Abstract
Our objective was to determine if 4-methylumbelliferyl-labelled enzyme substrates could be used to detect and quantify specific components of chitinase and cellulase activities as specific indicators of the presence and activity of fungal biomass. The fluorogenic substrates 4-methylumbelliferyl (MUF) N-acetyl-β-d-glucosaminide and MUF β-d-lactoside were used for the detection and quantification of β-N-acetylglucosaminidase (EC 3.2.1.30) (NAGase) and endo 1,4-β-glucanase (EC 3.2.1.4)/cellobiohydrolase (EC 3.2.1.91) (CELase), respectively. Culture screenings on solid media showed a widespread ability to produce NAGase among a taxonomically diverse selection of fungi on media with and without added chitin. NAGase activity was expressed only in a limited number of bacteria and on media supplemented with chitin. The CELase activity was observed only in a limited number of fungi and bacteria. Bacterial CELase activity was expressed on agar media containing a cellulose-derived substrate. In soil samples, NAGase activity was significantly correlated with estimates of fungal biomass, based on the content of two fungus-specific indicator molecules, 18:2ω6 phospholipid fatty acid (PLFA) and ergosterol. CELase activity was significantly correlated with the PLFA-based estimate of fungal biomass in the soil, but no correlation was found with ergosterol-based estimates of fungal biomass.
The determination of enzyme activities is a simple approach to the study of microbially mediated processes within the soil environment. Thus, soil enzyme activities have been interpreted as indirect measures of microbial biomass, rhizosphere effects, soil productivity, and mineralization potential of naturally occurring substrates or xenobiotics (4). However, few studies have attempted to correlate soil enzyme activities with the presence and activities of specific components of the microbial community. The ability of soil-inhabiting fungi to produce a range of enzymes capable of degrading complex litter substances could make the use of an enzymatic approach to study soil fungal populations possible. These enzymes must be specific for fungal presence and activity. In one study of chitinase in soil (24), chitinase activity and the number of fungal propagules in chitin-amended soils were strongly correlated. The same correlation was not found for actinomycetes or bacteria. Thus, chitinase activity appears to be a suitable indicator of actively growing fungi in the soil. The hydrolysis of cellulose requires the interaction of a number of hydrolases produced by cellulolytic microorganisms. A major role is played by the cellulase system, which consists of several distinct enzymes that are produced by a large number of microorganisms, including fungi, actinomycetes, and bacteria. However, fungi have been suggested as the predominant source of β-d-glucosidase (EC 3.2.1.21) (16, 17) and endo 1,4-β-glucanase (EC 3.2.1.4) (23) activity in soils.
Fluorogenic 4-methylumbelliferyl (MUF)-labelled enzyme substrates have been introduced for process-oriented studies in aquatic systems (3, 18) and, more recently, in peatlands (11). MUF substrates have been used to assay cell-bound activities in pure cultures of fungi, as the soluble substrate can enter the cell wall, making periplasmic enzyme activity detectable (15). These substrates have been used to detect fungal chitinolytic activities (17a) and cellulases (6) in vitro. The substrates may be added to environmental samples and, when hydrolyzed, release 4-methyl-umbelliferone (4-MU), which fluoresces and can be quantified in nanomolar concentrations (3).
A variety of methods to quantify fungi in soil have been described. The techniques include direct microscopic observation and extraction of fungus-specific indicator molecules such as glucosamine or ergosterol (9). More recently, the phospholipid fatty acid (PLFA) 18:2ω6 has been proposed as an indicator of fungal biomass (7, 12). Our objectives in the present study were to determine if (i) components of chitinase and cellulase activities could be used as indicators of the presence and activity of fungal biomass and (ii) enzyme activities detected with specific MUF substrates in soil samples were correlated with the content of the fungus-specific indicator molecules 18:2ω6 PLFA and ergosterol.
MATERIALS AND METHODS
Preparation, incubation, and sampling.
Soil samples were collected from Danish and Italian sites in September 1994 (Table 1). The soils were sent to the GSF-Forschungszentrum für Umwelt- und Gesundheit GmbH, Institute für Bodenökologie, Neuherberg, Germany, where the soils were incubated. Soils were incubated in containers 7 cm high and 10 cm in diameter. To each container was added 700 g of rewetted equilibrated soil with maize litter either incorporated or left on top. Samples were taken after 14, 30, 60, 120, 240, and 360 days of incubation. Ergosterol and enzymes were measured for each sample. PLFA analysis was performed on the 14-, 30-, 120-, and 360-day-old samples. Enzyme assays were performed at the University of Copenhagen, Copenhagen, Denmark, ergosterol determinations were performed at the University of Innsbruck, Innsbruck, Austria, and PLFA analyses were performed at the GSF Institute für Bodenökologie.
TABLE 1.
Main chemical and physical parameters of the experimental soils
| Soil | pH | Texture (%)
|
Corg (%)a | Nt (%)b | Density (g/cm3) | ||
|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | |||||
| Danish | 6.6 | 47 | 43 | 10 | 1.4 | 0.15 | 1.52 |
| Italian | 6.3 | 37 | 39 | 24 | 1.5 | 0.12 | 1.24 |
Corg, total soil organic C.
Nt, total soil N.
Enzyme activity screening on solid media.
4-Methylumbelliferyl (MUF) N-acetyl-β-d-glucosaminide and MUF β-d-lactoside (Sigma Chemical Co.) were used for the detection of β-N-acetylglucosaminidase (EC 3.2.1.30) (NAGase) (19) and endo 1,4-β-glucanase (EC 3.2.1.4)/cellobiohydrolase (CELase) (EC 3.2.1.91) (6), respectively. A diverse selection of fungi and bacteria were screened on soil extract agar (SEA) containing either MUF N-acetyl-β-d-glucosaminide or MUF β-d-lactoside, chitin agar containing MUF N-acetyl-β-d-glucosaminide, and carboxymethylcellulose (CMC) (Sigma Chemical Co.) agar containing MUF-β-d-lactoside.
The SEA contained (g liter−1 distilled water) soil extract, 400 ml; glucose, 1; peptone, 1; yeast extract, 1; K2HPO4, 1; and agar, 15. The CMC agar contained (g liter−1 distilled water) CMC, 10; asparagine, 0.5; yeast extract, 0.5; Mg2SO4 · 7H2O; (NH4)2SO4, 0.5; KH2PO4, 1; KCl, 0.5; CaCl2, 1; agar, 15. The chitin agar contained (g liter−1 distilled water) hydrolyzed chitin precipitate, 10; K2HPO4, 1; MgSO4 · 7H2O, 0.5; NaCl, 0.5; CaCl2, 0.1; Fe(NH4)2(SO4)2 · 6H2O, 0.05; NH4Cl, 0.1; agar, 15.
Hydrolyzed chitin precipitate was prepared by adding 10 g of chitin (Fluka BioChemika) to 200 ml of distilled water and was kept overnight at 4°C. Four hundred milliliters of 75% H2SO4 was added, and the resulting solution was left at 4°C for 24 h. The chitin was precipitated by mixing the solution with 9 liters of 4°C distilled water. After 48 h, the supernatant was decanted and the precipitate was filtered to remove large pieces of undissolved chitin. The precipitate was centrifuged and washed with 0.2% K2HPO4, and the supernatant was removed in 5 to 6 repeated cycles. The resulting hydrolyzed chitin precipitate was used in the preparation of the chitin agar.
After autoclaving, 2 ml of (200 μM) filter-sterilized (0.2-μm pore size) MUF-β-d-lactoside or MUF-β-d-glucosaminide was added to the agar media. Media were dispensed into microtiter plates (24-well plate; Greiner Labortechnik), and each well was inoculated with a pure fungal or bacterial culture. The microtiter plates were incubated at room temperature (21 to 23°C) and were visually examined daily under UV light (366 nm) for 8 days. Activity against specific MUF substrates was indicated by fluorescence. All screenings were done in triplicate.
Enzyme activity on soil samples.
NAGase and CELase activities in soil samples were determined as follows. Bulk soil was weighed into 10-ml plastic tubes. One hundred milligrams of soil in four replicates was found to be representative of the soils in this experiment (coefficient of variation < 15%); however, the amount of soil and the number of replications necessary for the enzymatic assay to be representative of a given soil may vary with soil type. Soil samples were not physically treated prior to weighing. All assays were conducted at 25°C in 2 ml of 50 mM Tris-maleate buffer (pH 5). MUF N-acetyl-β-d-glucosaminide or MUF-β-d-lactoside was added to a final concentration of 20 μM. Reactions were incubated for 30 min (NAGase) and 180 min (CELase). Optimum pH was determined for each enzyme. Turnover rates were constant during the assay time (unpublished data). Controls for extract and substrate fluorescence were processed in parallel. An additional control was processed to correct for quenching agents in the soil sample (11); however, quenching was not detected in these experiments. The assay was terminated by adding 2 ml of ice-cold 96% ethanol. After centrifugation, 2.7 ml of supernatant was transferred to plastic cuvettes (10 by 10 by 48 mm; Sarstedt) containing 300 μl of 2.5 M Tris buffer at pH 10. MUF substrate turnover was calculated by reference to a standard curve. Fluorescence derived from liberated 4-MU was determined with an LS50 luminescence spectrometer (Perkin-Elmer, Buckinghamshire, United Kingdom) at 446-nm emission and 377-nm excitation. Enzyme activities in soil samples were expressed as nanomoles of 4-MU per hour per gram (dru weight) of soil.
Extraction and quantification of ergosterol and PLFA.
Ergosterol analysis was performed as described by Roessner (25) with minor modifications: 2 g of moist soil was placed in 100-ml Schott flasks and treated with 20 ml of methanol, 5 ml of ethanol and 2 g of KOH and saponified for 40 min at 70°C. After cooling, 5 ml of distilled water was added. The suspension was filtered through a paper filter (AGF 606, 125 mm; Frisenette) into a separatory funnel. Soil residues were washed twice with 20 ml of methanol. n-Hexane was added (30 ml), and the suspension was shaken for 2 min. The n-hexane phase containing the ergosterol fraction was evaporated to dryness at 40°C with a rotary evaporator. The dried extract was resuspended in 2 ml of methanol and filtered (0.02-μm pore size, Anotrop; Whatman Ltd). This final extract (20 μl) was analyzed by HPLC with an RFC-18 column (Merck no. 50829, LiChrospher 60, RP-select B, 5 μm diameter). The mobile phase was water-methanol in a ratio of 5:95 with a flow rate of 1.5 ml min−1. Column eluant was monitored for absorbance at 282 nm. The retention time of ergosterol under these conditions was approximately 3 min.
The amount of PLFA 18:2ω6 was determined by the method of Frostegård et al. (13) with minor modifications. Lipids were extracted from 2 g (wet weight) of soil with a one-phase mixture of chloroform, methanol, and citrate buffer (0.15 M, pH 4) at a 1:2:0.8 ratio. Methyl nonadeconoate was added to the phospholipid fractions as an internal standard. The fatty acid methyl esters were separated and quantified on a gas chromatograph (Varian) equipped with a flame ionization detector and a 50-m nonpolar phenylmethyl silicone capillary column (Hewlett-Packard, Palo Alto, Calif.). Identification of the fatty acid methyl ester was done by mass spectrometry (Hewlett-Packard gas chromatography-mass spectometry system). Linear regression analysis and analysis of variance with STATISTICA software (Statsoft, 1994) were used to investigate the relationship between the variables examined.
RESULTS
Solid media screenings.
Eighty-six percent of the fungi tested had NAGase activity, but only 14% had CELase activity when they were inoculated onto SEA agar (Table 2). The fungi were inoculated on chitin agar containing MUF N-acetyl-β-d-glucosaminide and CMC agar containing MUF-β-d-lactoside to determine if NAGase and CELase activity were inducible. CELase activity could be induced in three fungi, and NAGase activity could be induced in two of three previously negative fungi (Table 2). When grown on SEA, all bacteria tested negative with regard to NAGase or CELase activity (Table 2); however, seven bacterial isolates produced NAGase or CELase when they were cultured on chitin and CMC agar.
TABLE 2.
Enzymatic activity detected with MUF-N-acetylglucosaminide or MUF-β-d-lactoside incorporated in agar media
| Speciesb | NAGase activitya
|
CELase activity
|
||
|---|---|---|---|---|
| SEA | Chitin agar | SEA | CMC agar | |
| Fungi | ||||
| Acremonium inflatum (Mk001) | + | + | − | + |
| Aspergillus flavus (AGMf19) | + | + | − | − |
| Alternaria sp. (AGMf4) | − | + | − | − |
| Beauveria sp. (AGMf35) | + | + | − | − |
| Botrytis sp. (AGMf10) | + | + | − | − |
| Candida albicans (AGMf11) | + | + | − | − |
| Cephalosporium sp. (AGMf13) | + | + | − | − |
| Cylindrocladium sp. (AGMf36) | + | + | − | − |
| Diplodia sp. (AGMf39) | + | + | − | − |
| Fusarium solani (AGMf15) | + | + | − | + |
| Geotrichum sp. (AGMf16) | + | + | − | − |
| Gliocladium sp. (AGMf17) | + | + | + | + |
| Humicola sp. (AGMf19) | + | + | − | + |
| Lylea tetracoila (Mk007) | + | + | − | − |
| Mucor sp. (AGMf20) | + | + | − | − |
| Penicillium sp. (AGMf22) | + | + | − | − |
| Paecilomyces sp. (AGMf21) | − | + | − | − |
| Rhodotorula sp. (AGMf26) | − | − | − | − |
| Trichoderma polysporum (AGMf31) | + | + | + | + |
| Trichurus sp. (AGMf30) | + | + | + | + |
| Verticillium clamydosporium (AGMf23) | + | + | − | − |
| Bacteria | ||||
| Alcaligenes faecalis (AGMb48) | − | − | − | − |
| Arthrobacter simplex (AGMb45) | − | − | − | − |
| Bacillus cereus (AGMb8) | − | − | − | − |
| Bacillus megaterium (AGMb134) | − | − | − | + |
| Bacillus subtilis (AGMb34) | − | − | − | − |
| Bacillus thuringiensis (AGMb18) | − | − | − | − |
| Comamonas acidovarans (AGMb56) | − | − | − | − |
| Enterobacter aerogenes (AGMb62) | − | − | − | − |
| Escherichia coli (AGMb30) | − | − | − | − |
| Klebsiella sp. (AGMb52) | − | + | − | − |
| Micrococcus albus (AGMb28) | − | − | − | − |
| Micrococcus luteus (AGMb5) | − | + | − | − |
| Phaffia rhodozyma (AGMb67) | − | − | − | − |
| Proteus vulgaris (AGMb59) | − | + | − | − |
| Pseudomonas aeruginosa (AGMb23) | − | − | − | + |
| Rhizobium meliloti (AGMb60) | − | − | − | − |
| Staphylococcus aureus (AGMb20) | − | − | − | − |
| Staphylococcus epidermis (AGMb64) | − | − | − | − |
| Staphylococcus faecalis (AGMb21) | − | + | − | − |
| Serratia marcescens (AGMb3) | − | + | − | − |
| Vibrio auguillarium (AGMb46) | − | − | − | − |
−, Agar not fluorescing; +, agar fluorescing (activity against MUF substrate).
The source of the culture collection was the Department of General Microbiology, University of Copenhagen.
Correlation with fungal biomass.
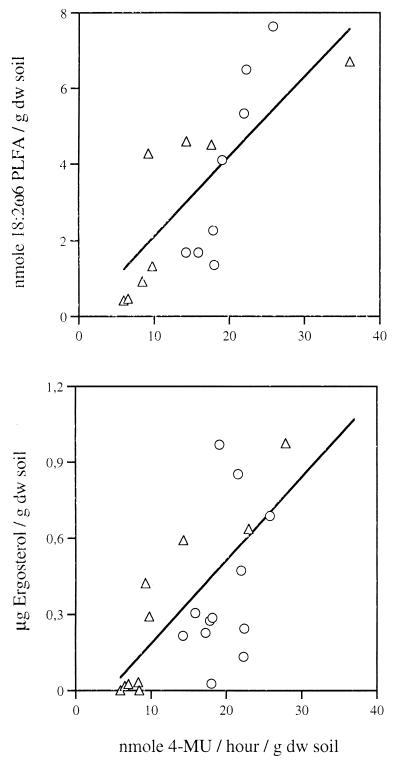
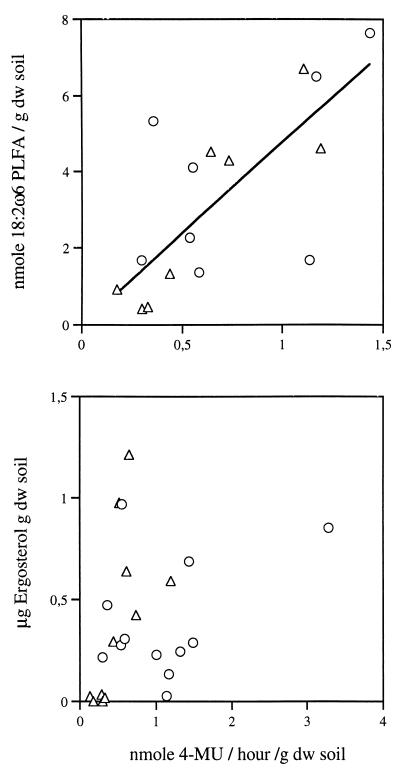
We evaluated the utility of MUF N-acetyl-β-d-glucosaminide and MUF β-d-lactoside substrates by correlating substrate turnover rates with the amount of 18:2ω6 PLFA and ergosterol. The enzyme activity determinations were reproducible (coefficient of variation < 15%). One-way analysis of variance identified differences between individual sampling times with respect to enzyme determinations (P < 0.001) and ergosterol (P < 0.01) and PLFA (P < 0.001). NAGase activity and PLFA were clearly correlated, and a weak but significant relationship between NAGase activity and ergosterol was detected (Fig. 1). CELase activity was correlated with PLFA but not with ergosterol (Fig. 2). Finally, there was a significant correlation between 18:2ω6 PLFA and ergosterol (r2 = 0.38, P < 0.01) and a weak but significant correlation between NAGase and CELase activities (r2 = 0.22, P < 0.05) (data not shown).
FIG. 1.
Scatter plots of NAGase activity and the content of the fungal biomass indicators 18:2ω6 PLFA (r2 = 0.61, P < 0.001) (top panel) and ergosterol (r2 = 0.36, P < 0.01) (bottom panel) in the Italian (▵) and Danish soil incubations (○).
FIG. 2.
Scatter plots of CELase activity and the content of the fungal biomass indicators 18:2ω6 PLFA (r2 = 0.52, P < 0.01) (top panel) and ergosterol (bottom panel) in the Italian (▵) and Danish soil incubations (○).
DISCUSSION
We found that NAGase activity was expressed by a diverse group of fungi. Furthermore, we detected NAGase activity in cultures growing on SEA agar containing glucose and peptone but no chitin, suggesting that NAGase is constitutive in these fungi. These results are consistent with those of other studies in which constitutive expression of NAGase activity in fungi was reported (5, 22). In contrast, CELase activity was restricted to a limited number of fungal species and the activity was predominantly expressed on CMC agar.
We used ergosterol and 18:2ω6 PLFA as indicators of fungal biomass in order to correlate NAGase and CELase activities with soil fungal biomass. The extraction of ergosterol is widely used for quantification of fungal biomass (9, 20). Potential for error exists in this method, which entails problems such as variability of specific ergosterol content in fungal tissue and in the relationship to living and nonliving fungal biomass (2, 27).
PLFA analysis has been used to describe soil microbial community structure and to detect changes in response to various disturbances (1, 7, 28). More recently, the 18:2ω6 PLFA was found to be highly correlated with ergosterol content in 15 types of soil (r2 = 0.85, P < 0.001) (12), consistent with the significant but weaker correlation we found in this study.
The correlations between NAGase activity and the soil content of ergosterol and 18:2ω6 PLFA, in combination with the constitutive expression of NAGase by a diverse group of fungi, support the use of this activity as an indicator of fungal biomass in soil samples. The fact that CELase activity was expressed in a limited number of fungal species excludes the use of this activity as a measure of fungal biomass. However, the correlation between 18:2ω6 PLFA and CELase activity is consistent with a predominant fungal origin (23).
In bacteria, NAGase and CELase activities were restricted to a limited number of species and expressed only on media containing chitin or CMC, suggesting a nutritional role for these enzymes. As fungi are considered to be the major important contributors to chitin turnover in soils (14), chitinolytic bacteria or actinomycetes are not likely to have a significant impact on the interpretation of NAGase activity as reflecting actively growing fungi in a soil sample. This conclusion is supported by our data and is in agreement with the significant, positive correlation found between chitinase activities, but not between bacteria or actinomycetes, and fungal propagules in chitin-amended soils (24).
The role of NAGase in fungal physiology is complex and has been linked to morphogenetics (15), N acquisition (14), and mycoparasitism (8). In an environmental sample, NAGase enzymes located in the periplasmic space will also contribute to a MUF substrate-derived signal, since MUF substrates have been used to measure periplasmic activities of fresh intact cells (15). Consequently, NAGase activity as determined by the enzymatic cleavage of MUF N-acetyl-β-d-glucosaminide in soil samples may reflect a nutritional as well as a possible morphogenetic aspect of fungal growth. Furthermore, NAGase activity in a soil sample may comprise enzyme activities of a recent biological origin as well as activities derived from enzymes that have retained their activity in nonliving soil compartments, e.g., dead cells, cell debris, and enzymes immobilized in the soil matrix (26). Further study has been initiated to investigate the relationship between NAGase activity and fungal growth and to determine the extent to which NAGase activity reflects enzyme activities of a recent biological origin, in order to evaluate the relationship to living and nonliving fungal biomass.
No definitive method to quantify fungal biomass in soil exists. Direct microscopic techniques are laborious and tend to underestimate the amount of fungi (10, 21), and calculations of fungal biomass with pure culture-derived conversion factors and the soil ergosterol content tend to overestimate fungal biomass in relation to total microbial biomass (12). The determination of NAGase activity is simple, i.e., no laborious physical or chemical treatment of the sample is necessary and the assay is rapid and reproducible.
In conclusion, we have shown that (i) NAGase activity is present in a diverse group of fungi, (ii) the activity appears to be constitutive, (iii) the activity is correlated with two independent indicators of fungal biomass, and (iv) the activity can be quantified in soil samples with a fluorogenic substrate. The data in the present study suggest that NAGase activity as determined by the turnover of MUF-N-acetyl-β-d-glucosaminide may provide a simple, sensitive, and rapid measure of soil fungal biomass.
ACKNOWLEDGMENTS
This project was supported by the EU Environment and Climate Program project no. EV5V-CT940-034.
We thank Kirsten Jensen for technical assistance and Bo Jensen and Jacob Møller for useful discussions about the manuscript.
REFERENCES
- 1.Bååth E, Frostegård Å, Fritze H. Soil bacterial biomass, activity, phospholipid fatty acid pattern, and pH tolerance in an area polluted with alkaline dust deposition. Appl Environ Microbiol. 1992;58:4026–4031. doi: 10.1128/aem.58.12.4026-4031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermingham S, Maltby L, Cook R C. A critical assessment of the validity of ergosterol as an indicator of fungal biomass. Mycol Res. 1995;99:479–484. [Google Scholar]
- 3.Boschker H T S, Cappenberg T E. A sensitive method using 4-methylumbelliferyl-β-cellobiose as a substrate to measure (1,4)-β-glucanase activity in sediments. Appl Environ Microbiol. 1994;60:3592–3596. doi: 10.1128/aem.60.10.3592-3596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns R G. Enzyme activity in soils: location and a possible role in microbial ecology. Soil Biol Biochem. 1982;14:423–427. [Google Scholar]
- 5.Cirano J U, Peberdy J F. Effect of carbon source on chitobiase production by Trichoderma harzianum. Mycol Res. 1993;97:45–48. [Google Scholar]
- 6.Claeyssens M, Van Tilbeurgh H, Tomme P, Wood T M. Fungal cellulase systems: comparison of the specificities of the cellobiohydrolases isolated from Penicillium pinophilum and Trichoderma reesei. Biochem J. 1989;261:819–825. doi: 10.1042/bj2610819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federle T W. Microbial distribution in soil—new techniques. In: Megusar F, Gantar M, editors. Perspectives in microbial ecology. Ljubljana, Yugoslavia: Slovene Society for Microbiology; 1986. pp. 493–498. [Google Scholar]
- 8.Flach J, Pilet P E, Jollés P. What’s new in chitinase research. Experientia. 1992;48:701–716. doi: 10.1007/BF02124285. [DOI] [PubMed] [Google Scholar]
- 9.Frankland J C, Dighton J, Boddy L. Methods for studying fungi in soil and forest litter. In: Grigorova R, Norris J R, editors. Methods in microbiology. Vol. 22. San Diego, Calif: Academic Press Limited; 1990. pp. 343–404. [Google Scholar]
- 10.Frankland J C, Dighton J, Swift M J. A comparison of two methods for the estimation of mycelial biomass in leaf litter. Soil Biol Biochem. 1978;10:468–471. [Google Scholar]
- 11.Freeman C, Liska G, Jones S E, Lock M A. The use of fluorogenic substrates for measuring enzyme activity in peatlands. Plant Soil. 1995;175:147–152. [Google Scholar]
- 12.Frostegård A, Bååth E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils. 1996;22:59–65. [Google Scholar]
- 13.Frostegård A, Bååth E, Tunlid A. Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol Biochem. 1993;25:723–730. [Google Scholar]
- 14.Gooday G W. The ecology of chitin degradation. In: Marshall K C, editor. Advances in microbial ecology. Vol. 11. New York, N.Y: Plenum Press; 1990. pp. 387–430. [Google Scholar]
- 15.Gooday G W, Zhu W, O’Donnel R W. What are the roles of chitinases in the growing fungus? FEMS Microbiol Lett. 1992;100:387–392. [Google Scholar]
- 16.Hayano K, Katami A. Extraction of β-glucosidase activity from pea field soil. Soil Biol Biochem. 1977;9:349–351. [Google Scholar]
- 17.Hayano K, Tubaki K. Origin and properties of β-glucosidase activity of tomato-field soil. Soil Biol Biochem. 1985;17:553–557. [Google Scholar]
- 17a.Hodge A, Alexander I J, Gooday G W. Chitinolytic enzymes of pathogenic and ectomycorrhizal fungi. Mycol Res. 1995;99:935–941. [Google Scholar]
- 18.Hoppe H G. Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl substrates. Mar Ecol Prog Ser. 1983;11:299–308. [Google Scholar]
- 19.McCreath K J, Gooday G W. A rapid and sensitive microassay for determination of chitinolytic activity. J Microbiol Methods. 1992;14:229–237. [Google Scholar]
- 20.Newell S Y. Estimating fungal biomass and productivity in decomposing litter. In: Caroll G C, Wicklow D T, editors. The fungal community: its organization and role in the ecosystem. New York, N.Y: Marcel Dekker; 1992. pp. 521–561. [Google Scholar]
- 21.Parkinson D. Filamentous fungi, 949–968. In: Page A L, Miller R H, Keeney D A, editors. Methods of soil analysis. Madison, Wis: American Society of Agronomy–Soil Science Society of American; 1982. [Google Scholar]
- 22.Rast D M, Horsch M, Furter R, Gooday G W. A complex chitinolytic system in exponentially growing mycelium of Mucor rouxii: properties and function. J Gen Microbiol. 1991;137:2797–2810. doi: 10.1099/00221287-137-12-2797. [DOI] [PubMed] [Google Scholar]
- 23.Rhee Y H, Hah Y C, Hong W. Relative contribution of fungi and bacteria to soil carboxymethylcellulase activity. Soil Biol Biochem. 1987;19:479–481. [Google Scholar]
- 24.Rodriguez-Kabana R, Godoy G, Morgan-Jones G, Shelby R A. The determination of soil chitinase activity: conditions for assay and ecological studies. Plant Soil. 1983;75:95–106. [Google Scholar]
- 25.Roessner H. Fungal biomass by ergosterol content. In: Schinner F, Oehlinger R, Kandeler E, Margesin R, editors. Methods in soil biology. Heidelberg, Germany: Springer-Verlag; 1996. pp. 49–51. [Google Scholar]
- 26.Skujins J. Extracellular enzymes in soil. Crit Rev Microbiol. 1976;4:383–421. doi: 10.3109/10408417609102304. [DOI] [PubMed] [Google Scholar]
- 27.Stahl D S, Parkin T B. Relationship of soil ergosterol concentration and fungal biomass. Soil Biol Biochem. 1996;28:847–855. [Google Scholar]
- 28.Zelles L, Bai Q Y. Fatty acid patterns of phospholipids and lipopolysaccharides in environmental samples. Chemosphere. 1994;28:391–411. [Google Scholar]