Abstract
A low-specificity l-threonine aldolase (l-TA) gene from Pseudomonas sp. strain NCIMB 10558 was cloned and sequenced. The gene contains an open reading frame consisting of 1,041 nucleotides corresponding to 346 amino acid residues. The gene was overexpressed in Escherichia coli cells, and the recombinant enzyme was purified and characterized. The enzyme, requiring pyridoxal 5′-phosphate as a coenzyme, is strictly l specific at the α position, whereas it cannot distinguish between threo and erythro forms at the β position. In addition to threonine, the enzyme also acts on various other l-β-hydroxy-α-amino acids, including l-β-3,4-dihydroxyphenylserine, l-β-3,4-methylenedioxyphenylserine, and l-β-phenylserine. The predicted amino acid sequence displayed less than 20% identity with those of low-specificity l-TA from Saccharomyces cerevisiae, l-allo-threonine aldolase from Aeromonas jandaei, and four relevant hypothetical proteins from other microorganisms. However, lysine 207 of low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558 was found to be completely conserved in these proteins. Site-directed mutagenesis experiments showed that substitution of Lys207 with Ala or Arg resulted in a significant loss of enzyme activity, with the corresponding disappearance of the absorption maximum at 420 nm. Thus, Lys207 of the l-TA probably functions as an essential catalytic residue, forming an internal Schiff base with the pyridoxal 5′-phosphate of the enzyme to catalyze the reversible aldol reaction.
β-Hydroxy-α-amino acids constitute an important class of compounds. They are natural products in their own right and are components of a range of antibiotics, for example, cyclosporin A, lysobactin, and vancomycin (30) and bouvardin and deoxybouvardin (6). 4-Hydroxy-l-threonine is a precursor of rizobitoxine, a potent inhibitor of pyridoxal 5′-phosphate (PLP)-dependent enzymes (32). 3,4,5-Trihydroxyl-l-aminopentanoic acid is a key component of polyoxins (32). l-threo-3,4-Dihydroxyphenylserine is a new drug for Parkinson’s disease therapy (13). However, the industrial production of β-hydroxy-α-amino acids has been limited to chemical synthesis processes, which need multiple steps to isolate the four isomers (l-threo form, d-threo form, l-erythro form, and d-erythro form). Threonine aldolase (EC 4.1.2.5), which stereospecifically catalyzes the retro-aldol cleavage of threonine, is a potentially useful catalyst for the synthesis of substituted amino acids from aldehyde and glycine (27, 31, 32).
Two different types of threonine aldolases are known so far. l-allo-Threonine aldolase (l-allo-TA), isolated and purified from Aeromonas jandaei DK-39 (8), stereospecifically catalyzes the reversible interconversion of l-allo-threonine and glycine. Low-specificity l-threonine aldolase (l-TA) catalyzes the cleavage of both l-threonine and l-allo-threonine to glycine and acetaldehyde, as well as the reverse reaction, aldol condensation. The enzymes have been purified and characterized from Candida humicola (9, 34) and Saccharomyces cerevisiae (12). Low-specificity l-TA activity has also been shown to exist in mammals (7, 23, 26) and a variety of other microbial species (2, 4, 17, 35). The enzyme is physiologically important for the synthesis of cellular glycine in yeast (12, 15, 16). Threonine aldolases with distinct stereospecificities are ideal targets for enzymology studies on structural and functional relationships. However, information on the primary structures of threonine aldolases was limited to our recent studies (11, 12). The construction of an overproduction system for threonine aldolase will be indispensable for the industrial biosyntheses of β-hydroxy-α-amino acids.
The present work focuses on the cloning, sequencing, and overexpression in Escherichia coli cells of the low-specificity l-TA gene from Pseudomonas sp. strain NCIMB 10558, the purification and characterization of the recombinant enzyme, and the identification of the active-site lysine residue of the enzyme by site-directed mutagenesis. Evidence is presented that Lys207 of low-specificity l-TA probably functions as a catalytic residue, forming an internal Schiff base with the PLP of the enzyme to catalyze the reversible aldol reaction. This is the first report showing a purified enzyme with l-β-3,4-dihydroxyphenylserine aldolase and l-β-3,4-methylenedioxyphenylserine aldolase activities, providing a new route for the industrial production of these important unnatural amino acids.
MATERIALS AND METHODS
Materials.
DEAE-Toyopearl 650M and Butyl-Toyopearl 650M were purchased from Tosoh (Tokyo, Japan); HiLoad Superdex 200 was obtained from Pharmacia Biotech (Uppsala, Sweden). PLP was obtained from Nacalai Tesque (Kyoto, Japan). dl-erythro-Phenylserine was a generous gift from Hideyuki Hayashi and Hiroyuki Kagamiyama, Department of Biochemistry, Osaka Medical College, Osaka, Japan. dl-β-3,4-Methylenedioxyphenylserine was prepared according to the method of Ohashi et al. (19). The other chemicals were all analytical grade.
Bacterial strains, plasmids, and culture conditions.
Pseudomonas sp. strain NCIMB 10558 was used as the source of chromosomal DNA (2). E. coli GS245 (pheA905 araD139 lacU169 ΔglyA::mu strA thi-1) (a generous gift from George V. Stauffer, University of Iowa, Iowa City) is a glycine-auxotrophic strain used as a host for gene cloning. E. coli XL1-Blue MRF′ (recA1 thi endA1 supE44 gyrA46 relA1 hsdR17 lac/F′ [proAB+ lacIq lacZΔM15::Tn10] Tetr (Toyobo, Osaka, Japan) was used for overexpression of the low-specificity l-TA gene. E. coli CJ236 (dut-1 ung-1 thi-1 relA1/pCJ105 F′ Camr) (Takara Shuzo, Kyoto, Japan) was used for the generation of a uracil-containing single-stranded DNA for site-directed mutagenesis. E. coli BMH71-18mutS [Δ(lac-proAB) supE thi mutS215::Tn10 Tetr/F′ traD36 proAB+ lacIq lacZΔM15] (Takara Shuzo) was used as a host strain for site-directed mutagenesis. Plasmid pUC118 (Takara Shuzo) was used as a cloning vector. Pseudomonas sp. strain NCIMB 10558 was grown in a medium comprising 1% peptone, 1% yeast extract, and 0.5% NaCl (pH 7.2). Recombinant E. coli cells were cultivated at 37°C in Luria-Bertani (LB) medium (1% peptone, 0.5% yeast extract, 1% NaCl [pH 7.2]) containing 0.1 mg of ampicillin per ml unless otherwise noted. For induction of the gene under the control of the lac promoter, 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to LB medium.
General recombinant DNA techniques.
Plasmid DNA was isolated by the alkaline sodium dodecyl sulfate (SDS) method. Plasmid DNA from large-scale preparations was purified with a plasmid purification kit from Qiagen Inc. (Chatsworth, Calif.). Restriction enzymes and DNA-modifying enzymes were purchased from Takara Shuzo and Toyobo and used according to the manufacturers’ protocols. Transformation of E. coli with plasmid DNA by electroporation was performed under standard conditions with a BTX ECM 600 electroporation system (Biotechnologies and Experimental Research, Inc., San Diego, Calif.). Other general procedures were performed as described by Sambrook et al. (25).
Cloning of the low-specificity l-TA gene (ltaP).
Chromosomal DNA isolated from Pseudomonas sp. strain NCIMB 10558 cells was partially digested with Sau3AI (24). Fragments in the molecular size range of 1 to 6 kb were separated by agarose gel electrophoresis and then purified with a GeneClean kit (Bio 101, Inc., Vista, Calif.). The fragments were ligated into BamHI-restricted pUC118, and the plasmids were introduced into E. coli XL1-Blue MRF′ cells to construct a genomic library of Pseudomonas sp. strain NCIMB 10558. For screening of ltaP-harboring clones, plasmids extracted from the established gene library were introduced into E. coli GS245 cells, and the recombinant E. coli cells were cultivated at 37°C for 24 h in M9 minimal medium supplemented with 50 μg of phenyalanine per ml, 10 μg of vitamin B1 per ml, and 100 μg of ampicillin per ml. Colonies grown on the plates were picked as positive clones for further study.
Sequence analysis.
pLTA was used as a sequencing template. The nucleotide sequence was determined by the dideoxy nucleotide chain termination method with Cy5 AutoCycle sequencing kits and a Pharmacia LKB ALFred DNA sequencer. A homology search was performed by means of the sequence similarity searching programs Fasta (1) and Blast (21). The Clustal V method was used to align the sequences (5).
Overexpression of the ltaP gene in E. coli.
To obtain the entire gene without excessive flanking parts, PCR amplification was carried out with 50 μl of 10 mM Tris-HCl (pH 8.3)–50 mM KCl–1.5 mM MgCl2–0.1 mM (each) deoxynucleotide triphosphate–100 pmol of each primer–1 μg of chromosomal DNA of Pseudomonas sp. strain NCIMB 10558–0.5 U of Ex Taq DNA polymerase (Takara Shuzo) at 94°C for 1 min, 65°C for 2 min, and 72°C for 3 min for a total of 30 cycles. The 5′ primer containing a Shine-Dalgarno sequence (lowercase letters) and an ATG initiation codon (bold letters) and the 3′ primer with the complement of the TGA termination codon (bold letters) had the respective sequences 5′-GCCGAATTCTTCaggaCAGAACCATGACCG-3′ and 5′-CCGCTGCAGTAGCCGCTGATGGTGTCAGG-3′, which were designed on the basis of the nucleotide sequence of the ltaP gene; to facilitate the cloning, an additional restriction site (underlined sequence) was incorporated into both primers. The amplified PCR product was digested with EcoRI and PstI, separated by agarose gel electrophoresis, and then purified with a GeneClean kit. The amplified DNA of approximately 1.1 kb, which contained the complete coding sequences of the ltaP gene, was inserted downstream of the lac promoter in pUC118 and then used to transform E. coli XL1-Blue MRF′ cells. The constructed plasmid was designated pLTA1.
Feasible purification of the low-specificity l-TA.
All enzyme purification operations were carried out at 0 to 5°C. Potassium phosphate (50 mM; pH 7.0) containing 10 μM PLP was used as the buffer throughout the purification procedures unless otherwise noted.
(i) Step 1: preparation of cell extract.
Cells of the E. coli transformant harboring overexpression plasmid pLTA1 were grown aerobically at 37°C for 14 h in 6 liters of LB medium containing 0.1 mg of ampicillin per ml and 0.2 mM IPTG. The cells were harvested and rinsed with buffer. After being suspended in 50 ml of buffer, the cells were disrupted by ultrasonic oscillation at 4°C for 20 min with a model 201M ultrasonic oscillator (Kubota, Tokyo, Japan). The cell debris was removed by centrifugation at 25,000 × g for 30 min.
(ii) Step 2: Butyl-Toyopearl column chromatography.
The supernatant solution, brought to 30% saturation with ammonium sulfate, was applied to a Butyl-Toyopearl 650M column (2.5 by 40 cm). Elution was carried out with a 1,200-ml linear gradient of 30 to 0% saturated ammonium sulfate in buffer at a flow rate of 5 ml/min. The active fractions were pooled and concentrated by ultrafiltration with a Centriprep-30 apparatus (Amicon, Inc., Beverly, Mass.).
(iii) Step 3: DEAE-Toyopearl column chromatography.
The enzyme solution was dialyzed against 1,000 volumes of buffer and applied to a DEAE-Toyopearl 650M column (2.5 by 20 cm) equilibrated with buffer. After the column was washed thoroughly with buffer containing 50 mM NaCl, linear gradient elution was performed with buffer supplemented with NaCl by increasing the concentration from 50 to 200 mM. The flow rate was maintained at 5 ml/min. The active fractions were pooled and concentrated by ultrafiltration with a Centriprep-30 apparatus. The purified enzyme was stored in buffer containing 20% (wt/vol) glycerol at −30°C.
Site-directed mutagenesis.
Mutants of the low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558 were prepared according to the method of Kunkel et al. (10). The mutant enzymes and synthetic mutagenic primers were as follows (the underlining indicates the mutagenized nucleotides): K207A, 5′-CATGCCGTTTGCGGTGCCG-3′, and K207R, 5′-CCATGCCGTTTCTGGTGC-3′. The substitutions were confirmed by DNA sequencing with AutoCycle sequencing kits and a Pharmacia LKB ALFred DNA sequencer. All of the mutant enzymes were produced by E. coli XL1-Blue MRF′ cells. The mutant enzymes were purified by the same procedure as that used for the wild type, except that the purification was monitored by SDS-polyacrylamide gel electrophoresis (PAGE) on a slab gel.
Molecular mass determination.
The molecular mass of the enzyme was determined by gel filtration on a HiLoad Superdex 200 column (1.6 by 60 cm).
The subunit molecular mass of the enzyme was determined by SDS-PAGE with the following marker proteins: phosphorylase b (94 kDa), albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa) (Pharmacia Biotech, Uppsala, Sweden).
PLP content.
The PLP content of the enzyme was determined with phenylhydrazine according to the method of Wada and Snell (33).
Spectrophotometric measurements.
The absorption spectra of the wild-type and mutant enzymes were measured at 20°C with 20 mM potassium phosphate buffer (pH 7.0) by use of a Beckman model DU 640 spectrophotometer.
Amino acid sequencing.
The NH2-terminal amino acid sequence was determined by the Edman degradation procedure with a model 476A protein sequencer (Perkin-Elmer, Norwalk, Conn.).
Enzyme assay.
Threonine aldolase activity was assayed with l-threonine as a substrate. The reaction mixture comprised 10 μmol of l-threonine, 0.01 μmol of PLP, 20 μmol of HEPES buffer (pH 8.0), and the enzyme in a total volume of 200 μl. The reaction was carried out at 30°C for 10 min and was terminated by the addition of 50 μl of 30% trichloroacetic acid. The acetaldehyde released was measured spectrophotometrically according to the method of Paz et al. (20). One unit of the enzyme was defined as the amount of enzyme which catalyzed the formation of 1 μmol of acetaldehyde per min under the assay conditions described. Threonine aldolase activity was also measured spectrophotometrically at 340 nm by coupling the reduction of acetaldehyde (oxidation of NADH) with yeast alcohol dehydrogenase (Wako, Osaka, Japan). The assay mixture comprised 100 μmol of HEPES buffer (pH 8.0), 0.05 μmol of PLP, 0.2 μmol of NADH, 30 U of yeast alcohol dehydrogenase, and appropriate amounts of the enzyme and substrate in a final volume of 1 ml. One unit of aldolase activity was the amount of enzyme that catalyzed the formation of 1 μmol of acetaldehyde (1 μmol of NADH oxidized) per min at 30°C; the molar extinction coefficient of NADH is 6.2 × 103 M−1 cm−1. The phenylserine aldolase, β-3,4-dihydroxyphenylserine aldolase, and β-3,4-methylenedioxyphenylserine aldolase activities were measured spectrophotometrically at 279, 350, and 320 nm, respectively, with molar extinction coefficients of 1.4 × 103 M−1 cm−1 for benzaldehyde, 16 × 103 M−1 cm−1 for protocatechualdehyde, and 9.0 × 103 M−1 cm−1 for piperonal, respectively. Quantitative assaying of the released glycine was carried out with an automatic amino acid analyzer (L-8500; Hitachi, Tokyo, Japan).
Chromatographic optical resolution of amino acid enantiomers.
The isomers of phenylserine and β-3,4-methylenedioxyphenylserine were analyzed by high-performance liquid chromatography as follows: column, Sumichiral OA-5000 (0.46 by 15 cm; Sumitomo, Tokyo, Japan); solvent, 2 mM copper sulfate containing 15% methanol; flow rate, 1.0 ml/min; detection, 254 nm; and temperature, 30°C. The isomers of β-3,4-dihydroxyphenylserine were also analyzed by high-performance liquid chromatography as follows: column, Crownpak CR (−) (0.4 by 15 cm; Daicel, Osaka, Japan); solvent, distilled water adjusted to pH 1.0 with perchloric acid; flow rate, 0.4 ml/min; detection, 220 nm; and temperature, 4°C.
Protein determination.
Protein concentration was determined with a Bio-Rad protein assay kit.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB001577.
RESULTS
Cloning of the ltaP gene.
A genomic library of Pseudomonas sp. strain NCIMB 10558 was constructed in pUC118 with E. coli XL1-Blue MRF′ as a host. From this library, we isolated three types of plasmids which were able to complement the glycine-requiring auxotroph E. coli GS245. Subsequently, these plasmids were separately introduced into E. coli XL1-Blue MRF′ cells, and the clone harboring plasmid pLTA showed obvious threonine aldolase activity.
Sequence analysis of the ltaP gene.
Nucleotide sequence analysis revealed that the open reading frame (ORF) of the ltaP gene consists of 1,041 bp starting with an initiation codon, ATG, and ending with a termination codon, TGA, at position 1219. A probable ribosome-binding sequence, AGGA, is present 7 bases upstream of the putative translational start codon (28). However, sequences similar to the E. coli −35 sequence (TTGACA) and the −10 sequence (TATAAT) were not found. The ORF encodes a protein of 346 amino acid residues. The NH2-terminal amino acid sequence coincided with that of the purified enzyme determined by Edman degradation.
In addition, another, incomplete ORF (320 bp), with a translational direction opposite that of the ltaP gene, was found about 250 bp upstream of the ltaP gene. In a search of protein amino acid sequence database SWISS-PROT by means of the sequence similarity searching program Fasta (1), the deduced amino acid sequence of this partial ORF was shown to have more than 75% identity with that of the serine hydroxymethyltransferase of E. coli (22). We thus supposed that this incomplete ORF might represent a partial serine hydroxymethyltransferase gene.
Overexpression of the ltaP gene in E. coli.
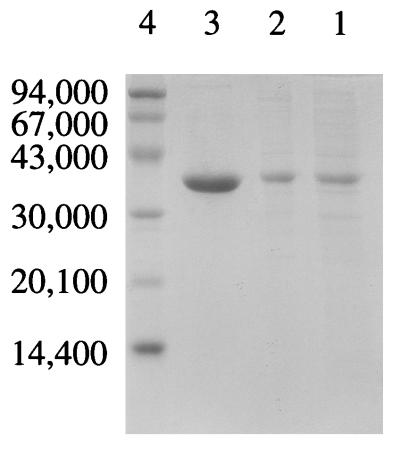
The whole ltaP gene amplified by PCR directly from Pseudomonas sp. chromosomal DNA, with a putative Shine-Dalgarno sequence (AGGA), an initiation codon (ATG), and a termination codon (TGA), was inserted into the EcoRI and PstI sites of pUC118. The resultant plasmid, named pLTA1, was introduced into E. coli XL1-Blue MRF′ cells. The nucleotide sequence of the whole amplified gene was further confirmed to have undergone no error matching during the PCR by sequencing both strands. Judging from the specific activity of the cell extract, low-specificity l-TA constitutes about 12% of the total soluble protein. The protein was produced only in the presence of IPTG (data not shown), indicating that the lac promoter is essential for overexpression. An attempt to further improve gene expression by inserting the ltaP gene downstream of either the tac promoter of pKK223-3 or the T7 promoter of pBluescript did not succeed. However, low-specificity l-TA produced by the overproducer pLTA1 was feasibly purified by two column chromatography steps, with a yield of 40% (Table 1 and Fig. 1).
TABLE 1.
Purification of the recombinant low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558a
| Step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 10,812 | 2,120 | 5.1 | 1.0 | 100 |
| Butyl-Toyopearl | 5,056 | 477 | 10.6 | 2.1 | 47 |
| DEAE-Toyopearl | 4,346 | 106 | 41.0 | 8.0 | 40 |
Threonine aldolase activity was determined with l-threonine as a substrate according to the method of Paz et al. (20) as described in Materials and Methods.
FIG. 1.
Purification of the recombinant low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558. Samples from each of the purification steps were loaded on an SDS–10% polyacrylamide gel and stained with Coomassie blue after electrophoresis. Lanes: 1, cell extract (10 μg); 2, Butyl-Toyopearl pool (10 μg); 3, DEAE-Toyopearl pool (20 μg); 4, molecular mass standards. The numbers to the left are the molecular masses of the standards.
Molecular mass.
The molecular mass of the recombinant enzyme was estimated to be about 145 kDa by gel filtration. The subunit molecular mass was determined by SDS-PAGE to be 38 kDa, the same as the value calculated from the deduced amino acid sequence (Fig. 1). These results suggest that the low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558 is composed of four subunits with identical molecular masses.
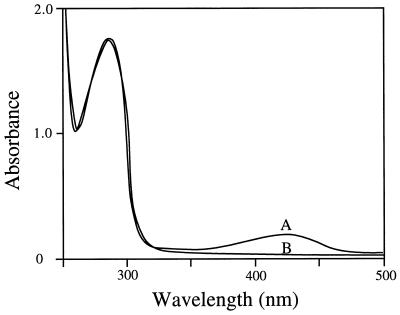
Cofactor requirement.
The enzyme exhibited absorption maxima at 280 and 420 nm, with an A280/A420 ratio of about 10 (Fig. 2). The absorption maximum at 420 nm suggests that the enzyme contains PLP as a cofactor. Reduction of the enzyme with sodium borohydride by the dialysis method of Matsuo and Greenberg (14) resulted in a loss of the enzyme activity, with a disappearance of the absorption maximum at 420 nm and a concomitant increase in the A330 (data not shown). The reduced enzyme was catalytically inert, and the addition of PLP did not restore the activity. This result suggests that sodium borohydride reduces the aldimine linkage of the internal Schiff base. The holoenzyme was converted to the apoenzyme by treatment with 1 mM hydroxylamine at 25°C for 30 min and then dialyzed against 10 mM potassium phosphate buffer (pH 7.0). The constructed apoenzyme did not show threonine aldolase activity. However, the activity was restored to 88% of that of the native enzyme on incubation with 0.1 mM PLP. All of these results show that PLP forms a Schiff base with a lysine residue of the low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558 to catalyze the reaction. The PLP content of the enzyme was determined to be 4 mol per 152 kg of protein, suggesting that the enzyme has the capacity to bind 1 mol of PLP as a cofactor/mol of 38-kDa subunits.
FIG. 2.
Absorption spectra of the wild-type (A) and K207R mutant (B) enzymes (the absorption spectrum of the K207A mutant enzyme is superimposed upon that of the K207R mutant enzyme, for which data are not shown). The absorption spectra were measured with 20 mM potassium phosphate buffer (pH 7.0) at a protein concentration of 1.5 mg/ml.
pH and temperature effects.
To examine the effect of pH on the enzyme activity, the initial reaction velocity was measured by the standard assay method with l-threonine as a substrate and with the following buffers of various pHs: 2-(N-morpholino)ethanesulfonic acid (pH 5 to 6.5), HEPES (pH 7.0 to 8.0), and 1,3-bis[tris(hydroxymethyl)methylamino]propane (pH 7.0 to 9.5). The maximum activity of the low-specificity l-TA was found to occur at pH 8.0 to 8.5. The enzyme was stable between pH 5.5 and 9.0 for 30 min at 30°C. The effect of temperature was also examined. The maximum activity of the low-specificity l-TA was observed at 25°C, and the enzyme retained 50% activity upon heating at 40°C for 15 min.
Substrate specificity.
The substrate specificity of the recombinant enzyme is shown in Table 2. The enzyme acted on both l-threonine and l-allo-threonine but not on d-threonine and d-allo-threonine. This bacterial enzyme showed a higher l-threonine specificity than the low-specificity l-TAs from S. cerevisiae and C. humicola (12, 34). Remarkably, l-threo-phenylserine, l-erythro-phenylserine, l-β-3,4-dihydroxyphenylserine, and l-β-3,4-methylenedioxyphenylserine were found to be substrates of the enzyme (Table 2).
TABLE 2.
Relative activities and steady-state kinetic constants of the recombinant low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558a
| Compound | Relative activityb (%) | Vmax (U mg−1) | Km (mM) | Vmax/Km (U mg−1 mM−1) |
|---|---|---|---|---|
| l-Threonine | 100 | 44.2 ± 1.0 | 14.7 ± 0.1 | 3.0 ± 0.1 |
| l-allo-Threonine | 70 | 31.1 ± 1.1 | 14.6 ± 0.8 | 2.1 ± 0.3 |
| d-Threonine | 0 | |||
| d-allo-Threonine | 0 | |||
| dl-threo-Phenylserine | ||||
| l-threo Form | 267 | 118.0 ± 5.6 | 7.3 ± 2.2 | 16.2 ± 0.8 |
| d-threo Form | 0 | |||
| dl-erythro-Phenylserine | ||||
| l-erythro Form | 514 | 227.1 ± 1.8 | 10.2 ± 0.5 | 22.3 ± 1.2 |
| d-erythro Form | 0 | |||
| dl-β-3,4-Dihydroxyphenylserine | ||||
| l-threo Form | 35 | 15.3 ± 5.6 | 8.3 ± 1.2 | 1.8 ± 0.6 |
| d-threo Form | 0 | |||
| dl-β-3,4-Methylenedioxyphenylserine | ||||
| l-threo Form | 360 | 159.0 ± 3.6 | 7.4 ± 2.5 | 21.5 ± 1.8 |
| d-threo Form | 0 | |||
| dl-Threonine hydroxamate | 0 | |||
| dl-Threonine methyl ester | 0 | |||
| dl-Threoninamide | 0 |
The stereospecific activity of the enzyme toward unresolved compounds was analyzed by high-performance liquid chromatography, and the enzyme activities were determined spectrophotometrically as described in Materials and Methods. The values of the kinetic constants are given as the means ± standard deviations of three different determinations.
Activity relative to that of the enzyme toward l-threonine.
Sequence similarity relative to other proteins.
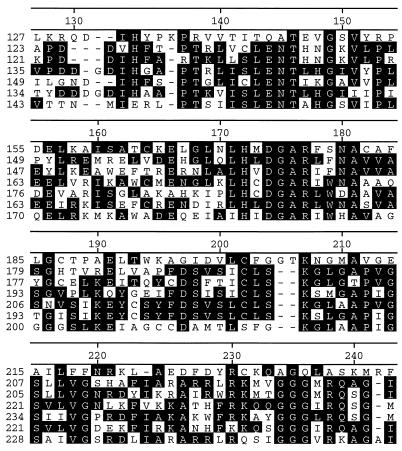
A search of protein amino acid sequence databases (GenBank, EMBL, PIR, and SWISS-PROT) by means of the sequence similarity searching programs Fasta (1) and Blast (21) revealed that the predicted amino acid sequence showed 19.4 and 19.2% identities to those of the low-specificity l-TA of S. cerevisiae (12) and l-allo-TA of A. jandaei DK-39 (11). In addition, we also found four hypothetical proteins from E. coli, Candida albicans, Schizosaccharomyces pombe, and Tolypocladium inflatum showing similarity in primary structure to threonine aldolases. The low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558 showed less than 20% identity to the other proteins, while the low-specificity l-TA of S. cerevisiae, the l-allo-TA of A. jandaei, and the four hypothetical proteins showed greater than 30% identity to one another. The phylogenetic tree of these proteins was constructed by the Clustal V method (data not shown) (5), and the low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558 was comparatively far from the other proteins. Figure 3 shows the segmental sequence alignment of the seven proteins. Notably, Lys207 of the low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558 was the sole lysine residue conserved in all of these proteins; this residue likely functions as the PLP-binding site of the enzyme.
FIG. 3.
Segmental sequence alignment of the low-specificity l-TA of Pseudomonas sp. strain NCIMB 10558 with other proteins. From top to bottom in each set, the proteins were the low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558, l-allo-TA from A. jandaei, hypothetical protein from E. coli, low-specificity l-TA from S. cerevisiae, and hypothetical proteins from S. pombe, C. albicans, and T. inflatum. Identical residues are boxed in black. The numbers on the left are the residue numbers for each amino acid sequence, and those on the top are the residue numbers for the Pseudomonas aldolase sequence.
Identification of the active-site lysine residue.
To identify the PLP-binding lysine residue of the low-specificity l-TA, two mutant enzymes were constructed and purified as described in Materials and Methods. The K207A mutant enzyme showed no detectable enzyme activity, and the K207R mutant enzyme showed only a remaining specific activity of 0.04 U/mg toward l-threonine; this activity is about 1,000 times lower than that of the wild-type enzyme. The two mutant enzymes showed the disappearance of the absorption maximum at 420 nm (Fig. 2), indicating that the Schiff base linkage between the ɛ-amino group of the active-site lysine residue and the PLP cofactor aldehyde group of the wild type is not present in the K207A and K207R mutant enzymes. To make certain that the isolated polypeptides are the mutant threonine aldolases, the N-terminal amino acid sequences of the two mutant proteins were confirmed by the Edman degradation procedure with a model 476A protein sequencer to be the same as that of the wild type.
DISCUSSION
This is the first report on gene cloning of a bacterial low-specificity l-TA that catalyzes the cleavage of both l-threonine and l-allo-threonine to glycine and acetaldehyde. The ltaP gene of Pseudomonas sp. strain NCIMB 10558 has a putative Shine-Dalgarno sequence but not an apparent ς70-type promoter. However, the enzyme was produced efficiently by recombinant E. coli cells under the regulation of the lac promoter in the presence of IPTG, leading to feasible purification of the enzyme by two column chromatography steps. This overexpression-purification system provided us with sufficient low-specificity l-TA to study the structural and functional relationships of the enzyme and its application.
Bruns and Fiedler reported the occurrence of phenylserine aldolase (EC 4.1.2.26) in the livers and kidneys of humans, rats, mice, and other animals (3). Naoi et al. found l-threo-3,4-dihydroxyphenylserine aldolase activity in the human brain (18). Neither of these enzymes, however, has been purified or studied extensively. To our knowledge, the recombinant low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558 is the first pure preparation showing l-β-3,4-dihydroxyphenylserine aldolase, l-β-3,4-methylenedioxyphenylserine aldolase, and phenylserine aldolase activities.
Low-specificity l-TA was recently shown to be the key enzyme for the synthesis of cellular glycine in S. cerevisiae (12, 15, 16). In contrast, serine hydroxymethyltransferase has been believed to be the sole enzyme responsible for cellular glycine in E. coli (29). However, in this study, on searching protein amino acid sequence databases, we found an anonymous hypothetical protein from E. coli that showed 52.3, 31.8, and 18% identities in primary structure with l-allo-TA from A. jandaei, low-specificity l-TA from S. cerevisiae, and low-specificity l-TA from a Pseudomonas sp., respectively. The significant similarity suggests that the hypothetical protein may be a threonine aldolase. To verify this idea, we amplified the gene from the genomic DNA of E. coli K-12 by PCR and further cloned and overexpressed the gene in E. coli cells. The purified enzyme was found to be a low-specificity l-TA with 30 times more activity toward l-allo-threonine than toward l-threonine (unpublished data). This result agrees with the sequence identity data, which showed that the E. coli aldolase had as much as 52.3% amino acid sequence identity with l-allo-TA from A. jandaei. To examine whether threonine aldolase is involved in the biosynthesis of cellular glycine in E. coli, a gene disruption study is under way. It is likely that the other three hypothetical proteins, from C. albicans, S. pombe, and T. inflatum, are also threonine aldolases. Here we present evidence based on primary structure similarity that threonine aldolase may be widespread in nature.
We showed that Lys207 of the low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558 is the PLP-binding site of the enzyme by site-directed mutagenesis; this finding was based on the fact that the two mutant enzymes significantly lost aldolase activity, with the corresponding disappearance of the absorption maximum at 420 nm (Fig. 2). However, the alteration may have resulted from global variations in protein structure and may not have been a specific effect of the side chains of the new amino acid. To clarify this point, two different aspects were considered. First, the similar circular dichroism spectra (200 to 300 nm) of the wild-type enzyme and the two mutant enzymes suggested that no drastic change in the secondary structure of the mutant molecule had occurred (data not shown). Second, the native molecular masses of the wild-type and K207A and K207R mutant proteins were determined by gel filtration on a HiLoad Superdex 200 column (1.6 by 60 cm) to be 145 kDa, a result which makes extensive conformational changes unlikely and which suggests that the tetrameric quaternary structure of the wild-type low-specificity l-TA is also characteristic of the mutant proteins.
We previously showed that Lys199 of l-allo-TA from A. jandaei, corresponding to Lys207 of the low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558, functions as the PLP-binding site of the enzyme (Fig. 3) (11). It is likely that l-allo-TA and low-specificity l-TA with different stereospecificities catalyze the reaction by the same reaction mechanism. In addition, a partial sequence, 172HXDGAR177, of the low-specificity l-TA from Pseudomonas sp. strain NCIMB 10558 is conserved (Fig. 3). It is worth examining the role of these residues in order to understand the structural and functional relationships of the enzyme.
ACKNOWLEDGMENTS
We are deeply indebted to G. V. Stauffer, who kindly provided us with E. coli GS245. Thanks are also due to H. Hayashi and H. Kagamiyama for their generous gift of dl-erythro-phenylserine.
This work was supported in part by grants-in-aid for scientific research (08760097) from the Ministry of Education, Science, Sports, and Culture of Japan and by RFTF (JSPS-RFTF 96I00301) from JSPS.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bell S C, Turner J M. Bacterial catabolism of threonine: threonine degradation initiated by l-threonine acetaldehyde-lyase (aldolase) in species of Pseudomonas. Biochem J. 1977;166:209–216. doi: 10.1042/bj1660209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns F H, Fiedler L. Enzymatic cleavage and synthesis of l-threo-β-phenylserine and l-erythreo-β-phenylserine. Nature. 1958;181:1533–1534. doi: 10.1038/1811533a0. [DOI] [PubMed] [Google Scholar]
- 4.Herbert, R. B., B. Wilkinson, G. J. Ellames, and E. K. Kunec. 1993. Stereospecific lysis of a range of β-hydroxy-α-amino acids catalysed by a novel aldolase from Streptomyces amakusaensis. J. Chem. Soc. Chem. Commun. 205–206.
- 5.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 6.Jolad S D, Hoffmann J J, Torrance S J, Wiedhopf R M, Cole J R, Arora S K, Bates R B, Gargiulo R L, Kriek G R. Bouvardin and deoxybouvardin, antitumour cyclic hexapeptides from Bouvardia ternifolia (Rubiaceae) J Am Chem Soc. 1977;99:8040–8044. doi: 10.1021/ja00466a043. [DOI] [PubMed] [Google Scholar]
- 7.Karasek M A, Greenberg D M. Studies on the properties of threonine aldolases. J Biol Chem. 1957;227:191–205. [PubMed] [Google Scholar]
- 8.Kataoka M, Wada M, Nishi K, Yamada H, Shimizu S. Purification and characterization of l-allo-threonine aldolase of Aeromonas jandaei DK-39. FEMS Microbiol Lett. 1997;151:245–248. doi: 10.1111/j.1574-6968.1997.tb12577.x. [DOI] [PubMed] [Google Scholar]
- 9.Kumagai H, Nagatae T, Yoshida H, Yamada H. Threonine aldolase from Candida humicola: purification, crystallization and properties. Biochim Biophys Acta. 1972;258:779–790. doi: 10.1016/0005-2744(72)90179-9. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 11.Liu J-Q, Dairi T, Kataoka M, Shimizu S, Yamada H. l-allo-Threonine aldolase from Aeromonas jandaei DK-39: gene cloning, nucleotide sequencing, and identification of the pyridoxal 5′-phosphate binding lysine residue by site-directed mutagenesis. J Bacteriol. 1997;179:3555–3560. doi: 10.1128/jb.179.11.3555-3560.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J-Q, Nakata S, Dairi T, Misono H, Shimizu S, Yamada H. GLY1 gene of Saccharomyces cerevisiae encodes a low-specific l-threonine aldolase that catalyzes cleavage of l-allo-threonine and l-threonine to glycine: expression of the gene in Escherichia coli and purification and characterization of the enzyme. Eur J Biochem. 1997;245:289–293. doi: 10.1111/j.1432-1033.1997.00289.x. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama W, Naoi M, Narabayashi H. The metabolism of l-DOPA and l-threo-3,4-dihydroxyphenylserine and their effects on monoamines in the human brain: analysis of the intraventricular fluid from parkinsonian patients. J Neurosci. 1996;139:141–148. [PubMed] [Google Scholar]
- 14.Matsuo Y, Greenberg D M. A crystalline enzyme that cleaves homoserine and cystathionine. J Biol Chem. 1959;234:507–515. [PubMed] [Google Scholar]
- 15.McNeil J B, Mcintosh E V, Taylor B V, Zhang F-R, Tang S, Bognar A L. Cloning and molecular characterization of three genes, including two genes encoding serine hydroxyltransferases, whose inactivation is required to render yeast auxotrophic for glycine. J Biol Chem. 1994;269:9155–9165. [PubMed] [Google Scholar]
- 16.Monschau N, Stahmann K P, Sahm H, McNeil J B, Bognar A L. Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis. FEMS Microbiol Lett. 1997;150:55–60. doi: 10.1111/j.1574-6968.1997.tb10349.x. [DOI] [PubMed] [Google Scholar]
- 17.Morris J G. Utilization of l-threonine by a Pseudomonad: a catabolic role for l-threonine aldolase. Biochem J. 1969;115:603–605. doi: 10.1042/bj1150603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naoi M, Takahashi T, Kuno N, Nagatsu T. l-threo-3,4-Dihydroxyphenylserine (DOPS) aldolase: a new enzyme cleaving DOPS into protocatechualdehyde and glycine. Biochem Biophys Res Commun. 1987;143:482–488. doi: 10.1016/0006-291x(87)91379-9. [DOI] [PubMed] [Google Scholar]
- 19.Ohashi, N., S. Nagata, K. Ishizumi, and K. Maeshima. April 1984. European patent 83,300,059.
- 20.Paz M A, Blumenfeld O O, Rojkind M, Henson E, Furfine C, Gallop P M. Determination of carbonyl compounds with N-methyl benzothiazolone hydrazone. Arch Biochem Biophys. 1964;109:548–559. doi: 10.1016/0003-9861(65)90400-5. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plamann M D, Stauffer G V. Characterization of the Escherichia coli gene for serine hydroxymethyltransferase. Gene. 1983;22:9–18. doi: 10.1016/0378-1119(83)90059-8. [DOI] [PubMed] [Google Scholar]
- 23.Roberto P, Roberto L, Lucia T, John C, Maria P, Enrico M. DL-allothreonine aldolase in rat liver. Biochem Soc Trans. 1991;19:346–347. [Google Scholar]
- 24.Saito H, Miura K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schirch L V, Gross T. Serine transhydroxymethylase: identification as the threonine and allothreonine aldolase. J Biol Chem. 1968;243:5651–5655. [PubMed] [Google Scholar]
- 27.Shibata K, Shingu K, Vassilev V P, Nishide K, Fujita T, Node M, Kajimoto T, Wong C-H. Kinetic and thermodynamic control of l-threonine aldolase catalyzed reaction and its application to the synthesis of mycestericin D. Tetrahedron Lett. 1996;37:2791–2794. [Google Scholar]
- 28.Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;154:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 29.Stauffer G V. Biosynthesis of serine and glycine. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 412–418. [Google Scholar]
- 30.Tymiak A, McCormick T J, Unger S E. Structure determination of lysobactin, a macrocyclic peptide lactone antibiotic. J Org Chem. 1989;54:1149–1157. [Google Scholar]
- 31.Vassilev V P, Uchiyama T, Kajimoto T, Wong C-H. l-Threonine aldolase in organic synthesis: preparation of novel β-hydroxy-α-amino acids. Tetrahedron Lett. 1995;36:4081–4084. [Google Scholar]
- 32.Vassilev V P, Uchiyama T, Kajimoto T, Wong C-H. An efficient chemo-enzymatic synthesis of α-amino-β-hydroxy-γ-butyrolactone. Tetrahedron Lett. 1995;36:5063–5064. [Google Scholar]
- 33.Wada H, Snell E E. The enzymatic oxidation of pyridoxine and pyridoxamine phosphates. J Biol Chem. 1961;236:2089–2095. [PubMed] [Google Scholar]
- 34.Yamada H, Gumagai H, Nagate T, Yoshida H. Crystalline threonine aldolase from Candida humicola. Biochem Biophys Res Commun. 1970;39:53–58. doi: 10.1016/0006-291x(70)90756-4. [DOI] [PubMed] [Google Scholar]
- 35.Yamada H, Kumagai H, Nagate T, Yoshida H. Formation of threonine aldolase by bacteria and yeasts. Agric Biol Chem. 1971;35:1340–1345. [Google Scholar]