Abstract
Manganese peroxidase (MnP) gene expression in the lignin-degrading fungus Phanerochaete chrysosporium is regulated by nutrient nitrogen levels and by Mn(II), the substrate for the enzyme, as well as by heat shock and other factors. Reverse transcription-PCR (RT-PCR) of total RNA can distinguish the mRNAs of each of the three sequenced P. chrysosporium mnp genes, i.e., mnp1, mnp2, and mnp3. Quantitative RT-PCR demonstrates that each of the three transcripts is present at a similar low basal level in nitrogen-sufficient cultures, with or without Mn, and in nitrogen-limited cultures lacking Mn. However, in 5-day-old, nitrogen-limited, stationary cultures supplemented with 180 μM Mn, the levels of the mnp1 and mnp2 transcripts increased approximately 100- and 1,700-fold, respectively, over basal levels. In contrast, under these conditions, the level of the mnp3 transcript did not increase significantly over the basal level. Quantitative RT-PCR of total RNA extracted from nitrogen-deficient, Mn-supplemented cultures on days 2 through 7 demonstrates that whereas the mnp1 transcript was present at relatively low levels on days 3 through 7, the mnp2 transcript level peaked on day 5 and the mnp3 transcript level peaked on day 3. Comparison of total RNA extracted on day 5 from nitrogen-deficient, Mn-supplemented stationary and agitated cultures indicates that in stationary cultures, mnp2 was the major expressed mnp gene, whereas in large agitated cultures, mnp1 was the major expressed mnp gene.
The white rot basidiomycete fungus Phanerochaete chrysosporium has been studied extensively for its ability to degrade lignin (19, 26) and a wide variety of aromatic pollutants (23, 25, 47). Two families of peroxidases, lignin peroxidase (LiP) and manganese peroxidase (MnP), and an H2O2-generating system are the major components of the extracellular lignin-degrading system of this organism (19, 22, 26). P. chrysosporium MnP isozyme 1 is a well-characterized H2O2-requiring heme glycoprotein with an Mr of ∼46,000 (16, 19, 22, 48, 49). The enzyme oxidizes Mn2+ to Mn3+; the latter, complexed with an organic acid chelator such as oxalate, which is secreted by the fungus, oxidizes the terminal phenolic substrate (16, 29, 30, 50) and also possibly nonphenolic substituents via a radical mediator (2, 51). X-Ray crystallographic (44) and site-directed mutagenesis (28, 31) studies have defined the Mn-binding site of MnP.
MnP occurs as a series of isozymes encoded by a family of closely related genes, and the sequences of cDNA (35, 37, 39, 40) and genomic clones (1, 18, 35) of three different mnp genes from P. chrysosporium have been determined. MnP expression is regulated at the level of gene transcription by the depletion of nutrient nitrogen (40). In addition, MnP activity is dependent on the presence of Mn2+ in the culture medium (5, 10), and mnp gene transcription is regulated by Mn2+ (9, 10, 14, 17). To date, this is the only system of Mn regulation of gene transcription to be studied at the molecular level. MnP also is regulated at the level of gene transcription by heat shock (11), H2O2, and other chemical stresses (33). However, there has been no previous study on the differential regulation of mnp gene transcription by Mn, nitrogen limitation, or culture agitation in defined medium.
Reverse transcription-PCR (RT-PCR) has facilitated the analysis of differential expression between and within families of genes involved in lignocellulose degradation, both in liquid cultures (6–8, 12, 41, 43, 45) and in pollutant-contaminated soil (3, 4, 32). In addition to its ability to distinguish among very similar mRNAs, RT-PCR has been shown to be 103- to 104-fold more sensitive than Northern blotting techniques (15, 42).
In this study, RT-PCR was used to further our investigations of mnp gene expression with greater sensitivity and specificity than was possible with our previous northern blot analyses (9–11, 33). Competitive RT-PCR was used to compare differential regulation by Mn of the transcription of genes encoding three MnP isozymes from P. chrysosporium, i.e., mnp1, mnp2, and mnp3, in stationary and agitated cultures.
MATERIALS AND METHODS
Culture conditions.
The P. chrysosporium wild-type strain OGC101 (ATCC 201542), a derivative of BKM-F-1767, was maintained as described previously (20). Stationary cultures were grown at 37°C from conidial inocula in 20 ml of medium in 250-ml Erlenmeyer flasks (10, 27). The medium contained mineral salts with trace elements lacking Mn, with 2% glucose as the carbon source, 1.2 mM (limiting nitrogen) or 12 mM (sufficient nitrogen) ammonium tartrate, and 20 mM sodium-2,2-dimethyl succinate (pH 4.5) as the buffer (10, 27). The cultures were grown in the presence or absence of 180 μM Mn as indicated. Cultures were incubated under air for 2 days and purged on day 3 with 100% O2 for 10 min. For the temporal expression experiments, the cultures were incubated under air for 2 days, after which they were purged daily with 100% O2 for 10 min. Agitated cultures were grown at 28°C from an inoculum of mycelial fragments in 1 liter of medium in 2-liter Erlenmeyer flasks, as described previously (16, 21). The medium was as above, except that it contained 6× trace elements (50), yielding a final Mn concentration of 180 μM, and 0.1% Tween 80. Benzyl alcohol (3 mM) was added on day 3 (50). The cultures were incubated under air for 3 days, then purged daily with 100% O2, and harvested on day 5.
RNA extraction.
The mycelia from 20-ml liquid cultures (∼100 mg/culture) were filtered through Miracloth (Calbiochem), dried between layers of paper towel, frozen rapidly in liquid nitrogen, and stored at −80°C. The frozen mycelia from each of two flasks were separately homogenized with acid-washed glass beads (1 gm) in a mini-bead beater in the presence of 580 μl of guanidinium thiocyanate (Fluka, Inc.) denaturing solution (24), together with 580 μl of diethylpyrocarbonate (DEPC)-treated water-saturated phenol (U.S. Biochemical) and 100 μl of 2 M sodium acetate (pH 4). The supernatants were extracted once with chloroform and once with phenol-chloroform, and the RNA was precipitated with isopropanol. An overnight 4 M LiCl wash of the precipitated RNA aided in the removal of carbohydrates, genomic DNA, and large rRNA. The precipitated RNA was washed with 75% ethanol and solubilized in DEPC-treated water. The absorbance was measured at 260 and 280 nm to quantitate the RNA (∼40 μg/culture).
RT-PCR.
RT-PCR amplifications were performed for RNA samples from each culture for the mnp1, mnp2, and mnp3 mRNAs. As a control, RT-PCRs also were carried out for the gpd mRNA, encoding glyceraldehyde-3-phosphate dehydrogenase (36). Oligonucleotide primers and probes were synthesized at the Oregon Regional Primate Research Center, Beaverton, Oreg. For the annealing reaction (10 μl in DEPC-treated water), 2 μg of total RNA and 3 pmol each of a conserved mnp 3′ primer and a 3′ primer from the gpd coding region (Table 1) were heated at 70°C for 5 min and then cooled slowly to 30°C. The RT reaction (25 μl) was initiated by the addition of 100 U of Moloney murine leukemia virus reverse transcriptase and buffer (GIBCO BRL) and 28 U of RNasin (Promega Biotech) in 10 mM dithiothreitol–1 mM deoxynucleoside triphosphates (U.S. Biochemical) (13), followed by extensions at 42°C for 60 min and 52°C for 30 min. Following the RT reaction, the volume was increased to 100 μl with DEPC-treated water. PCR mixtures (50 μl) contained 1 μl of cDNA, 1 U of Deep Vent polymerase and buffer (New England Biolabs), 30 pmol each of the appropriate gene-specific 5′ primer and the conserved mnp or gpd 3′ primer (Table 1), and deoxynucleoside triphosphates to a final concentration of 400 μM. The PCR temperature program was 94°C for 6 min, 54°C for 2 min, and 72°C for 40 min for 1 cycle followed by 94°C for 1 min, 54°C for 2 min, and 72°C for 5 min for 35 cycles and with a final 15-min extension at 72°C (43). Following amplification, 5 μl of each PCR product was analyzed by electrophoresis in a 1.5% agarose gel and stained with ethidium bromide. The gels were visualized with UV light and photographed.
TABLE 1.
Oligonucleotide primers and probes used in this studya
| Gene | Sequence of:
|
Size (bp)b:
|
Sequence of probe | ||
|---|---|---|---|---|---|
| 5′ Primer | 3′ Primer | cDNA | gDNA | ||
| mnp1 | CCGGTCAACGGCTTGGTATTC | CGTTGTTGG(C/G/A)(G/A)AGAAGTTGG | 336 | 512 | CGCTGTTCGTGCTGCCCCCACTGCGGT |
| mnp2 | AGCTCTCAAGGACATCCGCAC | CGTTGTTGG(C/G/A)(G/A)AGAAGTTGG | 346 | 565 | CATAACTCGCGCCGCCCCGACTGCGGA |
| mnp3 | CCGACGGTACCAAGGTCAACc | CGTTGTTGG(C/G/A)(G/A)AGAAGTTGG | 229 | 399 | CATTTCGCAAAGCAAGGGACCGAGCG |
| gpd | CGTATCGTCCTCCGTAATGC | ACTCGTTGTCGTACCAGGAG | 905 | 1106 | |
Sequences are based on previous studies as follows: mnp1 (18), mnp2 (35), mnp3 (1), and gpd (36). GenBank accession numbers are as follows: M77513 (mnp1), S69963 (mnp2), U70998 (mnp3), and M81754 (gpd).
Size predicted for PCR amplification products with cDNA and genomic (gDNA) templates.
Primer sequence from reference 3.
Competitive RT-PCR.
Competitive RT-PCRs were conducted as above, except that a series of known concentrations of plasmids containing the full-length genomic sequences for each gene (1, 18, 35, 36) were added as competitive templates to the appropriate amplification reaction mixtures (15). Introns within the competitive templates allowed the cDNAs and the genomic products to be separated on agarose gels (Fig. 1). For quantitation, photographs of the ethidium bromide-stained gels were scanned and densitometry was performed using IPLab Gel software (Signal Analytics Corp.). The transcript concentrations were estimated by determining the concentration of competitive DNA at which the intensities of the gDNA and cDNA targets (PCR fragments) were equal, taking into account the increased ethidium bromide staining of the larger gDNA target (15). Since transcript concentrations were expressed as picograms per 20 ng of total RNA, adjustments for the relative molar concentrations of the competitive genomic DNAs and the cDNAs and their respective targets were also made.
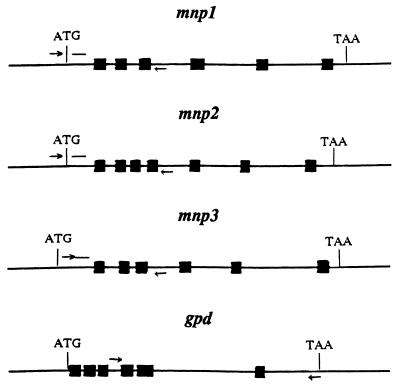
FIG. 1.
Strategy for PCR amplification of three mnp genes and the gpd gene from P. chrysosporium. The ATG translation start codons and the TAA stop codons are indicated. Solid boxes represent introns. Positions of the upstream primers (→), downstream primers (←) and gene-specific oligonucleotide probes (—) are indicated.
Southern blot analysis.
Southern blots of the mnp PCR products were analyzed with gene-specific oligonucleotide probes (Table 1) to ensure the specificity of each primer pair for its target sequence. PCRs were performed as described above, with each of the mnp primer pairs and P. chrysosporium genomic DNA (100 ng) as the template. RT-PCRs were performed as described above, with each of the mnp primer pairs and total RNA extracted from 5-day-old, nitrogen-limited, Mn-sufficient cultures. A 5-μl portion of each PCR product and 500 pg each of the EcoRI-linearized mnp genomic plasmids were separated on a 1.5% agarose gel and stained with ethidium bromide. The DNA was transferred to Magna NT membranes (Micron Separations, Inc.). Identical blots were hybridized with oligonucleotide probes corresponding to each mnp gene (Table 1). The probes (10 pmol) were end labeled with [γ-32P]dATP (Andotek Life Sciences), using T4 polynucleotide kinase (New England Biolabs, Inc.). Hybridizations were carried out at 55°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% sodium dodecyl sulfate–5× Denhardt’s solution–25 mM sodium phosphate (pH 7.0)–100 μg of herring sperm DNA per ml. The blots were washed in 6× SSC–1% sodium dodecyl sulfate three times at room temperature for 5 min each followed by twice at 50°C for 2 min each. The washed blots were exposed to Kodak XAR film.
RESULTS
Expression of individual mnp transcripts in stationary cultures.
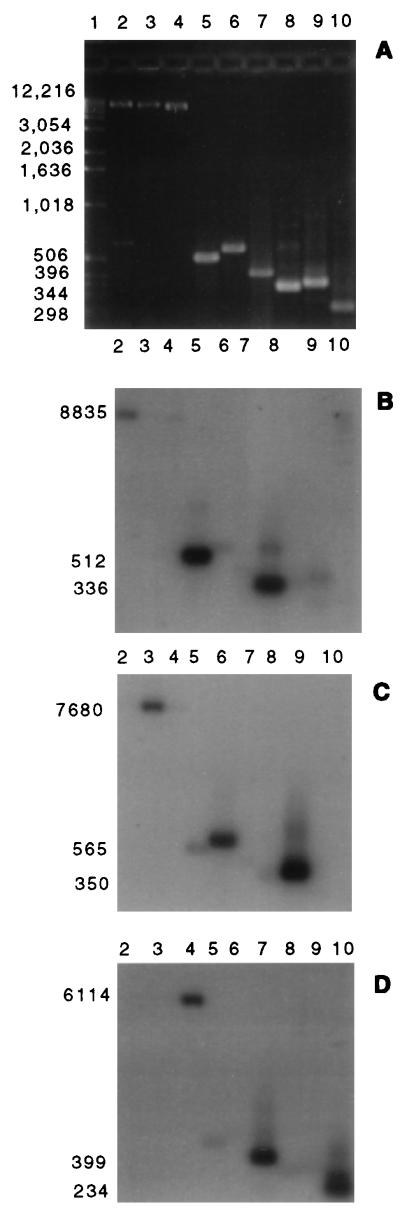
Nitrogen-sufficient and nitrogen-limited stationary cultures of P. chrysosporium OGC101 were grown at 37°C in the absence of Mn and in the presence of 180 μM Mn. On day 5, the cultures were harvested separately and the RNA was extracted. By using RT-PCR with mnp gene-specific primer pairs, the three mnp transcripts of the predicted sizes (Table 1; Fig. 1) were detected under all conditions tested. Southern blot hybridizations with mnp gene-specific oligonucleotide probes (Table 1) confirmed the identities of the PCR products and the specificities of the PCR primer pairs. No genomic DNA contamination was detectable among the PCR products derived from total RNA (Fig. 2).
FIG. 2.
Southern blots of mnp genomic plasmids, PCR amplification of P. chrysosporium genomic DNA, and RT-PCR of total RNA extracted from 5-day-old, nitrogen-limited, Mn-sufficient cultures probed with mnp gene-specific oligonucleotide probes. (A) Ethidium bromide-stained agarose gel. (B) Southern blot of the gel in panel A, probed with the mnp1-specific oligonucleotide. (C) Southern blot of the gel in panel A, probed with the mnp2-specific oligonucleotide. (D) Southern blot of the gel in panel A, probed with the mnp3-specific oligonucleotide. Lanes: 1, molecular size markers, indicated on the left of panel A in base pairs; 2 to 4, EcoRI-linearized plasmids containing the mnp1 gene (lane 2), the mnp2 gene (lane 3), or the mnp3 gene (lane 4); 5 to 7, PCR amplification of P. chrysosporium genomic DNA with the conserved mnp downstream primer and the mnp1 (lane 5), mnp2 (lane 6), and mnp3 (lane 7) upstream primers; 8 to 10, RT-PCR of total RNA with the conserved mnp downstream primer and the mnp1 (lane 8), mnp2 (lane 9), and mnp3 (lane 10) upstream primers. Sizes (in base pairs) of the probed plasmids and genomic and cDNA amplification products are indicated on the left of panel B through D. Oligonucleotide probes and primers are listed in Table 1.
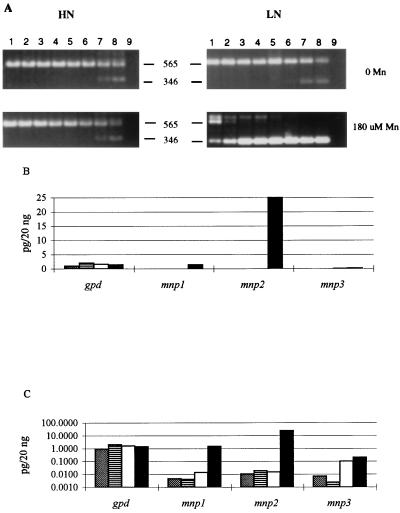
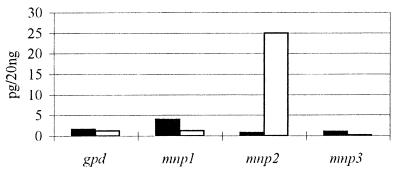
Competitive RT-PCR was used to quantitate the three mnp transcripts derived from duplicate cultures grown under each set of conditions. The RT reactions were as above, except that a 3′ gpd primer was included in addition to the conserved mnp 3′ primer (Table 1). For each of the four cDNA templates, eight identical PCR mixtures were spiked with serial dilutions of the appropriate competitive genomic template. A ninth reaction mixture lacked cDNA and was used as a control. The PCR products were analyzed by gel electrophoresis and ethidium bromide staining. The initial concentration of each cDNA was determined by observing the dilution of the genomic template at which the genomic and cDNA products were equivalent (Fig. 3A). After adjustments for the relative intensities of the larger genomic PCR products and for the relative molar concentrations of the genomic plasmids, the gDNA targets, the cDNAs, and the cDNA targets, the mnp transcript concentrations ranged from 10−3 to 25 pg/20 ng of total RNA. The gpd transcript concentration was between 1 and 2 pg/20 ng of total RNA under all conditions tested (Fig. 3B and C). The transcript concentrations in RNAs extracted from two identical cultures varied by less than 25%. The PCR target concentrations from two parallel PCRs of RNA from the same culture were essentially identical. The results with stationary cultures in Fig. 3B and C demonstrate that under nitrogen-sufficient conditions, the concentrations of the mnp transcripts were unaffected by the presence of Mn, with levels ranging from 10−2 pg for mnp2 to 2 × 10−3 pg for mnp3. However, under nitrogen-limited conditions, the addition of Mn to the culture medium resulted in an approximate 100-fold increase in the level of the mnp1 transcript and a 1,700-fold increase in the level of the mnp2 transcript. Less than a twofold increase of mnp3 transcript was observed on day 5 under nitrogen-limited conditions in the presence of Mn (Fig. 3B and C).
FIG. 3.
Competitive RT-PCR and quantitation of mnp and gpd transcripts in total RNA extracted from 5-day-old stationary cultures of P. chrysosporium. (A) Ethidium bromide-stained gels of competitive RT-PCR products (5 μl) obtained with the mnp2-specific upstream primer (Table 1). Cultures were nitrogen sufficient (HN) or nitrogen limited (LN) and contained 0 or 180 μM Mn, as indicated. RT-PCRs were performed as described in the text. Lanes: 1 to 8, PCR mixtures were spiked with 100, 50, 10, 5, 1, 0.1, and 0.05 pg, respectively, of plasmid containing the mnp2 gene; 9, the PCR mixture lacked both cDNA and plasmid DNA. The sizes of the genomic and cDNA PCR products are indicated. (B) Bar graph of transcript levels, determined from gels as shown for mnp2 in panel A. (C) Log scale bar graph of the transcript levels in panel B. Culture conditions: HN, no Mn (▧); HN, 180 μM Mn (▤); LN, no Mn (□); LN, 180 μM Mn (▪).
Temporal expression of mnp transcripts.
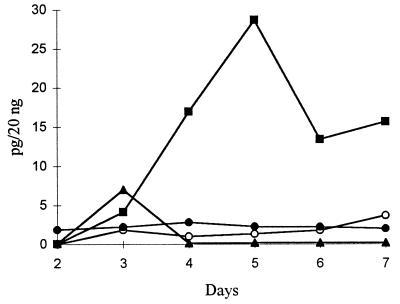
To determine the expression profiles over time of the mnp transcripts under nitrogen limitation, cultures of OGC101 were grown at 37°C in the presence of 180 μM Mn. cDNA was prepared from RNA isolated from each of two flasks on days 2 to 7 following inoculation, and competitive PCRs were performed as above. The transcript concentrations determined for RNAs extracted from duplicate cultures were averaged. There was less than a 25% variation between duplicate cultures. All three mnp transcripts were detectable at very low levels on day 2 (Fig. 4). The mnp3 transcript level, 0.1 pg/20 ng of RNA, was the highest level for the three transcripts on day 2. The mnp1 transcript level increased on day 3 by approximately 300-fold over day 2 and remained relatively constant through day 7. The mnp2 transcript level increased gradually on days 3 and 4, peaking on day 5 at approximately 1,000-fold higher than the level on day 2 and then declining on days 6 and 7. The mnp3 transcript level peaked on day 3 at approximately 75-fold higher than the level on day 2, returning to low levels on days 4 to 7. The levels of gpd transcript remained approximately constant on days 2 to 7 (Fig. 4). The compression of the basal-level data in Fig. 4 slightly misrepresents the increases in the mnp1 and mnp2 transcript levels described above.
FIG. 4.
Time course of mnp and gpd transcript levels. Nitrogen-limited, Mn-sufficient stationary cultures of P. chrysosporium were harvested and the RNA was extracted on the indicated days. Competitive RT-PCRs and quantitation were performed as described in the text. Symbols: ○, mnp1; ▪, mnp2; ▴, mnp3; •, gpd.
Expression of mnp transcripts in agitated cultures.
Competitive RT-PCR was performed on RNA extracted from 5-day-old, nitrogen-limited, Mn-sufficient cultures grown under agitated conditions, and the mnp transcript levels were compared with those from 5-day-old stationary cultures (Fig. 5). The mnp1 transcript level from agitated cultures was approximately threefold higher than that from stationary cultures. In contrast, the level of the mnp2 transcript from agitated cultures was approximately 34-fold lower than that from stationary cultures. Low levels of the mnp3 transcript were detected under both conditions. The levels of the gpd transcript were not affected (Fig. 5).
FIG. 5.
Bar graphs of mnp and gpd transcript levels in total RNA extracted from 5-day-old, nitrogen-limited, Mn-sufficient agitated (▪) or stationary (□) cultures of P. chrysosporium. Competitive RT-PCRs and quantitation were performed as described in the text.
DISCUSSION
The lignin-degradative system of P. chrysosporium is expressed during secondary metabolic (idiophasic) growth, whose onset is triggered by limiting nutrient nitrogen (19, 26). Likewise, LiP and MnP activities are detectable in the extracellular culture medium only during the secondary metabolic phase of growth (19, 26), and Northern blot analysis demonstrates that LiP and MnP expression is controlled at the level of gene transcription by nutrient nitrogen (34, 40). It has been reported that lip genes are differentially regulated in response to carbon and nitrogen (43). In addition, enzyme assays and Western blotting results have suggested that the MnP protein isozymes may be differentially regulated by carbon and nitrogen (38). The latter study also suggested that MnP isozymes might be differentially regulated by Mn, although this was not quantitated. In contrast, a recent report suggests that mnp genes are coordinately expressed during the bioremediation of contaminated soil by P. chrysosporium (3). The results of the present study constitute the first report of differential regulation of mnp gene transcription by any culture factor including Mn.
The mnp1, mnp2, and mnp3 promoter regions contain multiple putative consensus metal response elements (MREs) (1, 18, 35) that are identical to the cis-acting sequences responsible for heavy metal induction of mouse and other metallothionein genes (46). The mnp1, mnp2, and mnp3 promoters also contain putative heat shock elements (1, 18, 35). In this study, we used competitive RT-PCR to examine nitrogen and Mn regulation of the expression of individual mnp genes with greater sensitivity than is possible by Northern blot analysis.
Our earlier studies indicated that MnP activity and mnp RNA are observed only in nitrogen-limited cultures in the presence of Mn and reach a maximum in 5-day-old cultures (9, 10, 16). Our quantitative RT-PCR results in Fig. 3 demonstrate that all three mnp transcripts are present at very low levels on day 5 under nitrogen-sufficient conditions in the presence or absence of Mn, as well as under nitrogen-limited conditions in the absence of Mn. These levels (<0.02 pg/20 ng of total RNA) are below the usual limits of detection by Northern blot analysis. Furthermore, we did not detect MnP activity in the extracellular medium under these conditions (10). Thus, the three mnp genes are expressed at a basal level in the presence of sufficient nitrogen when the lignin-degradative system is not active, and this basal expression appears to be unaffected by Mn (Fig. 3), suggesting that the Mn-responsive MnP regulatory system is not functional under nitrogen-sufficient conditions. Low basal-level expression of a lip gene from P. chrysosporium also has been detected by RT-PCR under nitrogen-sufficient conditions (8).
The results in Fig. 3 also demonstrate that in the presence of 180 μM Mn, the levels of mnp1 and mnp2 transcripts from 5-day-old nitrogen-limited stationary cultures increase approximately 100- and 1,700-fold, respectively, over the basal levels. These transcript levels correlate well with previous estimates, indicating that under secondary metabolic conditions, MnP constitutes at least 2% of the total protein in P. chrysosporium (40). Under these conditions, the level of mnp3 transcript on day 5 is not increased significantly over the basal levels (Fig. 3B and C), suggesting that the mnp3 gene may not be regulated by Mn. The mnp3 promoter lacks paired putative MREs (1). In contrast, the mnp1 promoter has two pairs of putative MREs (18) and the mnp2 promoter has one pair (35).
Our earlier results with Northern blots (9, 10) indicate that mnp gene transcription in nitrogen-limited, Mn-sufficient cultures is first detectable on day 3 and peaks on day 5. The results in Fig. 4 confirm that total mnp mRNA level is highest on day 5. However, Fig. 4 demonstrates that this peak is caused almost entirely by mnp2 mRNA; mnp1 mRNA is present at relatively low levels from days 3 through 7, and the mnp3 mRNA level peaks on day 3. In contrast to these results, RT-PCR of RNA isolated from soil-grown P. chrysosporium indicated the coordinate regulation of these mnp genes (3). However, as demonstrated above, other culture conditions appear to significantly affect the regulation of individual mnp genes.
The above experiments yielded consistently higher levels of mnp2 transcript, compared with mnp1 transcript, in stationary cultures. In contrast, MnP1 is the primary MnP isozyme isolated from large agitated cultures of P. chrysosporium, which are grown at 28°C to maximize MnP1 protein production (16, 19). Therefore, quantitative RT-PCR was performed on RNA extracted from 5-day-old cultures grown under both conditions. The results shown in Fig. 5 demonstrate that the levels of mnp1 transcript are approximately threefold higher in large agitated cultures than in small stationary cultures. Furthermore, mnp2 transcript levels are approximately 34-fold lower in agitated cultures. Therefore, in agitated cultures, the level of the mnp1 transcript is approximately 5-fold higher than the level of the mnp2 transcript whereas in stationary cultures, the level of the mnp2 transcript is approximately 17-fold higher than the level of the mnp1 transcript. The levels of the mnp3 transcript apparently are unaffected by these culture conditions. Furthermore, the low levels of mnp3 do not appear to be regulated by Mn in agitated or stationary cultures. Stationary and agitated cultures differ in several aspects, including the type of inoculum and the growth temperature. However, mycelium grown either at 28 or at 37°C in agitated culture yields similar levels of the mnp transcripts (data not shown). The mycelial mat that forms under stationary conditions may differ from the mycelial pellets that form under agitated conditions in uptake of O2 or nutrients.
In conclusion, the results demonstrate that the mnp1 and mnp2 genes from P. chrysosporium are regulated by Mn in nitrogen-limited, 5-day-old cultures whereas the mnp3 gene does not appear to be significantly regulated by Mn. The mnp3 transcript peaks on day 3 and then returns to basal levels. In contrast, the mnp2 transcript predominates in day 5 stationary cultures and the mnp1 transcript predominates in agitated cultures. These results indicate that the P. chrysosporium mnp genes are differentially regulated at the transcriptional level in response to Mn and other culture conditions. These results support the hypothesis that multiple mnp genes are required to ensure expression under different environmental conditions. The results also demonstrate that the mnp3 gene is not strongly regulated, if at all, by Mn. The exact physiological conditions under which mnp3 is expressed are under investigation. The dramatic difference between mnp1 and mnp2 transcription in agitated and stationary cultures may be significant in attempts to overexpress single isozymes in large fermentors.
ACKNOWLEDGMENTS
This work was supported by grants MCB-9405978 and MCB-9723725 from the National Science Foundation.
REFERENCES
- 1.Alic M, Akileswaran L, Gold M H. Characterization of the gene encoding manganese peroxidase isozyme 3 from Phanerochaete chrysosporium. Biochim Biophys Acta. 1997;1338:1–7. doi: 10.1016/s0167-4838(96)00235-x. [DOI] [PubMed] [Google Scholar]
- 2.Bao W, Fukushima Y, Jensen K A, Jr, Moen M A, Hammel K E. Oxidative degradation of non-phenolic lignin during lipid peroxidation by fungal manganese peroxidase. FEBS Lett. 1994;354:297–300. doi: 10.1016/0014-5793(94)01146-x. [DOI] [PubMed] [Google Scholar]
- 3.Bogan B W, Schoenike B, Lamar R T, Cullen D. Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogan B W, Schoenike B, Lamar R T, Cullen D. Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:3697–3703. doi: 10.1128/aem.62.10.3697-3703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnarme P, Jeffries T W. Mn(II) regulation of lignin peroxidases and manganese-dependent peroxidases from lignin-degrading white-rot fungi. Appl Environ Microbiol. 1990;56:210–217. doi: 10.1128/aem.56.1.210-217.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broda P, Birch P R, Brooks P R, Sims P F. PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl Environ Microbiol. 1995;61:2358–2364. doi: 10.1128/aem.61.6.2358-2364.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broda P, Birch P R, Brooks P R, Sims P F. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol Microbiol. 1996;19:923–932. doi: 10.1046/j.1365-2958.1996.474966.x. [DOI] [PubMed] [Google Scholar]
- 8.Brooks P, Sims P, Broda P. Isozyme specific polymerase chain reaction analysis of differential gene expression: a general method applied to lignin peroxidase genes of Phanerochaete chrysosporium. Bio/Technology. 1993;11:830–834. doi: 10.1038/nbt0793-830. [DOI] [PubMed] [Google Scholar]
- 9.Brown J A, Alic M, Gold M H. Manganese peroxidase gene transcription in Phanerochaete chrysosporium: activation by manganese. J Bacteriol. 1991;173:4101–4106. doi: 10.1128/jb.173.13.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown J A, Glenn J K, Gold M H. Manganese regulates expression of manganese peroxidase by Phanerochaete chrysosporium. J Bacteriol. 1990;172:3125–3130. doi: 10.1128/jb.172.6.3125-3130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown J A, Li D, Alic M, Gold M H. Heat shock induction of manganese peroxidase gene transcription in Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:4295–4299. doi: 10.1128/aem.59.12.4295-4299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Covert S F, Vanden Wymelenberg A, Cullen D. Structure, organization, and transcription of a cellobiohydrolase gene cluster from Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:2168–2175. doi: 10.1128/aem.58.7.2168-2175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gettemy J M, Li D, Alic M, Gold M H. Truncated gene reporter system for studying the regulation of manganese peroxidase. Curr Genet. 1997;31:519–524. doi: 10.1007/s002940050239. [DOI] [PubMed] [Google Scholar]
- 15.Gilliland G, Perrin S, Bunn H F. Competitive PCR for quantitation of mRNA. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 60–69. [Google Scholar]
- 16.Glenn J K, Gold M H. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 17.Godfrey B J, Akileswaran L, Gold M H. A reporter gene construct for studying the regulation of manganese peroxidase gene expression. Appl Environ Microbiol. 1994;60:1353–1358. doi: 10.1128/aem.60.4.1353-1358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godfrey B J, Mayfield M B, Brown J A, Gold M H. Characterization of a gene encoding a manganese peroxidase from Phanerochaete chrysosporium. Gene. 1990;93:119–124. doi: 10.1016/0378-1119(90)90144-g. [DOI] [PubMed] [Google Scholar]
- 19.Gold M H, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold M H, Cheng T M, Mayfield M B. Isolation and complementation studies of auxotrophic mutants of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1982;44:996–1000. doi: 10.1128/aem.44.4.996-1000.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold M H, Kuwahara M, Chiu A A, Glenn J K. Purification and characterization of an extracellular H2O2-requiring diarylpropane oxygenase from the white rot basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys. 1984;234:353–362. doi: 10.1016/0003-9861(84)90280-7. [DOI] [PubMed] [Google Scholar]
- 22.Gold M H, Wariishi H, Valli K. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. ACS Symp Ser. 1989;389:127–140. [Google Scholar]
- 23.Hammel K E. Organopollutant degradation by ligninolytic fungi. Enzyme Microb Technol. 1989;11:776–777. [Google Scholar]
- 24.Janssen K. Single-step RNA isolation from cultured cells or tissues. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Brooklyn, N.Y: Greene Publishing Associates; 1987. pp. 4.2.4–4.2.6. [Google Scholar]
- 25.Joshi D K, Gold M H. Degradation of 2,4,5-trichlorophenol by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:1779–1785. doi: 10.1128/aem.59.6.1779-1785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirk T K, Farrell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 27.Kirk T K, Schultz E, Connors W J, Lorenz L F, Zeikus J G. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol. 1978;117:277–285. [Google Scholar]
- 28.Kishi K, Kusters-van Someren M, Mayfield M B, Sun J, Loehr T M, Gold M H. Characterization of manganese(II) binding site mutants of manganese peroxidase. Biochemistry. 1996;35:8986–8994. doi: 10.1021/bi960679c. [DOI] [PubMed] [Google Scholar]
- 29.Kishi K, Wariishi H, Marquez L, Dunford H B, Gold M H. Mechanism of manganese peroxidase compound II reduction. Effect of organic acid chelators and pH. Biochemistry. 1994;33:8694–8701. doi: 10.1021/bi00195a010. [DOI] [PubMed] [Google Scholar]
- 30.Kuan I C, Tien M. Stimulation of Mn peroxidase activity: a possible role for oxalate in lignin biodegradation. Proc Natl Acad Sci USA. 1993;90:1242–1246. doi: 10.1073/pnas.90.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusters-van Someren M, Kishi K, Lundell T, Gold M H. The manganese binding site of manganese peroxidase: characterization of an Asp179Asn site-directed mutant protein. Biochemistry. 1995;34:10620–10627. doi: 10.1021/bi00033a037. [DOI] [PubMed] [Google Scholar]
- 32.Lamar R T, Schoenike B, Vanden Wymelenberg A, Stewart P, Dietrich D M, Cullen D. Quantitation of fungal mRNAs in complex substrates by reverse transcription PCR and its application to Phanerochaete chrysosporium-colonized soil. Appl Environ Microbiol. 1995;61:2122–2126. doi: 10.1128/aem.61.6.2122-2126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Alic M, Brown J A, Gold M H. Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Appl Environ Microbiol. 1995;61:341–345. doi: 10.1128/aem.61.1.341-345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, Alic M, Gold M H. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayfield M B, Godfrey B J, Gold M H. Characterization of the mnp2 gene encoding manganese peroxidase isozyme 2 from the basidiomycete Phanerochaete chrysosporium. Gene. 1994;142:231–235. doi: 10.1016/0378-1119(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 36.Mayfield M B, Kishi K, Alic M, Gold M H. Homologous expression of recombinant manganese peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:4303–4309. doi: 10.1128/aem.60.12.4303-4309.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orth A B, Rzhetskaya M, Cullen D, Tien M. Characterization of a cDNA encoding a manganese peroxidase from Phanerochaete chrysosporium: genomic organization of lignin and manganese peroxidase-encoding genes. Gene. 1994;148:161–165. doi: 10.1016/0378-1119(94)90251-8. [DOI] [PubMed] [Google Scholar]
- 38.Pease E A, Tien M. Heterogeneity and regulation of manganese peroxidase from Phanerochaete chrysosporium. J Bacteriol. 1992;174:3532–3540. doi: 10.1128/jb.174.11.3532-3540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pease E A, Andrawis A, Tien M. Manganese-dependent peroxidase from Phanerochaete chrysosporium. Primary structure deduced from cDNA sequence. J Biol Chem. 1989;264:13531–13535. [PubMed] [Google Scholar]
- 40.Pribnow D G, Mayfield M B, Nipper V J, Brown J A, Gold M H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989;264:5036–5040. [PubMed] [Google Scholar]
- 41.Reiser J, Walther I S, Fraefel C, Fiechter A. Methods to investigate the expression of lignin peroxidase genes by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:2897–2903. doi: 10.1128/aem.59.9.2897-2903.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sooknanan R, Malek L, Wang X H, Siebert T, Keating A. Detection and direct sequence identification of BCR-ABL mRNA in Ph+ chronic myeloid leukemia. Exp Hematol. 1993;21:1719–1724. [PubMed] [Google Scholar]
- 43.Stewart P, Kersten P, Vanden Wymelenberg A, Gaskell J, Cullen D. Lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation and identification of a second dimorphic chromosome. J Bacteriol. 1992;174:5036–5042. doi: 10.1128/jb.174.15.5036-5042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sundaramoorthy M, Kishi K, Gold M H, Poulos T L. The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06-Å resolution. J Biol Chem. 1994;269:32759–32767. [PubMed] [Google Scholar]
- 45.Tempelaars C A, Birch P R, Sims P F, Broda P. Isolation, characterization, and analysis of the expression of the cbhII gene of Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:4387–4393. doi: 10.1128/aem.60.12.4387-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiele D J. Metal-regulated transcription in eukaryotes. Nucleic Acids Res. 1992;20:1183–1191. doi: 10.1093/nar/20.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valli K, Wariishi H, Gold M H. Degradation of 2,7-dichlorodibenzo-p-dioxin by the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Bacteriol. 1992;174:2131–2137. doi: 10.1128/jb.174.7.2131-2137.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wariishi H, Akileswaran L, Gold M H. Manganese peroxidase from the basidiomycete Phanerochaete chrysosporium: spectral characterization of the oxidized states and the catalytic cycle. Biochemistry. 1988;27:5365–5370. doi: 10.1021/bi00414a061. [DOI] [PubMed] [Google Scholar]
- 49.Wariishi H, Dunford H B, MacDonald I D, Gold M H. Manganese peroxidase from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Transient-state kinetics and reaction mechanism. J Biol Chem. 1989;264:3335–3340. [PubMed] [Google Scholar]
- 50.Wariishi H, Valli K, Gold M H. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium: kinetic mechanism and role of chelators. J Biol Chem. 1992;267:23688–23695. [PubMed] [Google Scholar]
- 51.Wariishi H, Valli K, Renganathan V, Gold M H. Thiol-mediated oxidation of nonphenolic lignin model compounds by manganese peroxidase of Phanerochaete chrysosporium. J Biol Chem. 1989;264:14185–14191. [PubMed] [Google Scholar]