Abstract
Numerous instances of reverse transcriptase (RT) inhibition of the PCR were observed while developing nonquantitative uncoupled RT-PCR techniques for detecting nitrogenase and ammonia monooxygenase gene expression in situ. The inhibitory effect of RT on the PCR was removed with increasing template concentrations beyond 105 to 106 copies. Including T4 gene 32 protein during the reverse transcription phase of the RT-PCR reaction increased the RT-PCR product yield by as much as 483%; if gene 32 protein was introduced after reverse transcription but prior to the PCR phase, no improvement in product yield was observed. Addition of 1 μg of exogenous calf thymus DNA or yeast tRNA did little to relieve RT inhibition of the PCR on both genomic DNA and mRNA templates. These results suggest that RT inhibition of the PCR is mediated through direct interaction with the specific primer-template combination (DNA and RNA) and point to specific assay modifications for estimating the extent of RT inhibition and counteracting some of the inhibitory effect. Furthermore, the working hypothesis of RT inhibition below a 105 to 106 copy threshold has important implications for quantitative RT-PCR studies. In particular, competitive, quantitative RT-PCR systems will consistently underestimate the actual RNA concentration. Hence, enumerations of RNA templates below 105 to 106 copies will be relative to an internal standard and will not be an absolute measure of RNA abundance in situ.
The field of microbial ecology is rapidly expanding due, in part, to the explosion of molecular microbial ecology and advances in nucleic acid techniques. Driven in large part by the availability of PCR, researchers are now able to recover and analyze nucleic acid sequences from microorganisms that remain uncultivated. Virtually every aspect of microbial ecology has been affected by these techniques. At the forefront of nucleic acid technological development are PCR and reverse transcriptase PCR (RT-PCR) assays for quantifying specific microorganisms, functional genes, and microbial activity in complex natural communities independently of culturability (7, 8, 13, 25, 27).
Nucleic acid techniques are particularly well suited to the study of nitrifying and denitrifying bacteria in situ, since these microorganisms are notoriously difficult to culture in the laboratory and grow very slowly under standard culture conditions (6). We have been studying the nitrogen-cycling processes in arid shrub-steppe ecosystems and are developing PCR-based techniques for quantifying microbial abundance and activity. During the course of RT-PCR development for nonquantitative detection of mRNA, we routinely observed that control DNA reaction mixtures containing RT did not give DNA amplification during PCR whereas control DNA reaction mixtures without RT showed no signs of PCR inhibition. This observation suggested that our ability to detect and amplify mRNA from environmental samples may be hindered by previously unknown interactions between the RT and the template. The purpose of this study was to investigate this phenomenon and gain a better understanding of the variables and limitations associated with the RT-PCR process. Our results suggest that the inhibitory effect of RT on the PCR is mediated through the RT interaction(s) with the specific mRNA or cDNA template and that the inhibitory effect is dependent upon template concentration (or copy number). Some of the inhibitory effect could be removed by including T4 gene 32 protein specifically during the reverse transcription reaction, which has unveiled some novel properties of T4 gene 32 protein that were not previously recognized or exploited. The implications of these findings apply to RT-PCR in general but also to quantitative RT-PCR studies where the abundance of specific mRNA may be relatively low.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Nostoc sp. strain 27895 was obtained from the American Type Culture Collection (Rockville, Md.), and Nitrosomonas europaea “freitag” was obtained from E. Bock, University of Hamburg (Hamburg, Germany). For DNA isolations, Nostoc was grown in ATCC medium 616 [containing, per liter, 1.5 g of NaNO3, 0.04 g of K2PO4, 0.075 g of MgSO4 · 7H2O, 0.036 g of CaCl2 · 2H2O, 0.006 g of citric acid, 0.006 g of ferric ammonium citrate, 0.001 g of disodium EDTA, 0.020 g of Na2CO3, 2.86 mg of H3BO3, 1.81 mg of MnCl2 · 4H2O, 0.222 mg of ZnSO4 · 7H2O, 0.39 mg of Na2MoO4 · 2H2O, 0.079 mg of CuSO4 · 5H2O, and 49.4 μg of Co(NO3)2 · 6H2O (pH 7.1)] for 3 weeks under constant illumination at 26°C. For total RNA isolations, Nostoc was grown in ATCC medium 616 without nitrate (NaNO3) for 3 weeks under constant illumination. N. europaea was cultured in ammonia oxidizer medium [containing, per liter, 0.5 g of (NH4)SO4, 1.34 mg of CaCl2 · 2H2O, 0.04 g of MgSO4 · 7H2O, 0.204 g of KH2PO4, 0.002 g of bromthymol blue, 0.1 mg of Na2MoO4 · 2H2O, 0.2 mg of MnCl2, 2.0 μg of CoCl2 · 6H2O, 100 μg of ZnSO4 · 7H2O, and 20 μg of CuSO4 · 5H2O] at room temperature, and the pH was adjusted with filter-sterilized 0.5 M K2CO3 as needed, for up to 2 months.
DNA isolation.
Cells from a 500-ml culture of Nostoc and N. europaea were collected by centrifugation and frozen at −20°C. The pellets were thawed and resuspended in 800 μl of lysis buffer (50 mM Tris, 50 mM disodium EDTA, 2% sodium dodecyl sulfate [pH 8.0]). Genomic DNA was liberated from the cells by ballistic disintegration with ∼1 g of sterile glass beads (0.1 mm in diameter) in an eight-place bead beater (BioSpec Products, Inc., Bartlesville, Okla.) at full speed for 1 min. Cellular debris and glass beads were removed by centrifugation at 13,600 × g at room temperature for 5 min, and the supernatant was transferred to a fresh tube. Beads and cell debris were extracted once more with 300 μl of TE (10 mM Tris, 1 mM disodium EDTA [pH 7.8]) and centrifuged as above. Like supernatants were combined, treated with 10 μl of RNase A (10 mg ml−1; Sigma, St. Louis, Mo.) for 30 min at 37°C, and extracted twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). Nucleic acids were precipitated with NaCl-isopropanol at room temperature for 10 min, collected by centrifugation for 8 min at 13,600 × g, washed once in 70% ethanol, dried under vacuum, and resuspended in sterile TE. DNA concentrations were determined by fluorometry with Hoescht 33258 stain and a TKO minifluorometer (Hoefer, San Francisco, Calif.).
Total-RNA isolation.
All glass, tubes, and plasticware were bleached, autoclaved, and treated with an RNase inhibitor (RNAseZap; Ambion Inc., Austin, Tex.), and all solutions were treated with diethylpyrocarbonate (DEPC) or prepared with DEPC-treated water. Nostoc culture (2 liters) and N. europaea culture (6 liters) were grown as described above and harvested by centrifugation. Cryptogamic crust (16 g [dry weight]) was rehydrated and incubated for 1 week at room temperature under natural lighting conditions. An equivalent of 2 liters of culture medium or 4-g equivalents of crust was lysed with 5 ml of GIPS solution (4.0 M guanidine isothiocyanate, 0.5% Sarkosyl, 0.25 M sodium citrate [pH 7.0]) and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The phases were separated by centrifugation at 6,000 × g for 10 min, and the aqueous phase was transferred to a new tube. Nucleic acids were precipitated with 2 volumes of ethanol overnight at room temperature and collected by centrifugation at 10,000 × g for 20 min. Nucleic acid pellets were washed once with 70% ethanol, dried briefly under vacuum, resuspended in DEPC-treated water, and stored at −80°C.
Isolation of mRNA and 16S rRNA.
Aliquots of total RNA were treated with amplification-grade DNase I as specified by the manufacturer (Life Technologies, Gaithersburg, Md.), and the DNase was removed by phenol-chloroform extraction. Purified RNA was then precipitated with sodium acetate-ethanol as described above, resuspended in DEPC-treated water, and quantified by UV absorption. Approximately 4.2 μg of Nostoc RNA and 13.5 μg of N. europaea RNA were used for affinity capture of nifH mRNA and 16S rRNA, respectively. The yield of cryptogamic crust total RNA could not be determined due to the coextraction of humic acids and other soil constituents that interfered with UV adsorption; in this case, 4-g equivalents of crust material in a 150-μl total volume were used for affinity capture of nifH mRNA. Biotinylated primers were synthesized by Keystone Laboratory Inc. (Menlo Park, Calif.) with the following sequences: Nostoc.I.MHC 5′-TTT TCT TCT AAG AAG TTR ATG GCG GTG AT-biotin for nifH transcripts (this study) and 1392.r 5′-ACG GGC GGT GTG TRC-biotin for 16S rRNA (47). The primers were reconstituted at 50 μM in DEPC-treated water. Total RNA was initially heat denatured at 65°C for 10 min and then subjected to affinity capture and purification with a PolyATtract mRNA isolation system as specified by the manufacturer (Promega Corp., Madison, Wis.), except that captured RNA was eluted in two washes of DEPC-treated water totaling 150 μl. After mRNA capture, residual DNA was removed by DNase I treatment and purified mRNA was recovered as outlined above for total RNA isolation; the final mRNA sample was resuspended in 150 μl of DEPC-treated water. RNA concentrations for N. europaea 16S rRNA were approximately 64 ng ml−1 as determined by UV absorption; the quantity of nifH mRNA was too small to be determined accurately by UV absorption.
Reverse transcription of RNA templates.
Purified nifH mRNA (1 to 10 μl) or N. europaea 16S rRNA (2 ng to 2 fg) and 2 pmol of reverse primer (see below) were heat denatured in 11 μl (total volume) at 70°C for 10 min. Where indicated, T4 gene 32 protein was introduced at 1.5 μg per sample during the initial 70°C template denaturation. After heat denaturation, reverse transcription reaction mixtures were assembled in 19.5 μl (total volume), which included an additional 0.5 μl of cloned RNase inhibitor, all as specified by the manufacturer of the RT (Life Technologies). The reverse transcription reaction mixtures were preheated to 42°C for 2 min, after which 1 μl of SuperScriptII RNase H− RT (200 U) was added. After a 50-min incubation at 42°C, the RT was heat inactivated at 100°C for 5 min and the reaction mixtures were quick-chilled on ice. Two microliters of each reverse transcription reaction mixture was then used as the template for PCR as described below. Control reverse transcription reaction mixtures included diluted genomic DNA with or without RT, no-template reactions with or without RT, and purified RNA templates with or without RT. Two additional RT enzymes, Moloney murine leukemia virus (MoMuLV) RT and avian myeloblastosis virus (AMV) RT, both from Life Technologies, were also tested.
PCR amplification of cDNA products.
Primers and PCR conditions for N. europaea 16S rRNA genes were as described elsewhere (9); a hot-start PCR and 40 amplification cycles were used. Control PCR mixtures included diluted genomic DNA that was not subjected to reverse transcription and all relevant controls described above for the RT reactions.
Specific primers for Nostoc nifH sequences were constructed with the sequence 5′ CAG AAC CCG GTG TAG GTT (forward) and 5′ GTA ACG ATG TAG ATT TCT TG (reverse) and are based upon a ClustalW alignment for Nostoc commune, Nostoc muscorum, Nostoc strain PCC6720, and Nostoc sp. nifH sequences retrieved from the Entrez database (accession numbers L23514, U04054, Z31716, and L15551, respectively). A 2-μl portion of each RT reaction mixture, primers, MgCl2, and water in 25 μl (total volume) were heat denatured at 80°C, after which PCR buffer, Taq polymerase (Perkin-Elmer, Foster City, Calif.), and deoxynucleoside triphosphates (Promega) were added. The final reaction conditions consisted of 2 μl of RT template, 10 mM Tris (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 0.2 μM each primer, and 2.5 U of Taq polymerase in 50 μl (total volume). Amplification consisted of 40 cycles at 94°C for 20 s, 50°C for 20 s, and 72°C for 1 min in 0.2-ml thin-walled tubes and a Perkin-Elmer 9600 thermal cycler. Control PCRs were similar to those described above.
After PCR amplification, 25 μl of each reaction mixture was resolved on 1% SeaKem GTG–1% NuSieve agarose (FMC Bioproducts, Rockland, Maine) gels in 1× Tris-acetate-EDTA running buffer, both containing ethidium bromide. Images of each gel were captured with a Gel Doc 1000 station and Molecular Analyst software package (Bio-Rad, Hercules, Calif.). Individual band intensities were quantified with the accompanying Analyst software package, and the localized background was subtracted from the computer-determined value of each band. We could therefore compare relative product yield between individual RT-PCRs and measure the effects of T4 gene 32 protein in a semiquantitative manner.
RESULTS
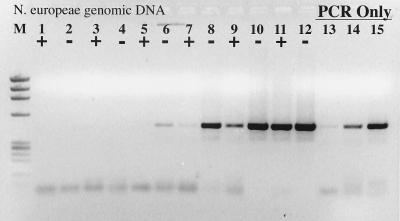
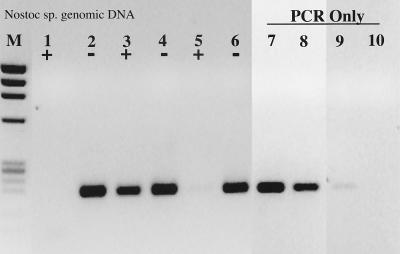
The initial observation prompting this study was that DNA in control (≤5 pg of genomic DNA) reaction mixtures containing RT did not amplify during PCR whereas control DNA reaction mixtures without RT showed no signs of PCR inhibition. This result was observed on at least six separate occasions with six different primer-template combinations and is not dependent upon the type or amount of RT (data not shown). The inhibitory effect of RT on the PCR, however, could be removed with increasing concentrations of genomic DNA (Fig. 1). An identical result and detection limit were obtained with the nifH PCR system and Nostoc sp. genomic DNA. Assuming an E. coli genome size of 4.5 fg cell−1, the PCR detection limit in both cases was approximately 45 cell equivalents of DNA, or 45 to 450 gene copies (assuming 1 to 10 copies per cell). RT inhibition of the PCR was overcome in both cases only when 4.5 × 105 cell equivalents of genomic DNA were present throughout the RT-PCR.
FIG. 1.
RT inhibition of the PCR. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1 and 2, no template; 3 and 4, 200 fg of N. europaea genomic DNA; 5 and 6, 2 pg; 7 and 8, 20 pg; 9 and 10, 20 pg; 11 and 12, 2 ng; 13, 200 fg of N. europaea genomic DNA amplified by PCR only; 14, 2 pg of N. europaea genomic DNA; 15, 20 pg of N. europaea genomic DNA.
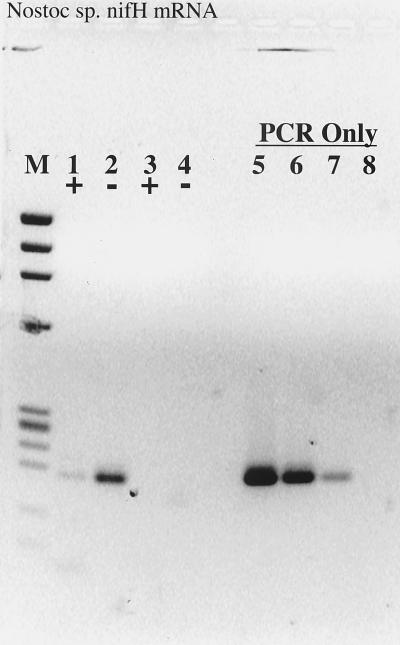
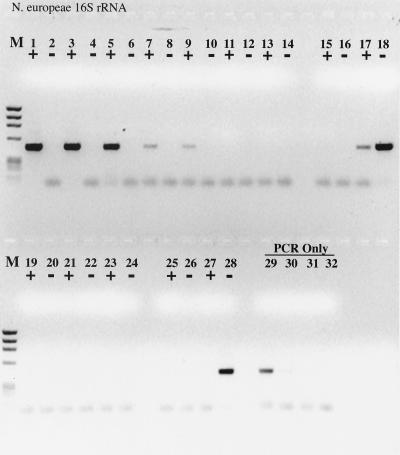
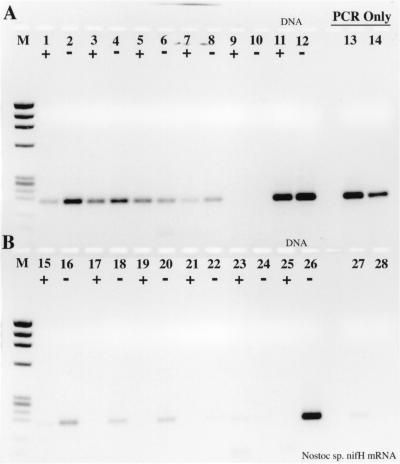
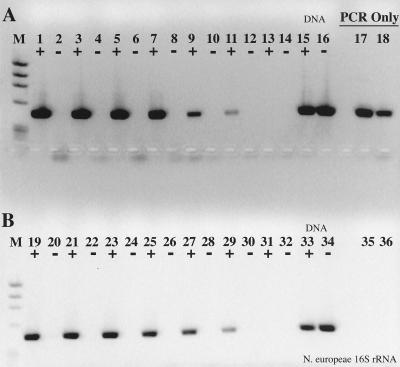
RT inhibition of the PCR was also observed when nifH mRNA was used as the template for RT-PCR (Fig. 2); in this case, Taq polymerase itself functioned as an RT, as indicated by the lack of PCR product when the template was pretreated with RNase (Fig. 2). Interestingly, the efficiency of reverse transcription and PCR was significantly greater with Taq as the sole polymerase than in the accompanying complete RT-PCR. When an identical experiment was performed with the N. europaea 16S rRNA, Taq polymerase was unable to reverse-transcribe the template and any inhibitory effects of RT could not be ascertained (Fig. 3). The detection limit for the 16S rRNA RT-PCR was approximately 0.4 amol (≤200 fg), or 4 × 105 copies, the same copy number at which obvious RT inhibition was overcome when amplifying from DNA templates.
FIG. 2.
RT inhibition of RT-PCR demonstrated on Nostoc sp. nifH mRNA. Lanes: M, φX174 × HaeIII molecular weight marker; 1, 5 μl of purified mRNA plus RT; 2, 5 μl of purified mRNA without RT; 3 and 4, same as 1 and 2, except that the mRNA sample was pretreated with RNase A prior to RT-PCR; 5, 20 pg of Nostoc sp. genomic DNA subject to PCR amplification only; 6, 2 pg of genomic DNA; 7, 200 fg of genomic DNA; 8, no-template control.
FIG. 3.
RT-PCR with a dilution series of purified N. europaea 16S rRNA as template. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1 and 2, 2 ng of 16S rRNA with or without RT; 3 and 4, 200 pg; 5 and 6, 20 pg; 7 and 8, 2 pg; 9 and 10, 200 fg; 11 and 12, 20 fg; 13 and 14, 2 fg; 15 and 16, no-template control; 17 and 18, 2 ng of N. europaea genomic DNA; 19 to 24, same as lanes 1 to 6, except that the sample was pretreated with RNase A prior to RT-PCR; 25 and 26, no-template controls pretreated with RNase A; 27 and 28, 2 ng of N. europaea genomic DNA pretreated with RNase A; 29, 20 pg of genomic DNA, PCR-only control; 30, 2 pg of genomic DNA; 31, 200 fg of genomic DNA; 32, no-template PCR-only control.
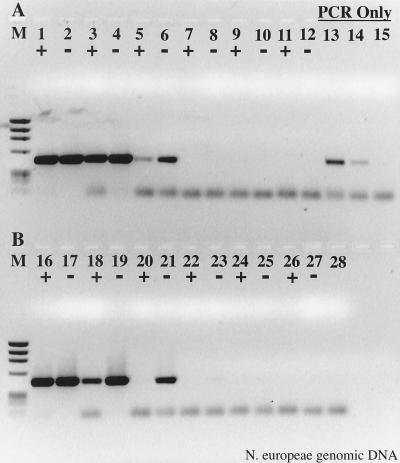
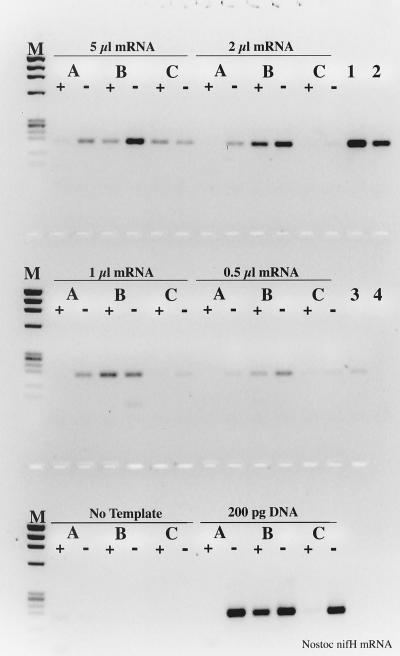
To further investigate the mechanism by which RT inhibits the PCR, T4 gene 32 protein was added during the reverse transcription step in an attempt to stabilize the DNA or displace RT from the genomic template. The addition of 1.5 μg of T4 gene 32 protein during RT-PCR resulted in a 50 to 142% improvement in PCR sensitivity when RT was also present (Fig. 4, compare lane 3 with lane 18 and lane 5 with lane 20). If T4 gene 32 protein was added after the RT step, RT inhibition of the PCR was again evident (Fig. 5). Extending these analyses to nifH mRNA templates clearly illustrated the improvement in RT-PCR sensitivity in the presence of T4 gene 32 protein (Fig. 6, compare lanes 1 to 8 with lanes 15 to 22). In the absence of T4 gene 32 protein, no amplification products were visible in the complete RT-PCR. Taq polymerase RT activity was augmented by 108 to 483% when T4 gene 32 protein was included in the nifH mRNA RT-PCR mixture. The improved RT-PCR performance was also evident with N. europaea 16S rRNA (Fig. 7). While the detection limit was identical whether or not T4 gene 32 protein was used in the RT-PCR (ca. 20 fg, or 40 zmol), increased product yield was again observed in reaction mixtures containing T4 gene 32 protein, ranging from a 20 to 80% increase when gene 32 protein was included during the initial 70°C template denaturation.
FIG. 4.
Relief of RT-PCR inhibition through the use of T4 gene 32 protein. N. europaea genomic DNA was used as template. (A) With T4 gene 32 protein; (B) without T4 gene 32 protein. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1 and 2, 2 ng of DNA with or without RT; 3 and 4, 200 pg; 5 and 6, 20 pg; 7 and 8, 2 pg; 9 and 10, 200 fg; 11 and 12, no-template RT-PCR controls; 13 to 15 = 20, 2, and 0.2 pg of genomic DNA PCR controls; 16 to 27, same as lanes 1 to 12, except without gene 32 protein; 28, no-template PCR control.
FIG. 5.
Relief of RT inhibition during the reverse transcription step in RT-PCR. Nostoc genomic DNA (200 pg) was used as the template. T4 gene 32 protein was added during the RT phase of RT-PCR or only for the PCR portion of the RT-PCR protocol. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1 and 2, no added T4 gene 32 protein; 3 and 4, gene 32 protein added during the RT step; 5 and 6, gene 32 protein added for the PCR phase only; 7, 20 pg of genomic DNA, PCR only; 8, 2 pg of DNA; 9, 200 fg of DNA; 10, no-template PCR control.
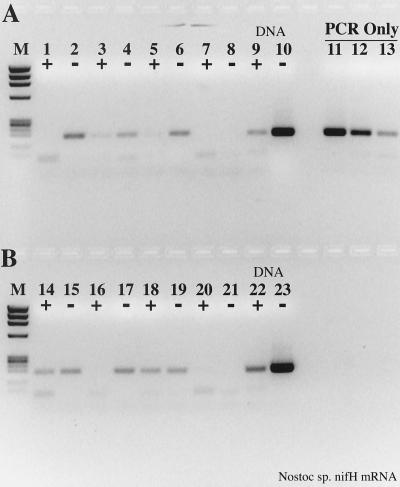
FIG. 6.
Effect of T4 gene 32 protein on RT-PCR amplification of Nostoc nifH mRNA. (A) Samples included 1.5 μg of gene 32 protein per RT-PCR; (B) no gene 32 protein present. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1, 2, 15, and 16, 5 μl of mRNA as template; 3, 4, 17, and 18, 2 μl of mRNA; 5, 6, 19, and 20, 1 μl of mRNA; 7, 8, 21, and 22, 0.5 μl of mRNA; 9, 10, 23, and 24, RT-PCR no-template controls; 11, 12, 25, and 26, RT-PCR 200 pg of Nostoc genomic DNA controls; 13, 14, 27, and 28, PCR-only controls; 13, 20 pg of genomic DNA; 14, 2 pg; 27, 200 fg, 28, no template.
FIG. 7.
Effect of T4 gene 32 protein on RT-PCR amplification of N. europaea 16S rRNA. (A) Samples included 1.5 μg of gene 32 protein per RT-PCR; (B) no gene 32 protein. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1, 2, 19, and 20, 2 ng of 16S rRNA as template; 3, 4, 21, and 22, 200 pg; 5, 6, 23, and 24, 20 pg; 7, 8, 25, and 26, 2 pg; 9, 10, 27, and 28, 200 fg; 11, 12, 29, and 30, 20 fg; 13, 14, 31, and 32, RT-PCR no-template controls; 15, 16, 33, and 34, 200 pg of N. europaea genomic DNA RT-PCR controls; 17, 18, 35, and 36, PCR-only controls; 17, 20 pg of N. europaea genomic DNA; 18, 2 pg of DNA; 35, 200 fg of DNA; 36, PCR-only no-template control.
T4 gene 32 protein is normally considered a single-stranded-DNA (ssDNA) binding protein and dsDNA helicase (28, 30, 31, 40, 44), but the genomic DNA results in Fig. 5 suggested that the effect of T4 gene 32 protein was mediated during reverse transcription rather than PCR amplification. To help understand the role of T4 gene 32 protein in the RT-PCR process, T4 gene 32 protein was added to the nifH mRNA RT-PCR mixtures during the RT step or during the PCR phase of the protocol. At all template concentrations, gene 32 protein added prior to reverse transcription significantly increased the PCR product yield relative to that in the standard RT-PCR (51 to 232% increase [Fig. 8]). If T4 gene 32 protein was added after reverse transcription but at the beginning of PCR, no increase in RT-PCR sensitivity was observed whether or not RT was one of the reaction components. In fact, the series of RT-PCRs without T4 gene 32 protein were more sensitive than if T4 gene 32 protein was introduced during the PCR phase of RT-PCR.
FIG. 8.
Specificity of T4 gene 32 protein interaction during RT-PCR amplification of Nostoc nifH mRNA. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker. Lanes were assigned based upon template identification, as indicated above each block of samples. Samples were also paired according to T4 gene 32 protein treatment; A, no-gene 32 protein; B, gene 32 protein added during the reverse transcription step of RT-PCR; C, gene 32 protein added after the reverse transcription step, and beginning of PCR. PCR-only controls were Nostoc genomic DNA; lane 1, 20 pg; lane 2, 2 pg; lane 3, 200 fg; lane 4, no template.
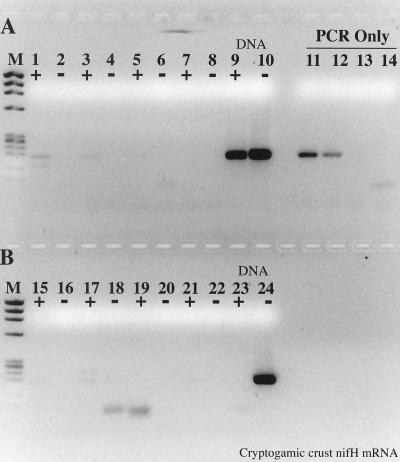
The importance of improved RT-PCR sensitivity was evident during an analysis of cryptogamic crust mRNA (Fig. 9). In the absence of T4 gene 32 protein, no amplification products were detected, whereas positive amplification was observed at the two highest sample volumes when T4 gene 32 protein was included in the RT-PCR mixture. Interestingly, Taq polymerase was unable to function as an RT on the nifH mRNA isolated from the crust material.
FIG. 9.
Effect of T4 gene 32 protein on RT-PCR amplification of Nostoc nifH mRNA isolated from cryptogamic crust. (A) Samples included 1.5 μg of gene 32 protein per RT-PCR; (B) no gene 32 protein. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1, 2, 15, and 16, 10 μl of mRNA as template; 3, 4, 17, and 18, 5 μl of mRNA; 5, 6, 19, and 20, 2 μl of mRNA; 7, 8, 21, and 22, no-template RT-PCR controls; 9, 10, 23, and 24, 200 pg of Nostoc genomic DNA RT-PCR controls; 11 to 14, PCR-only controls; 11, 20 pg of Nostoc genomic DNA; 12, 2 pg; 13, 200 fg; 14, no template.
Prior investigations have suggested that exogenous nucleic acids can relieve RT inhibition of the PCR (37). However, when either 1 μg of calf thymus DNA or 1 μg of yeast tRNA (5 × 102- to 5 × 106-fold excess) were included in the RT-PCR mixtures containing genomic DNA, the inhibitory effect of RT was not relieved (Fig. 10). An apparent 10-fold improvement in RT-PCR sensitivity was observed in the presence of exogenous calf thymus DNA, but the nonspecific amplification products indicate some level of primer cross-reactivity, which would serve to increase the effective copy number of the N. europaea 16S rDNA due to titration of the RT. Addition of exogenous DNA, then, did not significantly relieve RT inhibition of the PCR. Similarly, yeast tRNA did not appear to improve RT-PCR sensitivity from the nifH mRNA template (Fig. 11). While the data from Fig. 11B suggest some relief of RT inhibition for the full RT-PCR, the data from Fig. 11A indicate that RT-PCR was again more efficient in the absence of RT and without exogenous template. The value of exogenous template for improving RT-PCR sensitivity, then, is questionable. Since the RT is heat inactivated prior to PCR (5 min at 100°C) and neither calf thymus DNA nor yeast tRNA significantly improved RT-PCR performance at low template concentrations, we conclude that RT inhibition of the PCR is mediated through a specific interaction with the primer-template pair undergoing reverse transcription rather than with Taq polymerase itself.
FIG. 10.
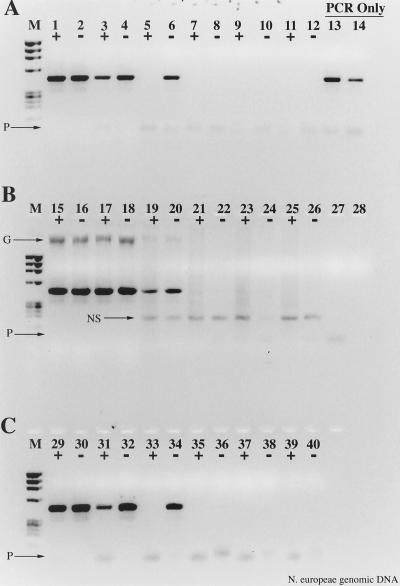
Effects of nonspecific, noncompetitive templates on RT inhibition of the PCR. N. europaea genomic DNA was used as the template. (A) No added competitive template; (B) 1 μg of calf thymus DNA added during the reverse transcription step; (C) 1 μg of yeast tRNA added during the reverse transcription step. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1 and 2, 2 ng of DNA; 3 and 4, 200 pg; 5 and 6, 20 pg; 7 and 8, 2 pg; 9 and 10, 200 fg; 11 and 12, no template; 13, 20 pg of DNA, PCR-only control; 14, 2 pg of DNA, PCR-only control; 15 to 26, same as lanes 1 to 12 except with calf thymus DNA as nonspecific template; 27, 200 fg of DNA PCR-only control; 28, no-template PCR-only control; 29 to 40, same as lanes 1 to 12 except with yeast tRNA as the nonspecific template. Arrows show the position of calf thymus genomic DNA (G), a nonspecific amplification product resulting from calf thymus genomic DNA (NS), and primer artifacts (P).
FIG. 11.
Effects of nonspecific template on RT-PCR from Nostoc nifH mRNA. (A) No added nonspecific template; (B) 1 μg of yeast tRNA added during the reverse transcription step. +, RT present; −, RT absent. Lanes: M, φX174 × HaeIII molecular weight marker; 1 and 2, 2 μl of nifH mRNA template; 3 and 4, 1 μl; 5 and 6, 0.5 μl; 7 and 8, no template; 9 and 10, 1 ng of Nostoc sp. genomic DNA, RT-PCR control; 11, 20 pg of genomic DNA, PCR-only control; 12, 2 pg of DNA; 13, 200 fg of DNA; 14, no-template PCR-only control; 15 to 23, same as lanes 1 to 10 except with yeast tRNA during the reverse transcription step.
DISCUSSION
Taq polymerase as an RT.
The initial observations of RT inhibition of the PCR were made on genomic DNA templates (ca. 1,000 copies) that were used as controls for RT-PCR (Fig. 1). Similar results, however, were also obtained with an mRNA template (Fig. 2). We initially viewed the mRNA data with suspicion, since Taq polymerase is not normally considered or used as an RT itself. However, pretreatment of the template with RNase abolished the signal (Fig. 2), indicating some level of Taq polymerase RT activity. This result is not new (21, 22, 38), although the efficiency of reverse transcription by Taq polymerase is very low relative to that of the true RT enzymes (<1% [21]). However, Taq polymerase was unable to function as an RT on mRNA isolated from cryptogamic crust (Fig. 8). We speculate that Taq RT activity is more sensitive to copurified contaminants associated with the nucleic acid or that Mg2+ ions are sequestered by copurified contaminants, so that the RT-PCR conditions applied to the crust mRNA were suboptimal for Taq RT activity relative to the control mRNA template. The differential activity of Taq on a control mRNA template and test mRNA template, however, may point to a qualitative difference between “environmental” and “control” nucleic acid targets that might also affect the true RT enzymes. This possibility warrants further study within the context of quantitative RT-PCR studies (see below).
RT inhibition of the PCR.
Several reports indicate that RT itself can interfere with PCR amplification of first-strand cDNA (1, 29, 37), but the results are mixed and confusing. Sellner et al. (37), for example, also observed that control DNA in RT-PCRs was not amplified during PCR whereas standard PCR would produce the desired product. An AMV RT-to-Taq ratio of ≥3:2 appeared to be the threshold for Taq inactivation, but addition of exogenous tRNA to the RT reaction mixture could alleviate the effect of RT inhibition. Aatsinki et al. (1) describe a one-tube RT-PCR system where the RT-PCR sensitivity was adversely affected by enzyme ratios only at limiting (100 ng) RNA concentrations. At higher RNA template concentrations, the ratio of RT to Taq was inconsequential. Mallet et al. (29), however, observed PCR inhibition at an AMV RT-to-Taq ratio of 20:2.5 when amplifying human immunodeficiency virus (HIV) mRNA from 1 of 100,000 infected cells.
These conflicting and confusing results suggest that the optimal RT-to-Taq enzyme ratio is determined by specific reaction conditions or primer-template pairs. Part of the confusion and disparate results also appear to be related to the abundance of specific target mRNA in the total RNA pool, where inhibitory effects become noticeable only at low (ca. 100-ng) total-RNA concentrations (1, 29). Much of the confusion and uncertainty surrounding PCR inhibition by RT can be removed if we consider a more useful barometer of template concentration, such as the template copy number. When previous studies are reevaluated with this normalized scale of template concentration, there is a clear contrast between previous investigations and the results presented here. In particular, our data suggest that at template concentrations of ≥105 to 106 copies in an uncoupled RT-PCR, all signs of RT inhibition disappear. Aatsinki et al. (1) used 1 pg of plasmid DNA as the lowest concentration of template; assuming a generous 10 kbp per plasmid, 1 pg is approximately 105 copies. Hence, the lowest concentration of template used in that study was at or above the threshold at which we would not expect to see PCR inhibition by the RT. An estimate of template copy number cannot be calculated from the study of Mallet et al. (29), but the concentration of HIV-positive cells (1 in 105) in their blood sample suggests that their specific template was probably present at ≤105 to 106 copies per PCR.
Sellner et al. (37) reported RT inhibition of the PCR at a plasmid DNA concentration of 100 pg (ca. ≥107 copies, assuming 10 kbp per plasmid) in a one-tube, coupled RT-PCR. One of the conclusions in this work was that RT interacts directly with Taq polymerase to inhibit the PCR. At elevated ratios of AMV to Taq (i.e., 14:2), some nonspecific protein-protein interaction may be inevitable and potentially lead to Taq inactivation. On the other hand, prolonged incubation of Taq polymerase during an RT incubation combined with a 5-min heat denaturation at 95°C may reduce Taq activity such that the amplification efficiency during the PCR would be less than under standard, optimal PCR conditions. Conclusions from other experiments were that primers, template, or MgCl2 alter RT enzyme conformation so that it is less susceptible to heat inactivation.
T4 gene 32 protein effects.
Several studies have shown that T4 gene 32 protein can improve PCR sensitivity during amplification of DNA derived from environmental samples (24, 39, 43). T4 gene 32 protein has also improved the PCR amplification of long templates (36), DNA-sequencing reactions (23), recovery of mutated DNA sequences with pronounced secondary structure (11), and PCR-based diagnostics in clinical material (33). T4 gene 32 protein increases the fidelity of DNA replication in vitro by interacting with DNA and proteins at the replication fork (31, 40) and greatly reduces the thermodynamic cost of helix melting in vivo (2). There is evidence for a direct interaction between T4 gene 32 protein and RNA (14), but this property has not been extensively studied or exploited. An analogous protein isolated from chicken fibroblast cells chronically infected with Rous sarcoma virus is also capable of binding to all forms of nucleic acid, including dsDNA, ssDNA, dsRNA, ssRNA, and DNA-RNA hybrids (19).
Our results suggests that T4 gene 32 protein is exerting its influence on the mRNA or RNA-DNA duplex during reverse transcription, in contrast to but consistent with the traditional notion and use of gene 32 protein as a ssDNA binding protein and helicase. Most compelling is the augmentation of Taq RT activity, as the inclusion of T4 gene 32 protein in the reverse transcription phase of the reaction consistently and significantly improved nifH product yield relative to that in standard RT-PCRs. Since all RT-PCRs were performed under identical conditions and the results were replicated on three separate occasions, we conclude that first-strand cDNA synthesis was more efficient in the presence of T4 gene 32 protein than without it, whether Taq or SuperScriptII RT was the primary source of RT activity. Improved N. europaea 16S rRNA RT-PCR product yield also conforms to this hypothesis, since 16S rRNA contains extensive secondary and tertiary structures (45), which are probably maintained at the reverse transcription reaction temperature of 42°C. The increased yield of nifH RT-PCR products from cryptogamic crust also support this hypothesis. The observation that Taq polymerase could not function as an RT on mRNA isolated from cryptogamic crust (Fig. 8), however, suggests that inhibitory effects may not be uniformly operative on control RNA relative to experimental RNA.
While we postulate that T4 gene 32 protein is involved in RNA binding and stabilization, a direct interaction with RT (MoMuLV or Taq) cannot be ruled out; other studies suggest that T4 gene 32 protein is multifunctional, including domains for interacting with DNA, DNA ligase, DNA polymerase, recombination nucleases, and membrane proteins (31, 44). Considering the relative inefficiency of Taq RT activity (21, 22) and the consistently superior performance of Taq over the SuperScriptII RT during amplification of nifH mRNA, a direct interaction between gene 32 protein and Taq DNA polymerase is plausible. In any case, our results suggest that T4 gene 32 protein is invaluable for improving RT-PCR sensitivity, and we therefore recommended its use in uncoupled RT-PCRs.
Possible mechanism of RT inhibition.
Our data suggest that interaction of RT with the template (DNA and RNA) is the mechanism for PCR inhibition at low template concentrations. Increasing template concentration alleviates the inhibition (Fig. 1 and 3), as does the addition of T4 gene 32 ssDNA binding protein during the RT incubation (Fig. 5 through 8). Altering the ratio of Superscript RT to Taq to 50:2.5 did nothing to alleviate PCR inhibition below 105 to 106 copies of template, and the inhibitory effect was evident with three different RT enzymes (AMV, MoMuLV, and Superscript M-MLV RNAse H−) used at or below the recommended levels (data not shown). In addition, 1 μg of exogenous yeast tRNA or calf thymus DNA was of limited value for improving RT-PCR sensitivity with the recommended levels of RT in the RT-PCR (Fig. 10 and 11).
Any RT has an inherent affinity for both DNA and RNA templates, since both forms of nucleic acid are bound by the enzyme during polymerization. Any nucleic acid present during an RT incubation, then, could serve as an RT substrate regardless of the sequence or source of nucleic acid. Since exogenous nucleic acid added during the reverse transcription step did not relieve PCR inhibition at standard RT-to-Taq ratios (Fig. 10 and 11), the binding of RT appears to be specific for the primer-template combination under investigation, perhaps in conjunction with primer extension on RNA and (at lower levels) DNA templates. In addition to our own data, many results presented by Sellner et al. (37) are also explained by this mechanism, and the reduced RT-to-Taq molar ratio required to alleviate PCR inhibition in the latter case may simply reflect titration of RT enzyme on abundant targets, Taq affinity for RNA templates (which may preclude RT binding to the target site), low-level Taq RT ability (see above and references 21, 22, and 38), or more efficient displacement of RT by Taq during repeated amplification cycles. Furthermore, heat inactivation of the RT is normally used to eliminate RT and RT nuclease activity. As such, the use of heat to inactivate the RT enzyme does not necessarily translate into the release of template (DNA or RNA) upon heat denaturation; the very existence of hyperthermophilic Archaea (18) demonstrates that extreme heat does not necessarily result in complete protein denaturation. Proteinase K digestion (37), however, will fragment the RT into smaller peptides and would therefore be expected to release substrates (template, primer, and Mg2+) otherwise sequestered by the RT. These results (37) also suggest that RT is binding specifically to the nucleic acid target and is not being released or displaced upon heat inactivation and subsequent cycles of PCR. Thus, most of the previous accounts of PCR inhibition by RT can be described by a copy number effect, and we therefore conclude that RT inhibition of the PCR at low template concentrations is an inevitable feature of uncoupled RT-PCRs.
Implications for quantitative RT-PCR studies.
The power of RT-PCR to rapidly amplify RNA templates with exquisite sensitivity and specificity has naturally led to the development of numerous quantitative RT-PCR systems (4, 7, 15, 17, 20, 27, 34, 35). Given the widespread use of quantitative RT-PCR in both clinical and environmental applications, it is surprising that RT inhibition of the PCR has not been more commonplace or widely recognized. As Sellner et al. note (37), many investigators may have unwittingly avoided this problem by using relatively high levels of template during RT-PCR; we observed this phenomenon only because we naively used small quantities of genomic DNA as a control template throughout the reverse transcription and PCR steps of RT-PCR.
On the surface, the finding that RT can interfere with PCR amplification appears to be a rather innocuous situation, one that can be easily overcome by performing RT-PCR at elevated template concentrations. The question then becomes one of whether ≤105 to 106 copies is an experimentally relevant concentration of target. In many cases, detection of ≤105 to 106 RNA templates in a given sample, such as the detection of HIV, food and public health viruses and pathogens, plant pathogens, or other infectious agents in both humans and animals, is extremely relevant (3, 10, 12, 16, 26, 41, 42). The detection of certain mRNA transcripts in complex environmental samples is equally relevant, since the relatively low recovery of nucleic acids from soils and sediments (27, 32) and the inherent instability of mRNA quickly deteriorate into problems of low-copy-number nucleic acid detection. For these reasons, RT-PCR has become a primary technique for detecting in situ gene expression and viral RNA genomes, and it is being used to enumerate RNAs in numerous clinical and environmental settings. However, small variations in amplification efficiency during early rounds of the PCR can result in disparate quantities of PCR product during the exponential phase of amplification, so the effect of RT inhibition on the PCR has important consequences for more quantitative attempts to detect ≤105 to 106 copies of RNA template.
Competitive, quantitative RT-PCR assays are normally assembled in one of two formats: reverse transcription of test RNA is carried out independently of a competitive template and the competitive template is added during the PCR process, or a copied RNA (cRNA) template is synthesized and subject to both the reverse transcription and PCR amplification in the presence of the test RNA. Our initial PCR experiments with DNA templates are analagous to a quantitative RT-PCR assay in which the competitive cDNA is added after reverse transcription. If RT inhibition of the PCR is true, as our data suggest, then this quantitative RT-PCR design will clearly give an underestimate of the actual template concentration in the RNA sample. That is, the competitive template will not interact with active RT and will therefore be amplified with greater efficiency than the test RNA sample. Therefore, less competitor will be required to obtain equivalence between test RNA and competitor PCR product concentrations. In this respect, quantitative RT-PCR systems that use an internal cRNA competitor to account for RT inefficiency and inhibition will be much more accurate indicators of the actual RNA copy number. However, our T4 gene 32 protein experiments show that reverse transcription of RNA targets may also be inhibited at lower template concentrations and that the extent or occurrence of inhibition may be different on mRNAs extracted from environmental samples. Since the deduced quantity of test RNA in these systems is always relative to the internal standard, the efficiency of reverse transcription of the test RNA cannot be determined directly, and nucleic acids extracted from environmental samples may contain additional inhibitors that do not act equally on the test RNA and cRNA templates, enumerations of RNA templates in the test sample should not be considered “absolute” (4, 5, 46).
Summary.
This study has established an inhibitory effect of RT on the PCR, which manifests itself as reduced RT-PCR efficiency in uncoupled RT-PCR systems below 105 to 106 copies of starting RNA. More importantly, however, these results imply that uncoupled, competitive quantitative RT-PCR assays used in both clinical and environmental settings will consistently underestimate the actual target RNA concentration in a sample if the starting concentration is below the 105- to 106-copy threshold. The extent of inhibition can be estimated by performing control RT-PCR amplifications of both DNA and RNA independently of the amplifications of the test sample but under otherwise identical reaction conditions. Some of this inhibition can also be relieved by incorporating 1.5 μg of T4 gene 32 protein during the initial template denaturation prior to reverse transcription, a property of gene 32 protein that has not been previously reported. However, because the outcome of quantitative RT-PCR can be significantly influenced by minor variations in amplification efficiency during the early rounds of PCR and because the extent of RT or PCR inhibition of test samples cannot be determined directly, this study suggests that quantitative RT-PCR enumerations of low-copy-number RNAs will underestimate mRNA levels in test samples. Consequently, a more thorough molecular and mechanistic understanding of RT-PCR and PCR at low template concentrations is required to accurately implement RT-PCR systems for quantifying low-abundance in situ RNA templates in both clinical and environmental samples.
ACKNOWLEDGMENTS
This research was supported by the Program for Ecosystem Research, Office of Health and Environmental Research, U.S. Department of Energy (DOE). Pacific Northwest National Laboratory is operated for the DOE by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
REFERENCES
- 1.Aatsinki J T, Lakkakorpi J T, Pietila E M, Rajaniemi H J. A coupled one-step reverse transcription PCR procedure for generation of full-length open reading frames. BioTechniques. 1994;16:282–288. [PubMed] [Google Scholar]
- 2.Alberts B, Sternglanz R. Recent excitement in the DNA replication problem. Nature. 1977;269:655–661. doi: 10.1038/269655a0. [DOI] [PubMed] [Google Scholar]
- 3.Atmar R L, Neill F H, Romalde J L, LeGuyader F, Woodley C M, Metcalf T G, Estes M K. Detection of norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl Environ Microbiol. 1995;61:3014–3018. doi: 10.1128/aem.61.8.3014-3018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker-André M. Absolute levels of mRNA by polymerase chain reaction-aided transcript titration assay. Methods Enzymol. 1993;218:420–445. doi: 10.1016/0076-6879(93)18034-a. [DOI] [PubMed] [Google Scholar]
- 5.Becker-André M. Evaluation of absolute mRNA levels by polymerase chain reaction-aided transcript titration assay (PATTY) In: Larrick J W, Siebert P D, editors. Reverse transcriptase PCR. New York, N.Y: Ellis Horwood; 1995. pp. 121–149. [Google Scholar]
- 6.Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 7.Bogan B W, Schoenike B, Lamar R T, Cullen D. Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler D P, Brockman F J. Estimating biodegradative gene numbers at a JP-5 contaminated site using PCR. Appl Biochem Biotechnol. 1996;57/58:971–982. doi: 10.1007/BF02941777. [DOI] [PubMed] [Google Scholar]
- 9.Chandler D P, Schreckhise R W, Smith J L, Bolton H., Jr Electroelution to remove humic compounds from soil DNA and RNA extracts. J Microbiol Methods. 1997;28:11–19. [Google Scholar]
- 10.Chiou P P, Drolet B S, Leong J C. Polymerase chain amplification of infectious hematopoietic necrosis virus RNA extracted from fixed and embedded fish tissue. J Aquat Anim Health. 1995;7:9–15. [Google Scholar]
- 11.Craik C S, Largman C, Fletcher T, Roczniak S, Barr P J, Fletterick R, Rutter W J. Redesigning trypsin: alteration of substrate specificity. Science. 1985;228:291–297. doi: 10.1126/science.3838593. [DOI] [PubMed] [Google Scholar]
- 12.Deacon E M, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson A B, Young L S. Epstein-Barr virus and Hodgkin’s disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degrange V, Bardom R. Detection and counting of Nitrobacter populations in soil by PCR. Appl Environ Microbiol. 1995;61:2093–2098. doi: 10.1128/aem.61.6.2093-2098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delius H, Duesberg P H, Mangel W F. Electron microscope measurements of Rous sarcoma virus RNA. Cold Spring Harbor Symp Quant Biol. 1975;39:835–843. doi: 10.1101/sqb.1974.039.01.097. [DOI] [PubMed] [Google Scholar]
- 15.Fleming J T, Sanseverino J, Sayler G S. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at Town Gas manufacturing sites. Environ Sci Technol. 1993;27:1068–1074. [Google Scholar]
- 16.Frerichs G N. Rhabdoviruses of fishes. In: Ahne W, Kurstak E, editors. Viruses of lower vertebrates. Berlin, Germany: Springer-Verlag KG; 1989. pp. 317–332. [Google Scholar]
- 17.Gilliland G, Perrin S, Bunn H F. Competitive PCR for quantitation of mRNA. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 60–69. [Google Scholar]
- 18.González J M, Kato C, Horikoshi K. Thermococcus peptonophilus sp. nov., a fast-growing, extremely thermophilic archaebacterium isolated from deep-sea hydrothermal vents. Arch Microbiol. 1995;164:159–164. [PubMed] [Google Scholar]
- 19.Hung P P, Lee S G. Isolation of nucleic acid-binding protein: stimulation of reverse transcription-catalysed DNA synthesis. Nature. 1976;259:499–502. doi: 10.1038/259499a0. [DOI] [PubMed] [Google Scholar]
- 20.Jessen-Eller K, Picozza E, Crivello J F. Quantitation of metallothionein mRNA by RT-PCR and chemiluminescence. BioTechniques. 1994;17:962–973. [PubMed] [Google Scholar]
- 21.Jones M D. Reverse transcription of mRNA by Thermus aquaticus DNA polymerase followed by polymerase chain reaction amplification. Methods Enzymol. 1993;218:413–419. doi: 10.1016/0076-6879(93)18033-9. [DOI] [PubMed] [Google Scholar]
- 22.Jones M D, Foulkes N S. Reverse transcription of mRNA by Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1989;17:8387–8388. doi: 10.1093/nar/17.20.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaspar P, Zadrazil S, Fabry M. An improved double stranded DNA sequencing method using gene 32 protein. Nucleic Acids Res. 1989;17:3616. doi: 10.1093/nar/17.9.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreader C A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamar R T, Schoenike B, Wymelenberg A V, Stewart P, Dietrich D M, Cullen D. Quantitation of fungal mRNAs in complex substrates by reverse transcription PCR and its application to Phanerochaete chrysosporium-colonized soil. Appl Environ Microbiol. 1995;61:2122–2126. doi: 10.1128/aem.61.6.2122-2126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laroche C, Drouet E B, Brousset P, Pain C, Boibieux A, Biron F, Icart J, Denoyel G A, Niveleau A. Measurement by the polymerase chain reaction of the epstein-barr virus load in infectious mononucleosis and AIDS-related non-hodgkin’s lymphomas. J Med Virol. 1995;46:66–74. doi: 10.1002/jmv.1890460115. [DOI] [PubMed] [Google Scholar]
- 27.Lee S Y, Bollinger J, Bezdicek D, Ogram A. Estimation of numbers of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C C, Burke R L, Hibner U, Barry J, Alberts B. Probing DNA replication mechanisms with the T4 bacteriophage in vitro system. Cold Spring Harbor Symp Quant Biol. 1979;43:469–487. doi: 10.1101/sqb.1979.043.01.053. [DOI] [PubMed] [Google Scholar]
- 29.Mallet F, Oriol G, Mary C, Verrier B, Mandrand B. Continuous RT-PCR using AMV-RT and Taq DNA polymerase: characterization and comparison to uncoupled procedures. BioTechniques. 1995;18:678–687. [PubMed] [Google Scholar]
- 30.Morris C F, Sinha N K, Alberts B M. Reconstruction of bacteriophage T4 DNA replication apparatus from purified components: rolling circle replication following de novo chain initiation on a single-stranded circular DNA template. Proc Natl Acad Sci USA. 1975;72:4800–4804. doi: 10.1073/pnas.72.12.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosig G, Luder A, Garcia G, Dannenberg R, Bock S. In vivo interactions of genes and proteins in DNA replication and recombination of phage T4. Cold Spring Harbor Symp Quant Biol. 1978;43:501–515. doi: 10.1101/sqb.1979.043.01.056. [DOI] [PubMed] [Google Scholar]
- 32.Ogram A V, Sayler G S. The use of gene probes in the rapid analysis of natural microbial communities. J Ind Microbiol. 1988;3:281–292. [Google Scholar]
- 33.Panaccio M, Lew A. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 1991;19:1151. doi: 10.1093/nar/19.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riedy M C, Timm E A J, Stewart C C. Quantitative RT-PCR for measuring gene expression. BioTechniques. 1995;18:70–76. [PubMed] [Google Scholar]
- 35.Scheuermann R H, Bauer S R. Polymerase chain reaction-based mRNA quantification using an internal standard: analysis of oncogene expression. Methods Enzymol. 1993;218:446–473. doi: 10.1016/0076-6879(93)18035-b. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz K, Hansen-Hagge T, Bartram C. Improved yields of long PCR products using gene 32 protein. Nucleic Acids Res. 1990;18:1079. doi: 10.1093/nar/18.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sellner L N, Coelen R J, Mackenzie J S. Reverse transcriptase inhibits Taq polymerase activity. Nucleic Acids Res. 1992;20:1487–1490. doi: 10.1093/nar/20.7.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaffer A L, Wojnar W, Nelson W. Amplification, detection, and automated sequencing of gibbon interleukin-2 mRNA by Thermus aquaticus DNA polymerase reverse transcription and polymerase chain reaction. Anal Biochem. 1990;190:292–296. doi: 10.1016/0003-2697(90)90196-g. [DOI] [PubMed] [Google Scholar]
- 39.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topal M D, Sinha N K. Products of bacteriophage T4 genes 32 and 45 improve the accuracy of DNA replication in vitro. J Biol Chem. 1983;258:12274–12279. [PubMed] [Google Scholar]
- 41.Tsai Y-L, Sobsey M D, Sangermano L R, Palmer C J. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl Environ Microbiol. 1993;59:3488–3491. doi: 10.1128/aem.59.10.3488-3491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai Y-L, Tran B, Sangermano L R, Palmer C J. Detection of poliovirus, hepatitis A virus, and rotavirus from sewage and ocean water by triplex reverse transcriptase PCR. Appl Environ Microbiol. 1994;60:2400–2407. doi: 10.1128/aem.60.7.2400-2407.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vahjen W, Tebbe C C. Enhanced detection of genetically engineered Corynebacterium glutamicum pUN1 in directly extracted DNA from soil, using the T4 gene 32 protein in the polymerase chain reaction. Eur J Soil Biol. 1994;30:93–98. [Google Scholar]
- 44.Williams K R, LoPresti M B, Setoguchi M. Primary structure of the bacteriophage T4 DNA helix-destabilizing protein. J Biol Chem. 1981;256:1754–1762. [PubMed] [Google Scholar]
- 45.Woese C R, Gutell R, Gupta R, Noller H F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zachar V, Thomas R A, Goustin A S. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;21:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]