Abstract
In water microcosm experiments, the survival times of Campylobacter isolates differed by up to twofold, as determined by culturing; this difference increased to fourfold when particular combinations of temperature and oxygenation were used. The mean survival times were much longer at 4 and 10°C (202 and 176 h, respectively) than at 22 and 37°C (43 and 22 h, respectively). The influence of anaerobiosis on survival time was less dramatic and differed considerably between isolates. In a two-stage water distribution model preparation containing a biofilm consisting of standardized autochthonous water microflora, Campylobacter isolates continued to differ in survival time. However, the survival times of cultures were considerably longer in the presence of the autochthonous water microflora (strains CH1 and 9752 survived 700 and 360 h, respectively, at 4°C) than in the sterile microcosms (strains CH1 and 9752 survived 230 and 157 h, respectively). Although increased temperature and oxygenation were generally detrimental to culturability, the interaction of these two factors influenced the two strains examined differently. When the organisms were grown aerobically at 30°C, the survival of the two strains was reversed; aerobiosis decreased the survival time of strain CH1 by 30%, but unexpectedly improved the persistence time of strain 9752 by more than threefold. Persistence times within biofilms were much longer when they were determined by detection methods not involving culturing. Immunofluorescent-antibody staining demonstrated that the pathogen persisted up to the termination of the experiments after 28 and 42 days of incubation at 30 and 4°C, respectively. The specificity of detection within intact biofilms was reduced because of high background fluorescence. However, preliminary studies with a Campylobacter-specific rRNA probe revealed the same extended persistence of the pathogen within the biofilms.
Campylobacter jejuni is the major cause of human bacterial gastroenteritis in the United Kingdom and probably worldwide (11, 36, 44). The four major sources of infection are raw meat (particularly poultry), untreated water, raw milk, and pets (1, 16, 36, 44). C. jejuni clearly survives in untreated and inadequately treated aquatic environments sufficiently well to directly cause human disease (3, 27, 37, 43, 45, 47). It has also been suggested that, although not universally the case (18, 21, 30), survival in the water systems of animal husbandry facilities and animal-processing units promotes infection in animals and cross-contamination of animal carcasses (2, 17, 23, 35). Thus, the survival of C. jejuni in aquatic environments is important both directly and indirectly in the causation of human disease.
The ability of C. jejuni to survive in aquatic, nutrient-poor environments may differ between strains (22, 48). Furthermore, this pathogen’s persistence may be enhanced or diminished by its interaction with, or incorporation into, natural biofilms. Biofilms are widely distributed in the environment and occur in most water supply and plumbing systems. They are composed of microcolonies of organisms, including bacteria, fungi, and protozoa, bound together by an extracellular matrix (9, 10, 13). Organisms in the biofilms create microenvironments which may provide a haven for other microorganisms and under low-nutrient and hostile conditions may provide protection against environmental extremes (9). There has been little systematic study of the comparative survival of different C. jejuni strains in water systems (22, 48), and no information is available at present on the ability of these organisms to integrate into biofilms, although it has been suggested that in simple experimental systems, predation by and competition with other microorganisms reduce the survival times of C. jejuni and Escherichia coli in water (25, 26). Conversely, for a number of other pathogens, interaction with biofilm microflora has been reported to promote survival (8, 29, 34, 40), and, in the case of C. jejuni, uptake by protozoans has been shown to protect against subsequent disinfection (24).
Survival of C. jejuni in simple water systems has been the subject of some previous studies. The experimental systems used, however, have varied, making systematic assessment of the results difficult. Survival in these “water microcosms” has been shown to be enhanced at low temperatures (6, 38, 41, 48), particularly at temperatures around 4°C. Dissolved oxygen tension may also influence survival in water. In one study aeration was shown to decrease survival time (41), while in a different study neither oxygen nor nutrient availability was particularly significant (38). Previously determined survival times for C. jejuni in water vary widely from a few days at ambient temperatures to weeks and even months at 4°C. The majority of the studies have put the long-term survival times of some strains of the organism at 4°C at about 2 to 4 weeks, although more extreme examples of survival for up to 4 months have been reported (41). These differences may reflect strain variations or merely differences in the experimental systems used. An additional confounding issue is the concept that organisms can adopt a viable but nonculturable form during prolonged exposure to nutrient-poor aquatic environments (42). Such organisms cannot be cultured on conventional laboratory media but still exhibit various degrees of metabolic activity and are thought to be capable of causing infection. Such a state has been proposed for campylobacters (14, 20, 22, 41, 46). The status of this condition, however, remains uncertain since resuscitation of putative viable but nonculturable forms in animals (22, 46) has not been reproducible (5, 33, 49).
Clearly, a number of important questions remain to be answered concerning the persistence of Campylobacter strains in aquatic systems and the risk that this poses to human health. This study addressed the effects of strain variation, competing microflora and biofilm involvement, and physical parameters, such as temperature and oxygen tension, on the persistence of C. jejuni.
MATERIALS AND METHODS
Bacterial isolates.
Campylobacter isolates were collected from a number of sources, including poultry, water, and human clinical cases (Table 1). On receipt, these isolates were minimally subcultured before stored at −70°C on MicroBank beads (ProLab) in accordance with the manufacturer’s instructions. Members of autochthonous water microfloras were isolated from potable water and were distinguished on the basis of distinct colonial characteristics following culture on R2A agar (39). These isolates were consistently recovered from a two-stage water model (see below). The isolates were stored frozen (−70°C) on MicroBank beads.
TABLE 1.
Sources and serotypes of Campylobacter strains
| Strain | Species | Sourcea | Other information | Serotypeb |
|---|---|---|---|---|
| CH1 | C. jejuni | Exeter PHL | Chicken skin isolate | 1 |
| CH5 | C. jejuni | Exeter PHL | Chicken skin isolate | 1 |
| C53 | C. jejuni | Exeter PHL | Chicken cecum isolate | 1 |
| W1 | C. jejuni | Exeter PHL | Water isolate | 5 |
| W2 | C. jejuni | Exeter PHL | Water isolate | 5 |
| W7 | C. jejuni | Exeter PHL | Water isolate | 54 |
| 206 | C. jejuni | Exeter PHL | Sporadic clinical isolate | 50 |
| 235 | C. jejuni | Exeter PHL | Sporadic clinical isolate | 11 |
| 247 | C. coli | Exeter PHL | Sporadic clinical isolate | 56 |
| 799 | C. coli | Exeter PHL | Sporadic clinical isolate | 56 |
| 9519 | C. jejuni | Exeter PHL | Sporadic clinical isolate | 59 |
| 9752 | C. jejuni | Exeter PHL | Sporadic clinical isolate | 50 |
| 10392 | C. jejuni | Exeter PHL | Sporadic clinical isolate | 6 |
| 81116 | C. jejuni | NCTC | NCTC 11828 | 6 |
| 11168 | C. jejuni | NCTC | NCTC 11168 | 2 |
| C677 | C. jejuni | NCTC | NCTC Letton Hall (outbreak) | 27 |
| 2172 | C. coli-C. upsaliensis | Southampton PHL | Sporadic clinical isolate | UT (45,58) |
PHL, Public Health Laboratory; NCTC, National Collection of Type Cultures.
The serotypes of the isolates were determined by using the Laboratory of Enteric Pathogens scheme (12). This is a modification of the Penner serotyping scheme for C. jejuni and C. coli based on the use of absorbed antisera to heat-stable antigens and utilizing direct whole-cell agglutination instead of passive hemagglutination as the detection system. UT, untypeable.
Culture of isolates.
Batch cultures of Campylobacter strains were prepared by harvesting cells from overnight Columbia blood agar (Oxoid) containing 5% defibrinated horse blood and placing these cells into 50-ml portions of CPMOD4 broth (28) (Bacto Tryptone [Difco], 10.00 g liter−1; proteose peptone [Oxoid], 10.00 g liter−1; glucose, 1.00 g liter−1; yeast extract [Difco], 2.00 g liter−1; sodium chloride, 5.00 g liter−1; pyruvic acid [sodium salt], 1.10 g liter−1) in 100-ml unbaffled Erlenmeyer flasks. The flasks were incubated overnight in an orbital shaker (37°C, 150 rpm, 5% O2–95% N2 atmosphere). Members of the autochthonous water microfloras from the two-stage model and from frozen bead storage were routinely cultured on R2A agar. Plates were incubated at ambient temperature for 7 days.
Water source and sterilization.
To provide water having a standardized chemical and bacteriological content, a single tap (bore hole source 192A1; Porton Down, Salisbury, United Kingdom) was used to supply water for all of the microcosm and two-stage continuous-culture water model experiments; this water was designated 192A1 water. Small volumes of water for microcosm experiments were routinely sterilized by autoclaving (15 lb/in2, 15 min), although for comparative purposes some duplicate volumes were also sterilized by filtration (0.2-μm-pore-size capsule filter; type TDSLK 2NFZP; Pall, Portsmouth, United Kingdom). Larger volumes (20 liters) of water for the two-stage continuous-culture experiments were also sterilized by filtration as described above.
Survival of Campylobacter strains in water microcosms.
The ability of Campylobacter strains to persist in batch water microcosms was determined over a range of temperatures (4 to 37°C) and at different levels of oxygenation (aerobic, microaerobic, and anaerobic).
The inocula for the water microcosms were grown in batch cultures in CPMOD4 broth. The optical density at 540 nm of each culture was recorded, 1 ml of the culture was centrifuged (13,000 × g, 3 min), and the pellet was resuspended. The cells were then washed twice in sterile water and were resuspended to an appropriate density for the inoculum (see below). Water from the designated source (source 192A1) was sterilized by autoclaving 10-ml aliquots (15 lb/in2, 15 min) or in some cases by filtration. After autoclaving, the bottles were allowed to cool to room temperature overnight. An appropriate volume (between 15 and 30 μl) of inoculum (washed) was dispensed into each McCartney bottle used so that the starting cell concentration was 2 × 106 cells ml−1. The bottles were then placed at the appropriate temperature and oxygen tension. Anaerobic conditions (80% N2, 10% H2, 10% CO2) were created in large anaerobic churns that were evacuated and filled by using a gas manifold. Microaerobic conditions were provided by a controlled-atmosphere incubator (model ASSAB; Don Whitley Scientific Ltd., Shipley, West Yorkshire, United Kingdom). Aerobic conditions were obtained by using standard incubators. Persistence was subsequently measured by quantitative plate culture counting. Serial 100-fold dilutions were prepared from samples of the microcosms in sterile water, and 50-μl samples of each dilution were spread onto Columbia blood agar plates. Colony counts were determined after 48 h of incubation at 37°C in a microaerobic incubator (ASSAB). Samples used to determine colony counts were taken from the water microcosms at 2-h intervals for 10 h and then daily until no further colonies were detected; the detection limit was 20 CFU · ml−1.
Two-stage continuous-mixed-culture aquatic biofilm model.
The two-stage model was comprised of two culture vessels, a first-stage (seed) vessel and a second-stage (experimental) vessel, essentially as described previously (40). The continuous-culture vessels were designed so that they could be interconnected when necessary, to allow flow of culture from the seed vessel to the experimental vessel. The seed vessel was inoculated with autochthonous water flora (see below). This vessel was used to supply a consistent inoculum consisting of autochthonous microflora to all subsequent experimental vessels. Once the experimental vessels were established with a consistent autochthonous population, they were deliberately inoculated with C. jejuni for the persistence studies. Each of the culture vessels was supplied constantly with filter-sterilized tap water (192A1 water) from separate reservoir bottles.
The continuous-culture equipment used in the water model experiments was essentially identical to the equipment described previously (28) and was based on LH Series 500 (Inceltech UK Ltd., Reading, United Kingdom) and Anglicon (Brighton Systems Ltd., Hove, United Kingdom) fermentation systems. The 1-liter culture vessel was short and wide in order to increase the ratio of surface area to volume of the culture and to maximize the formation of a biofilm. In addition to the probes and exit and entry ports, there were also four sets of glass coupons (10 by 12 by 1 mm) that were suspended (three coupons per set) in each culture vessel on titanium wires so that biofilm samples could be harvested. The top plate and sample port hood were also constructed of titanium, while all of the entry and exit ports were made of glass in order to eliminate extraneous iron from the culture. The culture vessel and its fittings were sterilized by autoclaving and were assembled aseptically.
The inoculum for the seed vessel was prepared by passing 10 liters of tap water (192A1 water) through a 0.2-μm-pore-size, 140-mm-diameter, nylon membrane disk filter (Pall) in a stainless steel housing (Sartorius) which had previously been autoclaved (15 lb/in2, 15 min). The filtered organisms were resuspended by shaking the filter overnight in a sterile 1-liter flask containing 100 ml of 192A1 water. The suspension was then centrifuged aseptically, and the pellet was resuspended in 10 ml of 192A1 water and used as the inoculum for the seed vessel.
After inoculation, the vessel was maintained in the batch mode for 21 days to allow the formation of a biofilm. After this, fresh sterile tap water was continuously added to the vessel with a peristaltic pump. The water used in all of these studies (192A1 water) was from the same source and was filter sterilized (0.2-μm-pore-size capsule filter; type TDSLK 2NFZP; Pall) directly into the reservoir bottle (10 liters; polypropylene; Nalgene). The working volume of the culture was maintained at 500 ml by continuously removing excess culture through a tube in the top plate set at a fixed height within the vessel. The waste was continuously removed and transferred into a sterile collection bottle by using a second peristaltic pump. The rate of flow of freshwater into the vessel was maintained at 12.5 ml · h−1 (dilution rate = 0.025 h−1; mean generation time = 27.7 h), and the temperature used was the ambient temperature.
The culture parameters (pH, Eh, dissolved oxygen tension, temperature) were routinely monitored. Planktonic-phase samples were regularly removed and appropriately diluted, and 100 μl of each dilution was spread onto R2A agar to determine autochthonous microflora counts. The progress of biofilm development was determined by carefully removing glass coupons and examining them by light microscopy. The biofilm population distribution was determined by scraping the surface deposits from duplicate glass coupons into sterile water with a sterile dental pick and plating appropriate dilutions onto R2A agar as described above. The viable cell count and autochthonous microflora population distribution in the seed vessel as determined by colonial and cell morphology stabilized 30 days postinoculation, after which the seed vessel was used to supply inocula for the experimental vessels.
The experimental vessels were assembled in a similar manner. An experimental vessel was inoculated by directing the entire flow of effluent from the seed vessel into it for 7 days. After this, the effluent flow from the seed vessel was redirected into its waste bottle. Then the experimental vessel received a continuous flow of fresh filter-sterilized water. The conditions established in the experimental vessels included different oxygen tensions (no O2, 4% O2, and ambient O2) and temperatures (4 and 30°C) on different occasions.
The experimental vessels were allowed to equilibrate for an additional 14 days before being challenged with C. jejuni CH1 or 9752 cells. These cells were harvested from overnight batch cultures as described above. The challenge inocula were prepared aseptically and washed twice in sterile 192A1 water. The cell concentrations of the washed suspensions were adjusted so that 10 ml of inoculum resulted in a starting cell concentration in the experimental vessel of 2 × 107 cells ml−1 (as determined by direct counting).
In every case the persistence of the challenge strain was determined by determining colony counts with serial 100-fold dilutions of the planktonic phase in sterile water. Each dilution (100 μl) was spread onto blood agar base (Oxoid) plates containing Skirrow selective supplement (Oxoid), and the plates were incubated microaerobically for 48 h. Persistence in the biofilms was also monitored directly by fluorescence microscopy of glass coupons removed from the cultures by using a Campylobacter-specific antibody and rRNA probes.
Monoclonal antibodies.
Monoclonal antibodies were developed in our laboratory by using standard techniques. The immunogen was an outer-membrane preparation of C. jejuni NCTC 11828. The cellular material for the outer-membrane preparation was obtained from cultures which had previously been grown in continuous culture (28) under defined conditions (iron-limited ABCD medium).
Immunofluorescence detection of Campylobacter strains.
Glass coupons coated with biofilm obtained from the water model experiment (see below) were washed gently with sterile water, fixed with acetone, and air dried. The coupons were then supported on microscope slides. A monoclonal antibody was appropriately diluted with fetal calf serum–phosphate-buffered saline (1:10, vol/vol), and 50-μl aliquots were dispensed onto each coupon. The slides were incubated in a humidified darkened box at 37°C with gentle shaking for 45 min. The slides were then gently washed in sterile water and placed in a heated cabinet (∼40°C) to air dry. Fifty microliters of appropriately diluted anti-immunoglobulin G- or anti-immunoglobulin M-specific goat anti-mouse fluorescein isothiocyanate-labelled conjugate (Sigma) was then placed on each slide, and the slides were incubated as previously described. The slides were again washed gently with sterile water and air dried. Coverslips were placed over each coupon along with approximately 5 to 10 μl of mountant and fluorescence enhancer Citifluor AFI (UKC Chem Lab, Canterbury, United Kingdom). The slides were examined by epifluorescence microscopy (Leitz model Dialux 20EB microscope) by using a water immersion objective (magnification, ×50), a 50-W mercury lamp, a type KP 490 blue excitation filter, a type K530 barrier filter, and a type TK510/K515 dichroic beam splitting mirror. Photographs were taken by using a Leitz Vario Orthomat camera and Ektachrome 320 tungsten color reversal film (Kodak).
rRNA probes: cell fixation, oligonucleotide hybridization, and microscopy.
Coupons were removed from the water model vessel (see below), air dried, placed in 4% paraformaldehyde for 3 h, washed with phosphate-buffered saline, and air dried again. The dry coupons were immersed in ice-cold filtered (pore size, 0.2 μm; Millipore) ethanol in 1-oz McCartney bottles for about 10 s. The biofilms on the coupons were then dehydrated by subjecting the coupons to a 50% ethanol–80% ethanol–98% ethanol series. The coupons were then each fixed to a glass slide by using a spot of cyanoacrylate adhesive, which was allowed to cure for 1 h. An oligonucleotide probe complementary to a region of the 16S rRNA of all known thermophilic Campylobacter species (R & D Systems, Abingdon, United Kingdom) was used to specifically detect the campylobacters, while a second probe complementary to a region of the 16S rRNA conserved in the domain Bacteria (4) was used as a positive control to detect all eubacteria. The Campylobacter probe was labelled with tetramethylrhodamine-5-isothiocyanate, and the eubacterial oligonucleotide probe was labelled with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester, which resulted in double labelling. For each hybridization, 8 μl of hybridization buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl [pH 7.2], 35% formamide) was mixed with 1 μl (50 ng) of the specific probe and 1 μl (50 ng) of the eubacterial probe. This mixture was added to each fixed biofilm. The coupon was then incubated at 46°C for 90 min in a humidity chamber. After hybridization, the excess probe was removed by washing the preparation in 50 ml of wash buffer (0.07 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl [pH 7.2], 5 mM EDTA) at 50°C for 30 min. The coupon was then rinsed with deionized H2O, allowed to air dry, and mounted in AFI. Probe-conferred fluorescence was visualized and photographed by using an Olympus model BX-FLA epifluorescence microscope fitted with Olympus filter cube sets U-MNB, U-MWG, and U-MWU.
RESULTS
Persistence of C. jejuni strains in batch water microcosms.
In contrast with previous studies, a relatively large number of isolates from a range of different sources (poultry, water, human clinical material) were compared in this study. Most of the isolates were C. jejuni isolates; the exceptions were Campylobacter coli 247 and 799 and C. coli-Campylobacter upsaliensis 2172. A number of different serotypes were represented, although all three chicken isolates were members of the same serotype (1).
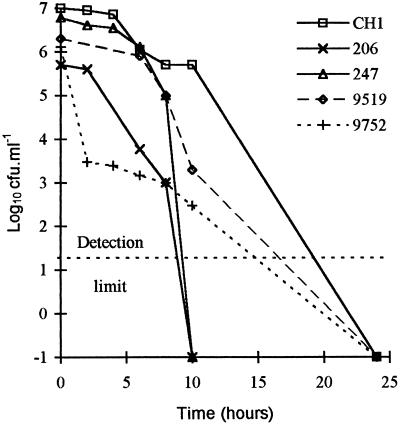
Culturability in the water microcosms declined progressively with time, and different strains displayed different death curve profiles. Figure 1 shows data for five strains with contrasting survival times. Survival was subsequently most reliably determined for comparative purposes by determining the time that it took for each cell population to cease to be culturable, since other parameters, such as the slope of the death curve, varied with time in many instances. Survival times varied considerably between strains; the survival times of some strains differed by more than twofold (Tables 2 and 3), and the survival times differed by fourfold when particular combinations of temperature and oxygenation were used. The differences between strains were statistically significant and independent of temperature when data were analyzed by using a multifactor analysis of variance procedure (P < 0.05, as determined by Statgraphics, version 5 [Manguistics]).
FIG. 1.
Decline in culturability of selected C. jejuni strains at 37°C in static aerobic microcosms. Points are means from duplicate determinations.
TABLE 2.
Comparison of the survival times of different Campylobacter strains in water microcosms over a range of temperatures under aerobic conditions
| Strain | Persistence time (h) ata:
|
Mean persistence time (h) | |||
|---|---|---|---|---|---|
| 4°C | 10°C | 22°Cb | 37°C | ||
| 9519 | 218 | 216 | 36 | 36 | 126.5 |
| 247 | 120 | 144 | 24 | 24 | 78.0 |
| 206 | 288 | 240 | 48 | 16 | 148.0 |
| 235 | 190 | 216 | 48 | 24 | 119.5 |
| 799 | 157 | 144 | 36 | 17 | 88.5 |
| 10392 | 170 | 168 | 29 | 24 | 97.8 |
| 2172 | 144 | 120 | 36 | 24 | 81.0 |
| 9752 | 157 | 108 | 48 | 14 | 81.8 |
| 81116 | 228 | 192 | 48 | 17 | 121.3 |
| 11168 | 289 | 168 | 48 | 24 | 132.3 |
| C677 | 276 | 192 | 48 | 17 | 133.3 |
| CH1 | 229.5 | 288 | 48 | 24 | 147.4 |
| CH5 | 229.5 | 240 | 48 | 24 | 135.4 |
| W1 | 190 | 120 | 48 | 9 | 91.8 |
| W2 | 168 | 120 | 48 | 17 | 88.3 |
| W7 | 156 | 120 | 36 | 36 | 87.0 |
| C53 | 218 | 192 | 48 | 24 | 120.5 |
| Mean | 201.6 | 175.8 | 42.6 | 21.8 | |
All survival times are means from duplicate determinations. The average coefficient of variation for the entire data set was 23%. A multifactor analysis of variance revealed statistically significant differences in survival times between strains (P < 0.05) and between temperatures (P < 0.0005); there was no interaction between these two factors.
Room temperature was nominally 22°C.
TABLE 3.
Comparison of the survival times of different Campylobacter strains in water microcosms at 10°C under aerobic and anaerobic conditions
| Strain | Persistence time (h)a
|
|
|---|---|---|
| Anaerobic | Aerobic | |
| 9519 | 168 | 216 |
| 247 | 96 | 144 |
| 206 | 144 | 240 |
| 235 | 144 | 216 |
| 799 | 216 | 144 |
| 10392 | 372 | 168 |
| 2172 | 72 | 120 |
| 9752 | 168 | 108 |
| 81116 | 240 | 192 |
| 11168 | 156 | 168 |
| C677 | 300 | 192 |
| CH1 | 408 | 288 |
| CH5 | 408 | 240 |
| W1 | 177 | 120 |
| W2 | 372 | 120 |
| W7 | 264 | 120 |
| C53 | 144 | 192 |
| Mean | 226.4 | 175.8 |
All survival times are means from duplicate determinations. The average coefficient of variation for the entire data set was 36%. There was no statistically significant difference in survival times between aerobic and anaerobic conditions.
The influence of temperature on survival time was also highly significant (P < 0.0005). The mean survival times were much longer at 4 and 10°C (202 and 176 h, respectively) than at 22 and 37°C (43 and 22 h, respectively), and the greatest change in survival times occurred between 22 and 10°C (Table 2).
Small differences in survival times were observed with the different levels of oxygenation. At 37°C the differences between the average survival times for all strains under aerobic conditions (22 h), microaerobic conditions (24 h), and anaerobic conditions (24 h) were not marked. At 10°C the difference between average survival times (Table 3) was greater, with the survival time under anaerobic conditions (226 h) being rather longer than the survival time under aerobic conditions (176 h), although the difference was not statistically significant.
The survival times of four isolates (9752, CH1, 81116, and 247) were also determined in filter-sterilized water under aerobic conditions at 10°C, room temperature, and 37°C. There were no differences in the survival times of the isolates in filter-sterilized water compared to the survival times in autoclaved water (data not shown).
Two-stage water model.
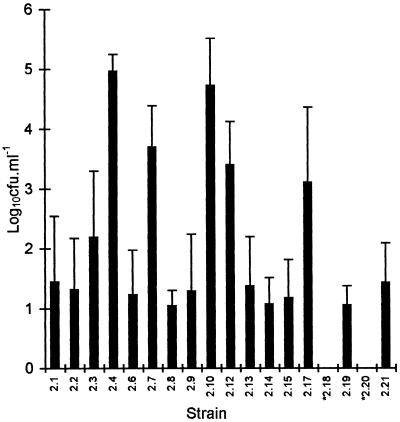
The mean viable cell count for the planktonic autochthonous microflora culturable on R2A in the seed and experimental vessels stabilized at 2.5 × 105 CFU · ml−1 after 21 days. The count fluctuated somewhat with time (standard deviation, 1.5 × 105 CFU · ml−1), and the maximum and minimum counts were 7.3 × 105 and 3.5 × 104 CFU · ml−1, respectively. The characteristics of the individual members of the culturable autochthonous microflora have been reported elsewhere (7). Although there were slight changes with temperature and oxygenation, the population distribution of the constituent bacteria remained largely constant in established cultures, with the individual counts varying in synchrony with fluctuations in the total population (Fig. 2).
FIG. 2.
Distribution of the culturable autochthonous water flora population. The solid bars show the means from 30 replicate determinations. The error bars indicate one standard deviation. Strains 2.18 and 2.20 were present intermittently at concentrations of ≥102 cells ml−1.
The mean viable count for autochthonous organisms in the biofilms in the seed and experimental vessels was 2.9 × 105 cells per cm2, and the viable counts ranged between 2.1 × 105 and 3.8 × 105 cells per cm2. The biofilm viable count remained essentially constant regardless of the changes in culture parameters. The population distribution of the constituent biofilm microflora also remained constant under the different culture conditions, paralleling that of the planktonic-phase population. Microscopic examination of the biofilms on the coupons demonstrated they had a heterogeneous structure composed of large numbers of microcolonies of bacteria interspersed with more sparsely populated areas (data not shown).
Challenge with C. jejuni.
The experimental vessels were challenged with either C. jejuni CH1 or C. jejuni 9752. These strains were selected as strains which had demonstrated good survival (CH1) and poor survival (9752) in the batch water microcosms.
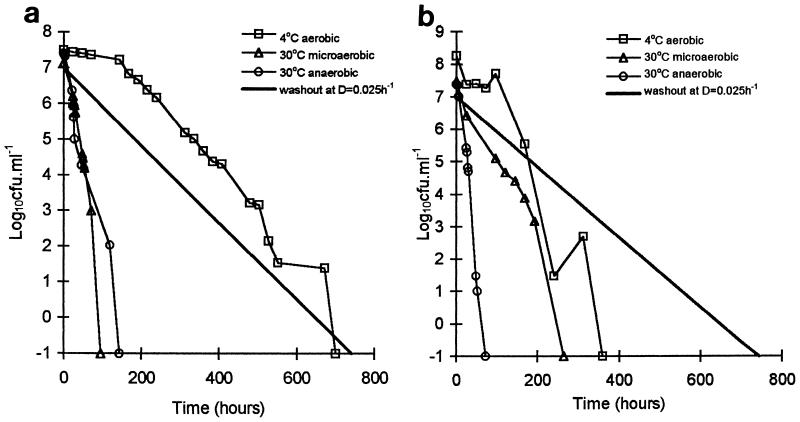
The culturability of C. jejuni CH1 and 9752 declined progressively after introduction into the experimental vessels under all oxygenation and temperature conditions examined (Fig. 3). The rate of decline was usually much faster than the rate of decline which would be expected to result from the culture dilution rate alone in the absence of cell division in the campylobacters; the only exception was the rate of decline of strain CH1 at 4°C, which was similar to the dilution rate. The survival times of both strains were always greater at 4°C than at 30°C. The level of oxygen also influenced survival. The survival time of strain CH1 at 30°C was slightly reduced when this organism was transferred from anaerobic conditions (144 h) to microaerobic conditions (96 h), whereas strain 9752 showed the reverse effect, with a more-than-threefold increase in survival time when it was transferred from anaerobic conditions (72 h) to microaerobic conditions (264 h). The interaction of aerobiosis and temperature influenced the survival of the two strains differently. At 30°C under anaerobic conditions and at 4°C under aerobic conditions, strain CH1 survived for approximately twice as long (144 and 700 h, respectively) as 9752 (72 and 360 h, respectively). At 30°C under microaerobic conditions, however, strain 9752 survived nearly threefold longer than CH1 (264 and 96 h, respectively). Thus, while anaerobiosis slightly increased the survival time of strain CH1 at 30°C, anaerobiosis markedly diminished the survival time of strain 9752.
FIG. 3.
Decline in culturability of C. jejuni CH1 (a) and C. jejuni 9752 (b) in the two-stage water model. Points are means from six replicate determinations; two separate cultures were analyzed in triplicate. D, dilution rate.
Survival times were greater in the water model than in the water microcosms. At 4°C strain 9752 exhibited a twofold increase (157 to 360 h) and strain CH1 exhibited a threefold increase (230 to 700 h) in survival time in the water model compared with the microcosms. Strain CH1 remained the more persistent of the two strains.
Indirect immunofluorescence.
Twenty-two different anti-Campylobacter monoclonal antibodies and six polyclonal rabbit antisera were evaluated. With several of these preparations, an indirect immunofluorescence analysis of biofilms on glass coupons presumptively revealed the presence of whole cells of C. jejuni CH1 and 9752 long after these organisms had ceased to be culturable. However, some nonspecific fluorescence was observed with the background autochthonous water microflora in the native biofilms; such nonspecific fluorescence had not previously been identified as a problem with any of the individual culturable biofilm components. This made more definitive identification of the Campylobacter cells in the biofilms problematic (data not shown).
rRNA probes.
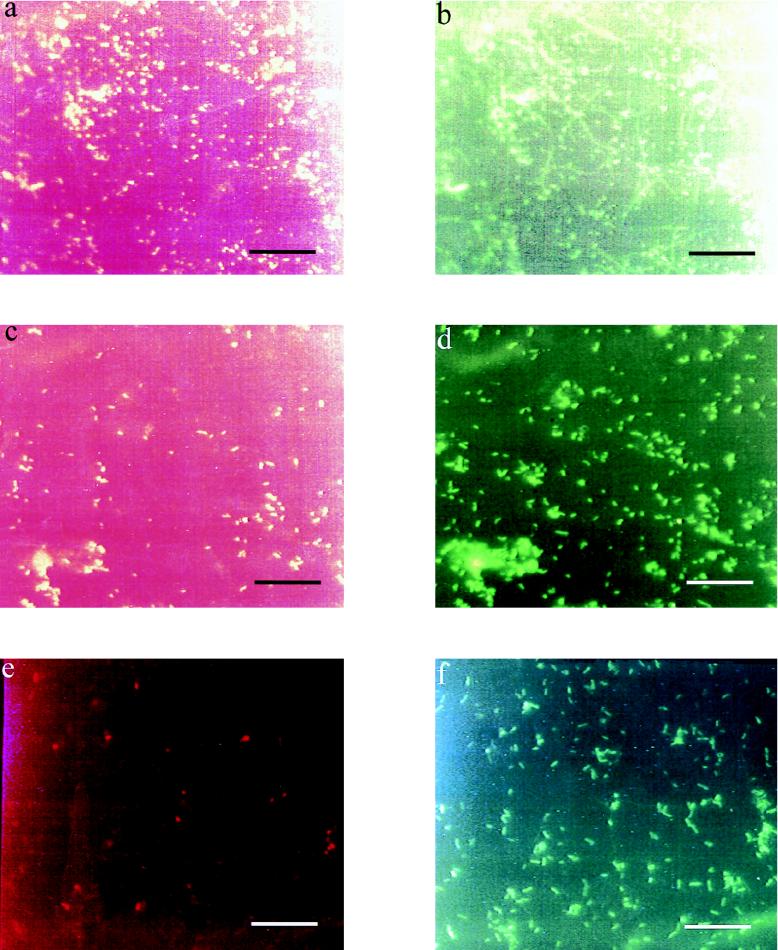
Nonspecific background fluorescence was much less problematic with the rRNA probes. Analysis of the biofilms by using Campylobacter-specific rRNA probes and fluorescence microscopy demonstrated that C. jejuni was present for at least 28 days at 30°C and for 42 days at 4°C following introduction into the experimental vessels. By this time the majority of the cells were spiral at 4°C but predominantly coccal at 30°C, as determined by epifluorescence microscopy. The rRNA probe signal also remained stronger at 4°C than at 30°C. At 4°C both CH1 and 9752 were detected equally after they had lost culturability at 700 and 360 h, respectively; this continued up to the point at which the experiments were terminated (42 days) (Fig. 4).
FIG. 4.
Micrographs of biofilm samples on glass coupons taken from the water model at two temperatures 42 days after contamination with strains of C. jejuni and stained with fluorescently labelled rRNA probes specific to the genus Campylobacter (red) and eubacteria (green). (a and b) Strain CH1 at 30°C. (c and d) Strain CH1 at 4°C. (e and f) Strain 9752 at 4°C. Bars = 20 μm.
DISCUSSION
In this study, we compared the survival of a range of Campylobacter isolates from a number of representative sources by using batch water microcosm experiments and a two-stage aquatic biofilm model. The former greatly extended the scope of the previously published experiments, while the latter system addressed novel factors related to potential associations with the autochthonous water microflora and biofilms.
In the water microcosms, the profiles of the death curves for the different Campylobacter isolates varied considerably. In most cases the initial decline in cell numbers was relatively slow and was followed by an increasingly rapid decline with time. However, in some instances either no initial lag occurred or the rate of decline decreased with time, which presumably reflected differences in the survival characteristics of strains.
Isolates differed significantly from one another in the length of time that they remained culturable in the microcosms; the survival times varied up to twofold when data for the various temperatures were averaged but up to fivefold when particular combinations of temperature and oxygenation were used. No significant differences were found (22, 48) in the small number of previous studies in which different C. jejuni isolates were compared. Previously, only differences between species have been reported, and C. coli apparently survived less well than C. jejuni in surface water (25, 26). By contrast, in our study, the survival times of the two C. coli isolates (strains 247 and 799) and the C. coli-C. upsaliensis isolate (strain 2172) were indistinguishable from the survival times of the C. jejuni isolates, and the variability between isolates was at least as great as the variability between species. Indeed, any correlation between species, serotype, or source of isolates and survival time was not obvious. Two of the strains (CH1 and CH5), however, which consistently exhibited the greatest survival times were members of the same serotype (1) and species (C. jejuni), and both were chicken isolates; more isolates will have to be examined to determine whether such a correlation is meaningful.
There has been little previously reported data concerning the direct effect of aerobiosis on the persistence of Campylobacter strains. In aquatic environments dissolved oxygen concentrations might be expected to be reduced by lower water flow rates, reduced turbulence, increased temperature, and the presence of competing microflora and organic matter, and it has been suggested previously that the latter two parameters reduce the survival of C. jejuni (48). On average, and in keeping with the microaerophilic nature of the pathogen, lower oxygen tensions in our study increased survival. However, the effect of anaerobiosis was not as consistent or as pronounced as might have been expected and was not statistically significant. Contrary to what might have been predicted, anaerobiosis actually appeared to be detrimental to the survival of certain isolates in some instances (for example, isolates 9519, 206, 235, 2172, and C53 at 10°C).
As expected from previous studies (22, 48), temperatures below 22°C were found to significantly increase survival. Some isolates remained culturable at 4 and 10°C for extended periods of time (more than 250 h), but they remained culturable for much shorter periods of time at 22 and 37°C, at which the maximum survival time was 48 h. The greatest change in survival time occurred between 10 and 22°C (the mean survival time under all conditions decreased from about 175 to 45 h). Although they used a different experimental system, Terzieva and McFeters (48) found that the survival times of C. jejuni in untreated stream water were similar at 6 and 16°C. This suggests that the threshold at which the survival time becomes markedly shorter is between 16 and 22°C, values which are closer to 22°C than to 10°C.
Based on the microcosm experiments, the survival of Campylobacter strains in nutrient-depleted aquatic systems appears to be dependent on a number of factors. Although lower temperatures generally promoted the survival of the pathogen, temperatures of 10 to 16°C and below appeared to especially encourage the survival of the organism, with survival continuing for up to 10 to 16 days for selected strains. The survival time at room temperature, by contrast, was always less than 2 days. The intrinsic variation between isolates was considerable, however, with some isolates surviving for comparatively short periods of time (3 to 4 days or less) even at 10°C. The degree of oxygenation influenced survival in an unpredictable manner, and the effect was particularly noticeable at the lower temperatures. The survival of some isolates improved at lower oxygen levels, whereas the survival of other isolates was reduced.
The two Campylobacter isolates (CH1 and 9752) which exhibited contrasting survival times in the water microcosms continued to show the same twofold disparity in survival times overall when they were introduced into the two-stage water biofilm model. Interestingly, however, the survival times, as measured by culturing, were considerably longer for both strains in this model than the survival times obtained previously in otherwise equivalent batch microcosm experiments. This is despite the fact that in this part of the study it was necessary to use selective plates for the recovery of Campylobacter strains in order to suppress the background autochthonous microflora. The use of selective plates has been shown to reduce the recovery of injured forms of Campylobacter strains (16, 19), and we expected that this method would reduce the apparent ability of the pathogen to persist. It seems that the background autochthonous water microflora and/or the presence of a biofilm enhances the survival of the pathogen considerably. The survival times were approximately doubled, up to 700 h, for CH1 and up to 350 h for 9752 at 4°C, for example. This observation is in agreement with studies on other pathogens which have demonstrated that integration into aquatic biofilms and interaction with the constituent microflora can enhance pathogen survival (8, 29, 34, 40). It will be interesting to determine whether mechanisms of interspecies specific interaction (coaggregation) are involved in the incorporation of Campylobacter strains and other pathogens into aquatic biofilms, since coaggregation appears to have a role in the interstrain binding of different members of the autochthonous water microflora (7).
In the absence of cell multiplication, cell death, or cell partitioning between the biofilm and the bulk water phase, the number of culturable Campylobacter cells in the experimental vessels was expected to decline at a rate which matched the dilution rate. The rates of loss of culturable cells from the experimental vessels in all but one case exceeded the dilution rate, revealing a contribution to the kinetics of either the loss of culturability of the cells and the adoption of a viable nonculturable state (or sublethal injury) or cell death. Only with isolate CH1 at 4°C did the cell number follow the dilution rate, indicating that much less significant cell death, if any, occurred over the entire 700 h (4 weeks) of the experiment. Loss of culturable cells in this case was most simply explicable in terms of cells being slowly washed out of the experimental vessel with the flow of water through it. This interpretation allows for no cell multiplication within the population of C. jejuni, but considering the low temperature, this is probably realistic.
As in the water microcosm experiments, the higher temperature in the two-stage water model was detrimental to the persistence of both isolates, despite the fact that at 30°C growth and division of C. jejuni are ordinarily supported in less hostile environments, such as laboratory batch media, and growth below this temperature is exceptional. Indeed, this temperature was chosen for this part of the study in order to maximize the potential for the pathogen to grow in the water model. Although increasing the temperature from 4 to 30°C reduced the survival time of CH1 considerably from 4 weeks to 4 to 5 days, isolate 9752 was much less affected, particularly under aerobic conditions, under which the survival time decreased from about 15 to 10 days. Nevertheless, the survival times of both isolates, even at this high temperature, were considerably greater than the survival times of the 1 to 2 days observed in the 22°C water microcosms in the absence of an autochthonous microflora.
The two Campylobacter isolates differed markedly in their responses to increased oxygen concentrations in the two-stage water model. At 30°C the survival time of isolate CH1 was marginally reduced by increased oxygen tension when it was transferred from anaerobic conditions to microaerobic conditions, whereas the survival time of isolate 9752 was increased by more than threefold. This paralleled the inconsistent effects of oxygenation on Campylobacter isolates obtained in the microcosm experiments. Although the survival times of isolates CH1 and 9752 were both reduced by an increase in oxygenation in the earlier experiments, there were a number of other isolates (e.g., strains 206 and 235) whose survival times were unexpectedly increased by higher dissolved oxygen tensions in the microcosms, as observed with isolate 9752 in the two-stage model. It seems that the background autochthonous water microflora and/or the presence of a biofilm modulates the influence of oxygen tension on the persistence of Campylobacter strains.
Problems with nonspecific fluorescence were encountered when we used monoclonal antibodies with natural biofilm samples from the two-stage water model. The high background fluorescence with the autochthonous microflora in the native biofilms may have been the result of several contributory factors. Species in the autochthonous microflora may express cross-reactive epitopes, and Campylobacter strains may condition the biofilm matrix (conditioning film) such that the surface becomes coated with the reactive epitope (either by cell lysis or by cell excretion). Although the results are unsatisfactory, Campylobacter strains could probably still be presumptively identified in the biofilm consortium on the basis of staining in conjunction with morphology. Although the numbers of Campylobacter cells appeared to decrease with time, particularly at the beginning of each experiment, Campylobacter cells persisted in the biofilm long after the cells had ceased to be culturable and right up to the termination of the experiment in all cases.
In order to improve the specificity of detection of the pathogen in the biofilm samples, a fluorescently labelled Campylobacter-specific rRNA probe was utilized in conjunction with a differentially labelled eubacterial probe which was used to simultaneously visualize the rest of the biofilm microflora. rRNA probes have been used similarly with success in many areas, including studies of biofilm structure, particularly with reference to cell-cell associations (32), and for the detection of pathogens in aquatic biofilms (31, 50). In our study, some nonspecific background was occasionally encountered with samples from the two-stage vessel; this background was probably due to difficulties with the hybridization conditions or detachment of the fluorescein label from the oligonucleotide during the hybridization process, which then bound to other constituent organisms in the biofilm. Nevertheless, the Campylobacter-specific rRNA probe was found to be more specific than the monoclonal antibody probes and not only was able to reveal the presence of Campylobacter strains in the biofilm samples but also highlighted cell morphology. Used in conjunction with the eubacterial probe, the Campylobacter-specific probe revealed an extended association of Campylobacter strains with the aquatic biofilm right up to the point at which the experiments were terminated (28 days at 30°C and 42 days at 4°C), long after Campylobacter strains had ceased to be culturable. In the majority of cases the Campylobacter strains were associated with microcolonies of autochthonous biofilm microflora, suggesting that the pathogen was incorporated within the biofilm matrix as opposed to being adherent to the surfaces of the coupons. The fluorescent signal from the Campylobacter cells was stronger with the biofilm samples at 4°C than at 30°C, and the cells tended to become coccal at 30°C but not at 4°C. The higher temperature, therefore, appeared to have promoted the transformation of the biofilm-associated Campylobacter cells into their coccal, possibly degenerate form; this correlated with an accelerated loss of rRNA signal compared to lower temperatures.
Despite the environmental sensitivity of Campylobacter cells, their survival in a culturable form in water can be sufficiently prolonged for this vehicle to be potentially important in the transmission of the pathogen, particularly at low temperatures and when the organism is associated with a biofilm, where survival can be measured in weeks. The considerable strain variation in survival could contribute to certain strains being of particular concern for human and animal infection. The significance of more extended persistence in a nonculturable form, as determined by specific rRNA probes in this study, for example, still remains to be addressed in relation to the concept of viable but nonculturable states. This could have significant consequences for further transmission of the organisms. Our studies in which potable water was used should be extended to determine the influence of water quality parameters, such as temperature and oxygen tension, on, for example, the water distribution systems in animal husbandry facilities.
ACKNOWLEDGMENTS
This work was funded by the Department of Health, London.
We are very grateful to Tom Humphrey of the Exeter Public Health Laboratory and Peter Hawtin of the Southampton Public Health Laboratory for supplying the Campylobacter strains. Typing of the isolates was kindly undertaken by Jenny Frost and Bernard Rowe of the Laboratory of Enteric Pathogens (Central Public Health Laboratory, Colindale, London, United Kingdom). The monoclonal antibodies were produced by the Animal Cell Technology Department, Centre for Applied Microbiology and Research Resources Division. We also thank Mark Linton and Geraldine Murphy for technical assistance in the rRNA probe work.
REFERENCES
- 1.Advisory Committee on the Microbiological Safety of Foods. Interim report on campylobacter. London, United Kingdom: Her Majesty’s Stationery Office; 1993. [Google Scholar]
- 2.Advisory Committee on the Microbiological Safety of Foods. Report on poultry meat. London, United Kingdom: Her Majesty’s Stationery Office; 1996. [Google Scholar]
- 3.Alary M, Nadeau D. An outbreak of Campylobacter enteritis associated with a community water supply. Can J Public Health. 1990;81:268–271. [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beumer R R, de Vries J, Rombouts F M. Campylobacter jejuni non-culturable coccoid cells. Int J Food Microbiol. 1992;15:153–163. doi: 10.1016/0168-1605(92)90144-r. [DOI] [PubMed] [Google Scholar]
- 6.Blaser M J, Hardesty H L, Powers B, Wang W L. Survival of Campylobacter fetus subsp. jejuni in biological milieus. J Clin Microbiol. 1980;11:309–313. doi: 10.1128/jcm.11.4.309-313.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buswell C M, Herlihy Y M, Marsh P D, Keevil C W, Leach S A. Coaggregation amongst aquatic biofilm bacteria. J Appl Microbiol. 1997;83:477–484. [Google Scholar]
- 8.Colbourne J S, Pratt D J, Smith M G, Fischer-Hock S P, Harper D. Water fittings as sources of Legionella pneumophila in a hospital plumbing system. Lancet. 1984;i:210–213. doi: 10.1016/s0140-6736(84)92126-3. [DOI] [PubMed] [Google Scholar]
- 9.Costerton J W, Cheng K-J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 10.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 11.Department of Health. On the state of the public health 1994. London, United Kingdom: Her Majesty’s Stationery Office; 1995. [Google Scholar]
- 12.Frost, J. A., A. N. Oza, R. T. Thwaites, and B. Rowe. A serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J. Clin. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 13.Gilbert P, Evans D J, Brown M R W. Formation and dispersal of bacterial biofilms in vivo and in situ. J Appl Bacteriol. 1993;74:67s–78s. doi: 10.1111/j.1365-2672.1993.tb04343.x. [DOI] [PubMed] [Google Scholar]
- 14.Hazeleger W C, Janse J D, Koenraad P M, Beumer R R, Rombouts F M, Abee T. Temperature-dependent membrane fatty acid and cell physiology changes in coccoid forms of Campylobacter jejuni. Appl Environ Microbiol. 1995;61:2713–2719. doi: 10.1128/aem.61.7.2713-2719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins R S, Olmsted R, Istre G R. Endemic Campylobacter jejuni infection in Colorado: identified risk factors. Am J Public Health. 1984;74:249–250. doi: 10.2105/ajph.74.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphrey T J. Techniques for the optimum recovery of cold injured Campylobacter jejuni from milk or water. J Appl Bacteriol. 1986;61:125–132. doi: 10.1111/j.1365-2672.1986.tb04265.x. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey T J, Beckett P. Campylobacter jejuni in dairy cows and raw milk. Epidemiol Infect. 1987;98:263–269. doi: 10.1017/s0950268800062014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphrey T J, Henley A, Lanning D G. The colonization of broiler chickens with Campylobacter jejuni: some epidemiological investigations. Epidemiol Infect. 1993;110:601–607. doi: 10.1017/s0950268800051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey T J, Martin K W, Mason M J. Isolation of Campylobacter species from non-clinical samples. Public Health Lab Serv Microbiol Digest. 1996;13:86–88. [Google Scholar]
- 20.Jacob J, Martin W, Holler C. Characterization of viable but nonculturable stage of C. coli, characterized with respect to electron microscopic findings, whole cell protein and lipooligosaccharide (LOS) patterns. Zentralbl Mikrobiol. 1993;148:3–10. [PubMed] [Google Scholar]
- 21.Jacobs-Reitsma W F, van de Giessen A W, Bolder N M, Mulder R W. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol Infect. 1995;114:413–421. doi: 10.1017/s0950268800052122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones D M, Sutcliffe E M, Curry A. Recovery of viable but non-culturable Campylobacter jejuni. J Gen Microbiol. 1991;137:2477–2482. doi: 10.1099/00221287-137-10-2477. [DOI] [PubMed] [Google Scholar]
- 23.Kazwala R R, Collins J D, Hannan J, Crinion R A, O’Mahony H. Factors responsible for the introduction and spread of Campylobacter jejuni infection in commercial poultry production. Vet Rec. 1990;126:305–306. [PubMed] [Google Scholar]
- 24.King C H, Shotts E B, Jr, Wooley R E, Porter K G. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl Environ Microbiol. 1988;54:3023–3033. doi: 10.1128/aem.54.12.3023-3033.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korhonen L K, Martikainen P J. Comparison of the survival of Campylobacter jejuni and Campylobacter coli in culturable form in surface water. Can J Microbiol. 1991;37:530–533. doi: 10.1139/m91-089. [DOI] [PubMed] [Google Scholar]
- 26.Korhonen L K, Martikainen P J. Survival of Escherichia coli and Campylobacter jejuni in untreated and filtered lake water. J Appl Bacteriol. 1991;71:379–382. doi: 10.1111/j.1365-2672.1991.tb03804.x. [DOI] [PubMed] [Google Scholar]
- 27.Kramer M H, Herwaldt B L, Craun G F, Calderon R L, Juranek D D. Surveillance for waterborne-disease outbreaks—United States, 1993–1994. Morbid Mortal Weekly Rep. 1996;45:1–33. [PubMed] [Google Scholar]
- 28.Leach S A, Harvey P C, Wait R. Changes with growth rate in the membrane lipid composition of and amino acid utilisation by continuous cultures of Campylobacter jejuni. J Appl Microbiol. 1997;82:631–640. doi: 10.1111/j.1365-2672.1997.tb02873.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee J V, West A A. Survival and growth of Legionella species in the environment. J Appl Bacteriol. 1991;70:121s–129s. [PubMed] [Google Scholar]
- 30.Lindblom G B, Sjorgren E, Kaijser B. Natural campylobacter colonization in chickens raised under different environmental conditions. J Hyg. 1986;96:385–391. doi: 10.1017/s0022172400066146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz W, Amann R, Szewzyk R, Szewzyk U, Stenstrom T A, Hutzler P, Schleifer K H. In situ identification of Legionellaceae using 16S rRNA-targeted oligonucleotide probes and confocal laser scanning microscopy. Microbiology. 1995;141:29–39. doi: 10.1099/00221287-141-1-29. [DOI] [PubMed] [Google Scholar]
- 32.Manz W, Szewzyk U, Ericsson P, Amann R, Schleifer K H, Stenstrom T A. In situ identification of bacteria in drinking water and adjoining biofilm by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl Environ Microbiol. 1993;59:2293–2298. doi: 10.1128/aem.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medema G J, Schets F M, van de Giessen A W, Havelaar A H. Lack of colonization of 1 day old chicks by viable, non-culturable Campylobacter jejuni. J Appl Bacteriol. 1992;72:512–516. doi: 10.1111/j.1365-2672.1992.tb01868.x. [DOI] [PubMed] [Google Scholar]
- 34.Payment P, Gamache F, Paquette G. Microbiological and virological analysis of water from two water filtration plants and their distribution systems. Can J Microbiol. 1988;34:1304–1309. doi: 10.1139/m88-228. [DOI] [PubMed] [Google Scholar]
- 35.Pearson A D, Greenwood M, Healing T D, Rollins D, Shahamat M, Donaldson J, Colwell R R. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl Environ Microbiol. 1993;59:987–996. doi: 10.1128/aem.59.4.987-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson A D, Healing T D. The surveillance and control of campylobacter infection, 2, R133–R139. Public Health Laboratory Service; 1992. Communicable Disease Report. [PubMed] [Google Scholar]
- 37.Penner J L, Pearson A D, Hennessy J N. Investigation of a waterborne outbreak of Campylobacter jejuni enteritis with a serotyping scheme based on thermostable antigens. J Clin Microbiol. 1983;18:1362–1365. doi: 10.1128/jcm.18.6.1362-1365.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickert A, Botzenhart K. Survival of Campylobacter jejuni in drinking water, river water and sewage. Zentralbl Bakteriol Mikrobiol Hyg Ser B. 1985;182:49–57. [PubMed] [Google Scholar]
- 39.Reasoner D J, Geldrich E E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers J, Dowsett A B, Dennis P J, Lee J V, Keevil C W. Influence of plumbing materials on biofilm formation and growth of Legionella pneumophila in potable water systems. Appl Environ Microbiol. 1994;60:1842–1851. doi: 10.1128/aem.60.6.1842-1851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rollins D M, Colwell R R. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl Environ Microbiol. 1986;52:531–538. doi: 10.1128/aem.52.3.531-538.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sacks J J, Lieb S, Baldy L M, Berta S, Patton C M, White M C, Bigler W J, Witte J J. Epidemic campylobacteriosis associated with a community water supply. Am J Public Health. 1986;76:424–428. doi: 10.2105/ajph.76.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skirrow M B. Epidemiology of Campylobacter enteritis. Int J Food Microbiol. 1991;12:9–16. doi: 10.1016/0168-1605(91)90044-p. [DOI] [PubMed] [Google Scholar]
- 45.Stehr-Green J K, Nicholls C, McEwan S, Payne A, Mitchell P. Waterborne outbreak of Campylobacter jejuni in Christchurch: the importance of a combined epidemiologic and microbiologic investigation. N Z Med J. 1991;104:356–358. [PubMed] [Google Scholar]
- 46.Stern N J, Jones D M, Wesley I V, Rollins D M. Colonisation of chicks by non-culturable Campylobacter spp. Lett Appl Microbiol. 1994;18:333–336. [Google Scholar]
- 47.Taylor D N, McDermott K T, Little J R, Wells J G, Blaser M J. Campylobacter enteritis from untreated water in the Rocky Mountains. Ann Intern Med. 1983;99:38–40. doi: 10.7326/0003-4819-99-1-38. [DOI] [PubMed] [Google Scholar]
- 48.Terzieva S I, McFeters G A. Survival and injury of Escherichia coli, Campylobacter jejuni, and Yersinia enterocolitica in stream water. Can J Microbiol. 1991;37:785–790. doi: 10.1139/m91-135. [DOI] [PubMed] [Google Scholar]
- 49.van de Giessen A W, Heuvelman C J, Abee T, Hazeleger W C. Experimental studies on the infectivity of non-culturable forms of Campylobacter spp. in chicks and mice. Epidemiol Infect. 1996;117:463–470. doi: 10.1017/s0950268800059124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]