Abstract
Time series incubations were conducted to provide estimates for the size selectivities and rates of protistan grazing that may be occurring in a sandy, contaminated aquifer. The experiments involved four size classes of fluorescently labeled groundwater bacteria (FLB) and 2- to 3-μm-long nanoflagellates, primarily Spumella guttula (Ehrenberg) Kent, that were isolated from contaminated aquifer sediments (Cape Cod, Mass.). The greatest uptake and clearance rates (0.77 bacteria · flagellate−1 · h−1 and 1.4 nl · flagellate−1 · h−1, respectively) were observed for 0.8- to 1.5-μm-long FLB (0.21-μm3 average cell volume), which represent the fastest growing bacteria within the pore fluids of the contaminated aquifer sediments. The 19:1 to 67:1 volume ratios of nanoflagellate predators to preferred bacterial prey were in the lower end of the range commonly reported for other aquatic habitats. The grazing data suggest that the aquifer nanoflagellates can consume as much as 12 to 74% of the unattached bacterial community in 1 day and are likely to have a substantive effect upon bacterial degradation of organic groundwater contaminants.
While heterotrophic protists have been found in pristine and contaminated aquifers (3, 29, 34, 37, 54–57), very little research has been performed to elucidate their role in the subsurface. In other environments (e.g., surface and marine waters, topsoil, and wastewater treatment plants), it is well documented that they typically consume bacteria (2, 11, 15, 41, 42, 47), although some have been observed to consume high-molecular-weight organics (48, 59) and even viruses (20, 39). Protists typically graze selectively, depending upon the size (1, 9, 17, 25, 52), growth condition (18, 53), species (16, 17, 35), and motility (18) of their prey. In carbon-limited environments, protists decrease bacterial competition, resulting in a greater bacterial uptake rate for organic substrate per unit of bacterial biomass (27). Based upon indirect field observations, it is also hypothesized that this may be the role they play in organically contaminated aquifers (31). In nutrient-limited environments, protists may release nitrogen or phosphorus needed by bacteria (10, 28, 44, 61).
Studies at the U.S. Geological Survey’s (USGS) Toxic Substances Hydrology Program research site at the Massachusetts Military Reservation (MMR) on Cape Cod, Mass., have shown that sandy aquifer sediments can harbor large protistan populations even at relatively low levels (≤2 mg/liter) of dissolved organic matter (30). Protistan abundances in the MMR aquifer plume range from 1 × 104 to 7 × 104 g (dry weight)−1 (30) and consist primarily of nanoflagellates (2 to 3 μm in length) (29) that belong to the genera Bodo, Cercomonas, Cryptaulax, Cyanthomonas, Goniomonas, and Spumella, along with some undescribed species (37). A few amoebae (63) and no ciliates (29) have been observed.
Results of a principal-component factor analysis of protistan and bacterial abundances and chemical constituents in the MMR plume suggested that the flagellates were preying upon unattached bacteria (30). Additional evidence of predation was obtained from flowthrough columns of aquifer sediment from which fluorescently labeled unattached bacteria eluted at much lower rates than they did from sterile (protist-free) controls (31). However, these results provide only indirect evidence of predation because no enumerations of the bacteria within the flagellates were performed.
The purpose of the research reported in this paper was to directly determine whether the MMR nanoflagellates can consume unattached bacteria in the plume and the extent to which they engage in size-selective grazing. Rates of bacterivory (grazing and clearance rates) were estimated in the laboratory by using fluorescently labeled, monodispersed bacteria (FLB) and nanoflagellates that had been isolated from the MMR aquifer plume. Although other methods exist (19, 26, 38, 43, 45, 62, 64–66), we chose to use fluorescent labeling to study flagellate bacterivory because this procedure requires shorter incubation times and relies upon direct visual observation of the prey within the predator. In addition, experiments could be designed with different sizes of FLB to determine if the nanoflagellates can discriminate between prey. This involved using FLB with cell lengths of 0.1 to 0.5, 0.5 to 0.8, 0.8 to 1.5, and >1.5 μm (average cell volumes of 0.06, 0.14, 0.21, and 0.87 μm3, respectively) in the grazing experiments.
MATERIALS AND METHODS
Study site.
The sand and gravel aquifer underlying the MMR contains a contaminant plume that is 5-km long, 1-km wide, and 23-km deep created by the discharge of 1,900 m3 of treated wastewater · day−1 onto rapid sand infiltration beds from 1936 until 1995 (33). The plume is characterized by 1 to 4 mg of dissolved organic carbon (DOC)/liter, 0 to 5 mg of dissolved oxygen/liter, ≤60 mg of nitrate/liter, 2 to 4 mg of alkylbenzene sulfonate detergents/liter in the distal reaches, and trace amounts of volatile organic compounds such as trichloroethylene (29).
Protists.
The nanoflagellates used in the experiments were cultured from aquifer sediments collected from the MMR plume in a contaminated zone 3 m below the water table at USGS well site F230 (0.12-km downgradient from the rapid-sand-infiltration beds). The sediments were recovered in the absence of drilling fluids by using a wireline piston corer in conjunction with a hollow-stem auger drill (67). The core was processed aseptically by the method developed by Bunn (6).
Culturing of the aquifer nanoflagellates occurred in covered, sterile 1-liter jars containing 500 g of sterile (15 min, 121°C, 15 lb/in2) sieved aquifer sediments (particle diameter, 0.5 to 1.0 mm) that were saturated with 125 ml of 4% sterile Cerophyl-Prescott (CP) medium (6, 21). The 4% CP medium, made from the extract of dehydrated cereal leaves (Sigma Chemical, St. Louis, Mo.), had a DOC content of ∼10 mg · liter−1 (similar to that of the groundwater near the infiltration beds) and was pH adjusted to 6.0 to match aquifer conditions. The grain size of the sediments was chosen because it represents the size of the predominant fraction in the MMR aquifer. The jars were inoculated with 1 g of the core material collected at F230 or 5 ml of liquid taken from another porous-medium culture at the peak biomass. There was ∼1 cm of freestanding liquid above the soil. The contents of the jars were swirled gently for 1 min after inoculation to ensure distribution of the microorganisms throughout the porous media. Bacteria that were present in the original core also grew in the porous-medium cultures and were primarily 0.5- to 2-μm-long rods and cocci (21). Nanoflagellates used in the grazing experiments were taken from 4- to 10-day-old porous-medium cultures grown at room temperature (20 ± 2°C). This time frame was used because it produces cultures with the greatest number of 2- to 3-μm-long highly active flagellates (as observed with inverted light microscopy). Video clips were taken of live flagellates from liquid culture (without porous media). (It is important to note that such cells are characteristically larger than those grown in porous-medium cultures.) These clips were obtained by converting analog clips recorded from an Olympus light microscope (fitted with a 40× objective and differential interference contrast) by using a JVC KY-F30 video camera. Analog clips were recorded on a Sony U-Matic SP recorder and converted to digital format by use of a Radius Video Vision high-resolution digital-film card and Adobe Premiere software on a Macintosh Quadra 840 AV computer. Single frames from the digital clips (frame duration, 1/25 s; frame resolution, 72 dots/in.) were exported to Adobe Photoshop 3.0 software. Each still image shown corresponds to a single frame. To enhance the clarity of the still images, the resolution was increased to 600 dots/in. by using software interpolation. After optimizing the brightness and contrast and reducing the image noise, all images were printed with a Tektronik Phaser II dye-sublimation printer.
Bacteria.
Four size classes of bacteria were used in separate grazing experiments (Table 1). The smallest size class was concentrated from 1 liter of contaminated groundwater obtained at MMR monitoring wells 0.05 to 0.08 km downgradient from infiltration beds (S318 and S314) at 13.8 and 6.6 m below the water table, respectively. The bacterial population was initially fractionated by filtration (bacteria passing through a 0.45-μm-pore-size Nuclepore filter) and captured on a 0.1-μm-pore-size filter. This sample was fixed with 3.7% (vol/vol) filter-sterilized formalin. To increase the number of bacteria in the 30-ml volume to ∼106 ml−1, the sample was concentrated to 15 ml by placing it in a 40°C oven for 6 days. (It is important to note that because the bacteria were fixed, lysis at 40°C was not a problem.)
TABLE 1.
Size classes of bacteriaa used in grazing experiments
| Size class (range of length [μm]) | Avg length (μm) | Aspect ratiob (μm) | Avg volc (μm3) |
|---|---|---|---|
| 0.1–0.5 | 0.5 | 1.3 ± 0.1 | 0.06 ± 0.01 |
| 0.5–0.8 | 0.7 | 1.2 ± 0.04 | 0.14 ± 0.02 |
| 0.8–1.5 | 1.1 | 2.0 ± 0.1 | 0.21 ± 0.02 |
| >1.5 | 1.8 | 2.1 ± 0.3 | 0.87 ± 0.05 |
Primarily rod-shaped bacteria.
Radius (r) = 0.5(L/aspect ratio), where L is cell length. Values are means ± standard errors.
Cell volume = [πr2(L − 2r)] + (4/3π(r3)], where r is radius. Values are means ± standard errors.
Bacteria for the three other size classes were obtained from the porous-medium cultures inoculated with core material. An aliquot of 4% CP solution was pipetted from beneath the soil surface of a 5-day-old culture and passed through a 0.8-μm-pore-size filter at 400 mm Hg (transmembrane pressure). The filtrate, free of large bacteria and protists, was inoculated into another jar containing sterile porous media (0.5- to 1.0-mm-diameter sieved sediments) along with sterile 4% CP and incubated at room temperature for 5 days. Bacteria for the different size fractions were obtained from this culture by differential filtration with 3.0-, 0.8-, and 0.45-μm-pore-size membrane filters. These bacteria were heat killed as outlined by Sherr and Sherr (49). Bacteria passing the 3.0-μm-pore-size filter and collected on the 0.8-μm-pore-size filter were not further subdivided. Rather, grazing experiments were conducted with this aggregate group of bacteria (0.8 to 3.0 μm in length). When samples were observed for nanoflagellates containing FLB, the sizes of the ingested bacteria and the nanoflagellates were measured by using a Whipple disk and an eyepiece micrometer (0.4-μm graduations at ×600 total magnification).
All bacteria were stained with the protein-binding fluorescent stain DTAF {5[(4,6-dichlorotriazin-2-yl)amino] fluorescein; Sigma Chemical} as outlined in Sherr and Sherr (49). In preliminary experiments, there were no significant differences (Student’s t test, P = 0.05) between the flagellates’ uptake of the DTAF-stained bacteria that had been frozen for ∼10 weeks and those that were used immediately after staining. Therefore, the DTAF-stained bacteria (FLB) were stored at 0°C for up to 3 months before use. Freezing ensured that a single stock containing the same FLB could be used in all replicate experiments.
Size frequency analyses were performed on each of the four operationally defined size classes of bacteria with an Optiphot II epifluorescence microscope (Nikon, Buffalo, N.Y.) and an image processor (Image Technology Corporation, Deer Park, N.Y.) connected to a personal computer, a Dage SIT66 black-and-white camera, and a Sony black-and-white monitor. The image system was optimized to analyze and calculate length, width, area, and perimeter of fluorescently stained bacteria in samples previously analyzed for bacterial abundances. Measurements from the image system were standardized by using fluorescently stained 0.95-, 1.07-, and 0.45-μm-diameter microspheres (Polysciences, Warrington, Pa.) to convert pixel measurements to micrometers. All analyses were performed at microscope magnifications of ×788 to ×1,260.
Grazing experiments.
The grazing experiments were modifications of the protocols outlined by Sherr and Sherr (49). Two separate experiments were run with each size class of FLB (Table 1). At the beginning of each experiment, 2.1 liters of the 4% CP medium was extracted by mild suction from several porous-medium cultures. After the medium was mixed to homogenize the extracts, 100-ml aliquots were dispensed into 21 sterile 125-ml screw-top micro-Fernbach flasks. The flasks sat undisturbed for 24 h in the dark at room temperature (20 ± 2°C) to allow the organisms to acclimate. Samples were then taken to determine protistan abundance. Seven replicate flasks were spiked with 1 ml of a specific size class of FLB (Table 1). The FLB constituted 16% of the total bacterial population in the experiment (Table 2). Immediately after the spiking, a 6-ml sample was taken from one of the replicate flasks from each size class to determine the initial (time [t] = 0) abundances of FLB, unstained bacteria from the porous-medium culture, and nanoflagellates. Subsequently, 6-ml samples were taken from one of the replicate flasks at t = 0.33, 0.67, 1, 2, 4, 10, and 20 h. Each flask was sampled only once to avoid changes in conditions due to reduction in volume over time.
TABLE 2.
Distribution of FLBa and unlabeled groundwater bacteria among operationally defined size classes in flagellate grazing experiments
| Size class (μm) | Bacterial abundance (105/ml)
|
FLB abundance
|
|||
|---|---|---|---|---|---|
| Unlabeled | Labeled | Unlabeled + labeled | % of totalb | % of specific size classc | |
| 0.1–0.5 | 4.9 | 1.0 | 5.9 | 6 | 17 |
| 0.5–0.8 | 3.0 | 0.9 | 3.9 | 5 | 23 |
| 0.8–1.5 | 4.9 | 0.5 | 5.4 | 3 | 9 |
| >1.5 | 1.4 | 0.3 | 1.7 | 2 | 16 |
| All size classes | 14.2 | 2.7 | 16.9 | 16 | 16 |
Labeling of bacteria was done with DATF.
Total bacterial abundance was 1.7 × 106 ml−1 (standard error = 1.5 × 105 ml−1).
Percent of specific size class = [(abundance of FLB in given size class)/(abundance of unlabeled bacteria + abundance of FLB in same size class)] × 100.
Each 6-ml sample was immediately fixed for 15 min with a spike of 10% filter-sterilized glutaraldehyde to achieve a 1% (vol/vol) final concentration. Two 1-ml aliquots were filtered onto 0.1-μm-pore-size black polycarbonate filters (Poretics Corp., Livermore, Calif.) to enumerate the FLB. The filters containing the FLB did not need to be stained before the bacteria were counted. Four 1-ml aliquots of the sample were stained with the nucleic acid-binding fluorescent stain DAPI (4′,6-diamidino-2-phenylindole; Sigma Chemical) to enumerate flagellates. These aliquots were filtered onto 0.8-μm-pore-size black polycarbonate filters (Poretics Corp.) by using a vacuum of ≤13 mm Hg. DAPI staining of flagellates followed the procedure outlined by Sherr and Sherr (49). All filters were air dried and placed on microscope slides that had one drop of low-fluorescence (Cargille A) immersion oil (VWR Scientific, Boston, Mass.). A second drop of immersion oil was added to the filter, and a glass coverslip was placed on top. Slides were kept refrigerated in the dark at 4°C for a maximum of 3 months prior to observation.
Controls.
For each experiment, initial FLB and flagellate concentrations were assessed prior to spiking the FLB into the CP extract. In addition, a negative control was run during every experiment for each size class of FLB. In the control, the nanoflagellates were fixed with glutaraldehyde (1% [vol/vol] final concentration) prior to the addition of the FLB. Any FLB observed associated with these protists were assumed to be surface associated because the flagellates were killed by the glutaraldehyde, which precluded feeding. The population of bacteria in the extract from the porous medium that contained flagellates was also enumerated to determine the total bacterial abundance. In these samples, the bacteria were stained with DTAF and processed as outlined above.
Enumeration.
All slides were observed with a Nikon (Garden City, N.Y.) Optiphot microscope equipped for epifluorescence. A 505-nm dichroic mirror was used for all stains. To observe the DAPI-stained nanoflagellates, the microscope was equipped with a 300- to 380-nm excitation filter and 420-nm barrier filter (UV-1A or UV-2A system). For the DTAF-stained bacteria, a 450- to 490-nm excitation filter was used in conjunction with a 520- to 560-nm barrier filter (a B2E filter system). In all cases, a 60× oil immersion Nikon objective was used with 10× oculars. Where needed, an Optitronics DEI-470 color video monitor camera and zoom lens system (Micro Video, Inc., Avon, Mass.) was employed to enhance the magnification, color, and/or contrast abilities of the microscope images. The DAPI-stained nanoflagellates were located first by using the UV-1A or UV-2A system. Once an individual cell was in focus, the B2E filter system was put in place to determine if FLB were present inside the flagellate. Resolution of the number of FLB associated with each nanoflagellate was sometimes difficult because of the proximity and overlapping of the FLB inside the 2- to 3-μm protists. Therefore, only the presence or absence of FLB inside each nanoflagellate was recorded.
Preliminary experiments were run to develop a counting array that would provide information regarding the combination of slides and fields per slide to be counted. These experiments used 0.1- and 1.0-μm-diameter, fluorescently labeled carboxylated microspheres (Polysciences) coated with bovine serum albumin. Previous experiments had shown that the statistical distributions of FLB and microspheres in the nanoflagellates were similar, so the counting protocol could be developed by use of the microspheres, which provided better contrast and were easier to work with than bacteria. In all of the experiments, there were ∼10 nanoflagellates per microscope field at ×600 magnification. This consistency, which resulted from using 4- to 10-day-old porous-medium cultures, was what enabled such a counting array to be readily developed. Preliminary experiments were conducted in the same way as the grazing experiments described above except that samples were taken at 10-, 20-, 60-, and 120-min intervals. For these experiments, the nanoflagellates in 20 microscope fields were observed on each of four replicate slides. The data from these preliminary experiments were used to develop a table (counting array) that predicted the number of observations (slides and fields per slide) needed to obtain an estimate of the fraction of the nanoflagellate population containing ≥1 FLB with a coefficient of variation of ≤20%.
Statistical analysis showed that error in estimating the proportion of nanoflagellates containing at least one FLB depended upon (i) p, the true proportion being estimated; (ii) N, the number of flagellates observed in each counting field; and (iii) m, the number of fields counted. A simulation model was developed to estimate the accuracy of possible counting procedures under these conditions. The model was based upon the assumption that N follows a Poisson distribution with mean μN and that X, the number of nanoflagellates containing at least one FLB, follows a binomial distribution with parameters N and p. This implies that X has a compound Poisson-binomial distribution which can be shown to be Poisson with mean pμN. The estimated proportion Pi from field i is, given Xi and Ni: Pi = Xi/Ni, where i = 1,…, m, leading to the estimator P = (P1 + P2 + … + Pm)/m to be used in estimating the proportion p.
While the variance of this estimator is not easily calculated, it can, for given values of p, m, and μN, be estimated by simulation. Once a table is constructed by use of this model and a wide range of values of the three parameters, an investigator can count a few fields to make preliminary estimates of p and μN and use the tabular values of the variance of P to determine the number of additional fields to be counted to reduce the variance to a level which will provide the accuracy required. The process can be facilitated by preparing samples at a common dilution to maintain an approximately constant value of μN.
The preliminary experiment indicated that the period of linear uptake occurred in the first few hours. Therefore, 0.33, 0.67, 1, 2, 4, 10, and 20 h were used as sampling times for all subsequent experiments. Additional sampling times of 10 and 20 h were added to monitor the longer-term fate of the FLB.
RESULTS
Populations.
During the 20 h of the FLB experiments, there was no significant change in the abundance of nanoflagellates, which ranged from 1 × 104 to 6 × 104 flagellates · ml−1 in the different grazing experiments. This is the typical concentration range seen in our porous-medium cultures at steady state and in the head of the MMR plume. The predominant flagellate in the cultures, Spumella guttula (Ehrenberg) Kent, was 2 to 3 μm in diameter (cell volume, ≅4 to 14 μm3; assuming a spherical shape). Fixation of the nanoflagellates with 1% (vol/vol) glutaraldehyde prior to enumeration did not markedly change their size or appearance compared to that of live cultures observed by using Hoffman interference optics and an inverted microscope. This contrasts with the twofold reductions in size resulting from fixation that are reported in the literature (5, 8).
For the grazing experiments, bacterial abundances in the interstitial liquid removed from the porous-medium cultures averaged 1.7 × 106 ml−1 (standard error = 1.5 × 105 ml−1). The FLB at the beginning of the experiments comprised 2 to 6% of that total bacterial population present in the flasks (Table 2). However, the percentages were 9 to 23% when each initial FLB abundance (C0) value was normalized to the total bacterial abundance present within its specific bacterial size class (Table 2). These percentages were within the range of 5 to 50% recommended in the literature (43, 48). The bacteria were primarily rods, and their size distributions in the porous-medium liquid (Table 2) were similar to what has been observed in some zones of the MMR plume.
FLB uptake.
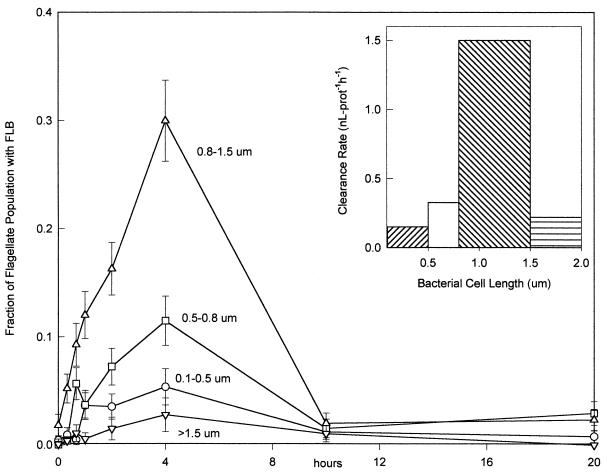
In the control preparations where glutaraldehyde was added to fix the nanoflagellates prior to inoculation with FLB, there were no FLB associated with them. Therefore, it was assumed that FLB associated with the live nanoflagellates in the experiments were being grazed. For all size classes of FLB, most of the uptake occurred within 4 h (Fig. 1). For all FLB sizes except 0.8 to 1.5 μm, there was a 0.6- to 1-h lag before appreciable uptake occurred. The fraction of flagellates containing FLB at 10 and 20 h was very low for all size classes of bacteria. Unlike the results obtained at ≤4 h, there were no significant differences in the fraction of protists containing the different sizes of FLB at these later times (analysis of variance, P = 0.05). This was probably due to the depletion and digestion of the FLB (16, 47) and is the reason why only initial uptake data (0 to 4 h) were used to calculate grazing rates.
FIG. 1.
Uptake of FLB by 2- to 3-μm aquifer nanoflagellates during the grazing experiments. Error bars represent standard errors of the mean and total experimental variability. Coefficients of determination (r2 values) for linear regressions fitted to 0- to 4-h data were 0.80 (for the 0.1- to 0.5-μm size class), 0.87 (0.5 to 0.8 μm), 0.98 (0.8 to 1.5 μm), and 0.93 (>1.5 μm). The inset shows clearance rates for each size class of FLB based on uptake rates observed during the first 4 h of the grazing experiments. prot, protist.
The data collected within the first 4 h for each size class of bacteria were used to generate a line of best fit. The slopes derived from these linear regressions were assumed to represent the uptake rates for each size class of bacteria (Fig. 1). Because only presence or absence of FLB was recorded due to the difficulty in resolving individual bacteria within the protists, it was assumed for these calculations that each observation was equivalent to one FLB per nanoflagellate. The coefficients of determination (r2 values) were significant for all bacterial size classes except the smallest (0.1 to 0.5 μm) (Fig. 1). With the exception of the slopes for the FLB size classes of 0.1 to 0.5 μm and >1.5 μm, all of the slopes were significantly different from each other (Student’s t test, P = 0.05). Clearance and uptake rates increased with bacterial cell length, except for those of the largest size class (>1.5 μm), which were significantly lower than those of the size class of 0.8 to 1.5 μm (Table 3). For the FLB of the size classes 0.1 to 0.5, 0.5 to 0.8, and 0.8 to 1.5 μm, a fitted linear relationship between clearance rate and cell length was significant (Fig. 1, inset) (r2 = 0.99, P = 0.05), indicating that flagellate grazing rates were dependent upon bacterial size. The x intercept for the linear relationship between clearance rate and cell volume (data not shown) corresponded to an average effective prey size of 0.028 μm3, equivalent to a bacterial cell of <0.1 μm in length.
TABLE 3.
Uptake and clearance rates of flagellates for the different size classes of FLB
| FLB source [reference(s)] | FLB cell length (μm) | FLB cell vol (μm3)a | Uptake rateb (bacteria · protist−1 · h−1) | Clearance rate (nl · protist−1 · h−1) |
|---|---|---|---|---|
| Contaminated aquifer (this study) | 0.1–0.5 | 0.06 ± 0.01 | 0.06 | 0.1 |
| 0.5–0.8 | 0.14 ± 0.02 | 0.13 | 0.33 | |
| 0.8–1.5 | 0.21 ± 0.02 | 0.77 | 1.4 | |
| >1.5 | 0.87 ± 0.05 | 0.04 | 0.2 | |
| Contaminated aquifer (columns) (31) | 1–2 | 3 | 12 | |
| Rivers (2, 16)c | 1.1–90.4 | 0.2–8.9 | ||
| Lakes (4, 25, 41)c | 2–181 | 0.2–44 | ||
| Marine (16,47)c | 5.2–27.4 | 1.4–4.3 | ||
| Estuarine (17, 42, 45)c | 0.32–3.2 |
Values are means ± standard errors.
Calculated by using the clearance rate of FLB and the total bacterial concentration of each size class present during grazing experiments.
The values from these references represent a range of experiments conducted by several researchers at various temperatures with various sizes of FLB and flagellates.
DISCUSSION
The results of the FLB uptake experiments demonstrated that the nanoflagellates that inhabit organically contaminated groundwater at the MMR site can consume unattached bacteria. This substantiates the hypothesis that the small groundwater protists are bacterivorous like flagellates in marine, freshwater, and topsoil environments (2, 15, 41, 42, 47, 51). In addition, the results show that the 2- to 3-μm-long flagellates in the MMR aquifer preferentially ingest fairly large bacteria (0.8 to 1.5 μm in length) in comparison to their own size. In contrast to observations made for surface-water habitats, our study shows that groundwater flagellates from the MMR aquifer can exhibit a predator/prey length ratio of ≤2:1. Their preference for the larger bacteria is also similar to trends observed for flagellates in other environments (1, 9, 17, 25, 52). Whether the preference is a function of mechanical processes (15), chemosensory detection (60), or the nature and species of the larger prey (16) is unknown.
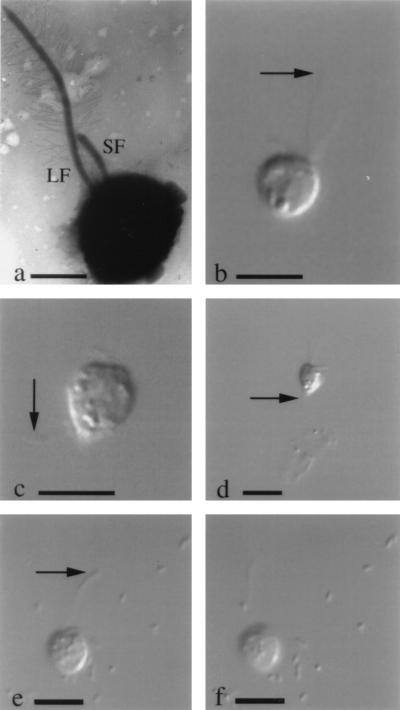
S. guttula, the dominant flagellate in the grazing experiments (Fig. 2), is a raptorial feeder that uses a direct-interception mode of feeding. In direct-interception feeding, protists generate critical flow lines (paths) along which there is a high probability that food particles (e.g., bacteria suspended in the medium) will be intercepted. S. guttula generates such flow lines as a result of the rapid beating action of the long flagellum (Fig. 2). Analogous to other flagellate species found in groundwaters (36), cells of S. guttula can swim actively in the medium (Fig. 2) but may also attach temporarily to sediment particles (Fig. 2). S. guttula attaches by means of a thin posterior protoplasmic filament (Fig. 2). Attachment of flagellates to surfaces may give rise to an increased probability of food particles being intercepted (13, 68). The transectional area of the flow along the critical lines past the cell and the specific clearance achieved are related to the radius of the flagellate (R) and the radius of the food particles (r) (15). Therefore, it is not surprising that the efficiency of feeding is dependent on the size of the prey. Using empirical data for flagellates and ciliates, Fenchel (14) has shown that if the r/R ratio is >0.1, protists are raptorial feeders. For our grazing experiments, which indicated that the 2- to 3-μm flagellates preferentially ingested 0.8- to 1.5-μm bacteria, the r/R ratio ranged from 0.27 to 0.75, corroborating the conclusion that S. guttula was feeding by direct interception.
FIG. 2.
Cells of S. guttula from liquid cultures from the MMR aquifer. Micrographs b to f are still images of live cells extracted from digital video clips. (a) Transmission electron micrograph of whole-mount shadow-cast preparation, showing the long flagellum (LF) with flagellar hairs and the short naked flagellum (SF). Scale bar, 2.5 μm. (b) Cell swimming to the rapid beat of the long flagellum; scale bar, 10 μm. (c) Cell which has just become detached from sediment particles (not visible); attachment was achieved by means of a thin posterior protoplasmic filament (arrow). Scale bar, 10 μm. (d) Cell attached to sediment particles. The attachment filament (not visible) arises from the pointed cell posterior (arrow). Scale bar, 10 μm. (e and f) Sequential frames of cell with actively beating long flagellum (arrow). Numerous food bacteria are visible in the background. Scale bar, 10 μm.
Unlike the data reported by Gonzalez et al. (17), the uptake and clearance rates of the groundwater nanoflagellates did not show a continuously increasing trend with bacterial size (Fig. 1). Indeed, the uptake rate for the largest bacteria (>1.5 μm in length) was low and not significantly different (P = 0.05) from that observed for the smallest FLB (0.1 to 0.5 μm in length). A similar trend was observed for a Spumella sp. by Holen and Boraas (25). However, when they accounted for the total volume of bacteria (cubic micrometers per protist per hour) ingested by Spumella, they found a monotonic increase with prey size. They suggested that Spumella became saturated with the larger bacteria as a function of their capability to regenerate cell membrane during formation of food vacuoles. Accounting for the total volume of the prey did not explain the decreased ingestion of the >1.5-μm bacteria by the groundwater nanoflagellates. Perhaps, bacteria with cell lengths of >1.5 μm are difficult for the 2- to 3-μm-long flagellates to ingest. Predator/prey volume ratios studied by other researchers (12, 17, 40) have ranged from 17:1 to >10,000:1, while in our study, this ratio was 5:1 to 233:1. For the largest size of bacteria we studied, the ratio was 5:1 to 16:1, and for the preferentially grazed 0.8- to 1.5-μm bacteria, it was 19:1 to 67:1.
The actual uptake rates observed in this study (Table 3) were lower than those reported in the literature for similarly sized flagellates (1.5 to 14 bacteria · protist−1 · h−1) (25, 47). This was due, in part, to the low concentrations of FLB in our experiments relative to the total bacterial population (2 to 6% of the total population). Clearance rates account for the differences in bacterial concentration and facilitate comparisons with other studies. These rates (Table 3) are more similar to those reported for flagellates from other environments (2, 4, 41, 42, 45, 47). For example, the rates (0.32 to 2.6 nl · protist−1 · h−1) obtained by Gonzalez et al. (16, 17) (with 0.03- to 0.32-μm3 bacteria and predominantly 3- to 4-μm-long marine and estuarine flagellates) are within the same range and may be slightly larger due to the greater cell volume of their flagellates. The rates are also very similar to those obtained by Holen and Boraas (25) (1.6 to 4.1 nl · protist−1 · h−1) with Spumella sp. (3 to 5 μm in length) isolated from Lake Michigan and fed 0.02 to 0.53 μm3 of bacteria. Our earlier flowthrough column studies (31) involving FLB advecting through aquifer sediments in the presence and absence of groundwater protists suggested higher clearance rates (12 to 23 nl · protist−1 · h−1) (Table 3) than those obtained in the present investigation. However, calculations of clearance rates from the column studies were based upon disappearance of the FLB relative to that observed for the protist-free control; direct uptake of FLB by flagellates was not monitored. It is likely that the true grazing and clearance rates in the aquifer may be somewhere between the values we have obtained in the laboratory.
The highest clearance rate (1.4 nl · protist−1 · h−1) was observed for the size class of unattached bacteria (length, 0.8 to 1.5 μm) that has the highest frequency of dividing cells in the MMR plume (34a). Selective grazing by protists on the bacteria which are most frequently dividing has been observed in a marine environment by Sherr et al. (46). By preferentially grazing on this size class, the flagellates have a more profound effect on bacterial production and, therefore, on the degradation of the organic contaminants by the bacteria.
The uptake rates calculated from the grazing experiments were used to estimate the impact of the groundwater nanoflagellates on the unattached bacterial standing crop in the aquifer (Table 4). The estimates were based on the assumption that the uptake rates observed over the 4-h incubation period would be maintained over 24 h. The percent consumption of the standing stock was computed by using concentrations of nanoflagellates and the different size classes of unattached bacteria in the MMR aquifer. For the 0.8- to 1.5-μm bacteria, 12 to 74% of the standing crop could be consumed per day by the groundwater flagellates. These estimates are generally in the range of 25 to >100% reported in the literature for other environments (50), and they are substantial when the modest bacterial growth rates reported for the MMR plume are considered (24). While they are only the first approximation of the impact of the nanoflagellates on the community of unattached bacteria that resides within the MMR plume, they demonstrate that protistan predation is probably significant and should be considered in models of in situ bioremediation.
TABLE 4.
Estimates of impact of groundwater nanoflagellates on the unattached bacterial standing crop
| Bacterial cell length (μm) | Uptake rate (bacteria · protist−1 · h−1) | Consumption ratea (bacteria · ml−1 · day−1) (104) | Avg unattached bacterial concnb (ml−1) | % of unattached bacterial standing crop consumedc (day−1) |
|---|---|---|---|---|
| 0.1–0.5 | 0.06 | 1.4–8.6 | 1.2 × 105 | 12–72 |
| 0.5–0.8 | 0.13 | 3.1–18.7 | 2.5 × 106 | 1.2–7.5 |
| 0.8–1.5 | 0.77 | 18.4–111 | 1.5 × 106 | 12–74 |
| >1.5 | 0.04 | 0.96–5.8 | 2.6 × 105 | 3.7–22 |
Assumes that the uptake rate from grazing experiments would be constant over 24 h.
In MMR aquifer near head of the plume.
Calculated by using the nanoflagellate concentration in the MMR aquifer near the head of the plume (1 × 104 to 6 × 104 ml−1).
Data collected on the growth and deposition for the unattached population of groundwater bacteria in the upgradient portion of the MMR plume clearly suggest that attachment to grain surfaces is insufficient to balance expected population increases due to growth. Although deposition is an important mechanism for removal of the unattached bacteria, injection and recovery investigations using DAPI-labeled groundwater bacteria (for an example, see reference 23) indicate that attachment to grain surfaces can explain, at most, a 1-log-unit removal for every 10 m (∼1 month) of travel through the MMR aquifer. However, growth rate estimates for this same population suggest an average generation time between 1 and 4 days for the upgradient portion of the plume (24). Because the abundance of the unattached bacterial population within the plume decreases steadily with increasing distance downgradient (22), there must be an additional removal mechanism(s), such as protistan grazing, that is capable of explaining the daily disappearance of significant fractions of the standing stock. Recent laboratory observations, comparing bacterial breakthrough in columns of sterile versus flagellate-containing aquifer sediment, suggest that under some conditions, predation by nanoflagellates can actually be more important than attachment to grain surfaces for removing unattached bacteria from the pore fluid of contaminated aquifer sediments (31). Mortality of the unattached bacterial community within the MMR plume, due to infection by bacteriophage, is a distinct possibility. Unfortunately, there are no data currently available on the effect of viruses on the population dynamics of bacteria in aquifers.
To improve these grazing estimates, we must further refine the bacterial uptake rates of the flagellates by conducting other experiments to estimate predation. One approach would be to use the method described by Starink et al. (58) that includes the impact of the protists on the bacteria associated with the sediment surface. This portion of the bacterial population, which can be active in biodegradation, was not considered in our liquid-phase experiments. Starink et al. (58) found a twofold-greater grazing rate when predation on surface-associated bacteria was included in their experiments with marine sediments. It is likely that some nanoflagellates in the MMR plume can preferentially graze on the more loosely attached bacteria. In fact, in our experiments, we observed one nanoflagellate that constituted ≤5% of the protistan population that rarely ingested FLB. It is possible that this organism grazes on surface-associated bacteria (7). While it is doubtful that the types of flagellates found in the MMR plume could ingest bacteria adhering to surfaces with a glycocalyx, bacteria that are weakly associated with surfaces in the so-called secondary minimum (<100 nm from the actual surface) may be a potential food source for surface-associated flagellates (e.g., Cercomonas) (37) and should be considered in future experimental designs. Ingestion of bacteria that are weakly (electrostatically) associated with grain surfaces may be particularly important for nanoflagellate populations inhabiting the distal portion of the MMR plume, which is characterized by low abundances of unattached bacteria (22).
The clearance rates measured for the groundwater nanoflagellates must be used cautiously in predicting what is occurring in situ. As noted by Gonzalez et al. (17), the rates calculated are “effective,” not absolute, grazing rates. In this study, we did not count the number of FLB per flagellate and made the assumption that each protist contained only one FLB. This resulted in a conservative estimate of clearance rates, because it appeared that many nanoflagellates contained more than one FLB, although it was difficult to resolve the exact number. In addition, preparation of the FLB may have altered the surface chemistry of the prey, affecting protistan uptake rates (17). In our experiments, the smallest bacteria (0.1 to 0.5 μm) were fixed with formalin, while all other size classes of FLB were heat killed. The changes to the prey’s surface caused by the type of fixation are not well understood and may have also affected the grazing rates. Sherr and Sherr (49, 51) noted that killing the bacteria before staining renders them nonmotile. This lowers the grazing rate because bacterivorous flagellates have higher grazing rates on motile bacteria (19, 32). In addition, the groundwater nanoflagellates are more likely to attach in situ, which will increase the probability of interception of food particles (13, 68). Conversely, higher temperature can increase flagellate grazing rates (47). Because our experiments were conducted at 20 ± 2°C, while the temperature of the MMR plume ranges from 8 to 10°C year round, the calculated rates would be higher than those in situ, offsetting some of the negative impacts described above.
Clearance rates are affected by the type (16, 35) and physiological state (18, 46) of the predator and the prey. The nanoflagellates that flourish in the porous-medium cultures represent only a few of the species present in cores taken from the MMR plume. More accurate grazing rates which are based on in situ populations will need to be calculated. In all of the experiments, the porous-medium cultures were sampled when they were at similar ages and population sizes. While this mitigated against interpretational problems resulting from comparisons of organisms with different physiological states, it makes it difficult to estimate what will happen in situ. Unfortunately, we know little about the in situ physiological states of either the bacteria or nanoflagellate populations occurring simultaneously in the MMR plume. Certainly, near the head of the plume, growth rates may be subject to considerable change due to temporal alterations in wastewater loading conditions.
These grazing experiments support our initial data (31) that indicated that nanoflagellates in the aquifer significantly impact the number of unattached bacteria present in the MMR contaminant plume. In the upgradient region of the plume, where the greatest biodegradation of the organic contamination is occurring, the unattached bacteria can comprise at least 30% of the total bacterial population (22). By ingesting the size class of unattached bacteria responsible for the greatest productivity, nanoflagellates probably enhance the rate of bacterial degradation of DOC in the MMR plume in a manner similar to what has been observed for other carbon-limited environments (for an example, see reference 27). We are conducting further laboratory and in situ experiments to determine groundwater bacterial and protistan population dynamics in response to controlled injections of readily degradable organic compounds as one indication of whether the nanoflagellates play such a role in a contaminated aquifer. The simplicity of the MMR plume’s ecosystem (no herbivores and no ciliates) makes it ideal for studying these relationships.
ACKNOWLEDGMENTS
This research was funded by the U.S. National Science Foundation (grant BSC 9312235) awarded to the University of New Hampshire.
The assistance of the Massachusetts/Rhode Island District of the USGS in obtaining aquifer sediments is gratefully acknowledged. We thank David W. Metge (USGS, Boulder, Colo.) for providing the aquifer bacteria. Laura Baumgartner (University of Colorado, Boulder) and David W. Metge are also thanked for their helpful reviews of the manuscript.
REFERENCES
- 1.Andersson A, Larsson U, Hagström Å. Size-selective grazing by a microflagellate on pelagic bacteria. Mar Ecol Prog Ser. 1986;33:51–57. [Google Scholar]
- 2.Barcina I, Ayo B, Muela A, Egea L, Iriberri J. Predation rates of flagellate and ciliated protozoa on bacterioplankton in a river. FEMS Microbiol Ecol. 1991;85:141–150. [Google Scholar]
- 3.Beloin R M, Sinclair J L, Ghiorse W C. Distribution and activity of microorganisms in subsurface sediments of a pristine study rate in Oklahoma. Microb Ecol. 1988;16:85–97. doi: 10.1007/BF02097407. [DOI] [PubMed] [Google Scholar]
- 4.Bloem J, Ellenbroek F M, Bär-Gilissen M-J B, Cappenberg T E. Protozoan grazing and bacterial production in stratified Lake Vechten estimated with fluorescently labeled bacteria and by thymidine incorporation. Appl Environ Microbiol. 1989;55:1787–1795. doi: 10.1128/aem.55.7.1787-1795.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloem J, Starink M, Bär-Gilissen M J B, Cappenberg T E. Protozoan grazing, bacterial activity, and mineralization in two-stage continuous cultures. Appl Environ Microbiol. 1988;54:3113–3121. doi: 10.1128/aem.54.12.3113-3121.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunn A L. Techniques for enumerating protozoa in saturated subsurface sediments. Ph.D. thesis. Durham: University of New Hampshire; 1992. [Google Scholar]
- 7.Caron D A. Grazing of attached bacteria by heterotrophic microflagellates. Microb Ecol. 1987;13:203–218. doi: 10.1007/BF02024998. [DOI] [PubMed] [Google Scholar]
- 8.Choi J W, Stoecker D K. Effects of fixation on cell volume of marine planktonic protozoa. Appl Environ Microbiol. 1989;55:1761–1765. doi: 10.1128/aem.55.7.1761-1765.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrzanowski T H, Simek K. Prey-size selection by freshwater flagellated protozoa. Limnol Oceanogr. 1990;35:1429–1436. [Google Scholar]
- 10.Clarholm M. Interactions of bacteria, protozoa and plants leading to mineralization of soil nitrogen. Soil Biol Biochem. 1985;17:181–187. [Google Scholar]
- 11.Curds C R, Cockburn A, Vandyke J M. An experimental study of the role of ciliated protozoa in the activated-sludge process. Water Pollut Control. 1968;67:312–329. [Google Scholar]
- 12.Cynar F J, Sieburth J M. Unambiguous detection and improved quantification of phagotrophy in apochlorotic nanoflagellates using fluorescent microspheres and concomitant phase contrast and epifluorescence microscopy. Mar Ecol Prog Ser. 1986;32:61–70. [Google Scholar]
- 13.Fenchel T. The ecology of heterotrophic microflagellates. Adv Microb Ecol. 1986;9:57–97. [Google Scholar]
- 14.Fenchel T. Protozoan filter feeding. Prog Protistol. 1986;1:65–113. [Google Scholar]
- 15.Fenchel T. Ecology of protozoa. The biology of free-living phagotrophic protists. New York, N.Y: Springer-Verlag; 1987. [Google Scholar]
- 16.Gonzalez J M, Iriberri J, Egea L, Barcina I. Differential rates of digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl Environ Microbiol. 1990;56:1851–1857. doi: 10.1128/aem.56.6.1851-1857.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez J M, Sherr E B, Sherr B F. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol. 1990;56:583–589. doi: 10.1128/aem.56.3.583-589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez J M, Sherr E B, Sherr B F. Differential feeding by marine flagellates on growing versus starving, and on motile versus nonmotile, bacterial prey. Mar Ecol Prog Ser. 1993;102:257–267. [Google Scholar]
- 19.Gonzalez J M, Sherr B F, Sherr E B. Digestive enzyme activity as a quantitative measure of protistan grazing: the acid lysozyme assay for bacterivory. Mar Ecol Prog Ser. 1993;100:197–206. [Google Scholar]
- 20.Gonzalez J M, Suttle C A. Grazing by marine nanoflagellates on viruses and virus-sized particles: ingestion and digestion. Mar Ecol Prog Ser. 1993;94:1–10. [Google Scholar]
- 21.Gruden C L. An investigation into the role of protozoa in an organically-contaminated aquifer. M.S. thesis. Durham: University of New Hampshire; 1993. [Google Scholar]
- 22.Harvey R W, Barber L B., II Associations of unattached bacteria and dissolved organic compounds in a plume of contaminated groundwater. J Contam Hydrol. 1992;9:91–103. [Google Scholar]
- 23.Harvey R W, Garbedian S P. Use of colloidal filtration theory in modeling movement of bacteria through a contaminated sandy aquifer. Environ Sci Technol. 1991;25:178–185. [Google Scholar]
- 24.Harvey R W, George L H. Growth determination for unattached bacteria in a contaminated aquifer. Appl Environ Microbiol. 1987;53:2992–2996. doi: 10.1128/aem.53.12.2992-2996.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holen D A, Boraas M E. The feeding behavior of Spumella sp., as a function of particle size: implications for bacterial size in pelagic systems. Hydrobiologia. 1991;220:73–88. [Google Scholar]
- 26.Hollibaugh J T, Fuhrman J A, Azam F. Radioactively labeling natural assemblages of bacterioplankton for use in trophic studies. Limnol Oceanogr. 1980;25:172–181. [Google Scholar]
- 27.Hunt H W, Cole C V, Klein D A, Coleman D C. A simulation model for the effect of predation on bacteria in continuous culture. Microb Ecol. 1977;3:259–278. doi: 10.1007/BF02010735. [DOI] [PubMed] [Google Scholar]
- 28.Johannes R E. Influence of marine protozoa on nutrient regeneration. Limnol Oceangr. 1965;10:434–442. [Google Scholar]
- 29.Kinner N E, Bunn A L, Harvey R W, Warren A, Meeker L D. Preliminary evaluation of relations among protozoa, bacteria, and chemical properties in sewage-contaminated ground water near Otis Air Base, Massachusetts. In: Mallard G E, Aaronson D A, editors. U.S. Geological Survey Water Resources Investigations report 91-4034. U.S. Reston, Va: Geological Survey; 1991. pp. 141–143. [Google Scholar]
- 30.Kinner N E, Harvey R W. Overview of research on the distribution and role of protozoa in an organically-contaminated aquifer at Cape Cod, MA. In: Morganwalp D W, Aaronson D A, editors. U.S. Geological Survey Water Resources Investigations report 94-4015. U.S. Reston, Va: Geological Survey; 1993. pp. 249–252. [Google Scholar]
- 31.Kinner N E, Harvey R W, Kazmierkiewicz-Tabaka M. Effect of flagellates on unattached bacterial abundance in an organically-contaminated aquifer. FEMS Microbiol Rev. 1997;20:249–259. doi: 10.1111/j.1574-6976.1997.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 32.Landry M R, Lehner-Fournier J M, Sundstrom J A, Fagerness V L, Selph K E. Discrimination between living and heat-killed prey by a marine zooflagellate, Paraphysomonas vestita (Stokes) J Exp Mar Biol Ecol. 1991;146:139–151. [Google Scholar]
- 33.LeBlanc D R, editor. Movement and fate of solutes in a plume of sewage-contaminated ground water. U.S. Geological Survey open-file report 84-475. U.S. Reston, Va: Geological Survey; 1984. [Google Scholar]
- 34.Madsen E L, Sinclair J L, Ghiorse W C. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science. 1991;252:830–833. doi: 10.1126/science.2028258. [DOI] [PubMed] [Google Scholar]
- 34a.Metge, D. W., N. E. Kinner, and R. W. Harvey. Unpublished data.
- 35.Mitchell G C, Baker J H, Sleigh M A. Feeding of a freshwater flagellate, Bodo saltans, on diverse bacteria. J Protozool. 1988;35:219–222. [Google Scholar]
- 36.Novarino G, Warren A, Butler H, Lambourne G, Boxshall A, Bateman J, Kinner N E, Harvey R W, Mosse R A, Teltsch B. Protistan communities in aquifers. A review. FEMS Microbiol Rev. 1997;20:261–276. doi: 10.1111/j.1574-6976.1997.tb00313.x. [DOI] [PubMed] [Google Scholar]
- 37.Novarino G, Warren A, Kinner N E, Harvey R W. Protists from a sewage-contaminated aquifer on Cape Cod, Massachusetts, USA. Geomicrobiol J. 1994;12:23–36. [Google Scholar]
- 38.Pace M L, Bailiff M D. Evaluation of a fluorescent microsphere technique for measuring grazing rates of phagotrophic microorganisms. Mar Ecol Prog Ser. 1987;40:185–193. [Google Scholar]
- 39.Pesan B F, Weinbauer M G, Peduzzi P. Significance of the virus-rich 2-200 nm size fraction of seawater for heterotrophic flagellates. I. Impact on growth. Mar Ecol. 1994;15:281–290. [Google Scholar]
- 40.Salonen K, Jokinen S. Flagellate grazing on bacteria in a small dystrophic lake. Hydrobiologia. 1988;161:203–209. [Google Scholar]
- 41.Sanders R W, Porter K G, Bennett S J, DiBiase A E. Seasonal patterns of bacterivory by flagellates, ciliates, rotifiers, and cladocerans in a freshwater planktonic community. Limnol Oceanogr. 1989;34:673–687. [Google Scholar]
- 42.Sherr B, Sherr E, Pedrós-Alió C. Simultaneous measurement of bacterioplankton production and protozoa bacterivory in estuarine water. Mar Ecol Prog Ser. 1989;54:209–219. [Google Scholar]
- 43.Sherr B F, Sherr E B, Andrew T L, Fallon R D, Newell S Y. Trophic interactions between heterotrophic protozoa and bacterioplankton in estuarine water analyzed with selective metabolic inhibitors. Mar Ecol Prog Ser. 1986;32:169–179. [Google Scholar]
- 44.Sherr B F, Sherr E B, Berman T. Grazing, growth and ammonia excretion rates of a heterotrophic microflagellate fed with four species of bacteria. Appl Environ Microbiol. 1983;45:1196–1201. doi: 10.1128/aem.45.4.1196-1201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherr B F, Sherr E B, Fallon R D. Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol. 1987;53:958–965. doi: 10.1128/aem.53.5.958-965.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherr B F, Sherr E B, McDaniel J. Effect of protistan grazing on the frequency of dividing cells in bacterioplankton assemblages. Appl Environ Microbiol. 1992;58:2381–2385. doi: 10.1128/aem.58.8.2381-2385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherr B F, Sherr E B, Rassoulzadegan F. Rates of digestion of bacteria by marine phagotrophic protozoa: temperature dependence. Appl Environ Microbiol. 1988;54:1091–1095. doi: 10.1128/aem.54.5.1091-1095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherr E B. Direct use of high molecular weight polysaccharides by heterotrophic flagellates. Nature. 1988;335:348–351. [Google Scholar]
- 49.Sherr E B, Sherr B F. Protistan grazing rates via uptake of fluorescently labeled prey. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Ann Arbor, Mich: Lewis Publishers; 1993. pp. 695–701. [Google Scholar]
- 50.Sherr E B, Sherr B F. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb Ecol. 1994;28:223–235. doi: 10.1007/BF00166812. [DOI] [PubMed] [Google Scholar]
- 51.Sherr E B, Sherr B F. Phagotrophy in aquatic microbial food webs. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C: ASM Press; 1997. pp. 309–315. [Google Scholar]
- 52.Simek K, Chrzanowski T H. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl Environ Microbiol. 1992;58:3715–3720. doi: 10.1128/aem.58.11.3715-3720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinclair J L, Alexander M. Effect of protozoan predation on relative abundance of fast- and slow-growing bacteria. Can J Microbiol. 1989;35:578–582. [Google Scholar]
- 54.Sinclair J L, Ghiorse W C. Distribution of protozoa in subsurface sediments of a pristine groundwater study site in Oklahoma. Appl Environ Microbiol. 1987;53:1157–1163. doi: 10.1128/aem.53.5.1157-1163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinclair J L, Ghiorse W C. Distribution of aerobic bacteria, protozoa algae and fungi in deep subsurface sediments. Geomicrobiol J. 1989;7:15–31. [Google Scholar]
- 56.Sinclair J L, Kampbell D H, Cook M L, Wilson J T. Protozoa in subsurface sediments from sites contaminated with aviation gasoline or jet fuel. Appl Environ Microbiol. 1993;59:467–472. doi: 10.1128/aem.59.2.467-472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinclair J L, Randke S J, Denne J E, Hathaway L R, Ghiorse W C. Survey of microbial populations in buried-valley aquifer sediments in northeastern Kansas. Ground Water. 1990;28:369–377. [Google Scholar]
- 58.Starink M, Krylova I N, Bär-Gilissen M J, Bak R P M, Cappenberg T E. Rates of benthic protozoan grazing on free and attached sediment bacteria measured with fluorescently stained sediment. Appl Environ Microbiol. 1994;60:2259–2264. doi: 10.1128/aem.60.7.2259-2264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tranvik L J, Sherr E B, Sherr B F. Uptake and utilization of colloidal DOM by heterotrophic flagellates in seawater. Mar Ecol Prog Ser. 1993;92:301–309. [Google Scholar]
- 60.Van Houten J, Hauser D C, Levandowski M. Chemosensory behavior in protozoa. In: Levandowsky M, Hunter S H, editors. Biochemistry and physiology of protozoa. 2nd ed. Vol. 4. New York, N.Y: Academic Press, Inc.; 1981. pp. 67–124. [Google Scholar]
- 61.Verhagen F J M, Duyts H, Laanbroek H J. Effects of grazing by flagellates on competition for ammonium between nitrifying and heterotrophic bacteria in soil columns. Appl Environ Microbiol. 1993;59:2099–2106. doi: 10.1128/aem.59.7.2099-2106.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vrba J, Simek K, Medoma J, Hartman P. 4-Methylumbelliferyl-β-N-acetyl glucosaminide hydrolysis by a high-affinity enzyme, a putative marker of protozoan bacterivory. Appl Environ Microbiol. 1993;59:3091–3101. doi: 10.1128/aem.59.9.3091-3101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warren A, Butler H. An investigation of amoebae from an organically-contaminated aquifer at Cape Cod, MA. Natural History Museum (London) Report SPC-94-4023. United Kingdom: Natural History Museum London; 1994. [Google Scholar]
- 64.Wikner J. Grazing rate of bacterioplankton via turnover of genetically marked minicells. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Ann Arbor, Mich: Lewis Publishers; 1993. pp. 703–714. [Google Scholar]
- 65.Wikner J, Anderson A, Normark S, Hagstrom A. Use of genetically marked minicells as a probe in measurement of predation on bacteria in aquatic environments. Appl Environ Microbiol. 1986;52:4–8. doi: 10.1128/aem.52.1.4-8.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright R T, Coffin R B. Measuring microzooplankton grazing on planktonic marine bacteria by its impact on bacterial production. Microb Ecol. 1984;10:137–149. doi: 10.1007/BF02011421. [DOI] [PubMed] [Google Scholar]
- 67.Zapico M M, Vales S, Cherry J A. A wireline piston core barrel for sampling cohesionless sand and gravel below the water table. Ground Water Monit Rev. 1987;7:74–87. [Google Scholar]
- 68.Zwart K B, Darbyshire J F. Growth and nitrogenous excretion of a common soil flagellate Spumella sp.—a laboratory experiment. Soil Sci. 1992;43:145–157. [Google Scholar]