Abstract
Microbial communities growing in laboratory-based flow chambers were investigated in order to study compartmentalization of specific gene expression. Among the community members studied, the focus was in particular on Pseudomonas putida and a strain of an Acinetobacter sp., and the genes studied are involved in the biodegradation of toluene and related aromatic compounds. The upper-pathway promoter (Pu) and the meta-pathway promoter (Pm) from the TOL plasmid were fused independently to the gene coding for the green fluorescent protein (GFP), and expression from these promoters was studied in P. putida, which was a dominant community member. Biofilms were cultured in flow chambers, which in combination with scanning confocal laser microscopy allowed direct monitoring of promoter activity with single-cell spatial resolution. Expression from the Pu promoter was homogeneously induced by benzyl alcohol in both community and pure-culture biofilms, while the Pm promoter was induced in the mixed community but not in a pure-culture biofilm. By sequentially adding community members, induction of Pm was shown to be a consequence of direct metabolic interactions between an Acinetobacter species and P. putida. Furthermore, in fixed biofilm samples organism identity was determined and gene expression was visualized at the same time by combining GFP expression with in situ hybridization with fluorescence-labeled 16S rRNA targeting probes. This combination of techniques is a powerful approach for investigating structure-function relationships in microbial communities.
In nature most bacteria do not exist as pure cultures, and significant proportions of all microorganisms are associated with surfaces in complex multispecies communities called biofilms (14). There are many examples of important microbial processes that cannot be performed by a pure culture. For example, anaerobic digestion of organic matter is believed to require juxtaposition of syntrophic H2-producing acetogens and H2-consuming methanogens (12, 33). Also, in the process of nitrification the combined action of ammonia- and nitrite-oxidizing bacteria (e.g., members of the genera Nitrosomonas and Nitrobacter) is needed to complete the oxidation of ammonia to nitrate (6, 28). The two-member consortium “Chlorochromatium aggregatum” is an elegant example of the sulfur cycle, in which sulfide-oxidizing phototrophic bacteria are directly attached to the surface of a single central sulfate- or sulfur-reducing bacterium. This association allows anoxic photosynthesis by the phototroph to occur (42). Furthermore, during degradation of many xenobiotic compounds, such as chlorinated herbicides (31, 60), nitrate esters (45), naphthalene derivatives (50), and alkylbenzene sulfonates (27), the combined action of several species present in bacterial communities enhances or is required for complete mineralization of the compounds. Processes which cannot be performed efficiently or which cannot be performed at all by a pure culture but depend on the joint action of two or more bacterial species are termed community level processes (7).
An important factor in understanding community level processes is the relationship between structure and function in microbial communities. Recent developments in bacterial rRNA-based phylogeny (57) have allowed workers to design phylogenetic stains (16) based on fluorescence-labeled rRNA targeting probes. rRNA probes can be used to identify and quantify phylogenetically defined groups of organisms in complex mixtures; thus, the key players can be identified and their population dynamics in a microbial community can be monitored (4, 49, 52, 56). In addition, by using hybridization with fluorescent probes, the cellular rRNA content can be quantified and the growth rate (16, 38, 44, 56) or even the metabolic activity (39) of a specific population in a community can be estimated. By using scanning confocal laser microscopy (SCLM) the three-dimensional structure or architecture (32) of a community can be described, and by using fluorescent molecular probes the extracellular polysaccharide composition and the heterogeneity with respect to oxygen or pH within the microenvironments of a community can be analyzed (8, 37). Furthermore, by using in situ hybridization in combination with SCLM the spatial distribution of organisms in bacterial communities has been visualized (39, 55). By using rRNA probes and SCLM in an investigation of the architecture of granular sludge, workers have visualized the close juxtaposition of syntrophic propionate-oxidizing bacteria and H2-consuming methanogens (21), and the results suggested that there is a structure-function relationship during the process of interspecies hydrogen transfer.
The next step in inferring structure-function relationships is to determine the function of the bacteria in situ. One approach has been to use microelectrodes to measure rates of microbial processes in combination with a description of community structure. In the study of Ramsing et al. (48), the presence of sulfate-reducing bacteria was found to be restricted to the anoxic layers. The use of microsensors for nitrate in combination with rRNA probes for ammonia- and nitrite-oxidizing bacteria showed that nitrification was restricted to the top layers of a trickling filter biofilm. In these layers ammonia and nitrite oxidizers were found in proximity to each other (51).
A more direct approach for determining the function of an organism in situ is to investigate its gene expression. This has been done by identifying specific mRNAs in the pool of extracted mRNA (43), and recently a method for detecting mRNAs in single bacterial cells by an in situ PCR has been developed (24). An alternative to mRNA detection is the use of genetic reporters (lux, luc, gus, or lacZ). In the field of environmental microbiology, such systems have been used to study bacterial infection of root hairs (25, 40) and surface regulation of alginate synthesis (15) and to monitor nitrogen or phosphate starvation (29). However, these reporters are not easily compatible with fluorescent in situ hybridization and SCLM. With the cloning and heterologous expression of the gene for green fluorescent protein (GFP) from Aequorea victoria (10) and subsequent improvements (13, 22), a nearly ideal reporter for fluorescence detection became available. GFP has been shown to be a useful tool for monitoring bacterial gene expression (18, 54), as well as plasmid transfer (11, 20). It is important to note that GFP is a very stable protein in the organisms studied so far (10), and GFP expression thus reflects the history of gene expression in the cell rather than expression at the time of observation.
We have studied induction of the genes for the well-characterized TOL pathway for toluene degradation (for a review see reference 35). In this pathway toluene degradation starts by oxidation of the C-1 methyl group to yield benzoic acid (the upper pathway), which is then further oxidized to catechol. The catechol then undergoes meta fission to produce a semialdehyde, which is further transformed to Krebs cycle intermediates (the meta pathway [61]). The genes for toluene degradation are organized into two separate operons, and transcription is driven from the Pu (upper) and Pm (meta) promoters. The upper-pathway operon is induced by toluene and xylenes and their alcohol derivatives (1), and this induction is mediated by the effector-activated XylR protein (26) together with the sigma factor RpoN (19). In Luria-Bertani medium “exponential silencing” (catabolite repression) of Pu has been observed, and RpoN is believed to function as the sensor of the cellular physiological state (9). The meta pathway is induced by benzoates (47), and induction is mediated by the substrate-activated XylS protein (46). In addition, expression from the Pm promoter can be switched on by upper-pathway substrates through a cascade regulatory system (46).
The strategy used in the present investigation of specific gene expression in a toluene-degrading mixed community biofilm was to create genetic fusions between the relevant promoters (for expression of catabolic genes) and the gfp gene, which allowed easy microscopic monitoring at the level of single cells, followed by introduction of the engineered strains into the microbial community.
MATERIALS AND METHODS
Strains, media, and cultivation of biofilms.
The biofilm community used consisted of seven bacterial strains (Table 1), all isolated from a biofilter treating toluene-containing waste gas (39). Biofilms were cultivated as mixtures of the seven isolates in rectangular four-channel flow cells (60) with individual channel dimensions of 1 by 4 by 40 mm and were maintained on FAB medium [1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM Fe-EDTA (catalog no. E6760; Sigma, St. Louis, Mo.), 0.15 mM (NH4)SO4, 0.33 mM Na2HPO4, 0.2 mM KH2PO4, 0.5 mM NaCl]; 0.26 mM benzyl alcohol (Merck, Darmstadt, Germany) was used as the sole carbon source. Before each experiment the tubing was autoclaved, and after the flow system was assembled, the system was sterilized by pumping a 0.5% (wt/vol) hypochlorite solution into the system and leaving it there overnight. The next day the system was flushed with 2 liters of sterile water before the medium was pumped into the system. During growth of biofilms the medium was pumped through the flow cells at a constant rate of 0.2 mm/s (approximately 2 ml/h) by using a peristaltic pump (model 205S; Watson Marlow, Calmouth, Cornwall, England). Flow cells were inoculated with mixtures of overnight cultures of the seven isolates grown in Luria-Bertani medium in the following ratios: Pseudomonas putida R1, 0.2 ml; Acinetobacter sp. strain C6, 2 ml; and D8, smo111, smo113, smo115, and smo127, 1 ml each. The mixtures were sonicated (Branson Ultrasonics Corp. [Danbury, Conn.] sonifier) for 1 min at an output control setting of 3 and a duty cycle setting of 40%. A 0.2-ml portion of the culture was injected into each channel, and the flow cell was turned upside down. After 1 h the flow was resumed.
TABLE 1.
Properties of the seven isolates of the biofilm community
| Isolate | Phylogenetic affiliation | Growth on toluened | Growth on benzyl alcoholf |
|---|---|---|---|
| P. putida R1 | γ Subgroup of Proteobacteriaa | + | + |
| Acinetobacter sp. strain C6 | γ Subgroup of Proteobacteriab | + | + |
| D8 | β Subgroup of Proteobacteriab | +e | + |
| smo127 | Gram-positive high-G+C-content bacteriac | − | − |
| smo111 | Gram-positive high-G+C content bacteriac | − | − |
| smo113 | α Subgroup of Proteobacteriac | − | − |
| smo115 | Gram-positive high-G+C-content bacteriac | − | − |
Plasmids.
The gene encoding the GFP was obtained as an enhanced version, gfp-mut3b (13). The gfp-mut3b gene was PCR amplified as a 0.7-kb SphI-HindIII fragment and inserted into a pUC18-NotI-based cloning vector, resulting in pJBA25. The sequence was changed during the PCR so that the gfp-mut3b contained an arginine residue instead of a serine residue at position 2. This was done to introduce a SphI site in the start codon of gfp-mut3b. The gfp-mut3b gene was inserted the optimal distance downstream from the ribosome binding site of phage T5 (RBSII), which ensured that efficient translation occurred. Translational stop codons in all three reading frames were inserted downstream of the HindIII site in pJBA25, and two strong transcriptional terminators, T0 (from phage lambda) and T1 (from the rrnB operon of Escherichia coli), were inserted downstream of the translational stop codons. The NotI fragment from pJBA25 containing RBSII, gfp-mut3b, the translational stop codons, and the transcriptional terminators were inserted into the NotI sites of pCNB1 and pCNB3 (17), creating the fusions Pm::gfp-mut3a and Pu::gfp-mut3a, respectively. pCNB1 and pCNB3 are based on universal mini-Tn5 delivery plasmid pUT described by Herrero et al. (23), and pCNB1 contains in addition to the Pm promoter the xylS activator gene, whereas pCNB3 contains in addition to the Pu promoter the xylR activator gene. In summary, the transposon carried on pJBA30 contained the regulator gene xylR, which is required for activation of Pu, and a npt gene which confers kanamycin resistance, while the transposon carried on pJBA26 contained the regulator gene xylS, which is required for activation of Pm, and the Sm/Sp gene, which confers streptomycin resistance. When this system was used, the transposons carried on pJBA30 (Pu::gfp) and pJBA26 (Pm::gfp) were capable of transposing into the chromosome of the recipient strain while not cotransposing the transposase gene, which stably integrated the reporter construct into the chromosome of the recipient. The Pu::gfp and Pm::gfp reporters were inserted into the chromosome of P. putida R1 by using triparental mating as described previously (11). The resulting insertants were screened on 5 mM benzyl alcohol–FAB medium plates containing 25 μg of streptomycin per ml (pJBA30) or on 2% sodium citrate–5 mM 3-methylbenzoate (3MB)–FAB plates containing 25 μg of kanamycin per ml (pJBA25). GFP-positive colonies were detected by using an Axioplan epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) equipped with a 100-W HBO bulb for excitation, and GFP-specific light was observed by using the filter for fluorescein (filter set 10; Carl Zeiss). GFP-positive clones grew on FAB medium supplemented with toluene vapor or 5 mM benzyl alcohol or on 5 mM sodium benzoate.
Sequencing of 16S rRNA.
Sequencing was performed with a model 373A automatic DNA sequencer (Applied Biosystems, Foster City, Calif.) directly by using PCR products generated from chromosomal DNA extracts and the manufacturer’s recommendations. The following primers were used: 11F (5′-GTTTGATC[A/C]TGGCTCAGATTG-3′), 344R (5′-CCCCACTGCTGCCTCCCGT-3′), 515R (5′-GTATTACCGCGGC[G/T]GCTGGCAC-3′), 922R (5′-GCTTGTGCGGGCCCCCGTC-3′), 1101R (5′-GACAAGGGTTGCGCTCGTT-3′),1389R (5′-GTGACGGGCGGTGTGTACAAG-3′), and 1465R (5′-CCCCAGTCATGAATCATAAAGTGGT-3′) (the suffix F indicates the forward direction, and the suffix R indicates the reverse direction). The initial phylogenetic analysis of the sequenced strains was performed by using the online services of the Ribosomal Database Project (SIMILARITY_RANK [34]). In addition, sequences were investigated for the phylogenetic signature sequences described by Woese (57) which allow differentiation of the major groups of the Proteobacteria.
Oligonucleotide probes.
For hybridizations probe EUB338, specific for the domain Bacteria (3), and probe PP986, specific for P. putida subgroup A 16S rRNA (39), were used. Probes for the toluene-degrading isolates Acinetobacter sp. strain C6 and strain D8 were designed on the basis of sequence information (Table 2). The specificities of ACN449 and D8_647 were tested by using previously published sequences and the CHECK_PROBE program from The Ribosomal Database Project (34). The probes were tested with the organisms in the community and were specific for their target organisms (data not shown). Oligonucleotide probes labeled with fluorescein isothiocyanate, CY3, or CY5 were purchased from Hobolth DNA Syntese (Hillerød, Denmark).
TABLE 2.
rRNA probes
Hybridization of hydrated biofilm samples with cells expressing GFP.
Biofilms were fixed in a freshly made 3% paraformaldehyde solution by pumping this solution through the flow cells with attached biofilms at a rate of 0.8 mm/s for about 5 min. The flow cells were then kept at 4°C for 1 h. The flow cells were then washed with phosphate-buffered saline for 10 min (flow rate, 0.5 mm/s) before each channel was embedded in 20% acrylamide (200:1 acrylamide-bisacrylamide; Sequagel; National Diagnostics, Atlanta, Ga.). The embedding was done in the following way. One milliliter of 20% (wt/vol) acrylamide was mixed with 8 μl of N,N,N′,N′-tetramethylethylendiamine and 20 μl of 1% ammonium persulfate (International Biotechnologies Inc., New Haven, Conn.). This allowed about 2 min before the acrylamide solidified, and approximately 0.5 ml was pumped into the channel at a flow rate of 0.8 mm/s. During this procedure the biofilm structure could be monitored microscopically, and when the procedure was performed carefully, we found that the method was nondestructive and preserved the biofilm in its native hydrated state.
After fixation and embedding the acrylamide block with biofilm was placed on a six-well hybridization slide (Novakemi ab, Enskede, Sweden) and equilibrated for 15 min with hybridization buffer containing 20 or 30% formamide (Table 2). Then 30 μl of the hybridization mixture (20 or 30% formamide [Table 2], 0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% [wt/v] sodium dodecyl sulfate) containing approximately 75 ng of probe was added to each hybridization well. The cells were incubated with hybridization solution for 3 h at 37°C in a moisture chamber. For washing, 50-μl portions of washing solutions were added to each well. First, the acrylamide blocks were washed with washing solution (20 or 30% formamide [Table 2], 0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% sodium dodecyl sulfate) for 40 min at 37°C, and then they were washed for another 40 min with washing solution II (0.1 M Tris [pH 7.2], 0.9 M NaCl) at 37°C, before they were finally rinsed twice with 50 μl of distilled water. The acrylamide blocks were mounted in 2× SlowFade phosphate-buffered saline-based antifade solution (Molecular Probes, Eugene, Oreg.). Fixation, embedding, hybridization, and the microscopic investigation were performed on the same day since we observed that the GFP signal degenerated rapidly.
Microscopy and image analysis.
All microscopic observations and image acquisition were performed with a model TCS4D confocal microscope (Leica Lasertechnik GmbH, Heidelberg, Germany) equipped with an argon-krypton laser and three detectors for simultaneous monitoring of fluorescein isothiocyanate, CY3, and CY5. In addition, a reflection detector for acquiring bright-field images was installed.
Multichannel simulated fluorescence projection (SFP) images were generated by using IMARIS software (Bitplane AG, Zürich, Switzerland) running on a Indigo 2 workstation (Silicon Graphics Inc., Mountain View, Calif.). Images were processed for display by using Photoshop (Adobe, Mountain View, Calif.).
Spatial profiles of organisms and gene expression were estimated by measuring the area covered by either hybridization or the GFP signal (represented by different colors) in a series of three-channel optical sections by using simple thresholding. Before thresholding, images were preprocessed in IMARIS; the background was removed by using lowpass filtering and background subtraction, and a final correction for loss of intensity from deeper layers was performed by using the emission attenuation or gamma correction algorithms of IMARIS. Thresholding was performed on the processed images by using the HIPS (30) image analysis package. Line profiles were generated by using NIH Image (software available at ftp site zippy.nimh.nih.gov).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. Y11464 (Acinetobacter sp. strain C6) and Y11465 (isolate D8).
RESULTS
Model community.
Previous studies of the kinetics of a laboratory scale waste gas biofilter indicated that P. putida, which constituted only 4% of the biofilm population, as judged by activity, was a dominant member of the biofilm community and was responsible for 65% of the toluene removed by the biofilter (39). Therefore, P. putida was chosen as the main organism to investigate gene expression in a more controlled model system. The toluene-degrading community used as a model system in the present study consisted of seven strains (Table 1), three toluene-degrading strains and four nondegrading strains. All of the organisms were isolated from the natural toluene-degrading biofilter community (39), and all of the strains were characterized by examining them repeatedly after plating. Moreover, we thought that it was important to include even strains unable to degrade toluene in the model system since they were present in the original bioreactor community. We chose benzyl alcohol as the substrate for ease of operation of the flow system. The P. putida strain used in the mixed community, P. putida R1, has been shown to degrade toluene through a pathway which is similar to that encoded by the TOL plasmid but which has restricted substrate specificity, so that m-xylene and p-xylene are not growth substrates due to blockage of the catabolic pathway at the level of the first enzyme of the meta-cleavage pathway (unpublished data). To study the expression of genes involved in toluene degradation in our artificial seven-strain community, we investigated the expression of two TOL plasmid-derived promoters, Pu (upper pathway) and Pm (meta pathway). We constructed two mini-Tn5-based transposons (see Materials and Methods) and inserted the fusions Pu::gfp and Pm::gfp together with the genes encoding their regulators (xylR and xylS, respectively) into the chromosome of P. putida R1. The resulting strains grew at rates identical to the rate of growth of the parent strain in minimal medium liquid cultures supplied with either citrate or benzyl alcohol as the carbon and energy source (data not shown). In citrate-supplemented medium there was no induced expression of fluorescence, whereas addition of benzyl alcohol resulted in induction of fluorescence in the Pu::gfp strain and addition of 3MB (a well-known inducer of the Pm promoter [1]) resulted in induction of fluorescence in the Pm::gfp strain; these observations showed that the two promoters behaved as expected with respect to their modes of gene expression control (the Pu promoter is inducible with toluene or benzyl alcohol but not with benzoate, and the Pm promoter in our construct is inducible only with benzoate [see below]).
Expression of Pu and Pm in pure-culture biofilms.
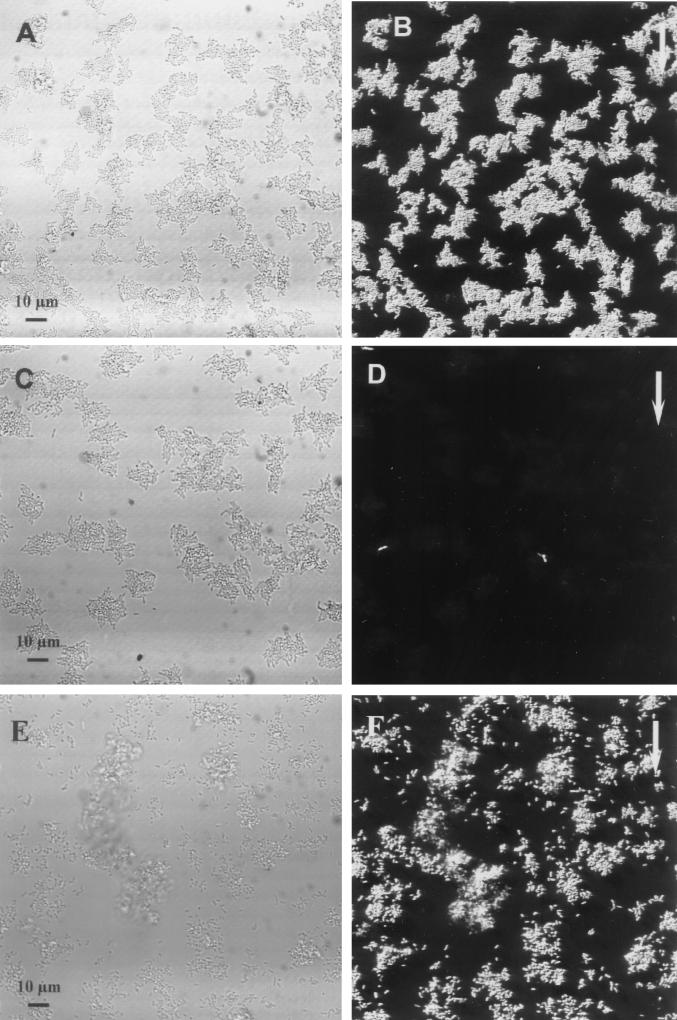
In order to analyze the gene expression patterns of the two P. putida R1 variants harboring the TOL promoters fused to the gfp gene, flow chambers with each of the variants in monoculture were established with a substrate based on benzyl alcohol as the only carbon source. Biofilms were grown on benzyl alcohol for 1 day, and monitoring of GFP expression showed that the Pu promoter was homogeneously induced in a pure-culture biofilm containing the relevant strain (Fig. 1A and B). In contrast, the GFP expression in a pure-culture biofilm containing P. putida R1 (Pm::gfp) grown on benzyl alcohol for 1 day showed that the Pm promoter was not induced in P. putida R1 except for strong induction in a few, very bright cells (Fig. 1C and D). Such bright cells were always present, but at a low frequency (<0.01%). The degrees of expression from the Pu::gfp and Pm::gfp fusions in the biofilm populations were quantified by performing an image analysis with series of optical sections that covered the thicknesses of the biofilms. The Pu promoter was induced in all cells, while the Pm promoter was not.
FIG. 1.
Online monitoring of Pu and Pm expression in pure-culture biofilms after 1 day of biofilm formation. (A and B) Biofilms formed by P. putida R1 (Pu::gfp). All cells are shown in the reflection image in panel A, and the GFP fluorescence in panel B shows that Pu expression was homogeneous. (C and D) Biofilms formed by P. putida R1 (Pm::gfp). All cells are shown in the reflection image in panel C, and the GFP fluorescence in panel D shows that there was no GFP expression except for that of a few bright cells, showing that Pm was not induced in the pure-culture biofilm. (E and F) Induction of Pm by 3MB. P. putida R1 (Pm::gfp) was grown for 1 day on benzyl alcohol, and then 5 mM 3MB was added to the medium. The reflection image (E) recorded on day 2 shows all cells, and the GFP signal from the same cells (F) reveals induction of Pm. The arrows indicate the direction of flow.
To further confirm these observations, a Pm::lacZ fusion was introduced into P. putida R1, and the β-galactosidase activity was monitored in cells growing with toluene, benzyl alcohol, or benzoate as the carbon source. High-level enzyme activity (8,000 Miller units) was expressed in cells cultivated with benzoate; however, in cells growing on toluene or benzyl alcohol the enzyme activity was about 30 times lower. These results suggest that in P. putida growing on toluene or benzyl alcohol, no significant accumulation of benzoate takes place. When biofilms of P. putida R1 (Pm::gfp) were grown on benzyl alcohol and 5 mM 3MB was added to the medium after 1 day of growth, induction of Pm::gfp was observed in all cells on day 2 (Fig. 1E and F). This showed that the Pm promoter was indeed inducible in P. putida R1 pure-culture biofilms.
Expression of Pu and Pm in mixed-culture biofilms.
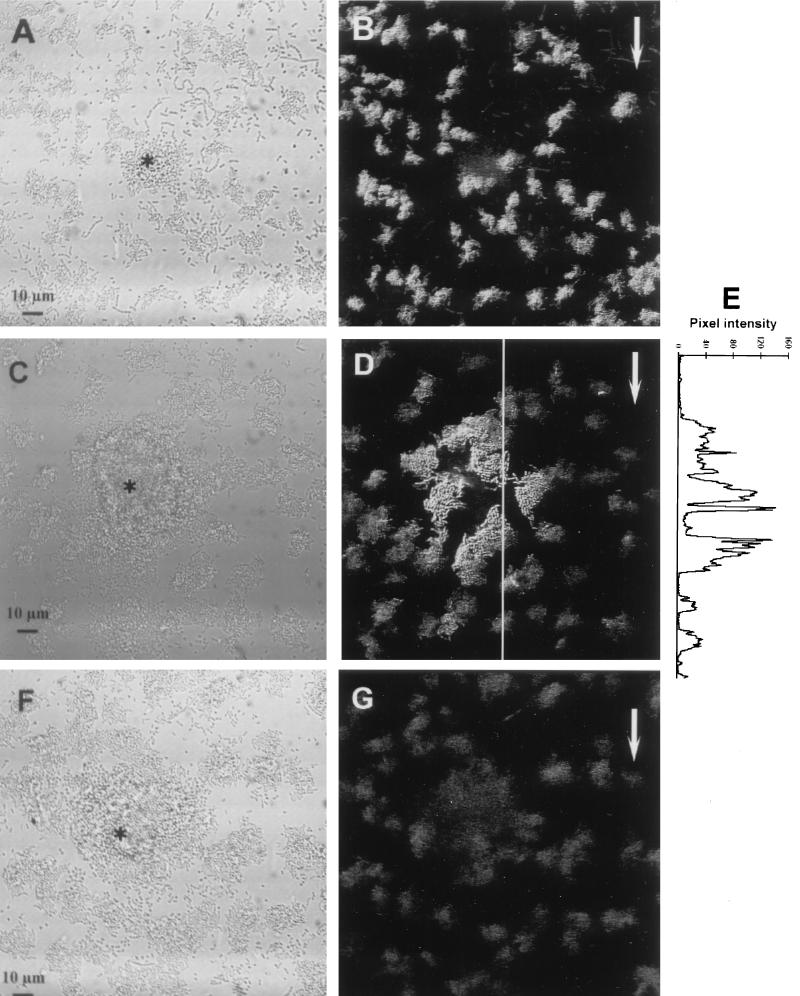
Expression from the Pu and Pm promoters was studied in the mixed community described above by monitoring the GFP levels in living biofilms. Flow cells were inoculated with the seven-member community or with communities in which wild-type P. putida R1 was replaced by either P. putida R1 (Pu::gfp) or P. putida R1 (Pm::gfp). GFP expression from Pu and GFP expression from Pm were monitored by using SCLM. Figures 2A and B illustrate GFP expression in a biofilm formed by the community containing P. putida R1 (Pu::gfp) after 1 day of growth and show that, as was observed with the single-strain biofilm described above, the Pu promoter was induced in the majority of the cells. The asterisk in Fig. 2A indicates a colony of cells that did not express GFP. The spherical microcolony morphology was typical of colonies formed by Acinetobacter sp. strain C6 (data not shown) and showed that both strain R1 and strain C6 were present in the biofilm.
FIG. 2.
Online monitoring of GFP expression from Pu and Pm in communities after 1 day of biofilm formation. (A and B) Pu expression in the biofilm formed by the community containing P. putida R1 (Pu::gfp). Note the spherical Acinetobacter sp. strain C6 colony (indicated by an asterisk) in the reflection image (A). The GFP fluorescence (B), visualized as an SFP, shows that Pu was constitutively expressed in the majority of the cells. (C through E) Expression of Pm in the biofilm formed by the community containing P. putida R1 (Pm::gfp). All cells (including the Acinetobacter sp. strain C6 colony indicated with an asterisk) are shown in the reflection image (C), and GFP fluorescence (D) shows the lack of homogeneous Pm expression on day 1. Panel E shows GFP expression quantified along the line shown in panel D and shows that GFP fluorescence increased as cells got closer to the Acinetobacter sp. strain C6 microcolony. The pixel intensities were measured from the maximum projection image (data not shown). (F and G) Biofilm formation by the wild-type community. The reflection image (F) and the SFP (G) show that no cells expressed GFP in the negative control. The asterisk indicates the spherical microcolonies of Acinetobacter sp. strain C6, and the arrows show the direction of flow.
P. putida R1 (Pm::gfp) was also grown with the six other strains in a mixed community, and expression from the Pm promoter was monitored (Fig. 2C to E). However, Pm expression was not homogeneous (as also observed in the monoculture biofilm), but very heterogeneous activity was observed after 1 day of biofilm formation. The reflection image in Fig. 2C shows a spherical Acinetobacter sp. strain C6 microcolony, and the SCLM image of the GFP signal (Fig. 2D) shows that the Acinetobacter sp. strain C6 colony was surrounded by P. putida R1 (Pm::gfp) cells expressing GFP. The GFP expression patterns indicated that cells close to the Acinetobacter sp. strain C6 microcolony were highly induced. In contrast, cells farther away from the Acinetobacter sp. strain C6 microcolony were only weakly induced or expressed background levels, results similar to the results observed in the single-strain biofilm (Fig. 1C and D). The gradient in gene expression could be further illustrated by quantifying the GFP signal along the line on the image in Fig. 2D. The plot in Fig. 2E shows that higher GFP expression levels (i.e., higher pixel intensities) occurred in cells located near the Acinetobacter sp. strain C6 microcolony. The background signal was recorded by using the biofilm formed by the wild-type community. A large Acinetobacter sp. strain C6 microcolony was observed in the reflection image (Fig. 2F), and only weak autofluorescence was detected in the SCLM image (Fig. 2G), showing that there was no green fluorescence from the negative control. Thus, the GFP signals observed in situ indicated that there was expression from the Pu and Pm promoters.
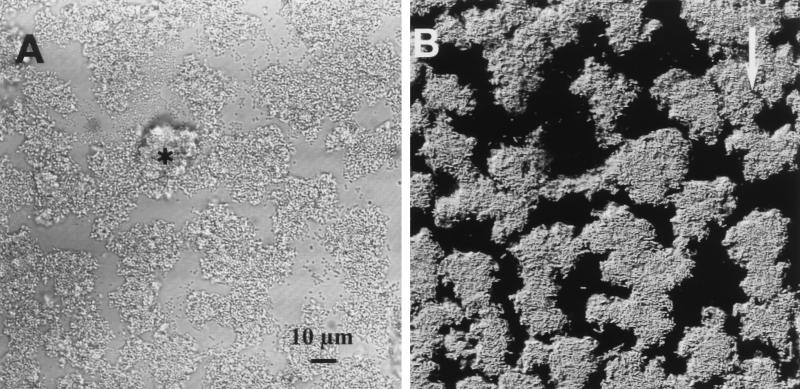
The pattern of induction for the Pm promoter in the presence of the other strains in the community was analyzed further. When P. putida R1 (Pm::gfp) pure-culture biofilms were grown for 1 day and then Acinetobacter sp. strain C6 was introduced, strong induction of the Pm promoter in P. putida R1 was observed on day 2 (Fig. 3). The other five community members became established in the P. putida R1 biofilms in very low numbers and were not able to cause induction of Pm (Table 3). This indicated that in the community studied, only Acinetobacter sp. strain C6 caused induction of the Pm promoter in P. putida R1. Our data also indicate (Table 3) that the interaction between P. putida R1 and Acinetobacter sp. strain C6 was not restricted to biofilm growth, but also occurred in liquid culture.
FIG. 3.
Addition of Acinetobacter sp. strain C6 to a P. putida R1 (Pm::gfp) biofilm. All cells are shown in the reflection image (A), and the GFP expression (B) shows that Pm was induced by the presence of Acinetobacter sp. strain C6. The asterisk indicates a spherical colony of Acinetobacter sp. strain C6, and the arrow indicates the direction of flow.
TABLE 3.
Metabolic interactions between community members
| Organism(s) added to P. putida R1 biofilms | Gene expression in biofilm
|
Gene expression in liquidb
|
||
|---|---|---|---|---|
| Pu::gfp | Pm::gfp | Pu::gfp | Pm::gfp | |
| None | + | − | + | − |
| Acinetobacter sp. strain C6 | + | + | + | + |
| D8a | + | − | + | − |
| smo111 + smo113 + smo115 + smo127a | + | − | + | − |
| Acinetobacter sp. strain C6 + D8 + smo111 + smo113 + smo 115 + smo127a | + | + | + | + |
All organisms except P. putida R1 and Acinetobacter sp. strain C6 became established in biofilms at very low concentrations (less than 1%).
Observed in an outgrown overnight culture.
Time course of Pm induction.
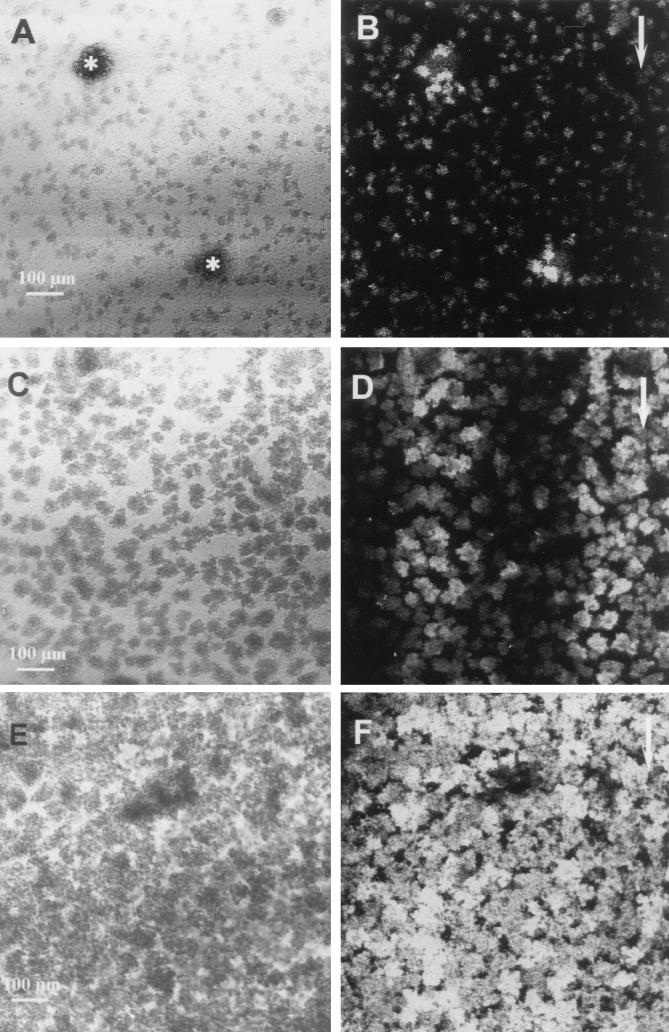
To illustrate the development of induction of the Pm promoter over time, a series of low-magnification images are shown in Fig. 4. At day 1 induction was very heterogeneous, and Pm was most strongly induced in P. putida R1 cells located close to Acinetobacter sp. strain C6 microcolonies (Fig. 4A and B), as shown above. The heterogeneous induction patterns observed on day 2 (Fig. 4C and D), with induced regions occurring as bands of induction parallel to the direction of flow, suggested that the inducing agent was able to diffuse in the biofilm. On day 3 the induction patterns for Pm in the biofilm had become more uniform (Fig. 4E and F), further suggesting that the inducing agent accumulated in the biofilm over time, which resulted in more complete induction of Pm in older biofilms.
FIG. 4.
Time course of GFP expression by P. putida R1 (Pm::gfp) in the community. (A and B) Lack of homogeneous expression of Pm::gfp on day 1. All cells are shown in the reflection image (A), and an optical section obtained 5 μm from the substratum shows the GFP expression (B). The asterisks indicate spherical microcolonies of Acinetobacter sp. strain C6. (C and D) Pm expression patterns on day 2. The reflection image (C) and an optical section of GFP expression obtained 5 μm from the substratum (D) show that induction occurred in bands parallel to the direction of flow. (E and F) Pm expression on day 3. The reflection image (E) shows all of the cells, and an optical section of the GFP fluorescence obtained 5 μm from the substratum (F) shows more homogeneous induction of Pm on day 3, indicating that the inducing agent accumulated in the biofilm. The arrows indicate the direction of flow.
Spatial distribution of organisms and gene expression.
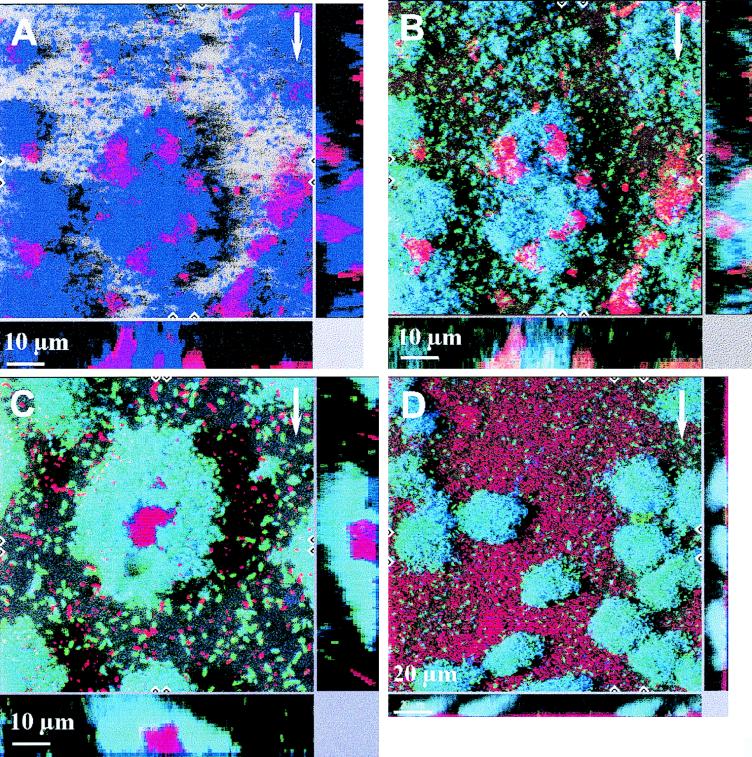
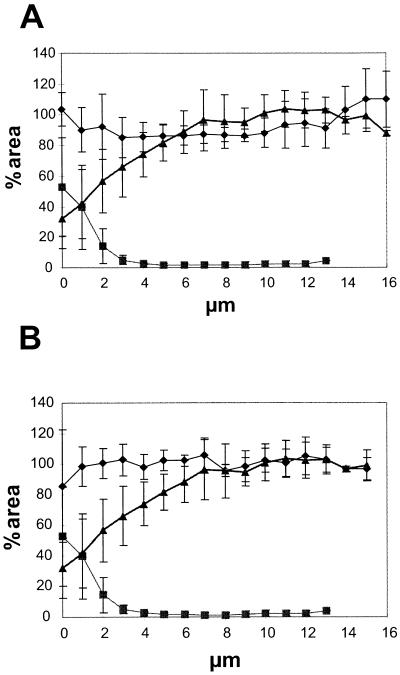
The spatial distribution of community members and simultaneous visualization of gene expression were studied by combining GFP expression and hybridization with 16S rRNA targeting probes. Figure 5A shows a three-dimensional representation of the organism distribution in a 3-day-old biofilm formed by the community containing P. putida R1 (Pu::gfp). Two probes were used, and P. putida R1 was hybridized with PP986 (blue) and Acinetobacter sp. strain C6 was hybridized with ACN449 (red). Figure 5B shows an overlay of the GFP signal that was simultaneously recorded in the green channel during scanning. The image is interpreted in the following way. P. putida R1 (Pu::gfp) expressing GFP is both blue and green and appears cyan. Acinetobacter sp. strain C6 not expressing GFP is unchanged and appears red. Expression of Pu::gfp was quantified by using image analysis, and Fig. 6A shows the spatial distribution of Pu expression in the community and shows that most P. putida R1 cells expressed GFP from the Pu promoter throughout the biofilm (between 80 and 100% of the area covered by the hybridization signal was also covered by GFP fluorescence [Fig. 6A]). The spatial distribution of the dominant organisms in the biofilm was also quantified. Figure 6A shows that Acinetobacter sp. strain C6 was the dominant organism at the substratum level (∼55% areal coverage), whereas P. putida R1 dominated the outermost layers of the biofilm (∼100% areal coverage).
FIG. 5.
Spatial distribution of organisms and gene expression in the community. The dominant organisms in the community were targeted by hybridization. (A) Dual hybridization of Acinetobacter sp. strain C6 (red) and P. putida R1 (blue) in the community containing P. putida R1 (Pu::gfp) on day 3. (B) Overlay of the GFP fluorescence (green) expressed from the Pu promoter. Cells that stained both blue (hybridization) and green (GFP expression) appear cyan. (C and D) Expression from Pm in the community containing P. putida R1 (Pm::gfp) grown for 3 days. Panel C shows a colony of Acinetobacter sp. strain C6 (red) surrounded by P. putida R1 (Pm::gfp) cells expressing GFP (cyan), and panel D shows surface coverage by Acinetobacter sp. strain C6 (red), with P. putida R1 distributed over the surface. These colonies expressed GFP (cyan). P. putida R1 was hybridized with PP986 labeled with CY5, Acinetobacter sp. strain C6 was hybridized with ACN449 labeled with CY3, and GFP was sampled in the green channel. The images shown are SFPs with x and z projections shown on the sides of the images; these projections provide extended focus images for the regions between the marks. The arrows indicate the direction of flow.
FIG. 6.
Profiles of organisms and gene expression in the biofilm. (A) Expression from the Pu promoter. Symbols: ⧫, percentage of the P. putida R1 population in the biofilm expressing Pu::gfp; ▴, percentage of cells in the biofilm that were P. putida R1 cells; ▪, percentage of cells that were Acinetobacter sp. strain C6 cells. (B) Expression from the Pm promoter. Symbols: ⧫, percentage of the P. putida R1 population in the biofilm expressing Pm::gfp; ▴, percentage of cells in the biofilm that were P. putida R1 cells; ▪, percentage of cells that were Acinetobacter sp. strain C6 cells. Percentages were calculated by determining the ratio between areas covered by the respective fluorescent signals in a series of multicolor optical sections. All bacteria were hybridized by using EUB338 labeled with CY5, P. putida R1 was hybridized with PP986 labeled with CY3 or CY5, Acinetobacter sp. strain C6 was hybridized with ACN449 labeled with CY3, and GFP was sampled in the green channel. Error bars indicate ±1 standard deviation. The profiles are the results obtained after we averaged the data for five independent positions in the biofilm.
The spatial distribution of Pm expression in the context of the community was also determined. Figure 5C shows a spherical microcolony of Acinetobacter sp. strain C6 entirely surrounded by P. putida R1 cells, all of which showed expression from the Pm promoter. Figure 5D shows a portion of the biofilm where the surface was entirely covered by Acinetobacter sp. strain C6 and colonies of P. putida R1 expressing Pm::gfp were distributed over the surface without any apparent spatial interaction with Acinetobacter sp. strain C6. This may indicate that direct contact between the two organisms (as suggested by Fig. 5B and C) is not necessary for induction of Pm. The distribution of Pm expression in the biofilm was quantified, and Fig. 6B shows Pm expression relative to the position of the dominant members of the community. On day 3 all of the P. putida R1 cells were expressing GFP from the Pm promoter throughout the biofilm (∼100% areal coverage) (Fig. 6B), and no spatial gradients in expression from Pm were observed.
The presence of D8 and the presence of the four non-toluene-degrading strains in the community could be confirmed by probing (by using D8_647 and EUB338, respectively). However, these organisms were present in only low numbers (data not shown). This confirms that P. putida R1 and Acinetobacter sp. strain C6 were the dominant organisms in the biofilm community.
DISCUSSION
Two of the seven organisms present in the community studied here, P. putida R1 and Acinetobacter sp. strain C6, were the focus of our study because they are present in fairly high numbers and because they interact. Both of these strains are able to degrade benzyl alcohol, and we have evidence (data not shown) that they both have degradative pathways similar to the degradative pathway located on the TOL plasmid. The two pathways are, however, not identical to the TOL plasmid pathway, and they are probably not identical in the two strains either. The differences occur at the level of substrate specificities and have been observed in hybridization experiments performed with probes derived from the TOL plasmid (unpublished data). The gene expression analysis described here took advantage of the existence of cloned promoters and regulatory genes derived from the TOL plasmid.
In biofilms expression from the Pu promoter was continuously induced in the mixed-strain community, as well as in pure culture. Benzyl alcohol is an inducer of the Pu promoter (1), and the homogeneous expression from Pu probably reflects the presence of the substrate in all microenvironments of the community during the spatial development of the biofilm, as well as during the temporal development of the biofilm.
In contrast, expression from the Pm promoter in the biofilms was more complex. In a pure-culture biofilm the Pm promoter was not induced by the benzyl alcohol added, probably because the cascade induction system could not operate since the xylS gene used in the designed fusions between the Pm promoter and gfp lacked the promoter that normally responds to benzyl alcohol coupled with XylR (17, 46) and because the benzoate formed in the cells from benzyl alcohol metabolism was quickly removed and did not accumulate in sufficient amounts to induce detectable levels of GFP production. In the early stages of biofilm development, induction of the Pm promoter was heterogeneous, and addition of the other community members to a P. putida R1 monoculture biofilm showed that Pm was induced only by the presence of Acinetobacter sp. strain C6. A simple explanation is that the Pm inducer, benzoate, leaked from Acinetobacter sp. strain C6 into the surrounding medium. Although both organisms carry the genes for a TOL-like degradation pathway for benzyl alcohol, the differences between them could easily result in different rates of conversion of the different metabolites, resulting in accumulation of benzoate in only one of them. Leakage of metabolites out of growing cells is known to occur; for example, during degradation of simple substrates, such as glucose, catabolites (i.e., acetate) can leak in significant amounts from E. coli (5). The excreted benzoate can then diffuse into the medium and be available as a substrate and/or inducer for other cells. This hypothesis is supported by the induction patterns observed. The Pm promoter was initially induced only in P. putida R1 cells located close to Acinetobacter colonies, whereas in later stages of biofilm development the Pm promoter was induced in all P. putida R1 (Pm::gfp) cells, which probably reflected accumulation of benzoate throughout the biofilm. Since Pu promoter expression was continuously induced, P. putida R1 cells in the community could be expected to grow on a mixture of benzyl alcohol and benzoate. Alternatively, the proximity of Acinetobacter cells could change the physiology of the P. putida R1 cells in such a way that intracellular benzoate would accumulate in adequate amounts to cause sufficient induction of the Pm promoter. In any case, the data present genetic evidence that metabolic interactions occur between community members in an aerobic degradative biofilm community.
Sequential metabolism of degradation products is well-known in many anaerobic community microbial processes. For example, the layered structure of granular sludge (21, 33) suggests that the spatial distribution of organisms reflects the positions of the organisms in the food chain. Thermodynamic considerations suggest that close juxtaposition of H2 producers and consumers occurs, and this suggestion has been supported by gas metabolism evidence (12, 53), as well as by direct microscopic observations (21, 53), which further support the significance of specific spatial arrangements during anaerobic degradation of organic matter. In addition, similar structures have been observed in granular sludge from different reactors (21, 33, 53), and this similarity is probably a consequence of tight coupling and strong interdependence in the anaerobic degradation process.
There are also many examples of aerobic processes in which different bacterial species are interdependent. As mentioned above, degradation of many xenobiotic compounds requires the action of a bacterial consortium and does not occur in a pure culture. Microbial associations in such degradative communities are less well-studied. Wolfaardt et al. (60) observed the development of structured consortia in a biofilm community growing on a chlorinated hydrocarbon (diclofob-methyl). The structure of the consortia rapidly degenerated when the biofilm was irrigated with labile carbon sources, indicating that there was a high degree of substrate-dependent coupling in the system (60). Furthermore, diclofob-methyl was shown to accumulate in the extracellular polysaccharide of certain community members (58) and to bind to sites of preferred chemistry (59), suggesting that there were regions where specific activities occurred in the community. In a separate investigation performed with a different community and a different hydrocarbon, Møller et al. (37) observed a similar substrate-dependent biofilm architecture. Taken together, these data suggest that the degradative biofilms studied are examples of tightly coupled and highly organized bacterial communities (36).
In contrast, the structural interactions in the benzyl alcohol-degrading biofilm investigated here seemed to be rather loose, although the architecture of the biofilm had characteristic features. The two dominant organisms appeared to have different niches in the biofilm, with Acinetobacter sp. strain C6 primarily attaching to the substratum and P. putida R1 being the dominant organism in the outermost layers of the biofilm (Fig. 6). SCLM images of the three-dimensional structure of the community revealed the heterogeneous architecture of the biofilm; in some regions Acinetobacter sp. strain C6 seemed to colonize P. putida R1 colonies (Fig. 5A), in other regions P. putida R1 seemed to colonize Acinetobacter sp. strain C6 colonies (Fig. 5C), and in yet other regions the two organisms seemed to grow independently (Fig. 5D). This suggests that close physical association between the two organisms was not essential for biofilm proliferation.
The loose architectural features described above probably reflect the fact that both P. putida R1 and Acinetobacter sp. strain C6 are able to degrade benzyl alcohol in pure culture (Table 3), which is not the case with the highly organized communities mentioned above. In spite of the loose organization, metabolic interaction between the two community members was observed and seemed to occur in all regions in the mature biofilm independent of the local architecture. In addition, the interaction was observed in liquid culture (Table 3). The importance of the metabolic interaction remains to be determined, but measurements of biofilm thickness indicated that there was synergy between community members; the seven-strain community formed biofilm thicker than the biofilms consisting only of P. putida R1 and Acinetobacter sp. strain C6, which were thicker than the pure-culture biofilms (39a). Our observations indicate that bacteria living in an apparently loosely organized microbial community may cooperatively interact with each other and consequently that community level processes (7) are not restricted to highly organized microbial communities.
ACKNOWLEDGMENTS
This work was supported by the Danish Biotechnology Program.
Palle Hobolth, Hobolth DNA Syntese (Hillerød, Denmark), is acknowledged for sequencing the 16S rRNA. Brendan Cormack, Rafael Valdivia, and Stanley Falkow are acknowledged for the gift of the gfp-mut3 allele used in this study.
REFERENCES
- 1.Abril M, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen K B, von Myenburg K. Are growth rates of Escherichia coli in batch cultures limited by respiration? J Bacteriol. 1980;144:114–123. doi: 10.1128/jb.144.1.114-123.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bock E, Koops H-P. The genus Nitrobacter and related genera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 2302–2309. [Google Scholar]
- 7.Caldwell D E, Costerton J W. Are bacterial biofilms constrained to Darwin’s concept of evolution through natural selection? Microbiol SEM. 1996;12:347–358. [PubMed] [Google Scholar]
- 8.Caldwell D E, Korber D R, Lawrence J R. Confocal laser microscopy and digital image analysis in microbial ecology. Adv Microb Ecol. 1992;12:1–67. [Google Scholar]
- 9.Cases I, de Lorenzo V, Pérez-Martin J. Involvement of sigma-54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol Microbiol. 1996;19:7–17. doi: 10.1046/j.1365-2958.1996.345873.x. [DOI] [PubMed] [Google Scholar]
- 10.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 11.Christensen B B, Sternberg C, Molin S. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (Gfp) from Aequorea victoria as a marker. Gene. 1996;173:59–65. doi: 10.1016/0378-1119(95)00707-5. [DOI] [PubMed] [Google Scholar]
- 12.Conrad R, Phelps T L, Zeikus J G. Gas metabolism evidence in support of the juxtaposition of hydrogen-producing and methanogenic bacteria in sewage sludge and lake sediments. Appl Environ Microbiol. 1985;50:595–601. doi: 10.1128/aem.50.3.595-601.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 14.Costerton J W, Cheng K J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie J T. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;42:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 15.Davies D G, Chakrabarty A M, Geesey G G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for identification of single cells. Science. 1989;243:1360–1362. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 17.de Lorenzo V, Fernández S, Herrero M, Jakubzik U, Timmis K N. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 1993;130:41–46. doi: 10.1016/0378-1119(93)90344-3. [DOI] [PubMed] [Google Scholar]
- 18.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 19.Dixon R. The xylABC promoter from Pseudomonas putida TOL plasmid is activated by nitrogen regulatory gene in Escherichia coli. Mol Gen Genet. 1986;203:129–136. doi: 10.1007/BF00330393. [DOI] [PubMed] [Google Scholar]
- 19a.Eberl, L. Personal communication.
- 20.Eberl L, Schulze R, Ammendola A, Geisenberger O, Erhart R, Sternberg C, Molin S, Amann R I. Use of green fluorescent protein as a marker for ecological studies of activated sludge communities. FEMS Microbiol Lett. 1997;149:77–83. [Google Scholar]
- 21.Harmsen H J M, Kengen H M P, Akkermans A D L, Stams A J M, De Vos W M. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl Environ Microbiol. 1996;62:1656–1663. doi: 10.1128/aem.62.5.1656-1663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 23.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodson R E, Dustman W A, Garg R P, Moran M A. In situ PCR for visualization of microscale distribution of specific genes and gene products in prokaryotic communities. Appl Environ Microbiol. 1995;61:4074–4082. doi: 10.1128/aem.61.11.4074-4082.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurek T, Reinhold-Hurek B, van Montagu M, Kellenberger E. Root colonization and systemic spreading of Azoarcus sp. strain BH71 in grasses. J Bacteriol. 1994;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inouye S, Nakazawa A, Nakazawa T. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEFG operons of the TOL plasmid. J Bacteriol. 1983;155:1192–1199. doi: 10.1128/jb.155.3.1192-1199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez L, Breen A, Thomas N, Federle T W, Sayler G S. Mineralization of linear alkylbenzene sulfonate by a four-member aerobic bacterial consortium. Appl Environ Microbiol. 1991;57:1566–1569. doi: 10.1128/aem.57.5.1566-1569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koops H-P, Möller U C. The lithotrophic ammonia-oxidizing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 2625–2637. [Google Scholar]
- 29.Kragelund L, Christoffersen B, de Bruijn F J, Nybroe O. Isolation of lux reporter gene fusions in Pseudomonas fluorescens DF57 inducible by nitrogen or phosphorus starvation. FEMS Microbiol Ecol. 1995;17:95–106. [Google Scholar]
- 30.Landy M S, Cohen Y, Sperling G. HIPS: a Unix-based image processing system. Comput Vision Graphics Image Process. 1984;25:331–347. [Google Scholar]
- 31.Lappin H M, Greaves M P, Slater J H. Degradation of the herbicide mecoprop [2-(2-methyl-4-chlorophenoxy)propionic acid] by a synergistic microbial community. Appl Environ Microbiol. 1985;49:429–433. doi: 10.1128/aem.49.2.429-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence J R, Korber D R, Hoyle B D, Costerton J W, Caldwell D E. Optical sectioning of microbial biofilms. J Bacteriol. 1991;173:6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLeod F A, Guiot S R, Costerton J W. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl Microbiol Biotechnol. 1990;56:1598–1607. doi: 10.1128/aem.56.6.1598-1607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marqués S, Ramos J L. Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathway. Mol Microbiol. 1993;9:923–929. doi: 10.1111/j.1365-2958.1993.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 36.Molin S, Molin J. CASE: complex adaptive systems ecology. Adv Microb Ecol. 1997;15:27–80. [Google Scholar]
- 37.Møller S, Korber D R, Wolfaardt G M, Molin S, Caldwell D E. Impact of nutrient composition on a degradative biofilm community. Appl Environ Microbiol. 1997;63:2432–2438. doi: 10.1128/aem.63.6.2432-2438.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Møller S, Kristensen C S, Poulsen L K, Carstensen J M, Molin S. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl Environ Microbiol. 1995;61:741–748. doi: 10.1128/aem.61.2.741-748.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Møller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Nielsen, A. T., and S. Møller. Unpublished data.
- 40.O’Kane D J, Lingle W L, Wampler J E, Legocki M, Legocki R P, Szalay A A. Visualization of bioluminescence as a marker of gene expression in rhizobium-infected soybean root nodules. Plant Mol Biol. 1988;10:387–399. doi: 10.1007/BF00014945. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen A R. Biological removal of toluene in waste gas reactors. Ph.D. thesis. Lyngby: Institute of Environmental Science and Engineering, The Technical University of Denmark; 1996. [Google Scholar]
- 42.Pfenning N. Syntrophic mixed cultures and symbiotic consortia with phototrophic bacteria: a review. In: Gottschalk G, Pfenning N, Werner H, editors. Anaerobes and anaerobic infections. Stuttgart, Germany: Gustav Fisher Verlag; 1980. pp. 127–131. [Google Scholar]
- 43.Pichard S L, Paul J H. Detection of gene expression in genetically engineered microorganisms and natural phytoplankton populations in the marine environment by mRNA analysis. Appl Environ Microbiol. 1991;57:1721–1727. doi: 10.1128/aem.57.6.1721-1727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos J L, Haïdour A, Duque E, Piñar G, Calvo V, Olivia J M. Metabolism of nitrate esters by a consortium of two bacteria. Nat Biotechnol. 1996;14:320–322. doi: 10.1038/nbt0396-320. [DOI] [PubMed] [Google Scholar]
- 46.Ramos J L, Mermod N, Timmis K T. Regulatory circuits controlling transcription of the TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987;1:293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 47.Ramos J L, Stolz A, Reineke W, Timmis K N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci USA. 1986;83:8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramsing N B, Kühl M, Jørgensen B B. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rozgaj R, Glancer-S̀oljan M. Total degradation of 6-aminonaphthalene-2-sulphonic acid by a mixed culture consisting of different bacterial genera. FEMS Microbiol Ecol. 1992;86:229–235. [Google Scholar]
- 51.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R I, Schleifer K H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiele J H, Chartrain M, Zeikus J G. Control of interspecies electron flow during anaerobic digestion: role of floc formation in syntrophic methanogenesis. Appl Environ Microbiol. 1988;54:10–19. doi: 10.1128/aem.54.1.10-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valdivia R H, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 55.Wagner M, Amann R I, Kämpfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K H. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst Appl Microbiol. 1995;17:405–417. [Google Scholar]
- 56.Wallner G, Erhart R, Amann R I. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl Environ Microbiol. 1995;61:1859–1866. doi: 10.1128/aem.61.5.1859-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfaardt G M, Lawrence J R, Headly J V, Robarts R D, Caldwell D E. Microbial exopolymers provide a mechanism for bioaccumulation of contaminants. Microb Ecol. 1994;27:279–291. doi: 10.1007/BF00182411. [DOI] [PubMed] [Google Scholar]
- 59.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, and D. E. Caldwell. Unpublished data.
- 60.Wolfaardt G M, Lawrence J R, Robarts R D, Caldwell S J, Caldwell D E. Multicellular organization in a degradative biofilm community. Appl Environ Microbiol. 1994;60:434–446. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]