3.47 | 20.4 | 75 | 4.0 | 16–18 | Integrated, SPF farm reinfected with EP |
| 7 | 1a | 2.44 | 21.2 | 72 | 13.0 | 12–15 | Integrated farm; acute respiratory problems in pigs |
| 8 | 1a | 2.90 | 21.4 | 55 | 10.0 | 12–16 | Growing farm; acute respiratory problems in pigs |
| 9 | 1 | 1.79 | 20.0 | 66 | 16.0 | 14 | Integrated farm; acute respiratory problems in pigs |
Room where positive air samples were collected.
NM, not measured.
On each farm, one to four rooms housing growing pigs were chosen on the basis of clinical signs (coughing) displayed within. The following parameters were recorded for each room: air volume per animal, air temperature and humidity (aspiration psychrometer; Haenni & Co. Ltd., Jegenstorf, Switzerland), clinical signs of EP (coughing), ages of animals, air quality (assessed subjectively), type of feed, housing system, and ventilation system. Coughs were counted for 10 min and then the counts were extrapolated to yield the number of coughs per 100 pigs per 10 min to make the figures comparable between rooms.
Each filter holder was positioned outside the pen, 30 to 50 cm above ground level and 10 to 20 cm from the animals. Air was pumped through the filter for 1, 10, 60, or 100 min. The sampled air volume was 18.3 to 20.0, 183 to 200, 1,200, or 1,830 to 2,000 liters, respectively. After sampling, the filter membranes were transferred into petri dishes for transport.
Three different sampling protocols were developed.
(i) Protocol 1.
Different sampling durations (1, 10, and 100 min) at one location in the room were used (farms 1 and 2).
(ii) Protocol 2:
Six samples, each taken over 10 min, were collected at six different locations in one room (farms 4, 5, and 6).
(iii) Protocol 3:
A cumulative sample was collected over 60 min at different locations in the room with one filter only (farms 4, 7, 8, and 9).
Additionally, on farms 4, 7, 8, and 9, dust samples were collected in the same room where air samples were collected. Dust was scratched from surfaces and ventilation ducts at different locations in the room and transferred into a sterile plastic container. From each sample, 100 mg of dust was suspended in 1 ml of TE buffer and subsequently extracted by the same protocols used for air sampling with the filters.
Statistical analysis.
Data were analyzed with the statistical package SPSS (version 7.5; SPSS Inc., Chicago, Ill.). To test for differences among groups, the χ2 test and the Mann-Whitney U test were used. Logistic regression techniques were used to explore farm and room variables influencing the PCR outcome.
RESULTS
Specificity and sensitivity of the nested PCR assay.
In order to analyze the specificities of the individual PCRs and in particular of the nested PCR assay, 2 ng of purified DNA from 15 different strains of M. hyopneumoniae, including the type strain, NCTC10110 (= ATCC 25934), as well as DNA from several closely related porcine Mycoplasma and Acholeplasma species, was used as a template in the reactions (Table 1). All strains of M. hyopneumoniae showed the expected DNA fragments in the individual PCRs and in the nested PCR, while none of the other Mycoplasma or Acholeplasma species showed any amplification product in either the single or the nested PCR assay (Table 1). DNA sequence analysis of the 808-bp fragment obtained from the inner PCR of a few selected strains with widely varying origins (Table 1) as well as from two field samplings revealed a 99.7% identity of the nucleotides to those in the corresponding fragment MHYP1-03-950 from the type strain, NCTC10110 (sequence AF004388). The fragments from strains S924, Hungary 1, and Beaufort, as well as those from the field samples, differed from sequence AF004388 at two nucleotide positions that were different for each strain, while strains BQ 14 and YZ differed from sequence AF004388 at the same three nucleotide positions. In control PCRs with the universal prokaryotic primers 16SUNI-L and 16SUNI-R, a 1.4-kb PCR product from the 16S rRNA gene rrs was amplified from all Mycoplasma and Acholeplasma species tested.
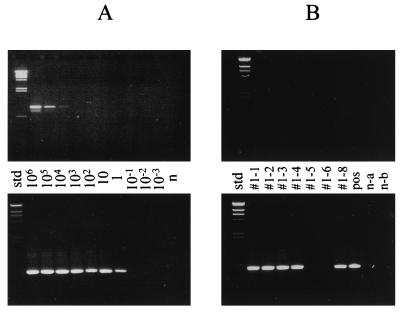
In order to analyze the suitability of the method for air sampling, nebulized, formaldehyde-inactivated M. hyopneumoniae cells were sampled in order to obtain filters with approximately 700, 4,000, and 24,000 CFU of M. hyopneumoniae. Nested PCR of the filter extracts (see Materials and Methods) revealed the presence of M. hyopneumoniae on filters from all three samplings. For analysis of the sensitivity, different filters artificially contaminated with various amounts of M. hyopneumoniae NCTC10110 containing seven copies of the target sequence were prepared as described above and subjected to nested PCR. Figure 1 shows the results of the electrophoretic analysis of the products of the first and second PCRs in the nested PCR assay. The detection limit of the first PCR with the primers MHP950-1L and MHP950-1R is in the range of 104 viable cells/filter, yielding a clearly visible 913-bp band. However, when the PCR products of this first reaction were used as templates for the second reaction in a nested PCR, the 808-bp fragment from the second step was clearly visible for samples containing as few as one viable cell/ml. No signal was detected for filters with averages of 10−1 viable cells/ml or fewer.
FIG. 1.
PCR analysis of air sampling with polyethersulfone membranes. The PCR amplification products from the first PCR, with primers MHP950-1L and MHP950-1R (upper photographs), and from the second PCR, with primers MHP950-2L and MHP950-2R (lower photographs), were analyzed by agarose gel electrophoresis on 0.7% gels. (A) Filters contaminated experimentally with known numbers of M. hyopneumoniae cells. The numbers between the two photographs are the average concentrations of viable cells/filter in the different samples. n, negative control sample; std, standard. Note that in some cases an additional PCR amplification product that was smaller than the 912-bp fragment was detected in small amounts in the outer PCR when high numbers of target organisms were used. This smaller PCR fragment, however, was not detected when purified M. hyopneumoniae genomic DNA was used. (B) Filters from field sampling on a farm with pigs with respiratory problems. Lanes #1-1, #1-3, #1-4, and #1-6, 18-liter samples; lanes #1-2 and #1-5, 200-liter samples; lane #1-8, 2,000-liter sample; lane n-a, negative control for buffers; lane n-b, negative control filter. std, standard; pos, positive control.
Field sampling.
The conditions under which air was sampled are described in Table 3, and PCR results are shown in Table 4. For farms where acute respiratory problems were present, the overall proportion of air filters with a positive PCR result was 53.6%, while none of the filters from SPF farms where reinfections occurred tested positive in the assay. Of the farms where acute respiratory problems were present, 80% had at least one positive filter result. All negative control samples tested negative.
TABLE 4.
Results for air samples analyzed with a nested PCR to detect DNA of M. hyopneumoniae
| Sample type and vol (liters) | No. positive/no. tested (%) in:
|
Total | No. of negative control samples positive/no. tested (%) | |
|---|---|---|---|---|
| Reinfected SPF herd without acute problems | Herd with acute respiratory problems | |||
| Air | ||||
| 18.3–20.0 | 0/1 (0) | 4/5 (80.0) | 6 | |
| 183.0–200.0 | 0/13 (0) | 7/16 (43.8) | 29 | 0/5 (0) |
| >1,000.0 | 0/2 (0) | 1/3 (33.3) | 5 | 0/1 (0) |
| Cumulative (1,098–1,200)a | 0/0 (0) | 3/4 (75.0) | 4 | |
| Total (by farm status) | 0/16 (0)c | 15/28 (53.6)d | 44 | |
| Dust | 0/0 (0) | 1/4 (25.0)b | 4 | |
The same filter was used at six different locations in one room, sampling 183 to 200 liters at each.
The result for one dust sample was not interpretable.
Zero percent of farms were positive.
Eighty percent of farms were positive.
It was observed that larger sample volumes were not necessarily associated with positive PCR results. The highest proportion of positive results was obtained with a short sampling period (1 min; 18.3 to 20.0 liters) and with the cumulative sampling protocol. In order to assess the potential inhibitory effect on the PCR of nontarget organisms and other substances that could have been sampled when collecting large volumes, we mixed equal volumes of extracts from filters used for long samplings (200 and 1,000 liters) and of extracts from filters that gave positive results and resubmitted the mixture to PCR. These samples also showed positive results without reduction of the signal intensity.
The results for the six samples collected at different locations in one room (protocol 2) showed that the distribution of mycoplasmas within a room did not seem to be uniform (data not shown). Also, air samples with positive PCR results and pens where coughing pigs were observed during the sampling session were not clearly associated.
Because no samples from reinfected SPF herds were positive, further analyses of factors influencing a positive PCR result were performed only for farms with pigs that displayed acute respiratory problems (28 filters). The likelihood of obtaining a positive air sample was influenced at the univariate level by the coughing intensity, ages of the pigs, and air humidity (Table 5). Positive PCR results were more likely if a sample was taken in a room with a higher coughing intensity, younger pigs, and a lower relative humidity. The likelihood of a positive test result was not significantly associated with the amount of air filtered, although the filters that yielded positive results had a slightly higher mean volume. In a multivariate logistic regression model with stepwise variable selection, only air humidity was marginally significant, with a P value of 0.07 (odds ratio, 0.93; 95% confidence interval, 0.84 to 1.01).
TABLE 5.
Factors associated with the outcome of a nested PCR assay to detect DNA from M. hyopneumoniae in air samplesa
| Result from filter | No. of coughs/100 pigs/10 min | % Relative humidity | Age of pigs (wk) | Filter vol (liters) |
|---|---|---|---|---|
| Negative (n = 13) | ||||
| Median | 10.0 | 75.0 | 14.0 | 200.0 |
| Mean | 9.62 | 75.69 | 15.38 | 416.92 |
| SE | 2.22 | 2.88 | 0.59 | 124.57 |
| Positive (n = 15) | ||||
| Median | 13.0 | 72.0 | 13.0 | 200.0 |
| Mean | 14.87 | 68.33 | 14.40 | 465.20 |
| SE | 2.43 | 2.54 | 0.57 | 156.16 |
P values obtained by comparing the means in the Mann-Whitney U test (exact, two-tailed) were 0.14, 0.06, 0.12, and 0.62 for the number of coughs/100 pigs/10 min, the relative humidity, the age of the pigs, and the filter volume, respectively.
Of four dust samples, three were negative and one yielded a smear on the agarose gel, which was not investigated further and which was not interpretable (Table 4).
DISCUSSION
This study documents the first successful attempt to definitely identify M. hyopneumoniae in samples from air in the vicinity of infected pigs. With an air sampling system using polyethersulfone membranes with a pore size of 0.2 μm, a simple method to recover DNA from the filters, and a nested PCR assay, it was possible to detect DNA of M. hyopneumoniae under both experimental and field conditions.
PCR amplification-based tests for the detection of microorganisms are generally highly sensitive, allowing in principle a single chromosome equivalent of a cell to be detected when pure DNA is used as a template. In practice, however, this sensitivity is only very rarely achieved due to inhibitory substances in the samples to be tested, dilutions of the original samples by preparative procedures, or loss of DNA during laborious extraction procedures. Typically for mycoplasmas, the sensitivity is on the order of 10,000 cells per reaction (20). This sensitivity of a one-step PCR is, however, not enough for the detection of microorganisms in the air. In order to increase the sensitivity, nested (two-step) PCRs have been developed for the detection of various microorganisms, including several mycoplasmas (20). We have developed a highly sensitive nested PCR method based on the DNA sequence of a repeated gene segment of M. hyopneumoniae and used it to detect M. hyopneumoniae on filters used for air samplings. The method includes rapid dissolution of polyethersulfone filters and simultaneous extraction of DNA to be used for the nested PCR. Using filters that contained known numbers of M. hyopneumoniae cells (titrated as live cells), we observed a sensitivity of 1 viable cell/filter with the nested PCR, compared with 104 viable cells/filter with one-step PCR. We had made a similar observation with a nested PCR used for the direct detection of Mycoplasma mycoides subsp. mycoides SC in clinical samples (17). Taking into account the dilution which is due to sample preparation and the small volume used for the first PCR step, the nested PCR was capable of detecting less than one viable cell per reaction. This is possible because prokaryotes contain more than one chromosome equivalent under exponential growth conditions. The gene target used for the PCR is the repetitive element MHYP1-03-950, one to seven copies of which are present in each chromosome of M. hyopneumoniae. This element is well conserved within the species M. hyopneumoniae, as shown by DNA sequence analysis, and it is absent from genetically and taxonomically related mycoplasmas. This explains the high detection limit and the high specificity of the nested PCR. Detection can be lower than 1 viable cell per reaction, which is of particular advantage in air sampling.
The field sampling showed that air sampling is most successful on farms with acute clinical problems (80% of these had at least one positive air sample). On farms with a higher rate of chronic disease associated with a lower prevalence of clinical signs, no M. hyopneumoniae DNA was found in air samples. This indicates a low concentration of the agent under these circumstances. It is well known that M. hyopneumoniae cannot be isolated consistently easily during the entire course of the disease (14, 23). Once the lung lesions start to regress at approximately 9 weeks after infection (14), the isolation of mycoplasmas becomes difficult as their concentration in tissue declines. It is therefore important to obtain samples from the right age group of pigs. In Switzerland, farmers wean pigs at the age of 4 to 6 weeks. At this stage, pigs will become infected if kept on a farm with EP problems, and the best time to sample for airborne mycoplasmas would be 6 to 8 weeks later (14), which is at an age of 10 to 14 weeks. In this study, success in isolating M. hyopneumoniae DNA from an air sample was also associated with the ages of the pigs. It appears that obtaining samples for somewhat younger pigs would have been advantageous. Also, in future studies, an assessment of the infection status of the pigs at the time of air sampling (for example, by analyzing nasal swabs) would be desirable (16, 23).
Coughing is considered the most reliable clinical indicator of EP infection (22). Coughing starts about 2 weeks after infection and peaks after 5 weeks before gradually declining (14, 23). Because mycoplasmas become airborne through coughing, the concentration of infectious aerosol particles will also be higher when coughing is most prevalent. This is consistent with the results of this study, in which more positive samples were obtained in rooms with a lot of coughing pigs.
The results also showed that the volume of air sampled is not the most important factor in the sampling protocol. Positive results were obtained with volumes as low as 20.0 liters, while some samples of >1,000 liters were negative. Although we could exclude in our assay an inhibitory effect on PCR amplifications due to samplings of large volumes, it has to be noted that samplings of large volumes, in particular from highly contaminated air, could give false-negative results due to the inhibitory action on PCR by nontarget DNA and other substances (1). The result therefore is probably related to the fact that the distribution of airborne M. hyopneumoniae is not uniform within a pig room. The complex air circulation patterns within a pig room make it hard to predict where positive samples may be obtained. It has therefore been suggested that air samples should be collected at a number of different sites within an animal room in order to obtain a representative result (27). The cumulative sampling protocol used in this study seems to be the most suitable approach in this situation.
In this study, air humidity was an influential factor as to whether infectious aerosols were detected in a pig room, with lower humidity being associated with positive sampling results. The relative humidity of the air influences particle aggregation and net water flow and therefore also particle size. The more hygroscopic a particle, the larger it gets in a humid environment and the higher its sedimentation rate (5). In a drier environment, particles will thus remain airborne over a longer time period, which increases the airborne particle concentration and the chance of particle capture in air filters.
The alternative method of sampling dust instead of air was not successful. Only one of four dust samples was suspected of being positive in the PCR assay. Dust samples may contain organic inhibitors which have a negative effect on the PCR assay.
The results of this study using a nested PCR assay strongly support the hypothesis of airborne transmission of M. hyopneumoniae. However, questions about the potential survival of airborne M. hyopneumoniae and maximum travel distances between farms remain to be investigated.
ACKNOWLEDGMENTS
We are thankful to Rudolf Meier for sharing with us his extensive air sampling experience and for making air sampling equipment available. We also thank Lazlo Stipkovits, Budapest, Hungary, for providing cultures of M. hyorhinis, M. hyosynoviae, Acholeplasma axanthum, Acholeplasma granularum, and A. laidlawii; Margrit Krawinkler for cultures of M. hyopneumoniae and M. flocculare; Marylin Kobisch, CNEVA, Ploufragan, France, and Steven Djordjevic, Camden, New South Wales, Australia, for M. hyopneumoniae strains; and Roger S. Morris for the critical review of the field sampling design. We thank Yvonne Schlatter for technical help and Marcus Spallek for helping with the field work.
This project was funded by the Swiss National Science Foundation (grant 823B-040072) and supported by the Institute of Veterinary Bacteriology, University of Berne.
REFERENCES
- 1.Alvarez A J, Buttner M P, Stetzenbach L D. PCR for bioaerosol monitoring: sensitivity and environmental interference. Appl Environ Microbiol. 1995;61:3639–3644. doi: 10.1128/aem.61.10.3639-3644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1990. [Google Scholar]
- 3.Blanchard B, Kobisch M, Bove J M, Saillard C. Polymerase chain reaction for Mycoplasma hyopneumoniae detection in tracheobronchiolar washings from pigs. Mol Cell Probes. 1996;10:15–22. doi: 10.1006/mcpr.1996.0003. [DOI] [PubMed] [Google Scholar]
- 4.Cox C S. The aerobiological pathway of microorganisms. Chichester, United Kingdom: John Wiley & Sons; 1987. [Google Scholar]
- 5.Cox C S. Physical aspects of bioaerosol particles. In: Cox C S, Wathes C M, editors. Bioaerosol handbook—1995. Boca Raton, Fla: CRC Press Lewis Publishers; 1995. pp. 15–25. [Google Scholar]
- 6.Djordjevic S P, Eamens G J, Romalis L F, Saunders M M. An improved enzyme linked immunosorbent assay (ELISA) for the detection of porcine serum antibodies against Mycoplasma hyopneumoniae. Vet Microbiol. 1994;39:261–273. doi: 10.1016/0378-1135(94)90163-5. [DOI] [PubMed] [Google Scholar]
- 7.Ernø H, Stipkovits L. Bovine mycoplasmas: cultural and biochemical studies. Acta Vet Scand. 1973;14:436–449. doi: 10.1186/BF03547431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Frey, J., and S. Djordjevic. Unpublished data.
- 8.Friis N F. Sensitivity of Mycoplasma suipneumoniae to penicillin-G. Acta Vet Scand. 1971;12:120–121. [PubMed] [Google Scholar]
- 9.Futo S, Seto Y, Okada M, Sato S, Suzuki T, Kawai K, Imada Y, Mori Y. Recombinant 46-kilodalton surface antigen (P46) of Mycoplasma hyopneumoniae expressed in Escherichia coli can be used for early specific diagnosis of mycoplasmal pneumonia of swine by enzyme-linked immunosorbent assay. J Clin Microbiol. 1995;33:680–683. doi: 10.1128/jcm.33.3.680-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giger T, Bruggmann S, Nicolet J. Immunologische Methoden zum Nachweis von Mycoplasma suipneumoniae in Gefrierschnitten und Bronchialabstrichen. Schweiz Arch Tierheilkd. 1977;119:125–134. [PubMed] [Google Scholar]
- 11.Goodwin R F W. Apparent re-infection of enzootic-pneumonia-free pig herds: search for possible causes. Vet Rec. 1985;116:690–694. doi: 10.1136/vr.116.26.690. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin R F W, Whittlestone P. The detection of enzootic pneumonia in pig herds. I. Eight years general experience with a pilot control scheme. Vet Rec. 1967;81:643–647. doi: 10.1136/vr.81.25.643. [DOI] [PubMed] [Google Scholar]
- 13.Keller H. Proceedings of the 10th International Pig Veterinary Society Congress 1988. 1988. The Swiss Pig Health Service (PHS) p. 334. [Google Scholar]
- 14.Kobisch M, Blanchard B, Le Potier M F. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and resistance to reinfection. Vet Res. 1993;24:67–77. [PubMed] [Google Scholar]
- 15.Kuhnert P, Capaul S E, Nicolet J, Frey J. Phylogenetic positions of Clostridium chauvoei and Clostridium septicum based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1996;46:1174–1176. doi: 10.1099/00207713-46-4-1174. [DOI] [PubMed] [Google Scholar]
- 16.Mattsson J G, Bergström K, Wallgren P, Johansson K-E. Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene. J Clin Microbiol. 1995;33:893–897. doi: 10.1128/jcm.33.4.893-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miserez R, Pilloud T, Cheng X, Nicolet J, Griot C, Frey J. Development of a sensitive nested PCR method for the specific detection of Mycoplasma mycoides subsp. mycoides SC. Mol Cell Probes. 1997;11:103–111. doi: 10.1006/mcpr.1996.0088. [DOI] [PubMed] [Google Scholar]
- 18.Nicolet J, Paroz P, Bruggmann S. Tween 20 soluble proteins of Mycoplasma hyopneumoniae as antigen for an enzyme linked immunosorbent assay. Res Vet Sci. 1980;29:305–309. [PubMed] [Google Scholar]
- 19.Olsson M, Sukura A, Lindberg L-A, Linder E. Detection of Pneumocystis carinii DNA by filtration of air. Scand J Infect Dis. 1996;28:279–282. doi: 10.3109/00365549609027173. [DOI] [PubMed] [Google Scholar]
- 20.Razin S. DNA probes and PCR in diagnosis of mycoplasma infections. Mol Cell Probes. 1994;8:497–511. doi: 10.1006/mcpr.1994.1071. [DOI] [PubMed] [Google Scholar]
- 21.Ross R F. Mycoplasmal diseases. In: Leman A D, Straw B E, Mengeling W L, D’Allaire S, Taylor D J, editors. Diseases of swine. 7th ed. Ames: Iowa State University Press; 1992. pp. 537–551. [Google Scholar]
- 22.Sørensen V, Barfod K, Feld N C, Vraa-Andersen L. Application of enzyme-linked immunosorbent assay for the surveillance of Mycoplasma hyopneumoniae infection in pigs. Rev Sci Tech Off Int Epizoot. 1993;12:593–604. doi: 10.20506/rst.12.2.699. [DOI] [PubMed] [Google Scholar]
- 23.Sørensen V, Ahrens P, Barfod K, Feenstra A A, Feld N C, Friis N F, Billehansen V, Jensen N E, Pedersen M W. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and evaluation of four diagnostic assays. Vet Microbiol. 1997;54:23–34. doi: 10.1016/s0378-1135(96)01266-7. [DOI] [PubMed] [Google Scholar]
- 24.Stärk K D C, Keller H, Eggenberger E. Risk factors for the reinfection of specific pathogen free pig breeding herds with enzootic pneumonia. Vet Rec. 1992;131:532–535. [PubMed] [Google Scholar]
- 25.Tamàsi G. Mycoplasma isolation from the air. In: Hers J F P, Winkler K C, editors. Airborne transmission and airborne infection. Utrecht, The Netherlands: Oosthoek Publishing Company; 1973. pp. 68–71. [Google Scholar]
- 26.Thomsen B L, Jorsal S E, Andersen S, Willeberg P. The Cox regression model applied to risk factor analysis of infections in the breeding and multiplying herds in the Danish SPF system. Prev Vet Med. 1992;12:287–297. [Google Scholar]
- 27.Wathes C M, Randall J M, editors. Aerosol sampling in animal houses. Proceedings of a workshop held at the University of Bristol, 26–29 July, 1988. EU report 11877. Luxembourg, Luxembourg: Commission of the European Communities; 1989. [Google Scholar]