Abstract
Secretion of proteolytic and chitinolytic enzymes is a hallmark of infection processes of Metarhizium anisopliae in response to host (insect) cuticular signals. The regulation of these enzymes (subtilisin-like proteases [Pr1a and Pr1b], trypsin-like proteases [Pr2], metalloproteases, aspartyl proteases, aminopeptidase, and chitinases) and a hydrophobin was investigated by Northern analysis and/or enzyme assay. The production of each enzyme showed a differential expression pattern in response to ambient pH; enzymes were synthesized only at pHs at which they function effectively, irrespective of whether the medium contained an inductive cuticle substrate. Three aspartyl proteases (pH optimum, 3), and chitinase (pH optimum, 5) showed maximal accumulation at acidic pHs. The highest level of aminopeptidase (pH optimum, 7) was detected at pH 7. The highest levels of five metalloproteases (pH optima, ca. 7) were detected over the pH range 6 to 8. Two trypsins and several subtilisin-like Pr1 isoforms with pH optima of ca. 8 were produced only under alkaline conditions. Northern analysis of RNA species corresponding to seven cDNA sequences encoding proteases and chitinase confirmed that the ambient pH played a major role in gene expression of secreted proteins. Hydrophobin was expressed almost equally at pHs 5 and 8 but was not expressed at pH 3. During fungal penetration, the pH of infected cuticle rises from about 6.3 to 7.7. Consistent with pH regulation of enzyme production, serine and metalloproteases were produced in situ during infection, but no production of aspartyl proteases was found. We propose that the alkalinity of infected cuticle represents a physiological signal that triggers the production of virulence factors.
Human, plant, and insect pathogenic fungi produce a complement of extracellular enzymes that degrade the integuments of their hosts (4, 11–13, 18, 24, 31–33). Elucidating the mechanisms regulating the secretion of these depolymerases is central to understanding pathogen growth and development in the host. The insect pathogen Metarhizium anisopliae has been the focus of studies of host-cuticle penetration and biocontrol of insect pests (32). This organism produces families of catalytically distinct extracellular subtilisin-like proteases (Pr1), trypsin-like proteases (Pr2), and metalloproteases, as well as several families of exo-acting peptidases that are believed to be important in insect cuticle degradation (19, 32). In addition, M. anisopliae produces several chitinolytic enzymes which act after the pathogen’s proteases have significantly digested the cuticle protein and unmasked the chitin component of the cuticle (18). Substantial knowledge of the physiology and biochemistry of these proteases and chitinases has been gained in recent years (15, 19, 29, 30). The cDNAs and genes encoding several cuticle-degrading enzymes have been cloned and sequenced (8, 9, 17, 26).
The regulation of these genes is complex, usually involving a combination of substrate induction and carbon and nitrogen repression (18). In M. anisopliae and other entomopathogens, chitinase is required for only a brief period during penetration of host cuticle and is tightly regulated by chitin degradation products (21). Proteases have an additional role in providing nutrients, before and after the cuticle is penetrated. Consequently, regulation is looser, with production being triggered in response to limitation for nutrients such as carbon and nitrogen (18). However, production is enhanced when the pathogen is grown on insect cuticle (15). Since many insect pathogens, including M. anisopliae, can grow in media over a wide pH range, 2.5 to 10.5 (6), it is also likely that they have a regulatory system to ensure that enzymes and other gene products that function beyond cell boundaries are synthesized only at pHs at which they can function effectively. Likewise, in Aspergillus nidulans and Yarrowia lipolytica, acidic phosphatases and proteases are secreted only in acidic environments and alkaline phosphatase is secreted only in alkaline environments (1, 35). Six regulatory genes that mediate this pH regulation in Aspergillus spp. have been identified and studied; the major mediator is the zinc finger transcription factor PacC, an activator for alkaline-expressed genes and a repressor for acid-expressed genes (10, 14). Similarly, mutations affecting the expression of pH-regulated genes in Y. lipolytica have also been described (3). These studies should eventually lead to an understanding of how these organisms sense ambient pH.
The study of the regulation of pathogen genes is of particular importance because pathogenic specialization may operate by way of regulatory controls that allow the expression of genes under conditions in which similar genes in nonpathogens are not expressed. Thus, a pH-regulated gene is involved in the morphological plasticity of Candida albicans, which is related to its pathogenicity to humans (16). Our objectives were (i) to determine if changes in ambient pH provide a physiological signal that triggers the production of putative virulence determinants such as Pr1 and Pr2, metalloproteases, aspartyl proteases, aminopeptidase, and chitinase and (ii) to determine if hydrophobins (34) were regulated by pH.
MATERIALS AND METHODS
Organism and growth.
M. anisopliae ARSEF strain 2575 (host: pecan weevil, Curculio caryae) was obtained from the U.S. Department of Agriculture Entomopathogenic Fungus Collection in Ithaca, N.Y. Fungal cultures were maintained on potato dextrose agar (20).
Preparation and analysis of culture filtrate.
Standardized mycelial inocula (5 g [wet weight] per 100 ml) from 24-h Sabouraud dextrose broth (SDB) cultures were incubated with shaking (at 100 rpm) at 25°C for up to 12 h in 100 ml of minimal medium [containing, per liter, 1 g of KH2PO4, 0.5 g of MgSO4, 0.7 mg of Na2B4O7 · 10H2O, 0.5 mg of (NH4)6Mo7O24 · 4H2O, 10 mg of Fe2(SO4)3 · 6H2O, and 0.3 mg of ZnSO4 · 7H2O] supplemented with cockroach cuticle at 1.0% (wt/vol). The medium’s pH was fixed and maintained throughout cultivation by using 0.1 M citric acid-sodium phosphate buffer (pH 3.0, 4.0, 5.0, 6.0, or 7.0) or 0.1 M HEPES buffer (pH 8.0). Measurements were made of the medium’s pH at five hourly intervals in order to confirm stability. Samples (15 ml) were filtered through Whatman no. 1 filter paper and then through a Millipore filter unit (pore size, 0.2 μm) and were desalted by using Centriprep-10 ultrafiltration units (Amicon).
Analytical isoelectric focusing (IEF).
Desalted samples were concentrated 50-fold by using Centricon-10 ultrafiltration units. Two-microliter aliquots of the concentrate were run on ultrathin polyacrylamide gels by using 1% ampholytes (Bio-Lyte 3/10; Bio-Rad) (28).
To characterize protease isoforms as to pH optima, gels were preincubated with 1.0 M citric acid-sodium phosphate buffer (pH 3.0, 4.0, 5.0, 6.0, or 7.0), 1.0 M HEPES buffer (pH 8.0 or 9.0) or 0.5 M NaHCO3–Na2CO3 (pH 10). After 30 s in buffer, protease activity was detected by gelatin zymography using gelatin-coated X-ray film (28). Membrane-gel sandwiches were incubated for 10 to 15 min.
The catalytic mechanisms of protease isoforms were characterized by using specific active-site inhibitors (28). In some experiments, gels were preincubated for 10 min in one of the following inhibitors dissolved in water: leupeptin (0.15 mM); pepstatin (0.2 mM); 1,10-phenanthroline and phosphoramidon (1 mM), and phenylmethylsulfonyl fluoride (PMSF) (0.2 mM). Alternatively, desalted extracts from infected cuticles were incubated directly with inhibitors for 10 min before being run on IEF gels.
Preparation of insect cuticle substrate.
Cuticle was obtained from the giant cockroach (Blaberus giganteus) by extraction of soft tissue from homogenized insects with potassium tetraborate (28).
Enzyme assays.
Subtilisin-like Pr1 activity [versus succinyl–(Ala)2–Pro–Phe–4-nitroanilide (NA) at pH 8], trypsin-like Pr2 activity (versus benzoyl-Phe-Val-Arg-NA at pH 8), and aminopeptidase activity (versus Ala-NA at pH 7) were determined as described previously (22). Activities are expressed as nanomoles of NA released per minute per milliliter. Aspartyl (acidic) protease was assayed against hide protein azure. Two-milliliter Eppendorf tubes were charged with 10 mg of substrate, 1.5 ml of 50 mM acetate buffer (pH 3.0), and 100 μl of enzyme. After incubation for 10 min at 30°C, reactions were terminated by the addition of trichloroacetic acid (0.25 ml; 500 g/liter). Following centrifugation (at 5,000 × g for 10 min) the absorbance was measured at 595 nm. Activities are expressed as change in optical density at 595 nm per 10 min per milliliter Assays of N-acetyl-β-d-glucosaminidase (NAGase) in microtitration trays contained 80 μl of 0.1 M citrate buffer (pH 5.0), 10 μl of 10 mM p-nitrophenol-N-acetyl-β-d-glucosaminide, and 10 μl of culture supernatant. Reaction mixtures were incubated at 37°C for 30 min, and reactions were terminated by the addition of 100 μl of 0.5 M NaHCO3–Na2CO3 buffer. The release of p-nitrophenyl (pNP) was determined at 405 nm in a Bio-Tek Instruments Microplate EL 309 autoreader. Activities are expressed as nanomoles of nitrophenyl (NP) released per minute per ml.
Northern blot analysis and determination of relative transcript levels.
Total RNA was isolated by using Tri-reagent, cDNA clones were radiolabelled, and Northern blots were performed as described previously (8). For the determination of transcript levels, signals were quantified by using a PhosphorImager (Molecular Dynamics). Relative abundance was calculated by dividing the given signal strength by the signal strength at pH 8.
The cDNA clones used for Northern blots were Pr1a (26), Pr1b (9), hydrophobin (ssg12), and tubulin (27). A genomic clone of Pr2 has been published (17). We isolated a cDNA clone (Pr2) from an expression library by using specific antibodies (30) as a probe. A cDNA clone of chitinase was also obtained by probing an expression library with specific antibodies (29) (GenBank accession no. U59484). A cDNA clone of carboxypeptidase (accession no. U76003) was obtained by using a differential-display technique (7a).
Intracellular pH determinations.
Standardized mycelial inocula (2.5 g [wet weight] per 50 ml) from 24-h SDB cultures were incubated with shaking (at 100 rpm) at 25°C for up to 3 h in 50 ml of basal medium buffered at pH 5.0 or pH 7.0 by using 0.1 M citric acid-sodium phosphate buffer or at pH 8.0 by using 0.1 M HEPES buffer. Following incubation, mycelia were filtered and were washed four times with 500 ml of distilled water, and intracellular pH was determined as described by Caddick et al. (1). Samples (2.5 g [wet weight]) were transferred to 20 ml of 0.1% (vol/vol) Triton X-100, frozen at −80°C overnight, and disrupted by rapid thawing to room temperature. The pH values were then determined immediately.
pH determinations of infected insect cuticle.
Cuticles from fifth-instar (3 days after ecdysis) Manduca sexta were dissected from other tissues, flash frozen in liquid nitrogen, and comminuted with a mortar and pestle. Samples (2 g [wet weight]) were transferred to 5 ml of distilled water, frozen at −80°C overnight, and thawed rapidly for pH determinations. Cuticles to be infected with fungal spores were soaked in 0.001% phenylthiourea (for 30 min), rinsed with four changes (5 min each) of sterile distilled water, and sterilized under an ethylene oxide atmosphere. Cuticles (about 3 by 2 cm) were placed on water agar (1.5%, wt/vol) plates and inoculated with 50 μl of distilled water containing about 5,000 conidia. Controls were inoculated with water alone. Following incubation (for 60 h) at 27.5°C, cuticles were ground under liquid nitrogen with a pestle and mortar, resuspended in distilled water, and frozen and thawed for pH determinations.
Extraction of enzymes from M. sexta cuticle.
Cuticles were infected with conidia, incubated as described above, and then extracted by vigorous shaking for 1 h in 0.2 M potassium phosphate buffer, pH 7.0, at 4°C (23). After centrifugation, extracts were desalted and concentrated 50-fold by using Amicon Centricon-10 ultrafiltration units before assaying for enzyme activities.
Materials.
Enzyme substrates and inhibitors were purchased from Sigma.
RESULTS
Influence of ambient pH on enzyme activities.
Exponentially growing mycelium of M. anisopliae was transferred to minimal medium with or without cockroach cuticle as the sole carbon source. Each flask received the same amount (5 g) of fungal biomass, reducing the dependence of enzyme production on total growth (27). Proteolytic and chitinolytic enzymes capable of degrading the protein and chitin components of insect cuticle were detected in cell-free culture supernatants, even in the absence of added cuticle. However, the levels of enzymes produced were higher when minimal medium was supplemented with insect cuticle (Table 1). The culture pH at which maximum activity of each enzyme was detected was close to its pH optimum. Thus, the highest levels of aspartyl proteinase (assayed at pH 3) were detected at culture pHs of 3 to 4. The highest level of NAGase (pH optimum, 5) was detected at pH 5. The highest level of aminopeptidase (pH optimum, 7) was detected at pH 7. The highest level of Pr2 (pH optimum, 8) was detected over a pH range of 6 to 8, and the highest level of Pr1 (pH optimum, 8) was detected at pH 8.
TABLE 1.
Secretion of enzymes as a function of growth pH in minimal medium alone or supplemented with cuticle
| pH | Enzyme (optimum pH) activitya
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NAGase (5)
|
Pr1 (8)
|
Pr2 (8)
|
Aminopeptidase (7)
|
Aspartyl protease (3)
|
||||||
| −C | +C | −C | +C | −C | +C | −C | +C | −C | +C | |
| 3 | 17 ± 3 (59) | 18 ± 4 (42) | 0 (0) | 0 (0) | 0 (0) | 0.1 (0) | 0 (0) | 0 (0) | 19 ± 3 (90) | 72 ± 5 (85) |
| 4 | 23 ± 4 (78) | 36 ± 5 (83) | 4 ± 1 (6) | 30 ± 3 (13) | 4 ± 1 (13) | 4 ± 1 (7) | 0 (0) | 0 (0) | 21 ± 2 (100) | 85 ± 4 (100) |
| 5 | 29 ± 4 (100) | 43 ± 6 (100) | 15 ± 2 (22) | 63 ± 5 (28) | 8 ± 1 (27) | 23 ± 3 (38) | 0 (0) | 0 (0) | 7 ± 1 (33) | 27 ± 2 (32) |
| 6 | 27 ± 3 (92) | 40 ± 4 (94) | 36 ± 4 (52) | 165 ± 8 (72) | 20 ± 2 (67) | 53 ± 3 (87) | 2.8 ± .5 (30) | 5.5 ± 1 (40) | 2 ± 0 (10) | 13 ± 1 (15) |
| 7 | 14 ± 3 (49) | 15 ± 2 (35) | 39 ± 6 (57) | 175 ± 8 (77) | 30 ± 4 (100) | 55 ± 4 (90) | 9.2 ± 1 (100) | 14 ± 1 (100) | 1 ± 0 (5) | 7 ± 1 (8) |
| 8 | 12 ± 2 (41) | 13 ± 2 (29) | 69 ± 7 (100) | 228 ± 11 (100) | 25 ± 3 (83) | 61 ± 5 (100) | 3.4 ± 1 (37) | 10 ± 2 (73) | 0 (0) | 3 ± 0.2 (4) |
Activity is expressed as nanomoles of NP per minute per milliliter (for NAGase), nanomoles of NA per minute per milliliter (for Pr1, Pr2, and aminopeptidase), or change in optical density per 10 min per milliliter (for aspartyl protease), followed by the percent of maximum activity of the enzyme (in parentheses). −C, minimal medium alone; +C, minimal medium supplemented with 1% cockroach cuticle.
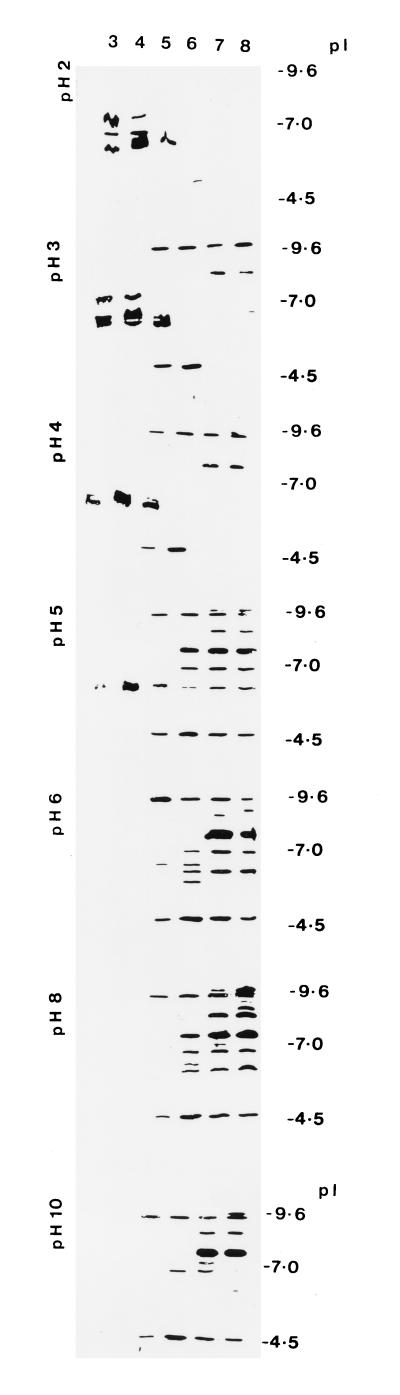
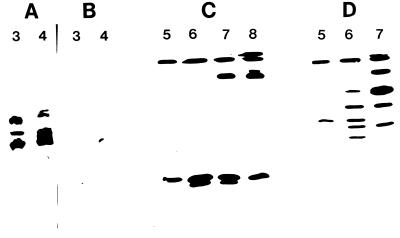
The profile of proteases produced at different pHs was assessed on IEF gels by using gelatin overlays to detect proteinases (Fig. 1). Mycelia incubated in cuticle-containing cultures at pHs 3, 4, and 5 produced at least three proteinases (pI 5.5 to 6.9) with pH optima of ca. 3 (Fig. 1). Pepstatin, a specific inhibitor of aspartyl (acidic) proteinases, inhibited all three activities (Fig. 2A and B). At higher culture pHs (pH 6, 7, and 8), 10 proteinases were produced (Fig. 1). Five of these bands (pI 5.8 to 7.6), active over a pH range of 5 to 8, were inhibited by 1, 10-phenanthroline, an inhibitor of zinc-containing metalloproteases (data not shown) and by phosphoramidon, a specific inhibitor of thermolysin-like metalloproteases (Fig. 2C). The greatest number of metalloproteinases was produced at pH 6 (five bands), but one of these bands (pI 7.6), with activity extending to pH 10, was produced at much higher amounts at pH 7 and pH 8 (Fig. 1 and 2). Two bands (pI 4 to 4.5), active over a pH range of 5 to 10, with optimal production at pH 6, were inhibited by leupeptin (Fig. 1 and 2D), identifying them as the trypsin-like Pr2 proteinases (28, 30). The most basic bands (pI greater than 9) are the subtilisin-like Pr1 isoforms previously characterized (28). The apparent continuation of Pr1 activity at pH 3 may be attributed to the basic pH of the surrounding gel, as longer (2-min) incubations in buffer at pH 3, but not at pH 4, eliminated this band.
FIG. 1.
pH regulation of proteinase production by mycelium of M. anisopliae transferred for 7 h to medium containing cockroach cuticle and buffered at pH 3, 4, 5, 6, 7, or 8 (top axis). Concentrated culture filtrates were run on IEF gels (see Materials and Methods). To investigate the pH optimum of each proteinase band produced at each pH, gels were incubated with buffer at pH 2, 3, 4, 5, 6, 8, or 10 (horizontal axis) and the proteinases were detected by gelatin zymography.
FIG. 2.
The effects of inhibitors on proteinases produced by M. anisopliae in medium containing cockroach cuticle. Experiments were carried out with cultures buffered at pH 3, 4, 5, 6, 7, or 8 (top axis). Shown are proteinases produced at pHs 3 and 4 without inhibitors (A) and gels incubated with pepstatin (B), phosphoramidon (C), and leupeptin (D).
Enzyme production during growth on infected M. sexta cuticle.
To our knowledge, direct measurements of cuticle pH have not as yet been accomplished. However, extracts of uninfected and infected (60 h) cuticles had pH values of 6.3 ± 0.2 (n = 5) and 7.7 ± 0.3 (n = 5), respectively, confirming that utilization of cuticle components during infection is accompanied by a rise in pH. To determine whether the cytosolic pH of M. anisopliae affects the bulk pH of infected cuticles, intracellular pH was measured as a function of extracellular pH. At ambient pH values of 5, 7, and 8, cytosolic pH values were 6.1 ± 0.1, 6.4 ± 0.1, and 6.5 ± 0.1, respectively, indicating that the rise in pH of infected cuticle does not result from changes in fungal cytosolic pH.
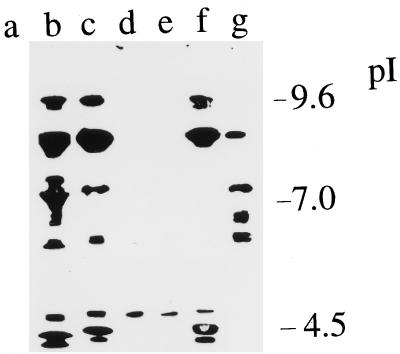
Extracts of infected cuticles were fractionated by analytical IEF (Fig. 3). Only one protease (pI ca. 4) could be detected in infected cuticles presoaked in 50 mM citric acid-sodium phosphate buffer (pH 3) (Fig. 3, lane d). This protease was not inhibited by pepstatin (Fig. 3, lane e) or phenanthroline but was inhibited by PMSF (an inhibitor of serine proteases) (Fig. 3, lane g). Extracts of unbuffered cuticles, or of cuticles buffered at pH 8 (50 mM HEPES), contained proteases with pI optima corresponding to the subtilisins, metalloproteases, and trypsins detected in culture (Fig. 3, lanes b and c). Consistent with this identification, the alkaline (pI above 8.5) and acidic bands were inhibited by PMSF (Fig. 3, lane g), and the neutral bands were inhibited by phenanthroline (Fig. 3, lane f).
FIG. 3.
Analytical IEF analysis of proteinases produced during penetration of M. sexta cuticle. Following 48 h of incubation, enzymes were extracted with 0.2 M potassium phosphate buffer. Shown are extracts from uninoculated cuticle (lane a), inoculated cuticle presoaked in water (lane b), inoculated cuticle presoaked in 0.1 M HEPES buffer (pH 8) (lane c), and inoculated cuticle presoaked in 50 mM citric acid-sodium phosphate buffer (pH 3) (lane d), extract from lane d treated with pepstatin for 10 min (lane e), extract from lane b incubated with 1 mM phenanthroline for 10 min (lane f), and extract from lane b incubated with PMSF for 10 min (lane g).
Expression patterns of the genes encoding secreted proteins.
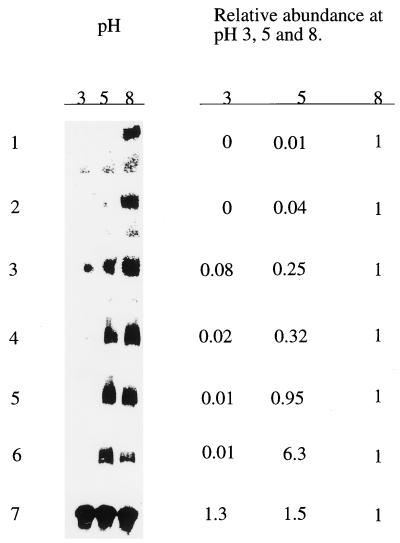
The pH-specific expression of transcripts encoding seven secreted proteins was analyzed by Northern blot analysis (Fig. 4). Exponentially growing mycelium was transferred to cuticle-containing cultures at pH 3.0, 5.0, or 8.0 and then grown for an additional 8 h. Total RNA was isolated from the harvested mycelium, blotted, and probed with cDNA clones. As a control we used the tubulin gene, which is expressed at similar levels at different pHs (Fig. 4). Pr1 mRNAs encoding the subtilisins Pr1a and Pr1b were most abundant at pH 8.0 and very much less at pH 5.0 and pH 3.0. Likewise, Pr2 and carboxypeptidase transcripts were most abundantly expressed at pH 8, but expression was also abundant at pH 5, with significant expression of Pr2 still detected at pH 3. Both hydrophobin and chitinase mRNAs were most abundantly expressed at pH 5 but were also produced at pH 8.
FIG. 4.
pH-specific expression of six genes encoding extracellular proteins. Total RNA (3 μg) from M. anisopliae grown on insect cuticle buffered at pH 3, 5, or 8 was subjected to Northern analysis using the complete cDNAs corresponding to Pr1a (row 1), Pr1b (row 2), Pr2 (row 3), carboxypeptidase (row 4), ssg12 (row 5), chitinase (row 6), and tubulin (row 7) as radiolabelled probes. Hybridization against tubulin mRNA was used to show the relative amounts of RNA present in each lane. The relative abundance was calculated by setting the value at pH 8.0 to 1.
DISCUSSION
In these experiments we studied the expression of several cuticle-degrading enzymes by M. anisopliae under strongly varying pH conditions. This study clarifies the situation with regard to the number and types of extracellular proteolytic enzymes produced by M. anisopliae. Analysis of pH regulation allowed us to identify aspartyl proteinases and several metalloproteinases, besides Pr1, Pr2, and the single metalloproteinase activities previously identified (28). Such analysis is important for the elucidation of the roles of these enzymes during infection and cuticle penetration.
Several hypotheses could explain the effects of pH on the extracellular enzyme activities of M. anisopliae, including transcription and translation activities, protein processing, enzyme stabilities, and the toxic effects of nonoptimal pH conditions on protein synthesis. M. anisopliae can grow over a wide pH range (pH 2.5 to 10.5) (6). The cytosolic pH was stable over the pH range 5 to 8, confirming efficient regulation of cytosolic pH and adaptation to survive in a broad array of environments. To facilitate nutritional versatility over a wide range of growth conditions, M. anisopliae produces several categories of proteases only at the pH values where they function effectively. In combination they enable exploitation of extracellular proteins over a pH range of 2 to 10. Stability measurements showed a gradual loss of Pr1 and Pr2 activities at pH values below 5 (22), so denaturation could be a factor in the low activities of these enzymes at pH 3. However, Northern analysis confirmed that the pH of the medium played a major role in gene expression: expression of both subtilisin genes and of Pr2 was turned off under acidic conditions. In contrast, a subtilisin-type serine protease produced by Aspergillus niger is expressed under native control at equally high levels at pHs 3 and 8, while the expression of aspartyl protease genes is completely turned off under alkaline conditions (7). When grown in the pH range 3 to 6, M. anisopliae produced three activities with acidic pH optima that were strongly inhibited by pepstatin, an inhibitor of aspartyl proteases. These activities were not detected in culture filtrates under alkaline conditions, indicating that regulation of these activities is similar to that of the extracellular aspartyl proteases of A. niger.
Only chitinase, with a pH optimum of 5 (28), showed maximal transcription of mRNAs at an acid pH; it is also expressed at pH 8 but not at pH 3. The pH within insect cuticle is unknown. However, growth over 60 h in isolated cuticles was accompanied by a rise in the pH of cuticular extracts from 6.3 to 7.7. Likewise, during growth by M. anisopliae in liquid culture containing cuticle, culture pH rises due to the release of ammoniacal by-products of cuticle protein degradation (25). Immunogold studies have shown that Pr1 and Pr2 are secreted by the fungus during cuticle penetration (5, 30). The pH-conditional expression of Pr1 and Pr2 observed in vitro suggests that the physiological pH in the infection site is alkaline. However, immunogold studies have shown that chitinase is secreted into the cuticle after the protease, during the later stages of infection (29). Limited production of chitinase at nonoptimal pH could be meaningful in terms of coordinated Pr1 and chitinase synthesis under alkaline conditions in infection sites. Pr1 production causes the degradation of cuticular proteins, exposing the chitin component of the cuticle and inducing the production of chitinase (18). The fact that chitinase is produced later and at nonoptimal pH supports other lines of evidence that chitinase plays a secondary role in infection processes compared with proteinases (18). Aspartyl proteases were not detected in extracts from cuticle, consistent with pH regulation of production, which suggests that this class of protease is not involved in the degradation of cuticle components. Hydrophobin was expressed at high levels over the pH range found in infection sites. The importance of this is that hydrophobins act as sensors of hydrophobic surfaces such as insect cuticles, which are conducive to infection (25, 34). Lack of transcription of hydrophobin at pH 3 is consistent with adaptation by M. anisopliae to being infectious at a neutral or basic pH.
These results indicate that regulation by pH is a general property of secreted M. anisopliae proteins. There is also evidence for a concerted action of pH and induction by cuticle on levels of enzyme production. Pr1 production is derepressed when the external pH is alkaline, even in the absence of cuticle. However, the presence of cuticle enhances Pr1 production threefold at pH 8 (Table 1), even though the inductive effects of cuticle do not override the negative effects on gene transcription of nonoptimum pH. The concerted action of induction by host cuticle and pH may provide a mechanism whereby environmental signals trigger the secretion of molecules capable of modifying the cuticle.
ACKNOWLEDGMENTS
This research was supported in part by the U.S. Department of Agriculture Competitive Research Grants Office (grant 9602033) and by a grant from the Park Foundation, Ithaca, N.Y.
REFERENCES
- 1.Caddick M X, Brownlee A G, Arst H N. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 2.Cooper R M, Wood R K S. Regulation of synthesis of cell wall degrading enzymes by Verticillium albo-atrum and Fusarium oxysporum f. sp. lycopersice. Physiol Plant Pathol. 1975;5:135–156. [Google Scholar]
- 3.Cordero R, Gaillardin C. Dominant mutations affecting expression of pH-regulated genes in Yarrowia lipolytica. Mol Gen Genet. 1996;252:311–319. doi: 10.1007/BF02173777. [DOI] [PubMed] [Google Scholar]
- 4.Cotty P J, Cleveland T E, Brown R L, Mellon J E. Variation in polygalacturonase production among Aspergillus flavus isolates. Appl Environ Microbiol. 1990;56:3885–3887. doi: 10.1128/aem.56.12.3885-3887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goettel M S, St. Leger R J, Rizzo N W, Staples R C, Roberts D W. Ultrastructural localization of a cuticle-degrading protease produced by the entomopathogenic fungus Metarhizium anisopliae during penetration of host (Manduca sexta) cuticle. J Gen Microbiol. 1989;135:2233–2239. [Google Scholar]
- 6.Hallsworth J E, Magan N. Culture age, temperature, and pH affect the polyol and trehalose contents of fungal propagules. Appl Environ Microbiol. 1996;62:2435–2442. doi: 10.1128/aem.62.7.2435-2442.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarai G, Buxton F. Nitrogen, carbon, and pH regulation of extracellular acidic proteases of Aspergillus niger. Curr Genet. 1994;26:238–244. doi: 10.1007/BF00309554. [DOI] [PubMed] [Google Scholar]
- 7a.Joshi, L., and R. J. St. Leger. Unpublished data.
- 8.Joshi L, St. Leger R J, Bidochka M J. Cloning of a cuticle-degrading protease from the entomopathogenic fungus, Beauveria bassiana. FEMS Microbiol Lett. 1995;125:211–218. doi: 10.1111/j.1574-6968.1995.tb07360.x. [DOI] [PubMed] [Google Scholar]
- 9.Joshi L, St. Leger R J, Roberts D W. Isolation of a cDNA encoding a novel subtilisin-like protease (Pr1B) from the entomopathogenic fungus, Metarhizium anisopliae using differential display-RT-PCR. Gene. 1997;197:1–8. doi: 10.1016/s0378-1119(97)00132-7. [DOI] [PubMed] [Google Scholar]
- 10.MacCabe A P, Johannes P T, van den Hombergh J T, Arst H N, Visser J. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol Gen Genet. 1996;250:367–374. doi: 10.1007/BF02174395. [DOI] [PubMed] [Google Scholar]
- 11.Mendgen K, Hahn M, Deising H. Morphogenesis and mechanisms of pathogenicity by plant pathogenic fungi. Annu Rev Phytopathol. 1996;34:367–386. doi: 10.1146/annurev.phyto.34.1.367. [DOI] [PubMed] [Google Scholar]
- 12.Monod, M., A. Fatih, K. Jaton-Ogay, S. Paris, and J. P. Latge. 1995. The secreted proteases of pathogenic species of Aspergillus and their possible role in virulence. Can. J. Bot. 73(Suppl. 1):S1087–S1091.
- 13.Murphy J M, Walton J D. Three extracellular proteases from Cochliobolus carbonum: cloning and targeted disruption of ALP1. Mol Plant-Microbe Interact. 1996;9:2290–2297. doi: 10.1094/mpmi-9-0290. [DOI] [PubMed] [Google Scholar]
- 14.Negrete-Urtasun S, Denison S H, Arst H N. Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paterson I C, Charnley A K, Cooper R M, Clarkson J M. Partial characterization of specific inducers of a cuticle-degrading protease from the insect pathogenic fungus Metarhizium anisopliae. Microbiology. 1994;140:3153–3159. doi: 10.1099/13500872-140-11-3153. [DOI] [PubMed] [Google Scholar]
- 16.Saporita-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smithson S L, Paterson I C, Bailey A M, Screen S E, Hunt B A, Cobb B D, Cooper R M, Charnley A K, Clarkson J M. Cloning and characterization of a gene encoding a cuticle-degrading protease from the insect pathogenic fungus Metarhizium anisopliae. Gene. 1995;166:161–165. doi: 10.1016/0378-1119(95)00609-3. [DOI] [PubMed] [Google Scholar]
- 18.St. Leger R J. Biology and mechanisms of invasion of deuteromycete fungal pathogens. In: Beckage N C, Thompson S N, Federici B A, editors. Parasites and pathogens of insects. Vol. 2. San Diego, Calif: Academic Press, Inc.; 1993. pp. 211–229. [Google Scholar]
- 19.St. Leger, R. J. 1995. The role of cuticle-degrading proteases in fungal pathogens of insects. Can. J. Bot. 73(Suppl. 1):S1119–S1125.
- 20.St. Leger R J, Cooper R M, Charnley A K. Cuticle-degrading enzymes of entomopathogenic fungi: synthesis in culture on cuticle. J Invertebr Pathol. 1986;48:85–95. [Google Scholar]
- 21.St. Leger R J, Cooper R M, Charnley A K. Cuticle-degrading enzymes of entomopathogenic fungi: regulation of production of chitinolytic enzymes. J Gen Microbiol. 1986;132:1509–1517. [Google Scholar]
- 22.St. Leger R J, Charnley A K, Cooper R M. Characterization of cuticle-degrading proteases by the entomopathogen Metarhizium anisopliae. Arch Biochem Biophys. 1987;253:221–232. doi: 10.1016/0003-9861(87)90655-2. [DOI] [PubMed] [Google Scholar]
- 23.St. Leger R J, Cooper R M, Charnley A K. Production of cuticle-degrading enzymes by the entomopathogen Metarhizium anisopliae during infection of cuticles from Calliphora vomitaria and Manduca sexta. J Gen Microbiol. 1987;133:1371–1382. [Google Scholar]
- 24.St. Leger R J, Durrands P K, Cooper R M, Charnley A K. Role of extracellular chymoelastase in the virulence of Metarhizium anisopliae for Manduca sexta. J Invertebr Pathol. 1988;52:285–293. [Google Scholar]
- 25.St. Leger R J, Butt T M, Staples R C, Roberts D W. Production in vitro of appressoria by the entomopathogenic fungus Metarhizium anisopliae. Exp Mycol. 1989;13:274–288. [Google Scholar]
- 26.St. Leger R J, Frank D C, Roberts D W, Staples R C. Molecular cloning and regulatory analysis of the cuticle-degrading protease structural gene from the entomopathogenic fungus Metarhizium anisopliae. Eur J Biochem. 1992;204:991–1001. doi: 10.1111/j.1432-1033.1992.tb16721.x. [DOI] [PubMed] [Google Scholar]
- 27.St. Leger R J, Staples R C, Roberts D W. Cloning and regulatory analysis of ssgA: a gene encoding a hydrophobin-like protein from the entomopathogenic fungus, Metarhizium anisopliae. Gene. 1992;120:119–124. doi: 10.1016/0378-1119(92)90019-l. [DOI] [PubMed] [Google Scholar]
- 28.St. Leger R J, Bidochka M J, Roberts D W. Isoforms of the cuticle-degrading Pr1 protease and production of a metalloproteinase by Metarhizium anisopliae. Arch Biochem Biophys. 1994;313:1–7. doi: 10.1006/abbi.1994.1350. [DOI] [PubMed] [Google Scholar]
- 29.St. Leger R J, Joshi L, Bidochka M J, Rizzo N W, Roberts D W. Characterization and ultrastructural localization of chitinases from Metarhizium anisopliae, M. flavoviride, and Beauveria bassiana during fungal invasion of host (Manduca sexta) cuticle. Appl Environ Microbiol. 1996;62:907–912. doi: 10.1128/aem.62.3.907-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St. Leger R J, Joshi L, Bidochka M J, Rizzo N W, Roberts D W. Biochemical characterization and ultrastructural localization of two extracellular trypsins produced by Metarhizium anisopliae in infected cuticles. Appl Environ Microbiol. 1996;62:1257–1264. doi: 10.1128/aem.62.4.1257-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. Leger R J, Joshi L, Bidochka M J, Roberts D W. Insecticidal properties of a genetically engineered entomopathogenic fungus constitutively expressing a toxic protease. Proc Natl Acad Sci USA. 1996;93:6349–6354. doi: 10.1073/pnas.93.13.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St. Leger R J, Bidochka M J. Insect-fungal interactions. In: Soderhall K, Vasta G, Iwanaga S, editors. Invertebrate immunology. Fair Haven, N.J: SOS Publications; 1996. pp. 441–478. [Google Scholar]
- 33.St. Leger R J, Joshi L, Roberts D W. Adaptation of proteases and carbohydrases of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology. 1997;143:1983–1992. doi: 10.1099/00221287-143-6-1983. [DOI] [PubMed] [Google Scholar]
- 34.Talbot N J. Fungal biology: growing into the air. Curr Biol. 1997;7:R78–R81. doi: 10.1016/s0960-9822(06)00041-8. [DOI] [PubMed] [Google Scholar]
- 35.Yamada T, Ogrydziak D M. Extracellular acid proteases produced by Saccharomycopsis lipolytica. J Bacteriol. 1983;154:23–31. doi: 10.1128/jb.154.1.23-31.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]