Abstract
Pulsed-field gel electrophoresis (PFGE) was optimized for genomic analyses of Clostridium botulinum (nonproteolytic) group II. DNA degradation problems caused by extracellular DNases were overcome by fixation of cells with formaldehyde prior to isolation. A rapid (4-h) in situ DNA isolation method was also assessed and gave indistinguishable results. Genomic DNA from 21 strains of various geographical and temporal origins was digested with 15 rare-cutting restriction enzymes. Of these, ApaI, MluI, NruI, SmaI, and XhoI gave the most revealing PFGE patterns, enabling strain differentiation. Twenty strains yielded PFGE patterns containing 13 pulsotypes. From summation of MluI, SmaI, and XhoI restriction fragments, the genome size of C. botulinum group II was estimated to be 3.6 to 4.1 Mb (mean ± standard deviation = 3,890 ± 170 kb). The results substantiate that after problems due to DNases are overcome, PFGE analysis will be a reproducible and highly discriminating epidemiological method for studying C. botulinum group II at the molecular level.
Throughout the world, with the exception of the continental United States, food-borne botulism is still the predominant disease form caused by Clostridium botulinum. Globally, approximately 450 outbreaks with 930 cases are recorded annually, of which more than 90% are caused by home-prepared or home-preserved foods (14). Most of these outbreaks occur in temperate regions, such as northern Europe, Canada, Alaska, and Japan, and they are caused mainly by the group II (nonproteolytic) strains (15). In the United States, infant botulism is the most common form of the disease, and in 1996 wound botulism also surpassed the classical food-borne infection for the first time (12). Despite its clinical importance, characterization studies of C. botulinum by other than serological methods have been lacking. Only one genomic analysis of group I C. botulinum 62 A and Hall A by pulsed-field gel electrophoresis (PFGE) has been published (21); to our knowledge, no studies involving other genotyping methods, such as plasmid profiling, restriction endonuclease analysis, ribotyping, or random amplification of polymorphic DNA, have been published.
The lack of substantial papers on the genotyping of C. botulinum might be due to difficulties in obtaining high-quality DNA for PFGE and other genotyping methods. This problem is encountered with many bacterial species, and it is often the result of DNA degradation during isolation. Within some bacterial genera, e.g., Campylobacter (13), Clostridium (4, 8, 18, 21, 29, 30), and Serratia (33), production of extracellular DNases can be very pronounced, turning the preparation of the intact in situ DNA needed for PFGE typing into a real challenge. Clostridial species and strains seem to have extensive differences in DNase production. We have found both C. perfringens and C. botulinum group I strains easy to type by PFGE but have encountered major problems in obtaining nondegraded DNA from group II strains (unpublished data). Samore et al. (28) were not able to type 70% of 33 C. difficile strains by PFGE, presumably due to the effect of particularly active DNases. Several approaches to overcoming this problem have been proposed. These include formaldehyde fixation of cells upon harvesting (13), heating of cells (18, 19, 21) or the use of lysis solution for the resuspension of cells prior to mixing with the insert gel (23), inclusion of hypertonic sucrose in the lysis solution (34), and shortening of the lysis and DNA plug wash steps (7, 13, 23).
This study was set out to evaluate the effect of different in situ preparation methods on the quality of DNA intended for use in PFGE typing of the presumably DNase-rich group II C. botulinum strains. The information obtained from the PFGE analysis of 21 strains was also evaluated in relation to their respective geographical and temporal origins, and the genome sizes of these C. botulinum group II strains were estimated.
MATERIALS AND METHODS
Bacterial strains and growth media.
The 21 C. botulinum group II strains characterized in this study are listed in Table 1. The cultures were grown for 3 days on anaerobic egg yolk agar (1) from which Trypticase (BBL Microbiology Systems)-peptone-glucose-yeast extract broth (11) was inoculated. All the cultures were incubated at 26°C in an anaerobic cabinet with an internal atmosphere of 85% N2, 10% CO2, and 5% H2 (MK III; Don Whitley Scientific Ltd., Shipley, England). The species and serotype of each colony grown for DNA extraction were ascertained by botulinum neurotoxin-specific PCR detection (16).
TABLE 1.
C. botulinum group II strains used in this study
| Strain | Origin | Yr of isolation | Location | Sourcea |
|---|---|---|---|---|
| 2 B | Marine sediment | 1960s | United States (Pacific coast) | Eklund/Lindrothb |
| 17 B | Marine sediment | 1960s | United States (Pacific coast) | Eklund/ATCC 25765 |
| 706 B | Salted salmon | 1977 | United States (Alaska) | Hatheway/Lindrothb |
| 1461 B | Rohschinken (dried ham) | 1980s | Germany | BF |
| 250 E | Canned salmon | 1978 | United States (Alaska) | Crowther/Lindrothb |
| Beluga E | Fermented white whale flippers | 1951 | United States (Alaska) | Dolman/Lindrothb |
| 211 E | Pickled herring | 1949 | Canada (Vancouver) | Dolman/Lindrothb |
| 92 E | Marine environment | 1960s | United States (Pacific coast) | Eklund/Lindrothb |
| 4062 E | Muktuk (fermented whale blubber) | 1981 | United States (Alaska) | Hatheway/Lindrothb |
| 31-2570 E | NKc | 1973 | United States | Hatheway/BF |
| 36208 E | Smoked salmon | 1934 | Canada (Nova Scotia) | Hazen/ATCC 9564 |
| R-90 E | Smoked whitefish | 1997 | Canada (Manitoba) | Our isolate |
| R-9087 E | Smoked rainbow trout | 1996 | Canada | Our isolate |
| RS-1 | Pacific red snapper | 1983 | United States (Pacific coast) | Lindrothb |
| KA-2 E | Seola Creek strain | NK | United States | Riemann/Lindrothb |
| C-51 E | Sealmeat | 1986 | Denmark (Greenland) | SSI |
| C-60 E | Dried mutton | 1989 | Denmark (Faeroes) | SSI |
| C-94 E | Sealmeat | 1990 | Denmark (Greenland) | SSI |
| 610B8-6 F | Salmon | 1966 | United States (Columbia River) | Craig/Lindrothb |
| 202 F | Marine sediment | 1965 | United States (Pacific coast) | Eklund/ATCC 23387 |
| FT 10 F | Herring | 1960s | United Kingdom (Inverness) | Hobbs/ATCC 27321 |
ATCC, American Type Culture Collection (Rockville, Md.); BF, Bundesinstitut für Fleischforschung (Kulmbach, Germany); SSI, Statens Serum Institut (Copenhagen, Denmark).
These strains have been collected from various sources by Seppo Lindroth (University of California, Davis). The preceding name denotes the original source, i.e., the person who presumably first isolated the strain.
NK, not known.
In situ DNA preparation.
As a reference to later results, DNA isolation was first performed as described by Maslow et al. (22). Subsequently, we modified this method essentially for the C. botulinum group II strains. An 8-ml volume of overnight culture in mid-log phase (absorbance at 540 nm, ≥1.0) was chilled on ice, and the cells were harvested by low-speed centrifugation (1,100 × g) at 4°C. To inactivate endogenous DNase activity, a modified formalin treatment step (13) was performed. The cells were resuspended in 3 ml of PIV (10 mM Tris [pH 7.5], 1 M NaCl) containing 3.5 to 4.0% (vol/vol) formaldehyde solution (Merck & Co., Darmstadt, Germany) and left on ice for 1 h. To obtain complete lysis, the cells were washed twice with PIV and resuspended in double-strength lysis solution (12 M Tris [pH 7.5], 2 M NaCl, 200 mM EDTA [pH 8.0], 1% Brij 58, 0.4% deoxycholate, 1% sodium lauroyl sarcosine, 40 μl of RNase per ml, 2 mg of lysozyme per ml, 40 U of mutanolysin per ml). The cell suspensions were mixed with an equal amount of 2% (wt/vol) low-melting-temperature agarose (InCert agarose; FMC Bioproducts, Rockland, Maine). Instead of insert molds, GelSyringe dispensers (New England Biolabs, Beverly, Mass.) were used as specified by the manufacturer. The syringe plugs were lysed overnight in single-strength lysis solution with gentle shaking at 37°C, and lysis was continued by three overnight ESP (0.5 M EDTA [pH 8.0], 10% sodium lauroyl sarcosine, 100 μg of proteinase K per ml) washes at 50°C. Phenylmethylsulfonyl fluoride inactivation of proteinase K and restriction endonuclease digestion of the agarose- embedded DNA were performed as described by New England Biolabs (24).
DNase inactivation experiments.
To further inactivate DNase activity, some additional procedures were tested: (i) cell suspensions were treated for 10 min at 50°C (21) or 65°C (18) before being mixed with the insert agarose; (ii) cells were lysed at 65°C before being mixed with the insert agarose, proceeding directly with the ESP wash (19); (iii) double-strength lysis solution was used for the overnight incubation step; (iv) sucrose (50%, wt/vol) was added to the lysis solution (34); (v) syringe plugs were cut into 1-mm slices prior to the lysis step; and (vi) the times for the regular lysis (overnight) and ESP wash (48-h) steps were progressively shortened, to as little as 60 min (30 plus 30 min).
Restriction enzyme digestions and electrophoresis.
Initially, 15 rare-cutting restriction enzymes (ApaI, AscI, AvrII, BssHII, ClaI, EagI, MluI, NaeI, NotI, NruI, RsrII, SacII, SmaI, XbaI, and XhoI [New England Biolabs]) were tested for cleavage of C. botulinum DNA. In some digestions, SacII was replaced by its isoschizomer KspI (Boehringer, Mannheim, Germany). Samples were electrophoresed at 10°C through a 1% (wt/vol) agarose gel (SeaKem Gold; FMC Bioproducts) in 0.5× TBE buffer (Amresco, Solon, Ohio) at 200 V for 16 to 22 h with a Gene Navigator system (Pharmacia, Uppsala, Sweden) with a hexagonal electrode. Different pulse time ramps were tested to find the most appropriate ones for each enzyme. Low Range, MidRange I, Lambda ladder, and Yeast chromosome PFG markers (New England Biolabs) were used for fragment size determination. The gels were stained for 30 min in 1 liter of used running buffer containing 0.5 mg of ethidium bromide and destained in running buffer until appropriate contrast was obtained for photography by standard procedures (27). DNA fragment sizes were estimated by measuring their respective running lengths in the gel in relation to the closest molecular weight markers. All DNA extractions and digestions were repeated at least three times.
RESULTS
DNase inactivation by different in situ DNA preparation methods.
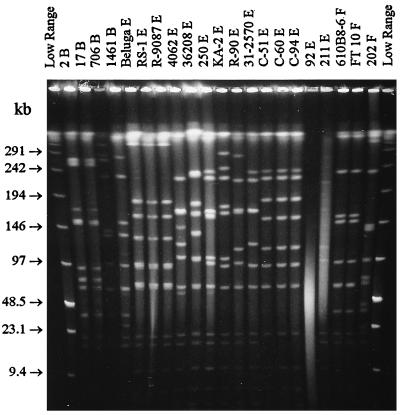
When a regular DNA isolation procedure was used (22), only 1 of the 21 strains yielded a visible PFGE pattern. The best results for the C. botulinum group II strains were achieved by using a large number of cells (8-ml cultures) and formaldehyde fixation on ice. For most strains, shortening of the isolation steps by following the protocol of Matushek et al. (23), i.e., a 2-h lysis at 37°C, a 1-h ESP wash at 50°C, and a 1-h TE wash at 50°C, had no effect on the outcome of PFGE. With this isolation procedure, lanes sometimes had less smearing due to degraded DNA but the restriction fragments lacked the intensity seen with DNA isolated by the 3-day procedure. The use of an ultrashort method (30 plus 30 min) gave lower DNA yields and resulted in reduced fragment visibility and increased smearing of PFGE lanes. The size and surface of the in situ DNA plug had no effect on the outcome of the results; neither did the use of double-strength lysis solution. The remaining DNase inactivation procedures that were tested (high lysis temperatures, omission of the 37°C lysis step, use of a sucrose-saturated lysis solution) did nothing to reduce DNase damage in affected strains and had a negative effect on the general outcome of the isolation; i.e., they resulted in increased smearing of PFGE lanes. One strain (92 E) was consistently untypeable by PFGE due to extensive DNase activity, and the genomic size of another strain (211 E) could not be estimated because of marked DNA degradation (Fig. 1).
FIG. 1.
SmaI digest of all C. botulinum group II strains from Table 1, including 1 nontypeable strain (92 E) and one strain with very faint PFGE patterns (211 E). The right- and left-hand lanes contain the low-range PFG marker. The pulse time was ramped from 1 to 18 s for 16 h at 200 V.
PFGE running conditions.
PFGE runs were mainly ramped from 1 to 18 s for 20 h. For the genome size determinations, three different electrophoretic ramps were used to move all the fragments to be sized into the linear range of the gel. For a good lower-molecular-size section (6 to 250 kb), pulses were ramped from 1 to 18 s for 16 h; in the middle-molecular-size section (50 to 500 kb), they were ramped from 1 to 40 s for 22 h; in the higher-molecular-size section (400 to 1,900 kb), they were ramped from 10 to 120 s for 22 h.
Typing of C. botulinum group II strains by PFGE.
Of the enzymes tested for DNA cleavage and PFGE typing of C. botulinum group II, ApaI, EagI, MluI, NaeI, NruI, SacII/KspI, SmaI, and XhoI produced convenient numbers of fragments (between 10 and 20) (Fig. 2 and 3). BssHII and RsrII generated only large fragments (one below 500 kb, and the rest above 500 kb), and although they were previously found practical for genome size estimations aided by transposon probing (21), they were unsuitable for basic PFGE interpretations. The reason for this is the stoichiometrically disproportionate ethidium bromide staining of large fragments, which causes band visualization problems when extremely rare-cutting enzymes, such as BssHII and RsrII, are used. AscI and NotI were altogether unsuitable, since they generated none or only one very large fragment under regular PFGE conditions. AvrII, ClaI, and XbaI generated too many small (<100-kb) fragments. Reproducibility of banding patterns between different DNA lots was excellent with all enzymes, although NaeI gave some problems with incomplete digestions (Fig. 2). For strain differentiation, ApaI, MluI, NruI, SmaI, and XhoI, gave the most revealing patterns, and these can be recommended for strain identification purposes in epidemiological studies (Fig. 3). MluI, SmaI, and XhoI were found to be the most suitable for genome size determination (Table 2).
FIG. 2.
PFGE separation of genome restriction enzyme digests of three C. botulinum group II strains belonging to different serotypes. DNA digested with AvrII (lanes 2 to 4), BssHII (lanes 5 to 7), EagI (lanes 8 to 10), KspI (lanes 11 to 13), and NaeI (lanes 14 to 16) is shown. Lanes: 1 and 17, lambda ladder; 2, 5, 8, 11, and 14, C. botulinum 17 B; 3, 6, 9, 12, and 15, C. botulinum 36208 E; 4, 7, 10, 13, and 16, C. botulinum FT10 F. The pulse time was ramped from 1 to 18 s for 20 h at 200 V.
FIG. 3.
Optimal PFGE separation and typing of C. botulinum group II strains was achieved by digestion with ApaI (lanes 2 to 4), MluI (lanes 5 to 7), NruI (lanes 8 to 10), SmaI (lanes 11 to 13), and XhoI (lanes 14 to 16). Lanes: 1 and 17, lambda ladder; 2, 5, 8, 11, and 14, C. botulinum 17 B; 3, 6, 9, 12, and 15, C. botulinum 36208 E; 4, 7, 10, 13, and 16, C. botulinum FT10 F. The pulse time was ramped from 1 to 18 s for 20 h at 200 V.
TABLE 2.
Genome size estimates and number of restriction fragments produced by endonuclease cleavage of C. botulinum group II genomic DNA
| Straina | Restriction fragment no.b and genome size (kb)c after cleavage with:
|
Mean size ± SD | |||||
|---|---|---|---|---|---|---|---|
|
MluI
|
SmaI
|
XhoI
|
|||||
| No. | Size | No. | Size | No. | Size | ||
| 2 BA | 9 | 3,528 | 15 | 3,581 | 12 | 3,654 | 3,588 ± 63 |
| 17 BA | 9 | 3,528 | 15 | 3,581 | 12 | 3,654 | 3,588 ± 63 |
| 706 B | 10 | 3,737 | 16 | 3,763 | 14 | 3,850 | 3,783 ± 59 |
| 1461 B | 17 | 3,605 | 15 | 3,590 | 11 | 3,623 | 3,606 ± 17 |
| 250 E | 15 | 4,158 | 18 | 4,184 | 13 | 4,105 | 4,149 ± 40 |
| Beluga EB | 20 | 3,939 | 17 | 3,990 | 18 | 4,035 | 3,988 ± 48 |
| RS-1B | 20 | 3,939 | 17 | 3,990 | 18 | 4,035 | 3,988 ± 48 |
| R-9087 EB | 20 | 3,939 | 17 | 3,990 | 18 | 4,035 | 3,988 ± 48 |
| 4062 E | 14 | 3,870 | 16 | 3,897 | 16 | 3,881 | 3,882 ± 13 |
| 31-2570 E | 22 | 3,767 | 15 | 3,762 | 17 | 3,773 | 3,767 ± 5 |
| 36208 E | 15 | 4,125 | 13 | 4,155 | 14 | 4,127 | 4,136 ± 17 |
| R-90 E | 16 | 3,965 | 15 | 4,094 | 14 | 4,055 | 4,038 ± 66 |
| KA-2 E | 14 | 3,849 | 16 | 3,878 | 12 | 3,861 | 3,863 ± 15 |
| C-51 EC | 19 | 3,873 | 15 | 3,777 | 15 | 3,878 | 3,806 ± 63 |
| C-60 EC | 19 | 3,873 | 15 | 3,777 | 15 | 3,878 | 3,806 ± 63 |
| C-94 EC | 19 | 3,873 | 15 | 3,777 | 15 | 3,878 | 3,806 ± 63 |
| 202 F | 14 | 3,958 | 16 | 3,998 | 14 | 4,034 | 3,996 ± 38 |
| 610B8-6 FD | 13 | 4,026 | 13 | 3,978 | 13 | 4,044 | 4,016 ± 34 |
| FT 10 FD | 13 | 4,026 | 13 | 3,978 | 13 | 4,044 | 4,016 ± 34 |
| Mean ± SD | 3,872 ± 176 | 3,881 ± 183 | 3,918 ± 157 | 3,890 ± 170 | |||
Different strains that have the same subscripts are seemingly clonal; i.e., they gave indistinguishable PFGE patterns with all enzymes used.
Only fragments larger than 6 kb.
Represents the average of at least three independent size determinations of each fragment separated under linear PFGE conditions.
Genome size determination.
As summarized in Table 2, the number of fragments generated by digestion of 19 C. botulinum group II genomes with MluI, SmaI, and XhoI ranged from 9 to 22, 13 to 18, and 11 to 18, respectively. With MluI, the smallest detected fragment was 24 kb and the largest was 1,192 kb; with SmaI, they were 6.7 and 1,664 kb; and with XhoI, they were 29 and 874 kb. The mean genome sizes obtained with the different restriction enzymes were 3,872 (MluI), 3,881 (SmaI), and 3,918 kb (XhoI), resulting in an overall genome size estimate of 3,890 ± 170 kb for C. botulinum group II.
DISCUSSION
When a regular in situ DNA isolation method is used (23), C. botulinum group II strains cannot be characterized by PFGE. The reason for this may be the low yields associated with DNA isolation in this group, further hampered by DNA degradation problems caused by extracellular DNases. This study shows that formaldehyde fixation of cells prior to lysis can to a large extent prevent DNase-related problems. With some strains, the results can be further improved by shortening the isolation steps at critical temperatures. On the other hand, some formaldehyde-treated cultures gave clearer PFGE patterns by the regular 3-day DNA isolation procedure. The additional modifications assessed (see Materials and Methods) did not limit DNase damage further with these C. botulinum group II strains and caused unnecessary smearing of PFGE lanes. Aside from the formaldehyde fixation of cells on ice, all other procedures tested had a very limited effect, be it positive or negative, on the outcome of the isolation. The small effects that could be seen seemed to have more to do with strain-specific differences in DNase production than with the procedure itself. We have found that optimal results can be obtained by performing both isolation procedures on the same agarose plug: after 2 h of lysis, we transfer half of the syringe plug into ESP for 1 h and TE buffer for 1 h, and we subject the other half to the 3-day protocol. This method has the benefit of rapid results while giving the researcher the option to choose the plug with the better isolation result for further PFGE studies. Of the 21 strains studied, only 1 (C. botulinum 92 E) could not be characterized by PFGE with any of the methods tested. Strain 211 E, also possessing DNA isolation problems, yielded PFGE patterns in the small-fragment range (<300 kb) when 16- or 24-ml cell cultures were used. However, its genome size calculation was impossible due to extensive large-fragment degradation. For these and other C. botulinum strains adversely affected by persistent DNases, further methodology studies are warranted.
The successful use of ultrashort lysing steps (30 min) and ESP washes (30 min) suggests that penetration of chemicals into the agarose plugs is not a problem. The indistinguishable outcomes with both agarose plug sizes, either 1-mm slices or whole syringe plugs, also warrant this conclusion. However, a surprisingly large number of nonproteolytic C. botulinum cells is needed to obtain sufficient amounts of DNA for PFGE analysis. To achieve proper visualization of the large (>600-kb) fragments needed in genome size calculations and the small (<50-kb) fragments that may possibly aid in species determination (5), at least 8 ml of TPGY broth at mid-log growth is needed for a 1-ml gel plug. This cell mass is about 5 to 10 times greater than the amount of other gram-positive bacteria used for PFGE in our laboratory (3, 25). Similarly low DNA yields are not as pronounced with other clostridia (6, 21, 25, 34). It seems to be a special feature of C. botulinum group II strains, possibly a result of DNases and/or the resistance of cell wall structures to lysis. This is supported by our experience with C. botulinum DNA isolation (unpublished results), in which group II strains consistently gave lower DNA yields than group I strains by normal and in situ isolation methods. The relatively unsmeared PFGE lanes of most strains in this study contradict the DNase hypothesis and suggest inadequate lysis as the cause of this particular problem.
This study clearly shows that when the DNase problems are overcome, PFGE is an efficient tool for epidemiological studies of nonproteolytic C. botulinum isolates. Regardless of the restriction enzyme used, the PFGE patterns of the different strains showed little resemblance to each other, sharing at most half of their bands. Except for the strains which always displayed identical patterns and apparently have a clonal origin, the different strains were easily distinguished (Fig. 1). It was difficult to assign strains to common lineages on the basis of shared fragments; instead, the patterns pointed at a large diversity within these group II strains. This is perhaps not very surprising, since the isolation of strains spanned more than 60 years throughout northern Europe and North America.
The common ancestry of some serotype B strains was evident, since both strains from Pacific Ocean marine sediments seemed clonal, regardless of the enzyme used for digestion. Strains 706 B isolated from salted salmon in Alaska and 1461 B isolated from ham in Germany shared 73% of their fragments but differed markedly from the two other type B strains. A closer relationship could be detected in the serotype F strains, where two strains seemed clonal although one was supposedly of Atlantic origin and the other was of Pacific origin. The third serotype F strain, also of Pacific origin, shared 62% of the bands found in the other two. The number of B and F serotyped strains in this study, however, is too small to make any far-reaching conclusions, either about strain diversity or about genome size. PFGE patterns of the 14 strains belonging to serotype E (Fig. 1) give a better example of the biodiversity in C. botulinum group II strains. Regarding clonality, it is interesting that strains Beluga, RS-1, and R-9087 showed identical patterns despite being isolated over a 45-year period and from three different marine species. Although all three originate in North America, it might be of epidemiological interest to point out that R-9087 was isolated from a product manufactured in Finland from imported Canadian fish. Three Arctic strains from Greenland and the Faeroe Islands (C-51, C-60, and C-94) also seemed clonal, although one strain was isolated from dried mutton and the two others were isolated from sealmeat over a period of 4 years. The PFGE patterns of the other serotype E strains in this study had less common features, which seems to denote a more distant common ancestry.
SmaI digests were performed more than 10 times for most strains and thus formed the basis of the observations regarding pulsotypes within the group II strains. The other enzymes used for closer scrutiny of the strains, ApaI, MluI, NruI, and XhoI, were not able to distinguish between the apparent clones revealed by SmaI digestion. Hence, their primary use was to aid in correct genome size determination (Table 2). An important feature of the SmaI digests was the four small bands (6.7, 7.9, 21, and 67 kb) shared by all the studied strains (Fig. 1). The common fragments might be group II species specific, since we have not seen them in the PFGE patterns of other C. botulinum strains (unpublished results). Similar species- or serovar-specific bands have been suspected in the low range of AscI digests of Listeria monocytogenes and L. innocua (5). In addition to these, a 19-kb SmaI restriction fragment was shared by all group II strains except the Arctic ones (C-51, C-60, and C-94), a 10-kb fragment was shared by all serotype E strains, and a 39-kb fragment was seen in all serotype B and F strains. The finding of shared fragments is essentially concordant with earlier small-subunit rRNA sequence studies on Clostridium (9, 17, 20), where the nonproteolytic C. botulinum serotypes were placed on their own phylogenetic branch.
One of the main PFGE applications is the calculation of bacterial genome size by adding the estimated sizes of resolved restriction fragments, cut preferably by a variety of suitable rare-cutting restriction enzymes (10). It is the most direct and accurate method available (21). The genome sizes of many species, including certain clostridia, have been estimated by this procedure: C. botulinum 62A, 4.04 Mb (21); C. perfringens CPN 50, 3.6 Mb (6); and C. acetobutylicum, 2.85 to 6.5 Mb (34). We were able to estimate the genome sizes of 19 of the 21 strains studied. The sizes of C. botulinum group II genomes ranged from 3.59 to 4.15 Mb, with a mean of 3.89 Mb (Table 2). When the fragment sizes were estimated, more accurate results were obtained by comparing the fragments in relation to the two closest marker fragments, as opposed to a calculation of fragment sizes through measurement of the migration distance from the well, plotted on a standard curve (31). Fragment size estimates generated through standard curves, even those based on a cubic spline formula (26), can easily be off by at least 10% at the top and bottom of the gel, where fragment migration is nonlinear.
Our results indicate that C. botulinum group II strains, although perhaps members of the same species, express a large genetic diversity. Differences in genome size between isolates of the same species has also been noted for other bacteria (2, 32), perhaps indicating the necessity for environmental bacteria to possess extensive genomic plasticity to cope with distinct ecological niches. For C. botulinum, this would be of assistance when it is transformed from its placid state as a soil bacterium into a food pathogen. The finding that clonal lineages of C. botulinum group II remain unchanged for decades is perhaps not very surprising, but it is intriguing that these clones are at the same time geographically so widespread. To further elucidate these results on C. botulinum group II biodiversity, more taxonomic and phylogenetic studies are warranted. When the remaining DNA extraction problems are resolved, other genotypic methods in addition to PFGE should also be applied.
ACKNOWLEDGMENTS
This work was supported by grants from the Academy of Finland, the Finnish Food Research Foundation, the Walter Ehrström Foundation, and the Finnish Veterinary Foundation.
We are grateful to Sirkka Ekström, Kirsi Ristkari, and Maria Stark for their invaluable technical assistance and to Tuula Johansson (National Veterinary and Food Research Institute, Helsinki, Finland) for having maintained the strain collection of the late Seppo Lindroth (University of California, Davis).
REFERENCES
- 1.American Public Health Association. Compendium of methods for the microbiological examination of foods. 3rd ed. Washington, D.C: American Public Health Association; 1992. p. 1108. [Google Scholar]
- 2.Bergthorson L, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björkroth K J, Ridell J, Korkeala H. Characterization of Lactobacillus sake strains associating with production of ropy slime by randomly amplified polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) patterns. Int J Food Microbiol. 1996;31:59–68. doi: 10.1016/0168-1605(96)00964-6. [DOI] [PubMed] [Google Scholar]
- 4.Blaschek H P, Klacik M A. Role of DNase in recovery of plasmid DNA from Clostridium perfringens. Appl Environ Microbiol. 1984;48:178–181. doi: 10.1128/aem.48.1.178-181.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch R, Chen J, Luchansky J B. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl Environ Microbiol. 1994;60:2584–2592. doi: 10.1128/aem.60.7.2584-2592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canard B, Cole S T. Genome organization of the anaerobic pathogen Clostridium perfringens. Proc Natl Acad Sci USA. 1989;86:6676–6680. doi: 10.1073/pnas.86.17.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chachaty E, Saulnier P, Martin A, Mario N, Andremont A. Comparison of ribotyping, pulsed-field gel electrophoresis and random amplified polymorphic DNA for typing of Clostridium difficile strains. FEMS Microbiol Lett. 1994;122:61–68. doi: 10.1111/j.1574-6968.1994.tb07144.x. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri P, Singh S D, Qari S H. DNase of Clostridium septicum—an obstacle in plasmid isolation. Indian J Anim Sci. 1996;66:653–656. [Google Scholar]
- 9.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall A, Shapiro L, Ely B. Analysis of bacterial genome organization and replication using pulsed-field gel electrophoresis. Methods Companion Methods Enzymol. 1990;1:160–168. [Google Scholar]
- 11.Food and Agricultural Organization. Manual of food quality control. Vol. 12. Rome, Italy: Food and Agricultural Organization of the United Nations; 1991. pp. 115–116. [Google Scholar]
- 12.Friedman C R, Hatheway C L. Proceedings of the 1996 Meeting of the Interagency Botulism Research Coordinating Committee. 1996. Laboratory-confirmed botulism in the United States in 1995; p. 34. [Google Scholar]
- 13.Gibson J R, Sutherland K, Owen R J. Inhibition of DNAse activity in PFGE analysis of DNA from Campylobacter jejuni. Lett Appl Microbiol. 1994;19:357–358. doi: 10.1111/j.1472-765x.1994.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 14.Hatheway C L. Botulism: the present status of the disease. Curr Top Microbiol Immunol. 1995;195:55–75. doi: 10.1007/978-3-642-85173-5_3. [DOI] [PubMed] [Google Scholar]
- 15.Hauschild A H W. Epidemiology of human foodborne botulism. In: Hauschild A H W, Dodds K L, editors. Clostridium botulinum—ecology and control in foods. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 68–104. [Google Scholar]
- 16.Hielm S, Hyytiä E, Ridell J, Korkeala H. Detection of Clostridium botulinum in fish and environmental samples using polymerase chain reaction. Int J Food Microbiol. 1996;31:357–365. doi: 10.1016/0168-1605(96)00984-1. [DOI] [PubMed] [Google Scholar]
- 17.Hutson R A, Thompson D E, Lawson P A, Schocken-Itturino R P, Bottger E C, Collins M D. Genetic interrelationships of proteolytic Clostridium botulinum types A, B and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Antonie Leeuwenhoek. 1993;64:273–283. doi: 10.1007/BF00873087. [DOI] [PubMed] [Google Scholar]
- 18.Kristjánsson M, Samore M H, Gerding D N, DeGirolami P C, Bettin K M, Karchmer A W, Arbeit R D. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J Clin Microbiol. 1994;32:1963–1969. doi: 10.1128/jcm.32.8.1963-1969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson P A, Gharbia S E, Shah H N, Clark D R. Recognition of Fusobacterium nucleatum subgroups Fn-1, Fn-2 and Fn-3 by ribosomal RNA gene restriction patterns. FEMS Microbiol Lett. 1989;65:41–46. doi: 10.1016/0378-1097(89)90363-7. [DOI] [PubMed] [Google Scholar]
- 20.Lawson P A, Llop-Perez P, Hutson R A, Hippe H, Collins M D. Towards a phylogeny of the clostridia based on 16S rRna sequences. FEMS Microbiol Lett. 1993;113:87–92. doi: 10.1111/j.1574-6968.1993.tb06493.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin W-J, Johnson E A. Genome analysis of Clostridium botulinum type A by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1995;61:4441–4447. doi: 10.1128/aem.61.12.4441-4447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed-field electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: ASM Press; 1993. pp. 563–572. [Google Scholar]
- 23.Matushek M G, Bonten M J M, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.New England Biolabs. ImBed Kit, reagents and protocols for embedding chromosomal DNA in agarose. Beverly, Mass: New England Biolabs; 1990. [Google Scholar]
- 25.Ridell, J., J. Björkroth, H. Eisgrüber, B. Schalch, A. Stolle, and H. Korkeala. Prevalence of the enterotoxin gene and clonality of Clostridium perfringens strains associated with food poisoning outbreaks. J. Food Prot., in press. [DOI] [PubMed]
- 26.Rodrigo A G, Borges K M, Bergquist P L. Pulsed-field gel electrophoresis of genomic digests of Thermus strains and its implications for taxonomic and evolutionary studies. Int J Syst Bacteriol. 1994;44:547–552. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 6.18–6.19. [Google Scholar]
- 28.Samore M H, Kristjánsson M, Venkataraman L, DeGirolami P C, Arbeit R D. Comparison of arbitrarily-primed polymerase chain reaction, restriction enzyme analysis and pulsed-field gel electrophoresis for typing Clostridium difficile. J Microbiol Methods. 1996;25:215–224. [Google Scholar]
- 29.Stern M, Warrack G H. The types of Clostridium perfringens. J Pathol Bacteriol. 1964;88:279–282. [PubMed] [Google Scholar]
- 30.Swiatek P J, Allen S D, Siders J A, Lee C H. DNase production by Clostridium septicum. J Clin Microbiol. 1987;25:437–438. doi: 10.1128/jcm.25.2.437-438.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thong K L, Puthucheary S D, Pang T. Genome size variation among recent human isolates of Salmonella typhi. Res Microbiol. 1997;148:229–235. doi: 10.1016/S0923-2508(97)85243-6. [DOI] [PubMed] [Google Scholar]
- 33.Timmis K, Winkler U. Isolation of covalently closed circular deoxyribonucleic acid from bacteria which produce exocellular nuclease. J Bacteriol. 1973;113:508–509. doi: 10.1128/jb.113.1.508-509.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson S R, Young M. Wide diversity of genome size among different strains of Clostridium acetobutylicum. J Gen Microbiol. 1993;139:1069–1076. [Google Scholar]