Abstract
We have isolated the genomic and cDNA clones encoding EG III (a low-molecular-mass endo-β-1,4-glucanase) gene from Trichoderma reesei QM9414. The nucleotide sequence of the cDNA fragment was verified to contain a 702-bp open reading frame that encodes a 234-amino-acid propeptide. The deduced protein sequence has significant homologies with family H endo-β-1,4-glucanases. The 16-amino-acid N-terminal sequence was shown to function as a leader peptide for possible secretion. Northern blot analysis showed that the EG III gene transcript, with a length of about 700 bp, was expressed markedly by cellulose but not by glucose. The protein has been expressed as a mature form in Escherichia coli and as secreted forms in Saccharomyces cerevisiae and Schizosaccharomyces pombe under the control of tac, alcohol dehydrogenase (ADH1), and human cytomegalovirus promoters, respectively. The S. cerevisiae and Schizosaccharomyces pombe recombinant strains showed strong cellulolytic activities on agar plates containing carboxymethyl cellulose. The E. coli strain expressed small amounts of EG III in an active form and large amounts of EG III in an inactive form. The molecular masses of the recombinant EG IIIs were estimated to be 25, 28, and 29 kDa for E. coli, S. cerevisiae, and Schizosaccharomyces pombe, respectively, by immunoblot analysis following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Parts of the yeast recombinant EG IIIs decreased their molecular masses to 25 kDa after treatment with endoglycosidase H and α-mannosidase, suggesting that they are N glycosylated at least partly.
Trichoderma reesei, a filamentous mesophilic fungus, is well-known to secrete the high cellulolytic activity required for a full spectrum of digestion of crystalline cellulose. For this fungus, two cellobiohydrolases (CBH I and II) and three endo-β-1,4-glucanases (EG I, II, and V), which belong to cellulase families C, B, C, A, and K, respectively (2, 9, 25, 31), have been identified and characterized (19, 25, 26, 29, 32, 34). All these cellulases have been shown to contain a cellulose-binding domain (CBD) and one or two linker regions comprised of proline and hydroxy amino acids (2, 31). Apart from these cellulases, the existence of a low-molecular-mass endoglucanase from T. reesei has been reported by several researchers (1, 6, 30), and we also have purified it (then named EG L and now renamed EG III) from a crude enzyme preparation of T. reesei PC-3-7 (12). EG III has a molecular mass of 25 kDa, which is smaller than those of other EGs (EG I and II) identified thus far, and was shown to play a role in degrading crystalline cellulose (Avicel) in concert with CBH I (12). It was presumed in a cellulose binding assay, however, that EG III has no CBD, in contrast to CBH I. We also showed that no EG III internal peptide sequences, which were partially examined, exist in the deduced amino acid sequences of other known T. reesei cellulases. On the other hand, the peptide sequences had significant homology with that of F1-carboxymethyl cellulase (F1-CMCase) from Aspergillus aculeatus (17), which belongs to cellulase family H. These results showed that EG III might not be a proteolytic artifact from the other cellulases but that it may be coded by another gene in the Trichoderma genome. To verify this speculation, we have isolated a clone of the EG III gene and deduced its protein sequence. Recently, Ward et al. (36) presented a preliminary report about the cloning and sequencing of a small, high-pI endoglucanase (named EG III). However, they showed only its amino acid sequence and not those of the cDNA and genomic clone.
To investigate the enzymatic characteristics of the individual cellulases, the cloned cellulase genes need to be expressed in cellulase-nonproducing microorganisms. The yeast Saccharomyces cerevisiae has been used as the host for the expression of the fungus cellulase genes. T. reesei cellulases (20, 21, 25, 34) were produced and effectively secreted into a growth medium by the yeast, although the yields depended on cellulase species and the secreted proteins were heterogeneously N glycosylated. Then we investigated whether or not the fission yeast Schizosaccharomyces pombe could be used for the heterologous expression of the fungus cellulases instead of S. cerevisiae. Consequently, it was found that recombinant Schizosaccharomyces pombe effectively secreted T. reesei CBH II to a level over 100 μg/ml in the growth medium, in which species of two molecular masses resulted from the difference in levels of glycosylation, and that the recombinant CBH IIs purified from the culture supernatant had almost the same enzymatic characteristics as those of the native one (14). Furthermore, T. reesei EG I, II, and V, as well as xylanases I and II, were also successfully secreted by the yeast (unpublished results).
Expression of nonglycosylated forms of fungal cellulases in a manner comparable to that of bacterial cellulases has been attempted for A. aculeatus F1-CMCase (16) and T. reesei CBH I (10) with Escherichia coli as the host. The inactive, aggregated CBH I protein was produced, and F1-CMCase could be expressed as an active form in E. coli cells.
In this paper we describe the cloning of the cDNA and genomic DNA encoding EG III from T. reesei QM9414 and discuss their molecular features and structural relationships with other β-glucanases. Furthermore, expression of the EG III cDNA in E. coli, S. cerevisiae, and Schizosaccharomyces pombe with corresponding high-expression vectors was conducted. The egl3 gene cloned by us provides a good example for evaluating which organism is suitable for expressing fungal cellulase genes.
MATERIALS AND METHODS
Strains and vectors.
The genotypes of the microbial strains and plasmids used in the present study are summarized in Table 1. T. reesei QM9414 was maintained on a potato dextrose agar slant. The T. reesei genomic DNA library was prepared in the lambda EMBL3 vector (Stratagene). Strains LE392 and P2392 (Stratagene) were used as hosts in the preparation of and screening from the genomic library. Plasmids pBluescript II KS(+) (Stratagene) and pT7Blue-T (Novagen) were used for subcloning and sequencing of restriction enzyme-treated DNA fragments and PCR products, respectively. Plasmids pKK223-3 (Pharmacia) and pUC18 (Takara Shuzo, Kyoto, Japan) were used for the construction of the E. coli expression vector. pUC18 was digested with PvuII to eliminate the fragment including the lac promoter, and the remaining fragment was ligated with the DNA fragment obtained after digestion of pKK223-3 with PvuII that contains the tac promoter and the rrn terminator, to lead to substitution of the multiple-cloning site derived from pKK223-3 for that from pUC18. The resulting plasmid, named pAG9-3, was used as an E. coli expression vector. S. cerevisiae expression vector pGAD10α was prepared by digesting pGAD10 (Clontech) with HindIII to remove the GAL4 activation domain and by religation. The Schizosaccharomyces pombe expression vector pCL2M was constructed by converting the AflIII site in pTL2M-2 to a BglII site (33) by site-directed mutagenesis. E. coli JM105 (Pharmacia), S. cerevisiae INVSC1 (Invitrogen), and the Schizosaccharomyces pombe leu1 mutant were used as hosts for EG III expression.
TABLE 1.
Microbial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| T. reesei QM9414 | Type culture | Kyowa Hakko |
| S. cerevisiae INVSC1 | MATα his3Δ1 leu2 trp1-289 ura3-52 | Invitrogen |
| S. pombe leu1 mutant | h− leu1-32 | ATCC 38399 |
| E. coli LE392 | hsdR514 supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 | Stratagene |
| P2392 | LE392 (P2 lysogen) | Stratagene |
| JM105 | thi rpsL endA sbcB15 hsdR4 supE Δ(lac-proAB) F′ (traD36 proAB+ lacIqlacZΔM15) | Pharmacia |
| Plasmids | ||
| pBluescript II KS(+) | Ampr | Stratagene |
| pT7Blue-T | Ampr | Novagen |
| pKK223-3 | AmprPtac-rrnBt | Pharmacia |
| pUC18 | Ampr | Takara |
| pAG9-3 | AmprPtac-rrnBt | This work |
| pAGegl3 | AmprPtac-egl3-rrnBt | This work |
| pGAD10 | AmprLEU2 ADH1pt GAL4 | Clontech |
| pGAD10α | AmprLEU2 ADH1pt | This work |
| pGADegl3 | AmprLEU2 ADH1p-egl3-ADH1t | This work |
| pTL2M-2 | AmprSV40p NeorSV40t hCMVp-SV40t | 14 |
| pCL2M | AmprSV40p NeorSV40t hCMVp-SV40t | This work |
| pCLegl3 | AmprSV40p NeorSV40t hCMVp-egl3-SV40t | This work |
| pAL7 | AmprLEU2 | 15 |
Cloning of the genomic egl3 DNA.
Construction of the genomic DNA library was carried out as follows. The genomic DNA isolated from T. reesei QM9414 was partially digested with Sau3AI. The digested DNA fragments were dephosphorylated with calf intestine alkaline phosphatase (Boehringer Mannheim) and ligated with the BamHI-digested EMBL3 vector. The DNAs were packaged in vitro, and E. coli P2392 was infected by the recombinant phages. The genomic library contained about 106 clones, with an average insert size of 15 kbp. Two internal amino acid sequences determined by sequencing lysylendopeptidase-digested peptides were used for the design of PCR oligonucleotide primers (Fig. 1) in light of the similarity of those amino acid sequences to that of the F1-CMCase from A. aculeatus (17). PCR was performed (7) by using primer 1 as a sense primer, primer 2 as an antisense primer (Fig. 1), and T. reesei QM9414 chromosomal DNA as a template. PCR conditions were 94°C for 1 min (denaturation), 42°C for 1 min (annealing), and 72°C for 2 min (extension). These conditions were repeated for 30 cycles. The amplified PCR fragment identified as an egl3 gene by nucleotide sequencing was labelled with a BcaBEST labelling kit (Takara Shuzo) and [α-32P]dCTP according to the supplier’s instructions. The labelled fragment was used for screening clones from the amplified chromosomal DNA library (8 × 104 clones) plated to a density of 1,000 PFU/87-mm-diameter plate. Plaques grown on plates were transferred to a Hybond-N nylon membrane (Amersham) and cross-linked to the surface of the membrane by UV light in a UV cross-linker (model; CL-1000; UVP, Inc.). The membrane was prehybridized in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10× Denhardt’s reagent–0.5% sodium dodecyl sulfate (SDS) for 2 h at 60°C and hybridized with the 32P-labelled PCR fragment for 16 h at 60°C. The membrane was washed with 6× SSC–0.1% SDS for 20 min at 60°C after being washed briefly with the same solution at room temperature and finally with 2× SSC–0.1% SDS for 20 min at 60°C. Positive clones were purified by a further two rounds of screening.
FIG. 1.
Partial amino acid sequences of the EG III purified from T. reesei and the design of the oligonucleotide primers used in PCR. (A) Amino acid sequences from the peptides obtained by lysylendopeptidase digestion. Fraction numbers indicate the peak numbers of the digested peptides eluted by high-performance liquid chromatography. The uncertain amino acids are in parentheses. (B) Designs of oligonucleotide primers based on the fraction 2 and fraction 3 peptides. The letter N denotes a mixture of all four bases. I, P, and Y denote an inosine, a mixture of adenine and guanine, and cytosine and thymine, respectively. Fr., fraction.
Isolation of the egl3 cDNA gene.
The egl3 cDNA clone was isolated from a T. reesei QM9414 first-strand cDNA library, which was prepared as described previously (24), by using the PCR method with the gene sequence-specific primers (sense, 5′-GGAGATCTATGAAGTTCCTTCAAGTCCTCC-3′, and antisense, 5′-CCAAGCTTAGTTGATAGATGCGGTCCAGGA-3′) corresponding, respectively, to the putative amino-terminal and carboxyl-terminal sequences of the product, deduced from the genomic egl3 DNA nucleotide sequence. The PCR conditions were the same as those for cloning of the genomic egl3 DNA except that annealing was carried out at 50°C. The amplified fragment was ligated with the pT7Blue-T vector. The egl3 genomic and cDNA fragments were subcloned into pBluescript.
Sequencing.
The nucleotide sequences were determined with an ALF automatic DNA sequencer by the Taq cycle sequencing method (Pharmacia) or with a BcaBEST sequencing kit (Takara Shuzo) and [α-32P]dCTP, according to the respective supplier’s instructions. The sequences obtained were characterized by using the Genetyx-Mac genetic information processing software, version 8 (Software Development Co., Ltd., Tokyo, Japan).
Southern and Northern blot analysis.
Standard methods described by Sambrook et al. (27) were followed. Genomic DNA (20 μg) was digested to completion with various restriction endonucleases purchased from New England Biolabs, Inc. (Beverly, Mass.). Digested DNA fragments were separated on a 1.0% (wt/vol) agarose gel in 1× Tris-acetate-EDTA buffer. HindIII-digested λ DNA fragments (from Nippon Gene, Toyama, Japan) were used as size markers.
Each 10 μg of total RNA samples and molecular mass standards (Promega) was separated on 1.0% (wt/vol) agarose gels in the presence of formaldehyde. DNA and RNA fragments in gels were blotted onto Hybond-N nylon membranes (Amersham). The membranes were probed with 32P-labelled egl3 cDNA. The labelled fragment was denatured by boiling for 5 min and used as a hybridization probe as described above for the screening of the genomic DNA library.
DNAs from the 10 λ clones recovered from the genomic library were digested with the restriction enzymes BamHI, EcoRI, HindIII, PstI, and SalI and separated on an agarose gel. The DNA was transferred to a nylon membrane and hybridized with the egl3 internal PCR fragment as described above.
Production of egl3 in yeasts.
Yeast expression plasmids were constructed as follows. The egl3 cDNA fragment was cut from pT7Blue-egl3 with BglII and HindIII and was inserted in BglII- and HindIII-digested pCL2M to give pCLegl3 (Fig. 2). The blunted fragment was also ligated with pGAD10α, which was digested with HindIII and blunted to generate pGADegl3 (Fig. 2). The obtained plasmids, pCLegl3 and pGADegl3, were used for EG III expression in Schizosaccharomyces pombe and S. cerevisiae, respectively. pCLegl3 was cotransformed with pAL7 into the Schizosaccharomyces pombe leu1 mutant as described previously (5, 15), resulting in Schizosaccharomyces pombe SP-cmv-egl3. pGADegl3 was transformed into S. cerevisiae INVSC1 by using the Li acetate method of Gietz et al. (4), resulting in S. cerevisiae SC1-adh-egl3.
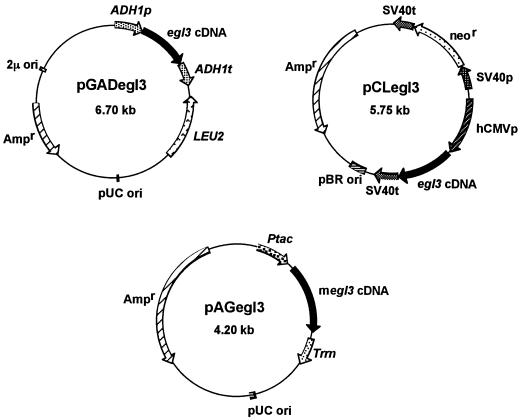
FIG. 2.
Plasmids pGADegl3, pCLegl3, and pAGegl3. Relevant gene locations are indicated. See the text for details on the construction of plasmids. ADH1p and ADH1t, promoter and terminator of the S. cerevisiae ADH1 gene, respectively; LEU2, LEU2 gene of S. cerevisiae; 2μ ori, origin of replication of the 2μm plasmid; pUC ori, replication origin of the E. coli pUC18 plasmid; hCMVp, promoter of the hCMV gene; SV40p and SV40t, promoter and terminator of the simian virus 40 (SV40) gene, respectively; neor, neomycin resistance gene of Tn5 conferring G418 resistance in Schizosaccharomyces pombe; pBR ori, origin of replication of the E. coli pBR322 plasmid; Ptac, tac promoter; Trrn, rrnB terminator; Ampr, ampicillin resistance gene.
The extracellular EG IIIs produced by transformed Schizosaccharomyces pombe and S. cerevisiae were detected by the plate assay as described by Farkas et al. (3) and by endoglucanase activity assay of the culture supernatant. The Schizosaccharomyces pombe and S. cerevisiae strains were grown on YEA-G41825 (0.5% yeast extract and 3% glucose supplemented with 25 μg of G418 per ml) plates for 7 days and leucine-deficient synthetic complete medium plates for 4 days, respectively, at 30°C. Both of the plates were supplemented with 0.1% carboxymethyl cellulose (CMC; Wako Pure Chemical Co., Osaka, Japan). The SC1-adh-egl3 and SP-cmv-egl3 strains were cultivated in 50 ml of YPDM medium (1% yeast extract, 2% peptone, 2% dextrose, 1% malt extract) and YPDM medium containing 25 μg of G418 per ml (YPDM-G41825), respectively, at 30°C in 300-ml shake flasks at 200 rpm. The culture supernatants were separated from the cells at the stationary phase (5.5 days for S. cerevisiae and 11 days for Schizosaccharomyces pombe) by centrifugation, concentrated by 80% saturated ammonium sulfate precipitation, and desalted by Bio-gel P-6 (Bio-Rad, Richmond, Calif.) column chromatography.
Production of mature EG III in E. coli.
For the construction of the mature EG III expression plasmid in E. coli, the DNA fragment which was comprised of the mature EG III-coding region framed with restriction enzyme sites at both ends was obtained by the PCR amplifying method with the sequence-specific primers and pT7Blue-egl3 as a template. The primers were 5′-GGCCATGGCACAAACCAGCTGTGACCAGTGGGC-3′ (sense) and the same sequence in the antisense direction as the carboxy-terminal primer for amplifying egl3 cDNA (italicized letters indicate the NcoI restriction site). Because of the addition of the NcoI site at the 5′ end, the protein expressed in E. coli was expected to have an extension of two extra amino acids at its amino terminus, Met-Ala-. The PCR fragment digested with NcoI and HindIII was blunt ended, ligated with pAG9-3 predigested with EcoRI, and blunted, generating pAG-megl3 (Fig. 2). pAG-megl3 was transformed into E. coli JM105 according to the method of Inoue et al. (8), resulting in E. coli 105-AG-megl3.
The intracellular EG IIIs produced by the recombinant E. coli strains were detected by the endoglucanase activity assay and immunoblotting. The transformants were grown for 2 h at 37°C in 2× TY medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl) supplemented with 50 μg of ampicillin per ml and, after the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), were further cultivated for 6 h. The cells harvested by centrifugation were suspended in 50 mM acetate buffer (pH 6.0) and disrupted with a sonicator, followed by centrifugation, and the resulting supernatant and pellet were analyzed for CMCase activity and proteins.
Preparation of antibodies.
Antiserum was prepared against the purified EG III of T. reesei PC-3-7 (12). Injection into rabbits, immunization, and collection of sera were done by Iwaki Glass.
Biochemical methods.
The endoglucanase activity was assayed as CMCase activity with CMC as a substrate in 50 mM acetate buffer (pH 6.0) at 50°C for 15 min. The amount of released reducing sugar was measured by the 3′,5′-dinitrosalicylic acid method described by Wood and Bhat (37). One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of glucose equivalent per min. SDS-polyacrylamide gel electrophoresis (PAGE) was done with 12.5% polyacrylamide gels with the Mini-PROTEAN II system (Bio-Rad) in accordance with the manufacturer’s instructions. Proteins were blotted onto polyvinylidene difluoride membranes (Bio-Rad) by using a TRANS-BLOT semidry transfer cell (Bio-Rad) and treated with the EG III antiserum. Endoglycosidase H (endo H; Seikagaku-Kogyo, Tokyo, Japan) and α-mannosidase (Wako Pure Chemical) treatment was carried out as described previously (14).
Nucleotide sequence accession number.
The DDBJ, EMBL, and GenBank accession number of the egl3 gene sequence is AB003694.
RESULTS
Isolation of genomic and cDNA clones and sequence analysis.
The N-terminal amino acid of purified low-molecular-mass EG III from T. reesei PC-3-7 was blocked, but its four digested peptide sequences were determined as shown in Fig. 1. In order to amplify a specific sequence corresponding to the egl3 gene, a PCR-based approach was taken. Under the experimental conditions described, a specific band of ca. 400 bp was amplified from T. reesei chromosomal DNA (data not shown). This band was then subcloned into the pT7Blue-T vector and checked by sequencing. The DNA fragment was identified as a part of the egl3 gene, since the internal peptide sequence (fraction 12 in Fig. 1) was found in the protein sequence deduced from the gene.
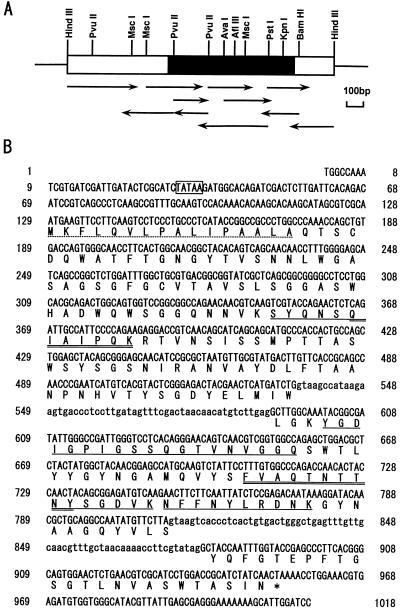
The PCR fragment was labelled and used as a probe to select the egl3 gene from a T. reesei QM9414 genomic library. Upon searching among ca. 80,000 phage plaques, 10 hybridizing clones (λL1 to λL10) were isolated. Only one clone (λL7) was chosen for further studies because all clones included the same egl3 gene. A partial restriction map of this genomic region is presented in Fig. 3A. The 2.0-kb HindIII-HindIII restriction fragment was sequenced, and the deduced protein sequence was found to include the four internal peptide sequences shown in Fig. 1 (Fig. 3B). By comparing them with the A. aculeatus F1-CMCase genomic sequence (18), putative N- and C-terminal amino acid sequences of EG III were presumed. Based on this assumption, egl3 cDNA was isolated by the PCR cloning method from a first-strand cDNA library and used as a template, which was prepared from T. reesei QM9414 grown on Avicel as a sole carbon source. The genomic and cDNA sequences along with the deduced protein sequence are shown in Fig. 3B. Two introns (55 and 66 bp) are present in positions identical to that of the F1-CMCase gene (18), and the suggested splicing signals showed homology with those of the T. reesei genes sequenced so far (data not shown). There is a putative TATA box located 96 bp upstream from the ATG of the initiation codon. Both the genomic and cDNA sequences were positively identified as EG III by the presence of all the partial amino acid sequences previously determined from the purified protein (Fig. 3B). Some minor differences between the peptide and DNA sequences are due to uncertain determinations made during the peptide sequencing. The mature protein presumably starts at amino acid 17, glutamine, which seemed to be pyroglutamylated, since the α-amino group of the N-terminal amino acid of the purified EG III was blocked like those of other T. reesei cellulases, all of which are pyroglutamic acid (12, 19, 26, 29, 32, 34). The peptide composed of 16 amino acids (Fig. 3B) showed the typical structure of a signal peptide with a high hydrophobic index following a positively charged amino acid (35). The protein deduced from the nucleotide sequence has 234 amino acids and a molecular mass of 25,158 Da for the unprocessed form, and if the initial 16 amino acids are excluded, the calculated molecular mass is 23,480 Da, in good agreement with the biochemical data (12).
FIG. 3.
Restriction map and sequencing strategy of the genomic DNA for T. reesei egl3 (A) and the complete nucleotide sequence of the gene and deduced amino acid sequence of the EG III protein (B). (A) The HindIII fragment of the egl3 genomic clones is shown as a bar, and the egl3 structural gene region is shown as a filled box. The orientations and lengths of coverage of sequencing primers are shown as horizontal arrows. (B) Intron sequences are in lowercase type. The standard one-letter amino acid code is used. The presumed signal sequence is indicated by the dotted underline. The internal amino acid sequences determined for the lysylendopeptidase-digested peptides of the purified T. reesei EG III are underlined. The amino acid sequences for the design of PCR primers are double underlined.
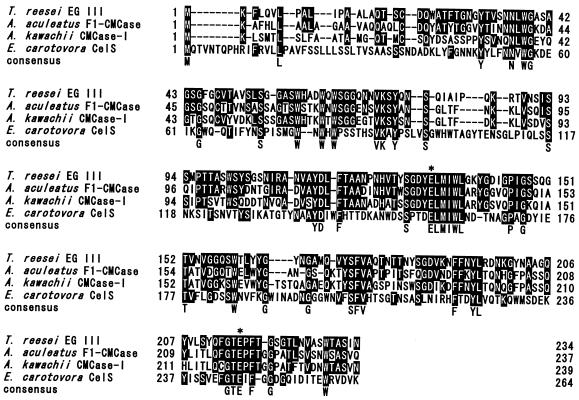
When the EG III sequence was compared with those available from the databases, F1-CMCase (17) from A. aculeatus, CMCase-I (24) from Aspergillus kawachii, and CelS (23) from Erwinia cartovora subsp. cartovora showed 56, 47, and 26% homology with EG III on the amino acid level, respectively (Fig. 4). These are all EGs which belong to the so-called family H cellulases.
FIG. 4.
Alignment of the EG III sequence with sequences of family H cellulases, namely, A. aculeatus F1-CMCase (29), A. kawachii CMCase-I (37), and Erwinia carotovora subsp. carotovora CelS (35). The standard one-letter amino acid code is used. Amino acid residues identical to those of EG III are indicated by white letters in black boxes, whereas the consensus indicates amino acid residues that are identical in all sequences. Hyphens indicate gaps. Putative catalytic amino acid residues are indicated by asterisks.
EG III gene and mRNA.
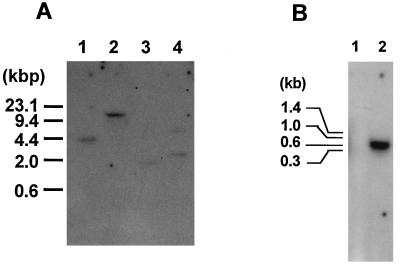
In order to test whether the egl3 gene is present in only one or multiple copies in the T. reesei QM9414 genome, Southern blotting was performed using total chromosomal DNA digested with several restriction enzymes and the egl3 cDNA as a probe. As shown in Fig. 5A, one hybridizing band is present in all the resulting DNA fragments except for those digested by PstI. The PstI-cut fragments appear to display two hybridizing bands because there is one PstI site in the egl3 gene (Fig. 3A). This result indicates that T. reesei may have one copy of the egl3 gene in its chromosomal DNA.
FIG. 5.
Southern hybridization analysis of T. reesei genomic DNA (A) and Northern hybridization analysis of T. reesei RNA (B). (A) Aliquots (20 μg) of T. reesei genomic DNA were digested with each of the following restriction enzymes: BamHI, EcoRI, HindIII, and PstI (lanes 1 to 4, respectively). The resulting fragments were fractionated by agarose gel electrophoresis and then transferred to a nylon membrane for hybridization. The probe used was the egl3 cDNA. The fragments of lambda DNA digested with HindIII were used as molecular size markers. (B) Total RNA samples (10 μg each) were isolated from cells grown in medium containing glucose (lane 1) and Avicel (lane 2). The positions of migration of RNA molecular standards are shown on the left.
To assess if egl3 is preferentially transcribed under the same conditions as other cellulase genes in T. reesei QM9414, Northern blotting was done with total RNA isolated from T. reesei grown on glucose or on Avicel as a sole source of carbon (Fig. 5B). The egl3 mRNA was detected in the Avicel-grown cells but not in the glucose-grown cells. This may be the same expression pattern as that of other T. reesei cellulase genes which are induced by cellulose and repressed by glucose. Thus, both the egl3 gene and the other cellulase genes have a common regulatory mechanism on the transcriptional level at least in part. The size of the mRNA was around 700 bp, in accordance with that of the egl3 cDNA coding sequence, indicating that 5′ and 3′ noncoding regions of egl3 mRNA are very short.
Expression of the egl3 gene in yeasts.
The egl3 cDNA was cloned into the S. cerevisiae multicopy expression vector pGAD10α under the control of the constitutive ADH1 promoter, and the resulting plasmid, pGADegl3, was transformed into S. cerevisiae INVSC1. In a similar manner, the egl3 expression vector for Schizosaccharomyces pombe, pCLegl3, was constructed by inserting the cDNA into the copy-number-controlled vector pCL2M under control of the human cytomegalovirus (hCMV) promoter. pCLegl3 was cotransformed into the Schizosaccharomyces pombe leu1 mutant with pAL7 harboring Schizosaccharomyces pombe ars and stb and S. cerevisiae LEU2 genes. The obtained recombinants, S. cerevisiae SC1-adh-egl3 and Schizosaccharomyces pombe SP-cmv-egl3, were analyzed for endoglucanase activity on CMC plates (Fig. 6). These strains produced clear halos, indicating that the Trichoderma enzyme was secreted in active forms by the yeasts. The control strains, S. cerevisiae INVSC1 transformed with the vector pGAD10α and the Schizosaccharomyces pombe leu1 mutant transformed with pCL2M, showed no endoglucanase activity in the plate assay.
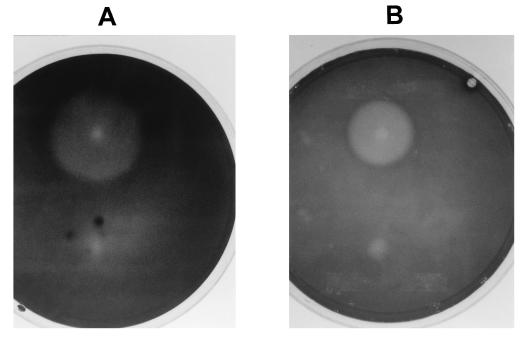
FIG. 6.
Hydrolysis halos produced on CMC plates by S. cerevisiae SC-adh-egl3 (A) and Schizosaccharomyces pombe SP-cmv-egl3 (B) expressing the egl3 gene of T. reesei. The control strains, S. cerevisiae and Schizosaccharomyces pombe containing only the pGAD10α (A) and pCL2M (B) vectors, respectively, were on the lower halves of the plates.
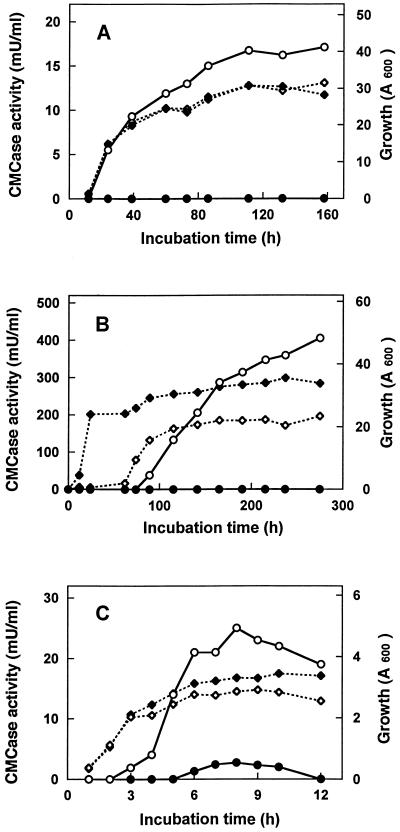
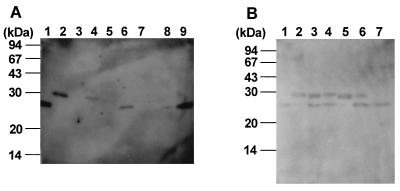
S. cerevisiae SC1-adh-egl3, Schizosaccharomyces pombe SP-cmv-egl3, and the control yeast strains were cultivated in shake flasks with YPDM medium for S. cerevisiae and with the medium containing 25 μg of G418 per ml for Schizosaccharomyces pombe. The S. cerevisiae SC-adh-egl3 strain had the same growth rate as the control strain transformed with the vector pGAD10α. The CMCase activity of the SC-adh-egl3 strain was observed from the beginning of growth and reached as much as 17 mU/ml after 158 h (Fig. 7A). On the other hand, the growth rate of the Schizosaccharomyces pombe SP-cmv-egl3 strain was very low compared with that of the control strain and the time taken to reach the stationary phase was about 5 days. Furthermore, the endoglucanase activity of the supernatant gradually increased beyond the stationary phase and reached 400 mU/ml after 275 h (Fig. 7B). The concentrated culture supernatants were analyzed for EG III protein by SDS-PAGE followed by immunoblotting (Fig. 8A). The apparent molecular masses of the extracellular EG IIIs produced by the yeasts were approximately 28 kDa in S. cerevisiae and 29 kDa in Schizosaccharomyces pombe, which are larger than that of the native enzyme. Treatments of the culture fluids with endo H altered the apparent molecular mass positions of parts of the EG IIIs to about 25 kDa, the same position as that of the purified enzyme from T. reesei (Fig. 8B). Additional α-mannosidase treatment gave a complete shift of the upper band to the 25 kDa band for S. cerevisiae EG III, but no more change from the SDS-PAGE pattern was observed in the Schizosaccharomyces pombe enzyme (Fig. 8B). This result indicates that the extracellular EG IIIs produced in the yeasts are heterogeneously N glycosylated, as has been shown for many other extracellular heterologous proteins expressed in S. cerevisiae and for the T. reesei CBH II expressed in Schizosaccharomyces pombe (14). The native enzyme appeared to remain significantly unaltered by endo H treatment and further α-mannosidase treatment (data not shown). This result is consistent with our previous data (12) and indicates that the T. reesei EG III is not glycosylated.
FIG. 7.
Time courses of cell growth (◊ and ⧫) and CMCase activity (○ and •) with S. cerevisiae (A), Schizosaccharomyces pombe (B), and E. coli (C) transformants. (A) ○ and ◊, S. cerevisiae SC-adh-egl3; • and ⧫, the control S. cerevisiae; (B) ○ and ◊, S. pombe SP-cmv-egl3; • and ⧫, the control Schizosaccharomyces pombe; (C) E. coli 105-AG-egl3 with IPTG addition (○ and ◊) and no addition (• and ⧫).
FIG. 8.
SDS-PAGE and immunostaining of extracellular EG IIIs secreted by S. cerevisiae and Schizosaccharomyces pombe and intracellular EG IIIs produced by E. coli. (A) Purified T. reesei EG III (lane 1), culture supernatants of Schizosaccharomyces pombe SP-cmv-egl3 (lane 2), the control Schizosaccharomyces pombe (lane 3), S. cerevisiae SC-adh-egl3 (lane 4), the control S. cerevisiae (lane 5), crude extracts of E. coli 105-AG-egl3 (lane 6), the control E. coli (lane 7), and the soluble fraction (lane 8) and the cell debris fraction (lane 9) of the crude extract of E. coli 105-AG-egl3. (B) Purified T. reesei EG III (lane 1), recombinant EG III from Schizosaccharomyces pombe (lanes 2 to 4), and recombinant EG III from S. cerevisiae (lanes 5 to 7). Lanes 2 and 5 were untreated, lanes 3 and 6 were treated with endo H, and lanes 4 and 7 were treated with endo H and α-mannosidase.
Expression of mature EG III in E. coli.
The mature form of EG III cDNA was made by PCR amplification to delete the putative signal sequence (see Materials and Methods). Thus, the amplified fragment was introduced into pAG9-3 under the control of the tac promoter and the resulting plasmid, pAGegl3, was transformed into a lacIq strain, JM105. The transformant obtained, 105-AG-egl3, was cultured, and IPTG was added to induce the recombinant EG IIIs. The cell extract of strain 105-AG-egl3 had endoglucanase activity of 25 mU/ml of the medium (Fig. 7C). Most of the produced protein was, however, detected in the cell debris as an inactive form (Fig. 8A), suggesting that the expressed EG III mainly formed an inclusion body without enzyme activity, and the slight amount of the soluble enzyme revealed endoglucanase activity as mentioned above. The EG IIIs in both the supernatant and the cell debris of the 105-AG-egl3 cell lysate had the same molecular size as that of the native enzyme.
DISCUSSION
In this paper we report the sequence and analysis of cDNA and genomic clones encoding low-molecular-mass EG III from T. reesei. For the cloning of egl3 cDNA by the method of PCR, N-terminal and C-terminal sequences of the protein were predicted from the similarity of the deduced sequence from the genomic egl3 gene to that of F1-CMCase from A. aculeatus (17). This prediction was consequently judged to be right after a determination of the amino acid sequence of EG III. The EG III protein sequence was identical to that of the small, high-pI endoglucanase reported by Ward et al. (36). EG III showed significant protein sequence homology with family H cellulases. When those sequences are aligned with EG III, structural homologies are evident, especially at certain conserved domains (Fig. 4). In addition, when the hydropathy profiles are compared, a clear pattern is conserved among these four endoglucanase enzymes (data not shown). Moreover, the putative catalytic site in F1-CMCase (13) is conserved in all these proteins, including EG III (Glu132 and Glu216 in Fig. 4). Hydrophobic amino acids with an aromatic side chain, especially tryptophans, are highly conserved throughout the overall sequence. The importance of their existence is not clear, but they may play a role in substrate recognition, as can be seen in the CBD of CBH I, which stacks with cellulose through the three conserved tyrosines (22). Most of the fungus cellulases studied thus far have the CBD, whereas the family H cellulases containing EG III do not. In our previous report we showed that purified EG III does not adhere to microcrystalline cellulose (Avicel), although the purified CBH I almost attached to the cellulose under those experimental conditions. From these results, EG III is shown to be the first enzyme without the CBD and the linker region among T. reesei cellulases. EG III was also induced by cellulose, as shown in Fig. 5B, possibly in harmony with other cellulases of T. reesei. It was demonstrated in our previous report that EG III synergistically degrades Avicel with T. reesei CBH I. These results suggest that EG III, a cellulase without a CBD, may play an important role in crystalline cellulose degradation by T. reesei. To determine whether the egl3 gene has an essential role in cellulose digestion will require testing of T. reesei with a genetic disruption in egl3. Furthermore, construction of the fusion enzymes by addition of the CBD to EG III will open an alternative way to elucidate the function of the CBD in cellulose degradation.
To study the enzymatic characteristics of EG III without other cellulase activity, we have attempted to produce the recombinant EG III in the heterologous, cellulase-nonproducing hosts S. cerevisiae, Schizosaccharomyces pombe, and E. coli. The yeast transformants secreted EG III enzymes in a catalytically active form, but the molecular masses were larger than that of the native enzyme from T. reesei. The enzymatic removal of the carbohydrate moieties demonstrated that the secreted enzymes were glycosylated. Treatment of the enzymes from S. cerevisiae and Schizosaccharomyces pombe with endo H reduced the molecular masses of a portion of the enzyme molecules to the molecular mass of the T. reesei enzyme, indicating that glycosylation might occur at least partly in the N-linked type. The low hydrolytic activity of endo H on the recombinant enzymes may thus be due to its substrate specificity for the high-mannose-type N-glycans of higher eukaryotes. It also cannot be ruled out that heterogeneous glycosylation took place in each yeast. The resistance of EG III glycans in Schizosaccharomyces pombe to α-mannosidase in contrast to the lack of resistance of EG III glycans in S. cerevisiae to α-mannosidase may be due to addition of galactose to the end of the oligomannosaccharide chains, which frequently occurs in Schizosaccharomyces pombe (11, 28). As shown in Fig. 7, S. cerevisiae SC-adh-egl3 and Schizosaccharomyces pombe SP-cmv-egl3 secreted 17 and 400 mU of endoglucanase activity per ml of culture medium, respectively, and these activities were estimated to represent 1.5 and 36 μg of the secreted protein per ml, respectively, on the basis of the specific activity of 11.2 μmol/min/mg of the purified enzyme from T. reesei (12). The amounts of EG IIIs secreted were rather less than those of CBH IIs in yeasts (14, 21). However, there is still much room for improvement in these systems, for example, the substitution of the EG III signal sequence for another sequence promising higher efficiencies of secretion in the respective yeasts of mating factor α, yeast killer toxin, or other T. reesei cellulases such as CBH II.
Although expression of the mature EG III in E. coli resulted mostly in the formation of enzymatically inactive inclusion bodies, a significant amount of the soluble, active enzyme was produced in the presence of IPTG (Fig. 7C). This study provides the first example showing that a T. reesei cellulase gene can be expressed in an active form as a mature enzyme without fusion in E. coli (10). Furthermore, we have succeeded in the recovery of the active form from the inclusion body by simple treatments (unpublished result).
At present the Schizosaccharomyces pombe expression system seems to be suitable for expressing fusion proteins such as EG III fused with the CBD. On the other hand, the E. coli system appears to be suited to the study of the catalytic function of EG III by site-directed mutagenesis. Further research in our laboratory will be directed toward studies of the contribution of EG III to crystalline cellulose degradation and its structure-function relationship.
ACKNOWLEDGMENTS
This work was partly supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan and a research grant from the Sapporo Bioscience Foundation.
We thank H. Watanabe for a critical reading of the manuscript.
REFERENCES
- 1.Beldman G, Leeuwen M F S-V, Rombouts F M, Voragen F G J. The cellulase of Trichoderma viride. Eur J Biochem. 1985;146:301–308. doi: 10.1111/j.1432-1033.1985.tb08653.x. [DOI] [PubMed] [Google Scholar]
- 2.Claeyssens M, Tomme P. Structure-function relationships of cellulolytic proteins from Trichoderma reesei. In: Kubicek C P, Eveleigh D E, Esterbauer H, Steiner W, Kubicek-Prnz E M, editors. Trichoderma reesei cellulases: biochemistry, genetics, physiology and application. Cambridge, United Kingdom: The Royal Society of Chemistry; 1990. pp. 1–11. [Google Scholar]
- 3.Farkas V, Liskova M, Biely P. Novel media for detection of microbial producers of cellulase and xylanase. FEMS Microbiol Lett. 1985;28:137–140. [Google Scholar]
- 4.Gietz D, St. Jean A, Wood R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giga-Hama Y, Tohda H, Okada H, Owada M K, Okayama H, Kumagai H. High-level expression of human lipocortin I in the fission yeast Schizosaccharomyces pombe using a novel expression vector. Bio/Technology. 1994;12:400–404. doi: 10.1038/nbt0494-400. [DOI] [PubMed] [Google Scholar]
- 6.Hakansson U L, Fagestam L, Pettersson G, Andersson L. Purification and characterization of a low molecular weight 1,4-β-glucan glucanohydrolase from the cellulolytic fungus Trichoderma viride QM9414. Biochim Biophys Acta. 1978;524:385–392. doi: 10.1016/0005-2744(78)90175-4. [DOI] [PubMed] [Google Scholar]
- 7.Innis M A, Gelfand D H. Optimizing of PCRs. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 8.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 9.Knowles J, Lehtovaara P, Teeri T. Cellulase families and their genes. Trends Biotechnol. 1987;5:255–261. [Google Scholar]
- 10.Laymon R A, Adney W S, Mohagheghi A, Himmel M E, Thomas S R. Cloning and expression of full-length Trichoderma reesei cellobiohydrolase I cDNAs in Escherichia coli. Appl Biochem Biotechnol. 1996;57/58:389–397. doi: 10.1007/978-1-4612-0223-3_35. [DOI] [PubMed] [Google Scholar]
- 11.Moreno S, Sanchez Y, Rodriguez L. Purification and characterization of the invertase from Schizosaccharomyces pombe: a comparative analysis with the invertase from Saccharomyces cerevisiae. Biochem J. 1990;267:697–702. doi: 10.1042/bj2670697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morikawa Y, Takahashi A, Yano K, Yamasaki M, Okada H. A low molecular weight endoglucanase from Trichoderma reesei. In: Shimada K, Hoshino S, Ohmiya K, Sakka K, Kobayashi Y, Karita S, editors. Genetics, biochemistry and ecology of lignocellulose degradation. Tokyo, Japan: Uni Publisher Co., Ltd.; 1994. pp. 458–467. [Google Scholar]
- 13.Okada H. Comparisons of primary, secondary and tertiary structures of xylanase of Bacillus pumilus and cellulase of Aspergillus aculeatus. Microbial Utilization of Renewable Resources. 1991;7:1–7. [Google Scholar]
- 14.Okada, H., T. Sekiya, K. Yokoyama, H. Tohda, H. Kumagai, and Y. Morikawa. Efficient secretion of Trichoderma reesei cellobiohydrolase II in Schizosaccharomyces pombe and characterization of its products. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 15.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooi T, Minamiguchi K, Kawaguchi T, Okada H, Murao S, Arai M. Expression of the cellulase (FI-CMCase) gene of Aspergillus aculeatus in Escherichia coli. Biosci Biotechnol Biochem. 1993;57:1960–1961. doi: 10.1271/bbb.57.1960. [DOI] [PubMed] [Google Scholar]
- 17.Ooi T, Shinmyo A, Okada H, Hara S, Ikenaka T, Murao S, Arai M. Cloning and sequence analysis of a cDNA for cellulase (FI-CMCase) from Aspergillus aculeatus. Curr Genet. 1990;18:217–222. doi: 10.1007/BF00318384. [DOI] [PubMed] [Google Scholar]
- 18.Ooi T, Shinmyo A, Okada H, Murao S, Kawaguchi T, Arai M. Complete nucleotide sequence of a gene coding for Aspergillus aculeatus cellulase (FI-CMCase) Nucleic Acids Res. 1990;18:5884. doi: 10.1093/nar/18.19.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penttila M, Lehtovaara P, Nevalainen H, Bhikhabhai R, Knowles J. Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene. 1986;45:253–263. doi: 10.1016/0378-1119(86)90023-5. [DOI] [PubMed] [Google Scholar]
- 20.Penttila M, Andre L, Saloheimo M, Lehtovaara P, Knowles J K C. Expression of two Trichoderma reesei endoglucanases in yeast Saccharomyces cerevisiae. Yeast. 1987;3:175–185. doi: 10.1002/yea.320030305. [DOI] [PubMed] [Google Scholar]
- 21.Penttila M E, Andre L, Lehtovaara P, Bailey M, Teeri T T, Knowles J K C. Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene. 1988;63:103–112. doi: 10.1016/0378-1119(88)90549-5. [DOI] [PubMed] [Google Scholar]
- 22.Reinikainen T, Ruohonen L, Nevanen T, Laaksonen L, Kraulis P, Jones T A, Knowles J K C, Teeri T T. Investigation of the function of mutated cellulose-binding domains of Trichoderma reesei cellobiohydrolase I. Proteins Struct Funct Genet. 1992;14:475–482. doi: 10.1002/prot.340140408. [DOI] [PubMed] [Google Scholar]
- 23.Saarilahti H T, Henrissat B, Palva E T. CelS: a novel endoglucanase identified from Erwinia carotovora subsp. carotovora. Gene. 1990;90:9–14. doi: 10.1016/0378-1119(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto S, Tamura G, Ito K, Ishikawa T, Iwano K, Nishiya N. Cloning and sequencing of cellulase cDNA from Aspergillus kawachii and its expression in Saccharomyces cerevisiae. Curr Genet. 1995;27:435–439. doi: 10.1007/BF00311212. [DOI] [PubMed] [Google Scholar]
- 25.Saloheimo A, Henrissat B, Hoffren A-M, Teleman O, Penttila M. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol Microbiol. 1994;13:219–228. doi: 10.1111/j.1365-2958.1994.tb00417.x. [DOI] [PubMed] [Google Scholar]
- 26.Saloheimo M, Lehtovaara P, Penttila M, Teeri T T, Stahlberg J, Johansson G, Pettersson G, Claeyssens M, Tomme P, Knowles J K C. EG III, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 1988;63:11–21. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Schweingruber A-M, Schoenholzer F, Keller L, Schwaninger R, Trachsel H, Schweingruber M E. Glycosylation and secretion of acid phosphatase in Schizosaccharomyces pombe. Eur J Biochem. 1986;158:133–140. doi: 10.1111/j.1432-1033.1986.tb09730.x. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker S, Schweickaut V, Ladner M, Gelfand D, Kwok S, Myambo K, Innis M. Molecular cloning of exocellobiohydrolase I derived from Trichoderma reesei strain L27. Bio/Technology. 1983;1:691–696. [Google Scholar]
- 30.Sprey B, Uelker A. Isolation and properties of a low molecular mass endoglucanase from Trichoderma reesei. FEMS Microbiol Lett. 1992;92:253–258. doi: 10.1016/0378-1097(92)90718-4. [DOI] [PubMed] [Google Scholar]
- 31.Teeri T, Penttila M, Keranen S, Nevalainen H, Knowles J K C. Structure function and genetics of cellulases. In: Ball C, Finkelstein D, editors. Biotechnology of filamentous fungi. London, United Kingdom: Butterworth; 1991. pp. 419–447. [DOI] [PubMed] [Google Scholar]
- 32.Teeri T T, Lehtovaara P, Kauppinen S, Salovuori I, Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene. 1987;51:43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- 33.Tohda H, Okada H, Giga-Hama Y, Okayama H, Kumagai H. A copy-number-controlled expression vector for the fission yeast Schizosaccharomyces pombe. Gene. 1994;150:275–280. doi: 10.1016/0378-1119(94)90437-5. [DOI] [PubMed] [Google Scholar]
- 34.van Arsdell J N, Kwok S, Schweickart V L, Ladner M B, Gelfand D H, Innis M A. Cloning, characterization and expression in Saccharomyces cerevisiae of endoglucanase I from Trichoderma reesei. Bio/Technology. 1987;5:60–64. [Google Scholar]
- 35.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 36.Ward M, Wu S, Dauberman J, Weiss G, Larenas E, Bower B, Rey M, Clarkson K, Bott R. Cloning, sequence and preliminary structural analysis of a small, high pI endoglucanase (EGIII) from Trichoderma reesei. In: Suominen P, Reinikainen T, editors. Proceedings of the Second Tricel Symposium on Trichoderma Cellulases and Other Hydrolases, Espoo. Vol. 8. Helsinki, Finland: Foundation for Biotechnical and Industrial Fermentation Research; 1993. pp. 153–158. [Google Scholar]
- 37.Wood T M, Bhat K M. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112. [Google Scholar]