Abstract
We applied transmission electron microscopy and densitometric image analysis to measure the cell volume (V) and dry weight (DW) of single bacterial cells. The system was applied to measure the DW of Escherichia coli DSM 613 at different growth phases and of natural bacterial assemblages of two lakes, Piburger See and Gossenköllesee. We found a functional allometric relationship between DW (in femtograms) and V (in cubic micrometers) of bacteria (DW = 435 · V0.86); i.e., smaller bacteria had a higher ratio of DW to V than larger cells. The measured DW of E. coli cells ranged from 83 to 1,172 fg, and V ranged from 0.1 to 3.5 μm3 (n = 678). Bacterial cells from Piburger See and Gossenköllesee (n = 465) had DWs from 3 fg (V = 0.003 μm3) to 1,177 fg (V = 3.5 μm3). Between 40 and 50% of the cells had a DW of less than 20 fg. By assuming that carbon comprises 50% of the DW, the ratio of carbon content to V of individual cells varied from 466 fg of C μm−3 for Vs of 0.001 to 0.01 μm3 to 397 fg of C μm−3 (0.01 to 0.1 μm3) and 288 fg of C μm−3 (0.1 to 1 μm3). Exponentially growing and stationary cells of E. coli DSM 613 showed conversion factors of 254 fg of C μm−3 (0.1 to 1 μm3) and 211 fg of C μm−3 (1 to 4 μm3), respectively. Our data suggest that bacterial biomass in aquatic environments is higher and more variable than previously assumed from volume-based measurements.
Determination of morphological and physiological parameters at the level of single cells is a major challenge in microbial ecology, especially if one considers the heterogeneity of natural bacterial populations. The quantification of microbial biomass in freshwater and marine ecosystems becomes particularly important in the case of growth and production rate determinations, as well as for turnover measurements of carbon, nitrogen, phosphorus, and sulfur (4, 6). Different methods have been proposed for biomass estimation, such as epifluorescence direct counts (14, 23) in combination with volume measurements (5, 7) and quantification of macromolecular components such as DNA (21) or proteins (30). However, the assumptions are often ambiguous, and errors remain large. A microscopically determined cell volume (V) may be converted into biomass if the density and percent dry weight (DW) of the cell are known, but often only average values such as a density of 1.1 g cm−3 and a ratio of dry to fresh weight of 0.22 are used (for a review, see reference 6). Bratback and Dundas (8), however, suggested a ratio of DW to wet weight closer to 0.4, and Bakken and Olsen (4) published a value larger than 0.3. If the biovolume is determined, the biomass is usually estimated by assuming a constant ratio (constant ratio model) of carbon content (C) to V. Therefore, experimentally obtained conversion factors are applied, although some authors (16) assumed a constant carbon mass (constant biomass model) per cell for natural bacterial populations. Biovolume-to-biomass conversion factors reported in the literature vary up to fivefold with respect to the commonly cited value of 0.121 g of C μm−3 (for a review, see reference 6).
According to Norland et al. (20), an allometric relationship can be determined between the biomass and volume of bacteria. It is described by the following: B = c · Va, where B is biomass, c is the conversion factor between biomass and V for unity volume (V = 1), and a is the scaling factor. This allometric relationship may partly explain the wide variation of the reported conversion factors. However, conversion factors were generally determined from cultured cells grown in supplemented media or by using cultured aquatic bacteria.
Consequently, they may not be applicable to natural aquatic bacteria, which are usually much smaller and taxonomically not well defined (2). As a matter of fact, differences in the physiological state of the investigated bacterial cells are another reason for the variety of conversion factors cited in the literature. Hence, the determination of conversion factors should be made with natural bacteria and based on the measurement of single cells.
The aim of this study was to develop a new method for DW estimation of individual, natural bacteria by means of electron microscopy and densitometric image analysis. The method has then been applied to Escherichia coli cells and bacterial populations of two lakes.
MATERIALS AND METHODS
Bacteria and sample preparation.
E. coli DSM 613 was grown in batch culture (500 ml) with Bact Nutrient Broth medium (8 g liter−1) (Difco Laboratories, Detroit, Mich.) at 37°C on a rotatory shaker. During exponential, late exponential, and stationary phases, subsamples were taken and fixed with formaldehyde (2% vol/vol). Cells (>0.1 μm in diameter) were concentrated with a hollow-fiber filtration system (Amicon, Witten, Germany) to reach a dense bacterial suspension.
Samples of natural bacterial populations were taken from the surface of two mountain lakes in August 1995, the mesotrophic Piburger See (913 m above sea level) and the oligotrophic Gossenköllesee (2,417 m above sea level) in Tyrol, Austria. To get a dense bacterial suspension, 10 liters of sample was concentrated to about 40 ml with the hollow-fiber system and fixed with formaldehyde (2% vol/vol). For calibration, latex spheres with a density of 1.055 g cm−3 and diameters of 0.093, 0.282, and 0.415 μm (Molecular Probes, Eugene, Oreg.) were sprayed in microdroplets onto grids and air dried. For transmission electron microscopy (TEM), copper slotgrids (Gröpl, Tulln, Austria) supported with Formvar film and coated with 0.5% bovine serum albumin (Sigma, Vienna, Austria) were used. Drops of cultures were added directly onto the grids. After 1 min, the drops were carefully drained off with filter paper, and the remaining cells were air dried before analysis.
Physical background.
In the electron microscope the contrast of an image of an effectively amorphous object (i.e., one in which the effect of coherent scattering is negligible) is almost entirely due to the differential scattering of electrons by various parts of the object. Differential scattering prevents various fractions of the incident beam from passing through the objective aperture of the microscope and contributing to the image intensity (33). The intensity of a light beam that passes through an object decreases exponentially with thickness and is directly related to the mass per unit area. The exponential law of transmission for the conventional TEM brightfield mode and the scanning TEM mode can be used for quantitative determination of mass thickness of amorphous specimens, such as supporting films, biological sections, and microorganisms (3, 26, 33).
Mass determination.
The analysis was carried out with an electron microscope, model EM 902 (Zeiss, Oberkochen, Germany), operating at 80 kV, an electron intensity of 60 · 10−12 eV mm−2 s−1, and a magnification of ×4,400. In order to increase the image contrast, we used elastically scattered electrons emitted under small angles and nonscattered electrons, i.e., electrons with energy losses of zero. To convert opacity values into mass, an electronic device (Dage camera; Zeiss) in combination with an image analysis system (LUCIA S; Laboratory Imaging, Prague, Czech Republic) was used. The electron opacity of an object caused a characteristic, proportional grey value when the densitometric measuring mode of the image analysis system was used. The mass of an object was calculated from the integrated logarithmic grey value of the object under investigation after subtraction of the integrated logarithmic grey value of the uniform background of the same area as the object. This corresponds to the integrated optical density (IOD), which gives an estimate of the mass of the object. As shading effects occurred, two images were taken for each measurement, one with the objects of interest and the second containing only the uniform background as close as possible to the object itself.
Cell volume determination.
An edge detection operator (28) was applied to measure cell volumes. The system was calibrated with latex beads of known size. Bacteria were considered as cylinders with hemispherical ends at each side. The following formula was used to calculate V from the length (l) and width (w) of the cells: V = [(w2 · π/4) · (l − w)] + (π · w3/6). This formula works equally well for cocci and rods, as for cocci l − w becomes zero. Bacterial length was calculated by means of a Feret box enclosing the object measured at different angles. The maximal Feret’s diameter (femax) at a certain angle equals the length of the object, and the minimal Feret’s diameter represents the width of the object. For elongated and thin objects the parameter length, calculated from area and perimeter (lengthAP), gave a better approximation of bacterial length than femax (data not shown). Therefore, bacterial length was defined as femax if femax was greater than or equal to lengthAP, and the minimal Feret’s diameter represents the width of the objects. If femax is less than lengthAP, bacterial length is described by lengthAP and bacterial width is described as area divided by lengthAP.
Statistical analysis.
Linear regression analyses were applied after log-log transformation of the data (log Y = a + b log X). A functional regression was used in the case where neither axis was independent (19).
RESULTS
For determining bacterial DW by means of TEM in combination with densitometric image analysis, optimizations of microscope and camera properties were necessary. The signal produced in the electron microscope and processed by the image analysis system depends on the beam current and the properties of the camera. It was necessary to ensure a linear relationship between input light intensity and measured grey value and to ensure a large dynamic range of the camera while keeping the optimal settings of contrast, sensitivity, and brightness constant. An electron intensity of 60 · 10−12E mm−2 s−1 was selected as the beam current for DW measurements. At this beam current the image produced on the fluorescent screen of the electron microscope was dark for the observer, but the camera was sensitive enough to obtain a measurable picture. Working with this highly sensitive camera allowed us to keep the beam current as low as possible, thus reducing mass losses to a minimum. On the other hand, because of the low beam current the noise level was high, and averaging of 255 images was necessary.
The illumination was heterogeneous, showing a decrease in brightness of up to 60% from the center of the image to the edges. The subtraction of background led to an even signal across the field. However, if latex beads were shifted through the viewing field (512 by 512 pixels), different IODs for identical latex beads were measured, because of geometrical, pincushion-like distortions caused by the TEM and video camera system. These errors were compensated for by applying a linear-correction factor depending on the distance from the center. Apart from this correction, the area of measurement was limited to the central part of the viewing field. By using the above-mentioned corrections, the standard deviation of the IOD of small, medium, and large latex beads could be reduced to <10%, <5%, and <3%, respectively.
Small latex beads (diameter = 0.093 μm; DW = 0.85 fg) were close to the lower sensitivity limit of this method, because even small changes in the thickness of the film or flickering of the cathode had a measurable impact on the accuracy of IOD determinations. Variations in the thickness of the supporting film occurred within the viewing field, and therefore measurements of the background had to be made as close as possible to the object itself. Flickering of the cathode and therefore alterations of the electron beam were controlled by use of look-up tables. Look-up tables were used to derive colors for grey-scale values to create a pseudocolor display (27), so small or gradual changes in image brightness due to cathode flickering became clearly visible. By keeping all error sources under control during the whole measurement, we could achieve a standard deviation of <10% for small and <5% for medium-sized spheres. The upper limit of the method is set by the thickness of the objects. If objects are too thick, multiple scattering will occur and a higher transmission than predicted is observed (26), yielding to an underestimation of DW. According to Bahr and Zeitler (3), the method is independent of shape, inhomogeneities, and chemical composition of the objects within its working range of 10−11 to 10−18 g. Nevertheless, multiple scattering could be detected in bacterial cells with inclusions such as storage granules (data not shown). Furthermore, multiple scattering is very likely if cultured cells thicker than 1 μm are used.
The IOD of latex beads, based on mixed combinations of beads of different diameters, showed a linear relationship with DW (Fig. 1). For DW calculations we relied on the mean diameter, with standard deviations from 2.6 to 7.9% of the mean (n = 500) given by the manufacturers.
FIG. 1.
Relationship between IOD and DW of latex beads (mixed combinations of three different sizes). The linear regression model resulted in the following: IOD = 5.27 · DW − 1.08 (r2 = 0.996; n = 140).
The DW of cultured E. coli cells from different growth phases ranged from 83 to 1,172 fg (Fig. 2), with a median value of 222 fg. For natural bacteria from Piburger See the DW was between 4 and 326 fg (Fig. 2), with a median value of 19 fg (Table 1). The median cell volume in Gossenköllesee samples (0.04 μm3) was higher than in Piburger See samples (0.027 μm3), and the DW was between 3 and 1,177 fg (Fig. 2), with a median value of 36 fg (Table 1). Also, 12 very light objects with a DW of <3 fg and a diameter of <0.2 μm were detected in the Gossenköllesee samples.
FIG. 2.
Log-log plot of DW versus V with data from cultured and natural bacterial assemblages.
TABLE 1.
Quartiles and median values of DW of cultured E. coli DSM 613 cells from different growth phases and of natural bacteria from two different lakes
| Sample | n | DW (fg) in percentile
|
||
|---|---|---|---|---|
| 25% | Median | 75% | ||
| Lake water | ||||
| Piburger See | 283 | 11 | 19 | 46 |
| Gossenköllesee | 195 | 16 | 36 | 87 |
| E. coli DSM 613 | ||||
| Exponential | 136 | 358 | 489 | 622 |
| Stationary | 116 | 148 | 179 | 211 |
DISCUSSION
DW measurements.
Published average DWs for E. coli grown in batch culture with minimal medium were 278 fg in the late exponential phase of growth and 154 fg at the early stationary phase (13). Fagerbakke et al. (10) reported a value of 710 fg (n = 26) for E. coli B6 wild type in the exponential growth phase and 180 fg (n = 20) in the stationary phase by summing up the dry masses of all measured elements and assuming a hydrogen content of one-sixth of C. Large differences in DW during the exponential growth phase depend on nutrient conditions and the type of the strain, but these differences are small in the stationary phase (10). Contrary to E. coli B6, our strain was formaldehyde fixed, which may lead to a loss of potassium and chlorine while other constituents remain in the cell (13). Thus, artifacts caused by shrinkage due to fixation and air drying may have affected our mass and volume measurements. Natural bacteria were generally much smaller than cultured ones, although there was a size overlap between E. coli and bacteria from Gossenköllesee (Fig. 2). The existence of cells with DWs around or below 3 fg, however, is questionable, if one considers the minimum requirements for a cell, i.e., the minimum amount of DNA, ribosomes, enzymes, and lipids, etc., which make up an organism (25). Assuming a minimum genome size of ca. 0.6 · 106 base pairs (9) and a very large ratio of DNA to cell mass (15%), we calculated a minimum DW of ca. 7 fg. It is thus debatable if objects below 7 fg can be considered bacteria (29). However, cell masses between 3 and 10 fg were reported by other authors (10, 13).
Cell volume determination.
It has been observed that the volume of formaldehyde-fixed and air-dried E. coli cells was 40% smaller than that of air-dried cells without any fixatives (10). According to Heldal et al. (13), bacteria flatten during air drying, whereas width and average cell length remain unchanged compared with living cells. We observed heat-fixed E. coli cells with a scanning probe microscope (unpublished data) and found a considerable flattening, while width and length were less affected by the drying process. The median volume of E. coli DSM 613 measured by TEM was 54% smaller than that determined by phase-contrast light microscopy for exponentially growing cells and 64% lower for cells in the stationary phase. In both cases, it was not length but primarily the width of the cells that was affected. The volume distribution of natural bacterial populations from Piburger See and Gossenköllesee determined by TEM was similar to that determined by epifluorescence microscopy (EFM) and DAPI (4′,6-diamidino-2-phenylindole) staining (Fig. 3), despite measuring lower cell numbers by TEM (Gossenköllesee, n = 195; Piburger See, n = 283) than by EFM (Gossenköllesee, n = 954; Piburger See, n = 668). The median cell volume for Gossenköllesee was 0.04 μm3 measured by TEM and 0.035 μm3 measured by EFM. We found similar median cell volumes also for Piburger See (TEM, 0.027 μm3; EFM, 0.028 μm3), which suggests that we may use our mass calculation formula for cell volumes determined by EFM. One should consider, however, that it is very difficult to measure the real size of free-living bacteria; thus, our volume measurements by EFM—although comparable to values found by TEM—can be used only as a relative standard. An absolute calibration is still missing, but most morphometric data on aquatic bacteria are determined by EFM. Consequently, our formula can be applied to calculate dry mass for most standard applications.
FIG. 3.
Comparison of bacterial Vs in populations from Piburger See (A) and Gossenköllesee (B), determined by EFM and TEM.
DW:V.
To our knowledge, the only published formula for direct calculation of cellular DW from Vs is that of Norland et al. (20). Similarly, we found an allometric relationship between DW and V (Fig. 2). Our scaling factor was less than unity, suggesting that smaller cells tend to have a higher DW:V ratio than larger ones. Compared to a scaling factor of 1, i.e., a constant volume-to-mass ratio, our scaling factor gives about twice as much weight to very small cells (V = 0.01 μm3) and ca. 40% more weight to cells of 0.1 μm3. An allometric relationship was reported also by Simon and Azam (30) for the protein content of bacterial assemblages from seawater, and by Kroer (15) for carbon and nitrogen. Verity et al. (31) published an allometric relationship between cell volume and C and N content of unicellular algal cultures. Their correlation between C (in femtograms) and V (in cubic micrometers) (C = 433 · V0.863) was similar to our functional relation, although ours was for dry weight (DW = 435 · V0.86). According to Norland (19), the use of a predictive regression is not appropriate as none of the axes is independent; thus, a functional regression coefficient (scale factor) of 0.86 was used. In our case it was rather similar to the scaling factor of 0.85 resulting from a predictive regression with volume as the independent variable.
C:V.
For calculating C of individual cells it is generally agreed that carbon comprises approximately 50% of the DW (5, 10, 18). By using this conversion factor, we found an average C:V ratio of 382 fg of C μm−3 (range, 128 to 725 fg of C cm−3; n = 477) for natural bacterial populations, which is consistent with findings of high C:V values, such as 560 fg of C μm−3 (7), 354 fg of C m−3 (5), 380 fg of C μm−3 (16), and 720 fg of C μm−3 (15), but in contrast with recent results of Fagerbakke et al. (10), who found only 32 to 160 fg of C μm−3. In that study, however, the authors put more emphasis on the quantification of the C:N:P quotient of single cells than on the determination of absolute C:V ratios. Lower C:V ratios were also determined by Nagata and Watanabe (18) for freshwater bacterial populations grown in lake water filtrate or in water enriched with nutrients. Their C:V ratio of 140 fg of C μm−3 compares well with the commonly used C:V ratio of 121 fg of C μm−3 from Watson et al. (32). As mentioned above, one possible reason for finding higher conversion factors is the underestimation of cell volumes, caused by the shrinkage effects due to fixatives and air drying, although several conversion factors in the literature were established with fixed cells without correction for shrinkage effects.
Constant biomass model.
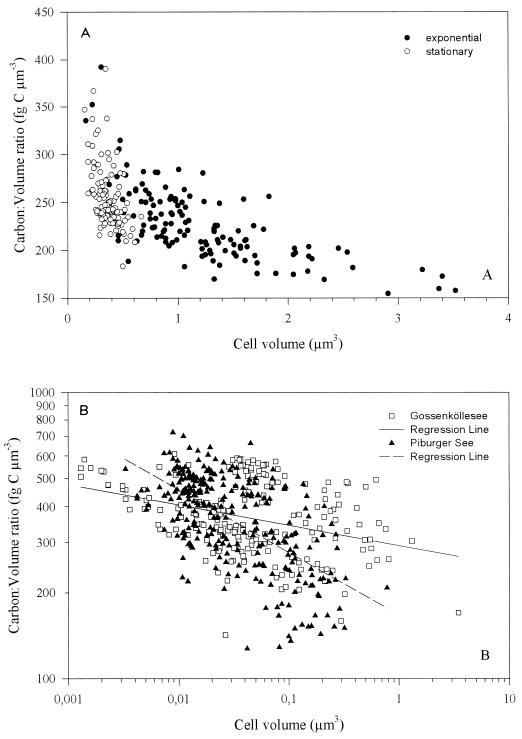
Besides the widely used constant ratio model, Lee and Fuhrman (16) proposed a constant biomass model. They found that bacteria with cell volumes of 0.036 and 0.073 μm3 both contained 20 ± 0.8 fg of C cell−1, whereas Simon and Azam (30) concluded that cells in the range of 0.036 to 0.07 μm3 would contain 13 to 19 fg of C cell−1, based on measurements of protein and macromolecular inventory. Cells in that size range from Piburger See and Gossenköllesee had a comparable mean C of 20 ± 8 fg of C cell−1 (range, 5 to 58 fg of C cell−1; n = 92) but with a much higher variation (40%), which shows that the assumption of a constant C per cell is not suitable. And yet a constant ratio model would not fit our data (Fig. 4), which show that C:V ratios for natural bacteria are between ca. 150 and 700 fg of C μm−3. Moreover, natural bacteria have a larger scatter than cultured E. coli cells (Fig. 2 and 4), but also for E. coli the conversion factor changed when the culture shifted from the exponential to the stationary phase (Table 2 and Fig. 4A). In addition, natural bacterial populations from different lakes may have different conversion factors, as suggested by Fig. 4B, but we still need more data to confirm this hypothesis. Kroer (15) showed that conversion factors were dependent not only on bacterial size but also on temporal and geographic variations, indicating possible influences of species composition, nutrition state, and growth rate, etc. He observed changes from 60 to 350 fg of C μm−3 within 3 weeks, and in cultures with approximately equal bacterial sizes in the early stationary phase C ranged from 350 to 1,350 fg of C μm−3 (15). Nevertheless, according to Nagata and Watanabe (18) the fivefold variability of the conversion factor found in the literature cannot be explained on the basis of possible differences in the nutritional stage of bacterial populations. Furthermore, differences in habitat (i.e., freshwater versus seawater) may also explain the large variability of the conversion factors found in the literature. Bjørnsen (5) found that the C:V ratio for freshwater bacteria (310 fg of C μm−3) was significantly lower than that for estuarine bacteria (410 fg of C μm−3). Whether different media (e.g., those with a different salt concentration [10]) or the presence of different phyla (for instance, the dominance of α versus β Proteobacteria in marine environments [11, 12]) is responsible for such differences is an open question. For small marine planktonic bacteria an increasing ratio of DW to wet weight with decreasing V was detectable, with the consequence that smaller cells gradually become dry and richer in carbon (30). Although physiologically not well understood, this phenomenon may have major ecological consequences.
FIG. 4.
Plot of C:V ratios versus V of E. coli in exponential and stationary growth phases (A) and natural bacterial populations (B). Panel A is a linear and panel B is a log-log plot.
TABLE 2.
C:V ratios of different size classes and means of natural bacterial populations and of E. coli during exponential and stationary growth phasesa
| Sample |
C:V ratio of bacteria in size class (fg of C μm−3)
|
||||
|---|---|---|---|---|---|
| 0.001–0.01 μm3 | 0.01–0.1 μm3 | 0.1–1 μm3 | 1–4 μm3 | Mean | |
| Lake water | 466 ± 88 (54) | 397 ± 117 (323) | 288 ± 93 (99) | — | 382 ± 121 (476) |
| E. coli DSM 613 | — | — | 254 ± 40 (179) | 211 ± 34 (73) | 242 ± 43 (252) |
Values are means ± standard deviations. Values in parentheses are numbers of measured cells. —, n = 0.
Conclusions.
Dry mass of natural bacterial cells varies over about 3 orders of magnitude, with a variable but high ratio of DW (or C) to V, which suggests that the importance of bacterioplankton in aquatic food webs may have been underestimated in previous studies. This has consequences for the calculation of biomass distribution, productivity, grazing, and element fluxes in aquatic ecosystems. To our knowledge, our approach is, with X-ray microanalysis, the only method available for the simultaneous determination of DW and size of single cells. Certainly, we need much more information about what happens when a living cell is stuck on surfaces such as microscope slides, membrane filters, and Formvar films, etc., to be observed in the epifluorescence, the transmission electron, or the atomic force microscope. We think, however, that our formula yields a correct mass estimation based on the size determination of fixed and DAPI-stained bacteria in the EFM while an absolute mass-to-volume ratio is still missing.
At present, the vast majority of the bacteria that exist in aquatic systems have been neither cultivated nor physiologically characterized (1, 2). Nonetheless, constant biomass or constant ratio models are widely used to estimate bacterial biomass, which is of little help for the understanding of functional relationships between bacteria and other components of food webs. Consequently, we do not know much about, e.g., biomass allocation within different size classes or phyla (22) or the relationship between activity and cell size (24). Correct estimates of biomass distribution, however, are a basic requirement in microbial ecology. Therefore, we need more measurements of dry mass and carbon, nitrogen, and phosphorus contents of aquatic bacteria combined with a reliable determination of size, physiological status (DNA and RNA, etc.), and taxonomic classification of single cells under natural conditions.
ACKNOWLEDGMENTS
We thank Konrad Eller and Willi Salvenmoser for support in the electron microscopic work and Arnulf Lochs for calculating the correction factor for geometrical distortions. We also thank Karl-Paul Witzel (Max-Planck-Institut für Limnologie, Plön, Germany) for providing E. coli DSM 613 harvested at different growth phases. We are indebted to Birgit Sattler, Albin Alfreider, Stefan Andreatta, Jakob Pernthaler, Ruben Sommaruga, Thomas Posch, and Ferdinand Hofer for critical comments and suggestions which improved the quality of the paper.
This work was supported by the Austrian Ministry of Research (GZ 45.335/4-IV/6/94) and the National Bank (project 4677).
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F-O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr G F, Zeitler E. The determination of the dry mass in populations of isolated particles. Lab Invest. 1965;14:955–977. [PubMed] [Google Scholar]
- 4.Bakken L R, Olsen R A. Buoyant densities and dry-matter contents of microorganisms: conversion of a measured biovolume into biomass. Appl Environ Microbiol. 1983;45:1188–1195. doi: 10.1128/aem.45.4.1188-1195.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjørnsen P K. Automatic determination of bacterioplankton biomass by image analysis. Appl Environ Microbiol. 1986;51:1199–1204. doi: 10.1128/aem.51.6.1199-1204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjørnsen P K, Riemann B. Towards a quantitative stage in the study of microbial processes in pelagic carbon flows. Arch Hydrobiol Suppl. 1988;31:185–193. [Google Scholar]
- 7.Bratbak G. Bacterial biovolume and biomass estimations. Appl Environ Microbiol. 1985;49:1488–1493. doi: 10.1128/aem.49.6.1488-1493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bratbak G, Dundas I. Bacterial dry matter content and biomass estimations. Appl Environ Microbiol. 1984;48:755–757. doi: 10.1128/aem.48.4.755-757.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen H, Bakken L R, Olsen R A. Soil bacterial DNA and biovolume profiles measured by flow-cytometry. FEMS Microbiol Ecol. 1993;102:129–140. [Google Scholar]
- 10.Fagerbakke K M, Heldal M, Norland S. Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat Microb Ecol. 1996;10:15–27. [Google Scholar]
- 11.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 12.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K H, Schleifer K H. An in situ hybridisation protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 13.Heldal M, Norland S, Tumyr O. X-ray microanalytic method for measurement of dry matter and elemental content of individual bacteria. Appl Environ Microbiol. 1985;50:1251–1257. doi: 10.1128/aem.50.5.1251-1257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroer N. Relationship between biovolume and carbon and nitrogen content of bacterioplankton. FEMS Microbiol Ecol. 1994;13:217–224. [Google Scholar]
- 16.Lee S, Fuhrman J A. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol. 1987;53:1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagata T. Carbon and nitrogen content of natural planktonic bacteria. Appl Environ Microbiol. 1986;52:28–32. doi: 10.1128/aem.52.1.28-32.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata T, Watanabe Y. Carbon- and nitrogen-to-volume ratios of bacterioplankton grown under different nutritional conditions. Appl Environ Microbiol. 1990;56:1303–1309. doi: 10.1128/aem.56.5.1303-1309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norland S. The relationship between biomass and volume of bacteria. In: Kemp P F, Sherr B F, Sherr E B, Cole J C, editors. Handbook of methods in aquatic microbial ecology. London, United Kingdom: Lewis Publishers; 1993. p. 303. [Google Scholar]
- 20.Norland S, Heldal M, Tumyr O. On the relation between dry matter and volume of bacteria. Microb Ecol. 1987;13:95–101. doi: 10.1007/BF02011246. [DOI] [PubMed] [Google Scholar]
- 21.Paul J H, Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pernthaler J, Sattler B, Simek K, Schwarzenbacher A, Psenner R. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat Microb Ecol. 1996;10:255–263. [Google Scholar]
- 23.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 24.Posch T, Pernthaler J, Alfreider A, Psenner R. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, Cyto-Clear slides, and image analysis. Appl Environ Microbiol. 1997;63:867–873. doi: 10.1128/aem.63.3.867-873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Psenner R, Loferer M. Nannobacteria: size limits and evidence. Science. 1997;276:1776–1777. [PubMed] [Google Scholar]
- 26.Reimer L, editor. Transmission electron microscopy. Berlin, Germany: Springer-Verlag; 1984. p. 185. [Google Scholar]
- 27.Russ J C, editor. The image processing handbook. London, United Kingdom: CRC Press; 1992. [Google Scholar]
- 28.Schröder D, Krambeck C, Krambeck H-J. How to count and size fluorescent microbial plankton with digital image filtering and segmentation. Acta Stereol. 1991;10:123–129. [Google Scholar]
- 29.Sieracki M E, Viles C H. Distributions and fluorochrome-staining properties of submicrometer particles and bacteria in the North Atlantic. Deep-Sea Res. 1992;39:1919–1929. [Google Scholar]
- 30.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 31.Verity P G, Robertson C Y, Tronzo C R, Andrews M G, Nelson J R, Sieracki M E. Relationship between cell volume and the carbon and nitrogen content of marine photosynthetic nanoplankton. Limnol Oceanogr. 1992;37:1434–1446. [Google Scholar]
- 32.Watson S W, Novitsky T J, Quinby H L, Valois F W. Determination of bacterial number and biomass in the marine environment. Appl Environ Microbiol. 1977;33:940–946. doi: 10.1128/aem.33.4.940-946.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeitler E, Bahr G F. A photometric procedure for weight determination of submicroscopic particles in quantitative electron microscopy. J Appl Physics. 1962;33:847–853. [Google Scholar]