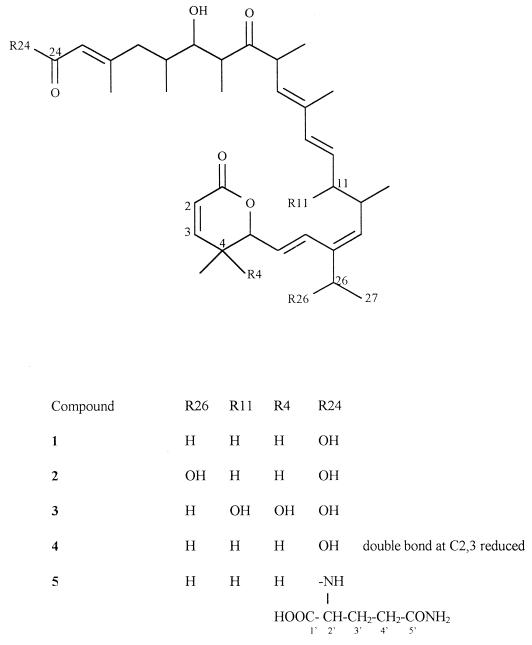
Abstract
Leptomycin B (LMB), a secondary metabolite produced by Streptomyces sp. strain ATS 1287, with known antifungal and antitumor effects, inhibits the nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 regulatory protein Rev and exhibits significant antiproliferative activity. Since LMB itself turned out to be distinctly cytotoxic, a bioconversion screening with a selected set of 29 bacterial and 72 fungal strains was performed in order to obtain metabolites of LMB with reduced antiproliferative effects. Several derivatives of LMB, more polar than the parent compound and produced in yields of >5%, were detected. Liquid chromatography-mass spectroscopy analysis indicated the type of bioconversion. Fermentations (1-liter scale) of those strains with high rates of transformation were suitable for isolation and characterization of the most prominent metabolites. Thus, bioconversion of LMB with Aspergillus flavus ATCC 9170 and Emericella unguis ATCC 13431 served for isolation of the novel derivatives 26-hydroxy-LMB (30% was the concentration of the metabolite [with respect to LMB] used for bioconversion) and LMB-24-glutaminamide (90%), respectively. Streptomyces rimosus ATCC 28893 converted LMB into 4,11-dihydroxy-LMB (13%) and 2,3-dihydro-LMB (55%). Although the antiproliferative effects of the LMB metabolites could be reduced through microbial conversion, none of these metabolites inhibited the nuclear export of Rev better than LMB itself.
Leptomycin B (LMB), produced by Streptomyces sp. strain ATS 1287, has attracted attention due to its antifungal (2–4) and antitumor (6, 7, 10, 18) effects. Recently it was found that, in addition to having antiproliferative activity, LMB inhibits the nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) regulatory protein Rev at low-nanomolar concentrations (16). Rev protein, which is responsible for the cytoplasmic accumulation of unspliced and singly spliced HIV-1 mRNA and thus for viral replication (1), must be translocated from the nucleus to the cytoplasm to exert its function (9, 15). Inhibitors of Rev protein translocation may, therefore, be useful for HIV therapy (16). However, due to its strong antiproliferative activity, LMB itself cannot be used therapeutically but, alternatively, may serve as a tool for dissecting nuclear transport pathways (16).
The aim of this study was to find LMB derivatives with reduced antiproliferative activities but preserved levels of inhibition of the nuclear export of Rev. Bioconversion reactions, such as hydroxylation and conjugation, are relevant detoxification reactions in living beings (14). Therefore, microbial conversion can serve as a tool to effect these useful derivation reactions with cellular enzyme equipment.
To identify novel metabolites of LMB, screening with a large variety of microorganisms selected on the basis of previous bioconversions was performed. Biotransformation products were detected by analyzing a combination of high-performance liquid chromatography (HPLC)-UV and liquid chromatography-mass spectroscopy (LC-MS) measurements. Fermentations (11) of those strains with high rates of transformation served for the isolation of derivatives, and their structures were elucidated mainly by nuclear magnetic resonance (NMR) techniques. All characterized metabolites of LMB were tested for their antiproliferative effects and inhibition of Rev translocation.
(A preliminary account of some of this work was presented at a poster session at the Biocat Screening ’96 Symposium, Ede, The Netherlands, 15 to 18 December 1996.)
MATERIALS AND METHODS
General experimental section.
For HPLC analysis, we used a model D-6000A interface; a model AS-2000A autosampler; a model L-6200A pump; a model L-4500 diode array detector; a model T-6300 column thermostat; software from Merck and Hitachi; a Lichrocart 125-4, Superspher 100, RP-18 column (4 μm); an RP-18 precolumn (5 μm); a linear gradient of 1% H3PO4–CH3CN (30 to 100% CH3CN) for 9 min, and a flow rate of 1.5 ml/min. Biotransformation products were detected at 220 nm by HPLC combined with diode array detection. For HPLC isolation, we used linear gradients of 0.098% H3PO4 (pH 4)–CH3CN (30 to 55% CH3CN) for 45 min and 0.098% H3PO4 (pH 4)–CH3CN (55 to 80% CH3CN) for 15 min, a Spherisorb RP-18 column (5 μm) (250 by 20 mm), UV detection at 220 nm, and a flow rate of 18 ml/min. To determine retention times for LC-MS, electrospray ionization MS was performed on a Finnigan TSQ 7000 triple-quadrupole MS. The instrument was controlled and data were analyzed with ICIS software (Finnigan). The electrostatic-spray ion source was operated at 4.5 kV, and the atmosphere-vacuum transfer capillary was heated at 220°C. The column effluent was split 5:1 with a Valco tee, allowing a flow rate of 0.05 ml/min into the electrospray nebulizer. Full-scan mass spectra were recorded from mass-to-charge ratios (m/z) of 500 to 1,400 in 1.8 s for the MS analysis of the derivatives. For LC analysis, we used a Spherisorb 5, RP-18 column (5 μm) and a gradient system with a column temperature of 50°C, UV detection at 220 nm, and a flow rate of 0.25 ml/min. All NMR spectra were recorded at 20°C on a Bruker AMX-400 spectrometer equipped with a 5-mm-diameter inverse triple-resonance probe. The spectra were acquired with CDCl3 by using trimethylsilyl (TMS) as the internal reference. Resonance assignments were obtained from a series of homonuclear and heteronuclear two-dimensional experiments. Chemical shifts are given in the δ scale; J values are given in hertz. MS spectra were recorded on an MS, model VC 70-SE (positive fast atom bombardment [FAB+]; Xe, 8 keV), with nitrobenzylic alcohol plus LiI as the matrix and an acceleration voltage of 8 kV.
Microorganisms and bioconversion conditions.
Strains (72 fungi, 29 bacteria) were purchased from the American Type Culture Collection, the National Collection of Industrial and Marine Bacteria, the Northern Utilization Research and Development Division, and the Centraalbureau voor Schimmelcultures.
Strains were stored on agar slants at −25°C. Spores and cells of one agar culture were suspended in 10 ml of 0.9% NaCl. Two-hundred-microliter Erlenmeyer flasks each containing 50 ml of seed culture medium (see below) were inoculated with 2 ml of these suspensions and incubated on a rotary shaker (200 rpm) at 24°C (fungi) or 27°C (bacteria) for 4 days. One-hundred-microliter Erlenmeyer flasks each containing 25 ml of main culture medium (see below) were inoculated with 2.5 ml of the seed culture and incubated on a rotary shaker (200 rpm) at 24°C. After 24 h, the parent compound dissolved in methanol (MeOH) was added to a final concentration of 50 mg/liter to the main cultures, which were harvested after 48, 72, or 96 h of incubation. All controls were treated under the same fermentation conditions as described for the bioconversion samples. Compound controls containing 50 mg of LMB/liter of main culture medium and strain controls lacking LMB were prepared. To stabilize LMB, main culture media that were adjusted to pHs of 2, 3, 4, 5, 6, 7, 8, 9, and 10 and contained 50 mg of LMB/liter of medium were prepared and treated as described above for the compound controls.
Compositions of the liquid seed media.
For bacteria, we used medium 231, which consists of 0.005% CaCO3, 0.7% glucose, 0.45% yeast extract (Gistex), 0.5% malt extract liquid (Wander), 1.0% soluble starch, 0.25% N-Z-Amine type A (Sheffield), and 0.1% (vol/vol) trace element solution a (pH 7.0). Trace element solution a contains 0.01% H3BO3, 0.5% FeSO4 · 7H2O, 0.005% KI, 0.2% CoCl2 · 6H2O, 0.02% CuSO4 · 5H2O, 0.2% MnCl2 · 4H2O, 0.4% ZnSO4 · 7H2O, and 0.1% (vol/vol) H2SO4 (97%). For fungi, we used medium SA, which consists of 0.1% Bacto Agar, 0.4% yeast extract (Gistex), and 2.0% malt extract liquid (Wander) (pH 5.0 to 5.5), and medium SB, which consists of 0.75% soy protein (Siber & Hegner), 2.0% dextrose (Difco), 0.1% malt extract liquid (Wander), 0.1% brewer’s yeast (Cenovis), 0.05% KH2PO4, 0.005% MgSO4 · 7H2O, 0.002% CaCl2 · 2H2O, 0.001% NaCl, 0.1% (vol/vol) trace element solution b, and 0.1% Bacto Agar (pH 6.0 to 6.2). Trace element solution b consists of 0.003% Na2MoO4 · 2H2O, 0.44% ZnSO4 · 7H2O, 0.55% FeSO4 · 7H2O, 0.008% CuSO4 · 5H2O, 0.018% MnCl2 · 4H2O, and 0.2% (vol/vol) H2SO4 (97%).
Compositions of the main media.
For bacteria, we used medium Act1, which consists of 0.5% glucose, 1.5% soluble starch, 1.0% N-Z-Amine type A (Sheffield), 0.2% brewer’s yeast (Cenovis), 0.06% K2HPO4–0.003% KH2PO4, 0.01% MgSO4 · 7H2O, 0.01% CaCl2 · 2H2O, 0.005% NaCl, and 0.1% (vol/vol) trace element solution a (pH 6.2 to 6.5), and medium Act3, which consists of 0.5% Bacto Tryptone, 0.3% Bacto yeast extract, and 1.0% glucose (pH 7.1). For fungi, we used medium MA, which consists of 2.0% glucose, 0.2% soy protein (Siber & Hegner) 0.2% malt extract liquid (Wander), 0.2% yeast extract (Gistex), 0.2% KH2PO4, 0.05% MgSO4 · 7H2O, 0.1% (vol/vol) trace element solution b, and 0.1% Bacto Agar (pH 5.1 to 5.4), medium MB, which consists of 1.0% soy protein (Siber & Hegner), 3.0% dextrose (Difco), 0.2% malt extract (Difco), 0.2% brewer’s yeast (Cenovis), 0.075% KH2PO4, 0.001% MgSO4 · 7H2O, 0.005% CaCl2 · 2H2O, 0.002% NaCl, and 0.15% (vol/vol) trace element solution b (pH 6.0 to 6.2), and medium MC, which consists of 0.6%, (NH4)2HPO4, 0.3% KH2PO4, 0.001% NaCl, 0.01% MgSO4 · 7H2O, 0.002% CaCl2 · 6H2O, and 0.05% (vol/vol) trace element solution b (pH 7.0 to 7.2). For agar slants, we used medium 330, which consists of 1.8% Bacto Agar, (vol/vol) 0.02% trace element solution a, 0.2% yeast extract (Bacto), and 1.0% soluble starch (pH 7.0).
Extraction and HPLC analysis.
Ethyl acetate (EtOAc; 20 ml) was added to 20 ml of the main culture, shaken for 20 min at 24°C at 200 rpm, and centrifuged at 4,500 rpm for 10 min. The EtOAc layer (15 ml) was evaporated to dryness and redissolved in 1.5 ml of MeOH. These extracts (10 μl) were then chromatographed by HPLC (for the conditions, see “General experimental section”).
Isolation of the LMB biotransformation products.
One-liter culture broths of the strains ATCC 9170, ATCC 28893, ATCC 13431, and ATCC 55060, to which 50 mg of LMB/liter was added, were filtered, and the filtrates were extracted three times with EtOAc (700, 500, and 400 ml), washed with demineralized water, and dried with Na2SO4. These extracts were each separated in one step by preparative HPLC (for the conditions, see “General experimental section”). Fractions (18 ml) were monitored for the desired leptomycin derivatives by analytical HPLC (for conditions, see “General experimental section”). The enriched fractions were extracted with equal volumes of EtOAc, and the extracts were dried under vaccuum. The extracts of the culture broths of ATCC 9170 (75 mg), ATCC 28893 (109.8 mg), and ATCC 13431 (81.3 mg) yielded compounds 2 (7 mg), 3 (6 mg), 4 (12 mg), and 5 (17 mg).
26-Hydroxy-LMB (compound 2).
UV (MeOH), c = 0.1 g/liter; λmax (in nanometers), 225. Infrared (IR) (film) ν max (per centimeter), 3,352, 2,966, 2,930, 1,707, 1,644, 1,455, 1,374, 1,252, 1,100, 1,046, 968, 885, 825, 736, 703. FAB-MS (positive mode), [M+Li-H]Li+, 569 (m/z 86); [M+Li]+, 563 (m/z 100), 551 (m/z 16), 425 (m/z 42), 413 (m/z 32), 397 (m/z 53), 377 (m/z 35). For 1H-NMR (CDCl3), see Table 2.
TABLE 2.
1H NMR data of conversion products 2 to 4 of LMB
| Hydrogen |
1H NMR data for LMB conversion producta:
|
||||
|---|---|---|---|---|---|
| 1(3) | 2 | 3 | 4 | 4 (log 1H-1H COSY)b | |
| H-2 | 6.0 (1H, d, J = 9.7) | 6.0 (1H, d, J = 9.8) | 5.95 (1H, d, J = 10.5) | 2.61 (2H, m) | H-3a, H-3b |
| H-3a | 6.95 (1H, dd, J = 9.7, 6.1) | 6.95 (1H, dd, J = 9.9, 5.5) | 6.85 (1H, d, J = 10.5) | 1.69 (1H, m) | H-2, H-3b, H-4 |
| H-3b | 1.98 (1H, m) | H-2, H-3a, H-4 | |||
| H-4 | 2.53 (1H, m, J = 6.8, 6.8, 6.1) | 2.55 (1H, m, J = 5.0, 5.5, 6.6) | 2.15 (1H, m) | H-3a, H-3b, H-5, H-25 | |
| H-5 | 5.0 (1H, dd, 7.4, 6.8) | 5.0 (1H, dd, 5.0, 7.2) | 4.85 (1H, d, J = 6.6) | 4.95 (1H, dd, 5.0, 7.2) | H-4, H-6 |
| H-6 | 5.72 (1H, dd, J = 15.6, 7.4) | 5.9 (1H, dd, J = 15.9, 7.2) | 5.79 (1H, dd, J = 15.4, 6.6) | 5.72 (1H, dd, J = 15.9, 7.2) | H-5, H-7 |
| H-7 | 6.65 (1H, d, J = 15.6) | 6.6 (1H, d, J = 15.9) | 6.78 (1H, d, J = 15.4) | 6.52 (1H, d, J = 15.9) | H-6 |
| H-9 | 5.23 (1H, d, J = 10.4) | 5.54 (1H, d, J = 10.5) | 5.36 (1H, d, J = 10.5) | 5.2 (1H, d, J = 10.5) | H-10 |
| H-10 | 2.67 (1H, m, J = 10.4, 7.4, 6.2) | 2.68 (1H, m, J = 10.5, 7.2, 6.1) | 2.83 (1H, m, J = 10.5, 7.2, 6.6) | 2.68 (1H, m, J = 10.5, 7.2, 6.1) | H-9, H-11, H-28 |
| H-11 | 2.09 (2H, t, J = 7.4) | 2.11 (2H, t, J = 7.2) | 3.9 (1H, dd, J = 8.3, 6.6) | 2.09 (2H, t, J = 7.2) | H-10, H-12 |
| H-12 | 5.59 (1H, m, J = 15.5, 7.5) | 5.6 (1H, m, J = 15.4, 7.2) | 5.67 (1H, dd, J = 14.9, 6.6) | 5.59 (1H, m, J = 15.4, 7.2) | H-11, H-13 |
| H-13 | 6.0 (1H, d, J = 15.5) | 6.1 (1H, d, J = 14.9) | 6.2 (1H, d, J = 14.9) | 6.02 (1H, d, J = 14.9) | H-11, H-12 |
| H-15 | 5.08 (1H, d, J = 10.4) | 5.12 (1H, d, J = 10.5) | 5.23 (1H, d, J = 10.5) | 5.1 (1H, d, J = 10.5) | H-16, H-29 |
| H-16 | 3.67 (1H, m, J = 10.4, 6.2) | 3.65 (1H, m, J = 10.5, 6.1) | 3.65 (1H, m, J = 10.5, 6.1) | 3.67 (1H, m, J = 10.5, 6.1) | H-15, H-30 |
| H-18 | 2.83 (1H, m, J = 7.2, 6.8) | 2.83 (1H, m, J = 7.2, 7.1) | 2.78 (1H, m, J = 7.2, 7.1) | 2.83 (1H, m, J = 7.2, 7.1) | H-19, H-31 |
| H-19 | 3.58 (1H, t, J = 6.8, 5.7) | 3.59 (1H, dd, J = 5.7, 7.1) | 3.59 (1H, dd, J = 5.7, 7.1) | 3.6 (1H, dd, J = 5.7, 7.1) | H-18, H-20 |
| H-20 | 1.75 (1H, m, J = 8.5, 7.2, 5.7) | 1.75 (1H, m, J = 7.2, 8.5, 7.2) | 1.68 (1H, m, J = 7.2, 8.5, 7.2) | 1.7 (1H, m, J = 7.2, 8.5, 7.2) | H-19, H-21a, H-21b, H-32 |
| H-21a | 1.9 (1H, dd, 12.5, 8.5) | 1.9 (1H, dd, J = 12.5, 8.5) | 1.9 (1H, dd, J = 12.5, 8.5) | 1.92 (1H, dd, J = 12.5, 8.5) | H-20, H-21b |
| H-21b | 2.21 (1H, dd, 12.5, 8.5) | 2.21 (1H, dd, J = 12.5, 8.5) | 2.20 (1H, dd, J = 12.5, 8.5) | 2.2 (1H, dd, J = 12.5, 8.5) | H-20, H-21a, H-23 |
| H-23 | 5.68 (1H, s) | 5.69 (1H, s) | 5.67 (1H, s) | 5.7 (1H, s) | H-21b |
| H-25 | 1.07 (3H, d, J = 6.8) | 1.08 (3H, d, J = 6.6) | 1.34 (3H, s) | 1.05 (3H, d, J = 6.6) | H-4 |
| H-26 | 2.20 (2H, m, J = 7.4) | 4.58 (1H, q, J = 5.5) | 2.3 (2H, m, J = 7.4) | 2.20 (2H, m, J = 7.4) | H-27 |
| H-27 | 1.05 (3H, t, J = 7.4) | 1.36 (3H, d, J = 5.5) | 1.1 (3H, t, J = 7.4) | 1.05 (3H, t, J = 7.4) | H-26 |
| H-28 | 0.97 (3H, d, J = 6.2) | 0.99 (3H, d, J = 6.1) | 0.99 (3H, d, J = 6.1) | 0.97 (3H, d, J = 6.1) | H-10 |
| H-29 | 1.82 (3H, s) | 1.82 (3H, s) | 1.82 (3H, s) | 1.82 (3H, s) | |
| H-30 | 1.14 (3H, d, J = 6.2) | 1.14 (3H, d, J = 6.1) | 1.14 (3H, d, J = 6.1) | 1.14 (3H, d, J = 6.1) | H-16 |
| H-31 | 1.15 (3H, d, J = 7.2) | 1.15 (3H, d, J = 7.2) | 1.15 (3H, d, J = 7.2) | 1.15 (3H, d, J = 7.2) | H-18 |
| H-32 | 0.79 (3H, d, J = 7.2) | 0.82 (3H, d, J = 7.2) | 0.82 (3H, d, J = 7.2) | 0.8 (3H, d, J = 7.2) | H-20 |
| H-33 | 2.13 (3H, s) | 2.1 (3H, s) | 2.11 (3H, s) | 2.12 (3H, s) | H-23 |
For 1H NMR analysis (400 MHz), δH values are relative to those of TMS and CDCl3. δH values are in parts per million, and J values are in hertz. Signals set in boldface type differ significantly from those of LMB (δH and/or J).
1H-1H COSY data are cross-peaks between signals.
4,11-Dihydroxy-LMB (compound 3).
UV (MeOH), c = 0.1 g/liter; λmax (in nanometers), 227. IR (film) ν max (per centimeter), 3,350, 2,963, 2,928, 1,708, 1,456, 1,376, 1,260, 1,099, 969, 870, 823, 736. FAB-MS (positive mode), [M+Li-H]Li+, 585 (m/z 63); [M+Li]+, 579 (m/z 47), 473 (m/z 24), 466 (m/z 100), 460 (m/z 27), 447 (m/z 72), 398 (m/z 35), 355 (m/z 35), 328 (m/z 82). For 1H-NMR (CDCl3), see Table 2.
2,3-Dihydro-LMB (compound 4).
IR (film) ν max (per centimeter), 2,964, 2,930, 1,706, 1,641, 1,455, 1,375, 1,250, 1,136, 966, 867, 702, 636, 620. FAB-MS (positive mode), [M+Li-H]Li+, 555 (m/z 83); [M+Li]+, 549 (m/z 63), 417 (m/z 22), 405 (m/z 31), 399 (m/z 100), 393 (m/z 34), 333 (m/z 20). For 1H-NMR and 1H-1H-COSY (CDCl3), see Table 2.
LMB-24-glutaminamide (compound 5).
IR (film) ν max (per centimeter), 3,348, 2,966, 2,931, 1,708, 1,663, 1,528, 1,455, 1,374, 1,286, 1,248, 1,480, 1,101, 1,046, 970, 825. FAB-MS (positive mode), [M+Li-H]Li+, 681, (m/z 100), [M+Li]+, 675 (m/z 76), 629 (m/z 11), 593, (m/z 11), 572, (m/z 12), 419 (m/z 13), 397 (m/z 27). For 1H-NMR, InvHCCORR, and 1H-1H COSY (CDCl3), see Table 3.
TABLE 3.
NMR data of LMB conversion product 5
| Position | NMR result(s)
|
||
|---|---|---|---|
| δHa | InvHCCORR (δc)b | 1H-1H COSYc | |
| H-C-2 | 6.0 (1H, d, J = 9.8) | 135.1 | H-3 |
| H-C-3 | 6.95 (1H, dd, J = 9.9, 5.5) | 151.4 | H-2, H-4 |
| H-C-4 | 2.55 (1H, m, J = 4.9, 5.5, 6.6) | 33.4 | H-3, H-5, H-25 |
| H-C-5 | 5.01 (1H, dd, J = 4.9, 7.2) | 81.4 | H-4, H-6 |
| H-C-6 | 5.72 (1H, dd, J = 15.9, 7.2) | 122.4 | H-5, H-7 |
| H-C-7 | 6.65 (1H, d, J = 15.9) | 130.0 | H-6 |
| H-C-9 | 5.23 (1H, d, J = 10.5) | 136.8 | H-10 |
| H-C-10 | 2.68 (1H, m, J = 10.5, 7.2, 6.1) | 31.7 | H-9, H-11, H-28 |
| H-C-11 | 2.09 (2H, t, J = 7.2) | 40.7 | H-10, H-12 |
| H-C-12 | 5.59 (1H, m, J = 15.4, 7.2) | 127.5 | H-11, H-13 |
| H-C-13 | 6.0 (1H, d, J = 14.9) | 120.0 | H-12 |
| H-C-15 | 5.07 (1H, d, J = 10.5) | 127.8 | H-16, H-29 long range |
| H-C-16 | 3.67 (1H, m, J = 10.5, 6.1) | 45.4 | H-15, H-30 |
| H-C-18 | 2.83 (1H, m, J = 7.2, 7.1) | 47.3 | H-19, H-31 |
| H-C-19 | 3.6 (1H, dd, J = 5.7, 7.2) | 73.1 | H-18 |
| H-C-20 | 1.68 (1H, m, J = 7.2, 8.5, 7.1) | 33.1 | H-19, H-21a, H-21b, H-32 |
| Ha-C-21 | 1.9 (1H, dd, J = 12.5, 8.5) | 45.4 | H-21b, H-20 |
| Hb-21 | 2.2 (1H, dd, J = 12.5, 8.5) | H-21a, H-20 | |
| H-C-23 | 5.7 (1H, s) | 119.8 | H-33 long range |
| H-C-25 | 1.05 (3H, d, J = 6.6) | 12.0 | H-4 |
| H-C-26 | 2.2 (2H, m, J = 7.4) | 26.1 | H-27 |
| H-C-27 | 1.05 (3H, t, J = 7.4) | 13.4 | H-26 |
| H-C-28 | 0.97 (3H, d, J = 6.1) | 20.7 | H-10 |
| H-C-29 | 1.82 (3H, s) | 13.1 | H-15 long range |
| H-C-30 | 1.14 (3H, d, J = 6.1) | 16.1 | H-16 |
| H-C-31 | 1.15 (3H, d, J = 7.2) | 13.7 | H-18 |
| H-C-32 | 0.76 (3H, d, J = 7.2) | 13.0 | H-20 |
| H-C-33 | 2.07 (3H, s) | 17.8 | H-23 long range |
| H-N-1′ | 7.3 (1H, br*) | H-2′ | |
| H-C-2′ | 4.5 (1H, br*, m) | 52.0 | H-1′, H-3a′ |
| Ha-C-3′ | 2.05 (1H, br*, m) | 27.1 | H-2, H-4 |
| Hb-3′ | 2.19 (1H, br*, m) | H-4 | |
| H-C-4′ | 2.39 (2H, br*, m) | 31.7 | H-3a′, H-3b′ |
| CO-NH2 | 6.45 (1H, br*) | ||
| 6.98 (1H, br*) | |||
For 1H NMR analysis (400 MHz), δH values are relative to those of TMS and CDCl3. δH values are in parts per million, and J values are in hertz. *, the broad signal (br) is due to rotation isomeric effects.
For InvHCCORR (400 MHz, 100 MHz) analysis, δH and δC values are relative to those of TMS and CDCl3.
1H-1H COSY data are cross-peaks between signals.
Biological assays.
Inhibition of Rev translocation and antiproliferative activity of LMB metabolites were determined by means of the Rev translocation assay (RTA) (16) and the sulforhodamine B staining assay for cellular protein (13), respectively.
RESULTS AND DISCUSSION
In the course of a screening of 29 bacterial and 72 fungal strains, various derivatives of LMB were detected (Table 1). Stability experiments with LMB under those fermentation conditions chosen for bioconversion cultures showed unambiguously that LMB remains stable from pH 4 to 8. As a consequence of this and with respect to strain and compound controls, which were treated exactly like the biotransformation samples, all products described in this paper are bioconversion metabolites.
TABLE 1.
Strains converting LMB
| Compound | Relative retention αa | Amt of derivative (%)b | Bacterium | Fungus | Medium | Incubation time (days) |
|---|---|---|---|---|---|---|
| 26-Hydroxy-LMB (2) | 7.06 | 12 | Streptomyces aureofaciens ATCC 13304 | Act3 | 2 | |
| 25 | Saccharopolyspora erythraea ATCC 11635 | Act1 | 2 | |||
| 30 | Aspergillus flavus ATCC 9170 | MB | 3 | |||
| 30 | Aspergillus fischeri ATCC 1020 | MB | 3 | |||
| 4,11-Dihydroxy-LMB (3) | 5.19 | 2 | Mucor hiemalis ATCC 20095 | MB | 4 | |
| 13 | Streptomyces rimosus ATCC 28893 | MC | 2 | |||
| 2,3-Dihydro-LMB (4) | 8.24 | 8 | Gliocladium catenulatum ATCC 10523 | MB | 4 | |
| 10 | Aspergillus alliaceus ATCC 10060 | MB | 4 | |||
| 55 | Streptomyces rimosus ATCC 28893 | Act1 | 2 | |||
| LMB-24-glutaminamide (5) | 8.01 | 2 | Streptomyces aureofaciens ATCC 13189 | Act1 | 2 | |
| 90 | Emericella unguis ATCC 13431 | MB | 3 | |||
| LMB (1) | 10 |
Relative retention α (HPLC) = Rt (metabolite) − Rt (mobile phase)/Rt (LMB) − Rt (mobile phase) × 10, where Rt is the retention time. For HPLC conditions (analysis), see “General experimental section.”
Area under the curve of the derivative × 100/area under the curve of LMB in the compound control.
One-liter fermentations of strains exhibiting high rates of transformation served for the isolation of the conversion derivatives 2 to 5 by preparative HPLC (Fig. 1).
FIG. 1.
LMB and conversion products.
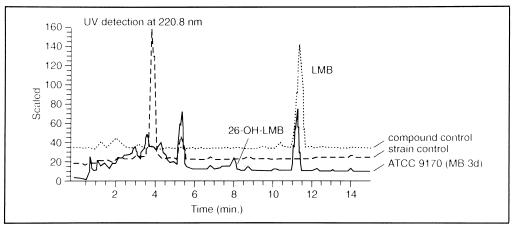
Through the screening, Streptomyces aureofaciens ATCC 13304 (5), Saccharopolyspora erythraea ATCC 11635 (12), Aspergillus fischeri ATCC 1020 (5), and Aspergillus flavus ATCC 9170 (5) were found to convert LMB into metabolite 2 (Table 1). Among these strains Aspergillus flavus ATCC 9170, an already known bioconversion strain exhibiting hydrolase activity (5), which produced predominantly metabolite 2 (Fig. 2), served for its isolation. The similarity between the UV spectrum of metabolite 2 and that of the parent compound confirmed the detection of an LMB derivative (Fig. 3).
FIG. 2.
HPLC chromatogram of LMB bioconversion with Aspergillus flavus ATCC 9170 (with medium MB and an incubation time of 3 days [MB/3d]).
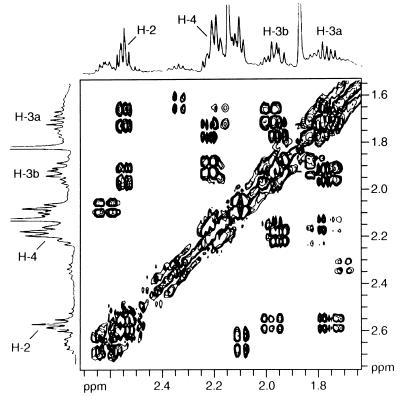
FIG. 3.
1H-1H COSY spectrum of 2,3-dihydro-LMB (compound 4) in CDCl3.
An m/z of 556 for compound 2 indicated monohydroxylation of LMB. Whereas most of the signals in the 1H-NMR analysis of compound 2 appeared almost identical to those of compound 1, a quadruplet of a single proton at δ 4.58 ppm was observed instead of the multiplet of the two H-26s at δ 2.2 ppm in the spectrum of compound 1 (Table 2). The loss of one proton at C-26 and the simultaneous significant downfield shift of the remaining H-26 is a strong hint for the introduction of a hydroxyl group. Concomitantly, the signal of the neighboring 27-CH3 changed from a triplet at δ 1.06 ppm in the spectrum of compound 1 to a doublet at δ 1.36 ppm, supporting this assumption. By these facts, compound 2 was identified as the novel 26-hydroxy-LMB (Fig. 1).
Streptomyces rimosus ATCC 28893 and Mucor hiemalis ATCC 20095, strains already known for their oxidation capabilities (8, 17), transformed compound 1 into the polar metabolite 3. An m/z of 572 indicated a dihydroxylation of compound 1. The 1H-NMR spectrum of compound 3 lacked the signal of H-4, and in contrast to the spectrum of compound 1 (4-CH3 δ = 0.96 ppm, d), the signal of the 4-CH3 appeared as a singlet at δ 1.34 ppm. This result indicated the presence of a hydroxyl group at C-4. The second hydroxyl group is located at C-11, since a multiplet of a single proton at δ 4.58 ppm was observed instead of the multiplet of the two protons of C-11 at δ 2.03 ppm (Table 2). Based on the evaluation of the 1H-NMR data, compound 3 was characterized as 4,11-dihydroxy-LMB (Fig. 1), a new metabolite. However, the relative stereochemistries of the newly generated hydroxyl groups bearing chiral centers in metabolites 2 and 3 were not determined.
Another metabolite of Streptomyces rimosus ATCC 28893 is the novel 2,3-dihydro-LMB (compound 4), which differs in mass from compound 1 by 2 U. This metabolite was also detected through bioconversion with Gliocladium catenulatum ATCC 10523 and Aspergillus alliaceus ATCC 10060. In the 1H NMR spectrum of compound 4, reduction of the olefin was clearly demonstrated by the disappearance of the significant olefinic signals of H-2 at δ 6.00 ppm and H-3 at δ 6.95 ppm and the appearance of H-2 and H-3 proton signals in a range typical for saturated hydrocarbons (H-2, [2H] δ 2.61 ppm, m; H-3a, [1H] δ 1.69 ppm, m; H-3b, [1H] δ 1.98 ppm, m). The assignment of the H-2 and H-3 protons was achieved by 1H-1H COSY (Fig. 3), showing a coupling of the H-3a and -3b with the H-2 (a and b) and the H-4. Based on these data, compound 4 was identified as the novel 2,3-dihydro-LMB (Fig. 1).
Finally, Emericella unguis ATCC 13431 (5) and Streptomyces aureofaciens ATCC 13189 (5) produced metabolite 5, an enzymatic conjugation product. First hints were obtained from the MS spectrum, which showed an increase in mass of 128 U with respect to that of compound 1. The proton signal pattern typical for LMB remained unchanged in the 1H NMR spectrum of compound 5, but additional signals were observed (Table 3). 1H-1H COSY (Table 3) and HCCORR spectra (Table 3) revealed that compound 5 is an amide formed by condensation of the free carboxyl group of LMB with the amino group of glutamine. The new broad signal at δ 7.28 ppm could be assigned to the amide proton, which showed a prominent coupling with the new H-2′. However, the relative stereochemistry of the chiral C-2′ in the new glutaminamide was not determined. According to these data, compound 5 was identified as LMB-24-glutaminamide.
Biological data.
The microbial transformation metabolites of LMB were evaluated in an RTA (16) and a 72-h proliferation assay with HeLa-Rev cells (16) (Table 4). Different types of enzymatic modifications (hydroxylation, reduction, and conjugation) at different sites of the molecule, achieved by microbial conversion, led to decreases in activities in both assays with respect to that of LMB. The loss of RTA activity was most significant for metabolite 4, indicating that the enone system has to be preserved for the inhibition of Rev translocation. All compounds described showed a close correlation in their activities in both the RTA and the proliferation assay. So far no derivative, either chemically (11) or enzymatically modified, with a therapeutic window between Rev translocation and cytotoxicity has been found.
TABLE 4.
Biological data of LMB and conversion products
| Compound | RTA activity [nM]a | IC50 proliferation [nM]b |
|---|---|---|
| LMB (1) | 0.8 | 0.1 |
| 26-Hydroxy-LMB (2) | 10.0 | 13.0 |
| 4,11-Dihydroxy-LMB (3) | 100.0 | 186.0 |
| 2,3-Dihydro-LMB (4) | 1,000.0 | 620.0 |
| LMB-24-glutaminamide (5) | 100.0 | 169.0 |
Minimum inhibitory concentration in an RTA (HeLa-Rev cells) determined after 7 h of incubation.
Fifty percent inhibitory concentration (IC50) in a cell proliferation assay with HeLa-Rev cells determined after 72 h of incubation by staining cellular protein with sulforhodamine B.
Microbial conversions can serve as models to mimic the mammalian metabolism (14). Drug metabolism in mammals, which is divided into phase I (i.e., hydroxylation) and phase II (i.e., conjugation) reactions, is associated with inactivation, activation, or termination of the biological activities of drugs (14). This may explain the reduced effects of the transformed derivatives on both cell proliferation and Rev nuclear export. On the other hand, the higher polarities of these derivatives may be advantageous in some cases for improved pharmacokinetic attributes. Furthermore, hydroxylated derivatives, which are more easily obtainable by bioconversion than by chemical methods, may serve as starting materials for the synthesis of new products.
ACKNOWLEDGMENTS
We thank L. Gschwind and M. Uhl for technical support and for preparing the large-scale fermentations and E. Bürgin for the LC-MS measurements. We are grateful to H. U. Naegeli for his advice in isolation of metabolites and to H. U. Gremlich and J. France for their help in the elucidation of the structures of the described bioconversion products. We finally thank E. Schreiner and M. Grassberger for critically reviewing the manuscript.
REFERENCES
- 1.Arya S, Guo C, Josephs S F, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III) Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- 2.Hamamoto T, Gunji S, Tsuji H, Beppu T. Leptomycins A and B, new antifungal antibiotics. I. Taxonomy of the producing strain and their fermentation, purification and characterization. J Antibiot. 1983;36:639–645. doi: 10.7164/antibiotics.36.639. [DOI] [PubMed] [Google Scholar]
- 3.Hamamoto T, Seto H, Beppu T. Leptomycins A and B, new antifungal antibiotics. II. Structure elucidation. J Antibiot. 1983;36:646–650. doi: 10.7164/antibiotics.36.646. [DOI] [PubMed] [Google Scholar]
- 4.Hamamoto T, Uozumi T, Beppu T. Leptomycins A and B, new antifungal antibiotics. III. Mode of action of Leptomycin B on Schizosaccharomyces pombe. J Antibiot. 1985;38:1573–1580. doi: 10.7164/antibiotics.38.1573. [DOI] [PubMed] [Google Scholar]
- 5.Kieslich K. Classification of the transformations according to reaction type. In: Kieslich K, editor. Microbial transformations of non-steroid cyclic compounds. Stuttgart, Germany: Georg Thieme Publishers; 1976. pp. 291–793. [Google Scholar]
- 6.Komiyama K, Okada K, Oka H, Tomisaka S, Miyano T, Funayama S, Umezawa T. Structural study of a new antitumor antibiotic, Kazusamycin. J Antibiot. 1985;38:220–223. doi: 10.7164/antibiotics.38.220. [DOI] [PubMed] [Google Scholar]
- 7.Komiyama K, Okada K, Hirokawa Y, Masuda K, Tomisaka S, Umezawa I. Antitumor activity of a new antibiotic, Kazusamycin. J Antibiot. 1985;38:224–230. doi: 10.7164/antibiotics.38.224. [DOI] [PubMed] [Google Scholar]
- 8.Kuhnt M, Bitsch F, France J, Hofmann H, Sanglier J J, Traber R. Microbial biotransformation products of cyclosporin A. J Antibiot. 1996;49:781–787. doi: 10.7164/antibiotics.49.781. [DOI] [PubMed] [Google Scholar]
- 9.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1548. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 10.Roberts B J, Hamelehle K L, Sebolt J S, Leopold W R. In vivo and in vitro anticancer activity of the structurally novel and highly potent antibiotic CI-940 and its hydroxy analog (PD 114,721) Cancer Chemother Pharmacol. 1986;16:95–101. doi: 10.1007/BF00256156. [DOI] [PubMed] [Google Scholar]
- 11.Schreiner, E., and B. Wolff. Unpublished data.
- 12.Schulman M, Doherty P, Zink D, Arison B. Microbial conversion of avermectins by Saccharopolyspora erythraea: hydroxylation at C28. J Antibiot. 1993;46:1016–1019. doi: 10.7164/antibiotics.46.1016. [DOI] [PubMed] [Google Scholar]
- 13.Skehan P, Storeng R, Scudiero D, Monks A, McMahan J, Vistica D, Warren J T, Bokesch H, Kenney S, Boyd M R. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 14.Smith R V, Rosazza J P. Microbial transformations as a means of preparing mammalian drug metabolites. In: Rosazza J P, editor. Microbial transformation of bioactive compounds. Vol. 2. 1982. pp. 1–42. , pt. 1. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 15.Wolff B, Cohen G, Hauber J, Meshcheryakova D, Rabeck C. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp Cell Res. 1995;217:31–41. doi: 10.1006/excr.1995.1060. [DOI] [PubMed] [Google Scholar]
- 16.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita H, Tsubokawa S, Endo A. Microbial hydroxylation of compactin (ML-236B) and monacolin K. J Antibiot. 1985;38:605–609. doi: 10.7164/antibiotics.38.605. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida E, Komiyama K, Naito K, Watanabe Y, Takamiya K, Okura A, Funaishi K, Kawamura K, Funayama S, Umezawa I. Antitumor effects of Kazusamycin B on experimental tumors. J Antibiot. 1987;40:1596–1604. doi: 10.7164/antibiotics.40.1596. [DOI] [PubMed] [Google Scholar]