Abstract
Bone morphogenic protein (BMP) signaling is critical for intestinal development, homeostasis, and function performance. Although the function of BMP signaling in the intestinal epithelium is well appreciated, the direct effect of BMP on intestinal stromal cells is poorly understood. Here, we show that disruption of BMP signaling by genetic ablation of Alk3 or Smad4 expands the stromal cell pool, the mucosa tumefaction, and colonic polyposis in the large intestine. Interleukin (IL) secretion by stromal cells is notably increased, including IL-1, IL-11, and IL-17. Specifically, IL-1 and IL-17a hyperactivate the mucin production by goblet cells through nuclear factor κB signaling, and abnormal mucin accumulation results in the morphological changes, epithelial barrier destruction, and polyposis development. Together, our results provide an insight into the role of BMP signaling in intestinal stromal cells to regulate epithelium function. This study further highlights the role of mucin-producing goblet cells in intestinal homeostasis and colitis development.
Stromal BMP signaling regulates intestinal mucin production via IL-1/17 for intestinal homeostasis and colitis development.
INTRODUCTION
The intestine is an important organ for food digestion, nutrient absorption, hormone secretion, immune regulation, and host defense (1). Its multiple functions are based on the cooperation between intestinal epithelium and mesenchyme. In addition to the immune cells, the intestinal mesenchyme also contains stromal cells, such as the telocytes for Wnt activation and bone morphogenic protein (BMP) inhibition (2, 3), Gli1+ cells for Wnt signals (4), and Pdgfralow/Cd34+/Cd81+ cells for BMP antagonists (5). The signaling cross-talk between stromal cells and the epithelium is fundamental to the balance maintenance between intestinal stem cells (ISCs) and mature epithelial cells, the basics of intestinal homeostasis (1).
Two types of cell surface receptors mediate BMP signaling: the type II receptor (BMPRII) and the type I receptors (BMPRIA, also called ALK3, and BMPRIB, also called ALK6). BMP binding enhances receptor complex formation and phosphorylation of type I receptors, which, in turn, phosphorylate and activate Smad1, Smad5, and Smad8 and promote a complex formation with Smad4. The Smad complex is accumulated in the nucleus to regulate gene expression. BMP receptors can also signal through non-Smad pathways, such as p38, c-Jun N-terminal kinase, and phosphatidylinositol 3-kinase to regulate multiple cellular functions (6). As an essential niche signal, BMP shows a gradually elevating gradient from the crypt bottom toward the villus compartment, which inhibits the proliferation of ISCs and transient-amplifying (TA) cells in the crypt (5, 7), and promotes the terminal differentiation of enteroendocrine cells along the crypt-to-villus as well as hormone switching (8). In addition, BMP drives the top villus cell states, including enterocytes and goblet cells, as loss of its type I receptor Bmpr1a reduces top villus gene expression (9). Impaired maturation of goblet cells, Paneth cells, and enteroendocrine cells are also observed in mice lacking Bmpr1a (10). These results demonstrate the vital function of BMP signaling in epithelial homeostasis.
Deregulation of BMP signaling contributes to intestine pathogenesis. Frequent mutations in BMP signaling components including BMPRIA/ALK3 and SMAD4 are identified in juvenile polyposis syndrome that is an inherited disease with a high risk of adenocarcinoma (11). Conditional deletion of Bmpr1a in mouse intestinal epithelium also induces expansion of ISC compartments, ultimately leading to intestinal adenomas (7). In line with these, juvenile polyposis-like phenotype and adenomatous polyps become obvious after ectopic expression of the BMP antagonist noggin (12). Moreover, the connection between BMP signaling and colorectal cancer development is evidenced on the treatment of this disease by Simvastatin (13).
Intestinal immune homeostasis, such as balanced secretion of interleukin (IL) signals, is critical for the functional performance of the intestinal epithelium, and its deregulation leads to pathological conditions (14, 15). In the homeostatic state, the cytokine IL-10 can support ISC renewal (16), while IL-13 can increase tuft cell differentiation and expand the goblet cell pool (17, 18). IL-22, a host defense factor (19), not only promotes ISC recovery following damage (20) but also protects ISC against DNA damage and malignant transformation (21). IL-17a also stimulates secretory cell differentiation, including enteroendocrine cells, Paneth cells, and goblet cells (19, 22). ILs are also involved in intestinal diseases, such as intestinal inflammation, colitis, and colorectal cancer. IL-33 shows a high level in the inflamed lesions of patients with inflammatory bowel disease (IBD) (23) and acts as either an inflammatory driver or an alarmin (24). IL-23 functions as a pathogenic factor in IBD (25), and its inhibition becomes a therapeutic strategy for colitis treatment (26). Colitis-induced IL-11 can promote colon carcinogenesis (27). IL-1 also promotes chemoradiotherapy resistance and disease progression in colorectal cancer through polarizing cancer-associated fibroblasts (28).
The intestinal epithelium is regulated by the underneath niche that contains stromal cells secreting various growth factors. For instance, FOXL1+ telocytes located near intestinal stem/TA cells provide a source of Wnt activation and BMP inhibition signals for ISC self-renewal (3, 29). CD90+ cells are important for colonic stem cell maintenance by secreting Rspo3, Wnt2b, and Grem1 (30). Pericyte-like cells, marked by Cspg4 and overlapped with the Foxl1+ population in the mouse stomach and small intestine, can produce Wnt signals (31). Stromal cells have been divided into three subpopulations: Pdgfrahigh/Cd34−, Pdgfralow/Cd34+/Cd81−, and Pdgfralow/Cd34+/Cd81+ (5). Pdgfrahigh/Cd34− cells are mainly located at the villus base and serve as a BMP reservoir, while Pdgfralow/Cd34+/Cd81+ cells are abundant just below the crypts and secrete BMP antagonists. Furthermore, Gli1+ mesenchymal cells may be critical for colonic homeostasis as blockage of Wnt secretion from these cells by deletion of Wntless leads to loss of stem cells in the colon but not in the small intestine (4). In addition, Twist+ stromal cells have been shown to participate in colitis pathogenesis by secreting prostaglandin E2 (32).
In this study, to investigate the function of BMP in intestinal stromal cells, we disrupted BMP signaling in Gli1+ stromal cells by ablating Alk3 and Smad4 and found that the large intestine showed abnormal morphology, accompanied with severe diarrhea and cyst-like polypus. Abundant accumulation of mucins and epithelial barrier destruction were observed in cyst-like polypus. Furthermore, BMP blockade increased the number of stromal cells and enhanced the expression of several ILs, including IL-1, IL-17, and IL-11. We demonstrated that IL-1 and IL-17 could induce goblet cell differentiation and mucin secretion through nuclear factor κB (NF-κB) signaling, which account for the observed cyst-like morphological changes. Together, our results uncover that BMP signaling in stromal cells is important to regulate goblet cell function and maintain epithelium homeostasis.
RESULTS
Gli1+ stromal cell–specific ablation of Alk3 and Smad4 causes mucosa tumefaction and promotes goblet cell differentiation in the large intestine
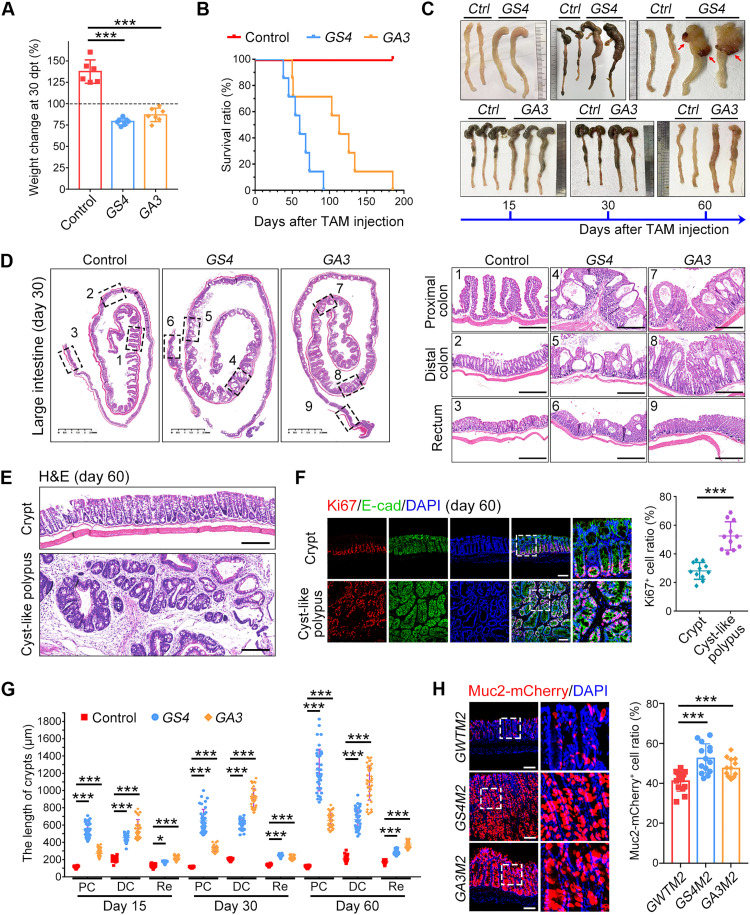
To explore the function of BMP signaling in stromal cells, we generated stromal-specific heterozygous mice: Gli1-CreERT2;Smad4wt/fl and Gli1-CreERT2;Alk3wt/fl. No notable difference was observed on the mouse body weight or the intestinal mucosa morphology in these mice 5 months later after tamoxifen-induced one-allele deletion (fig. S1, A and B). However, compared to control mice (Gli1-CreERT2), homozygous deletion of these genes in Gli1-CreERT2;Smad4fl/fl (GS4) and Gli1-CreERT2;Alk3fl/fl (GA3) mice resulted in a notably shortened life span, companied with marked body weight loss (Fig. 1, A and B, and fig. S1C). Along with the time, the large intestine was obviously distended, especially in the proximal colon (Fig. 1C). The large intestinal epithelium of BMP deficiency mice showed tumefaction morphology with a cavity structure (Fig. 1D and movies S1 to S3). GS4 mice also developed cyst-like polypus in 60 days after Smad4 knockout (KO) (Fig. 1, E and F) (33). Moreover, the crypt length of BMP-deleted mice was significantly increased in different segments of the large intestine (Fig. 1G). In addition, both GS4 and GA3 mice presented severe diarrhea (fig. S1D). These results suggest that the interruption of BMP signaling in stromal cells evokes morphological and functional disorder in the large intestine.
Fig. 1. Inactivation of Smad4 or Alk3 in Gli1+ stromal cells leads to intestinal mucosa tumefaction.
(A) Body weight changes of indicated mice compared with the starting body weight at day 30 post–tamoxifen administration (dpt). n = 6 mice for each group. (B) Survival rate of indicated mice after the last tamoxifen (TAM) administration. n = 8 mice for each group. (C) Representative images of large intestine at different time points after the last tamoxifen administration, and cyst-like polypus appeared at 60 dpt in GS4 mice (red arrow). Ctrl, control. (D) Representative images of hematoxylin and eosin (H&E)–stained large intestine sections and enlarged field of three large intestine segments at 30 dpt. Scale bars, 2.5 mm (left) and 500 μm (right). (E) Representative images of H&E-stained cyst-like polypus sections at 60 dpt. Scale bars, 200 μm. (F) Ki67 immunofluorescence staining in cyst-like polypus sections from GS4 mice at 60 dpt. The right panel shows the quantification of Ki67+ cells in the cyst-like polypus. Scale bars, 100 μm. DAPI, 4′,6-diamidino-2-phenylindole. (G) Crypt length quantification of three large intestine segments along with the time of conditional KO. PC, proximal colon; DC, distal colon; Re, rectum. (H) Representative images of Muc2-mCherry in vivo signals in distal colon sections at 30 dpt. The right panel shows the quantification of Muc2-mCherry+ cells along the crypt. Scale bars, 100 μm. *P < 0.05 and ***P < 0.001 by one-way analysis of variance (ANOVA) test in (A), (F), (G), and (H).
As Muc2+ goblet cells play a critical role in the large intestine by secreting mucus (34), we then generated Gli1-CreERT2;Muc2-mCherry (GWTM2), Gli1-CreERT2;Smad4fl/fl;Muc2-mCherry (GS4M2), and Gli1-CreERT2;Alk3fl/fl;Muc2-mCherry (GA3M2) mice to examine goblet cell function. As expected, goblet cells in the large intestine of GS4M2 and GA3M2 were significantly increased compared with GWTM2 mice (Fig. 1H). Similar results were obtained in GA3 and GS4 mice (fig. S1E), while Ki67+ cells and enteroendocrine cells were not changed after Alk3 or Smad4 KO (fig. S1, F and G). These results suggest that disruption of BMP signaling in Gli1+ stromal cells promotes goblet cell differentiation in the large intestine.
Ablation of Alk3 and Smad4 in Gli1+ cells had minimal effects on the small intestinal morphology in GA3 and GS4 mice (fig. S2A) but increased crypt length in the ileum (fig. S2, B and C). Disruption of BMP signaling in Gli1+ cells had no notable effect on Olfm4-marked ISCs (fig. S2D) or goblet cells in the small intestine (fig. S2E). To examine the possible effect on enterocytes, we established Gli1-Cre ERT2;Apoa1-mCherry (GWTA1), Gli1-Cre ERT2; Smad4fl/fl;Apoa1-mCherry (GS4A1), and Gli1-Cre ERT2; Alk3fl/fl;Apoa1-mCherry (GA3A1) mice and observed a reduction of enterocytes in the ileum by disruption of BMP signaling in Gli1+ cells (fig. S2F). However, Paneth cells showed raised location in ileum in GA3 mice but not in GS4 mice (fig. S2G). Abnormal location of Paneth cells in GA3 mice suggests that Smad4 may mediate other signals to influence Paneth cell maturation.
Disruption of BMP signaling in Gli1+ cells causes mucin accumulation in cavities and the squamous shape of epithelial cells
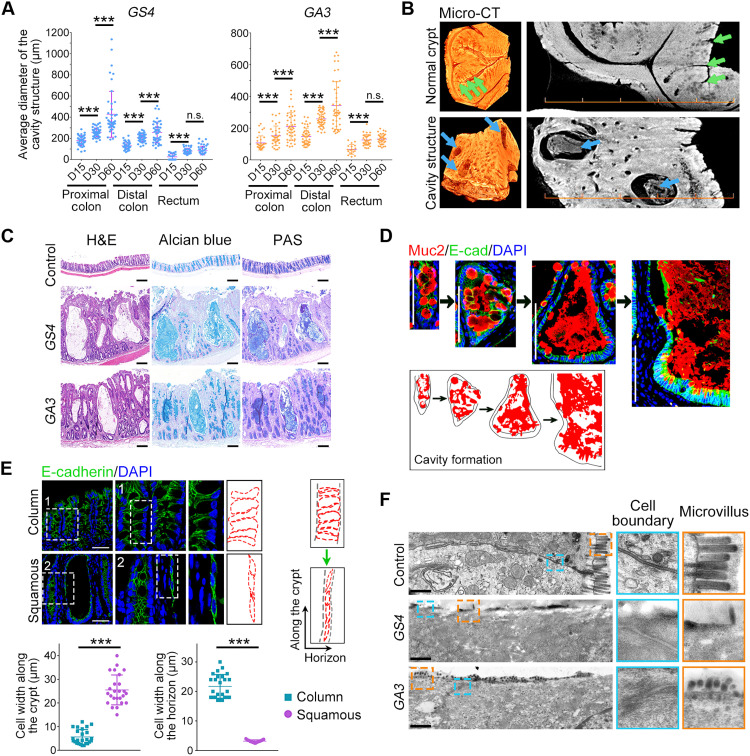
To further characterize the tumefaction phenotype and cavity structures in the large intestine upon BMP blockade in Gli1+ stromal cells, we quantified the cavity diameter in three large intestine segments along with the time and observed the cavity enlargement (Fig. 2A). Micro-scale computed tomography revealed that the cavities were filled by protein-like content (Fig. 2B and movies S1 and S2). Alcian blue and periodic acid–Schiff (PAS) staining further confirmed that the cavity was filled with mucins (Fig. 2C), which is consistent with the enrichment of goblet cells upon BMP blockade in Gli1+ stromal cells. Along the time, the cavities were filled with abnormal accumulated mucins (Fig. 2D).
Fig. 2. Mucin accumulation results in the cavity formation and the epithelial cell morphology change from column to squamous structure.
(A) The average diameter of the cavity structure in three large intestine segments of GS4 and GA3 mice at different KO times. (B) Representative micro-scale computed tomography (micro-CT) images of normal crypts (green arrows) and cavity structures (blue arrows) in the large intestine. Scale bars, 500 μm. (C) H&E, Alcian blue, and PAS staining of mucins in the cavities. Scale bars, 200 μm. (D) Muc2 and E-cadherin immunofluorescence staining during the cavity development. A sketch depicts an abnormal accumulation of mucins. Scale bars, 100 μm. (E) E-cadherin immunofluorescence staining in normal crypts showing the column epithelial cell structure and in cavities showing the squamous epithelial cell structure. The cell width along the crypt and the horizon was quantified (right and bottom). Scale bars, 50 μm. (F) Representative transmission electron microscopy images of the large intestinal epithelium and enlarged fields of microvillus and cell boundary. Scale bars, 1 μm. n.s., no significance; ***P < 0.001 by one-way ANOVA test in (A) and (E).
We also observed an epithelial morphology change from column to squamous structure accompanied with the cavity expansion based on cell junction staining (Fig. 2E and fig. S3A). In support of this observation, transmission electron microscopy revealed an elongated epithelium and reduced microvillus number along the cavity structure after Alk3 and Smad4 depletion (Fig. 2F). Less proliferative cells, goblet cells, and enteroendocrine cells were found in the cavity epithelium (fig. S3B). Together, these results indicate that disruption of BMP signaling in Gli1+ stromal cells results in abundant accumulation of mucins in the large intestine to form cavity structures and squamous epithelium with abnormal cell composition.
Inactivation of BMP signaling in Gli1+ cells enhances mucin production in goblet cells
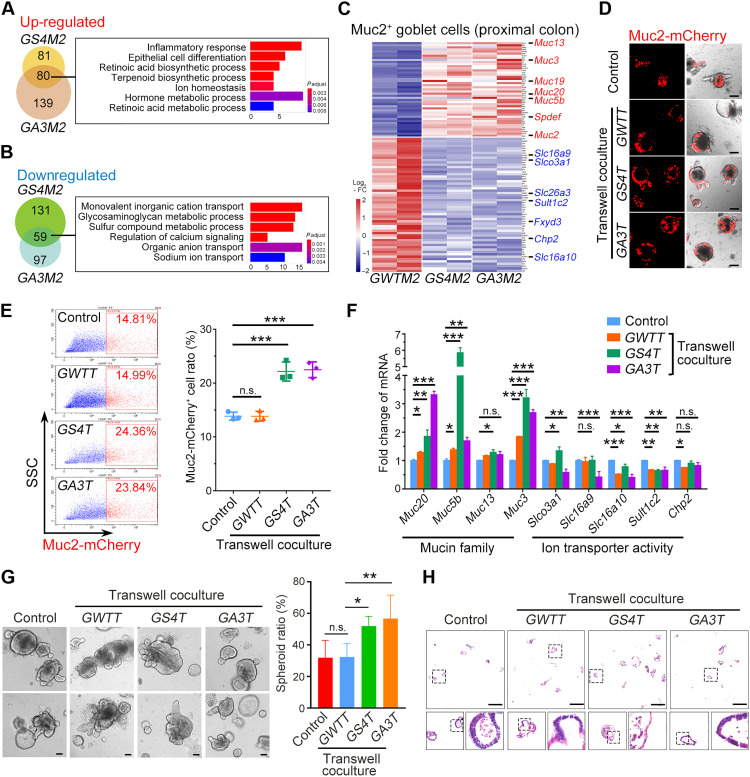
To explore the mechanism underlying Muc2+ cell enrichment after blocking BMP signaling in Gli1+ stromal cells, we examined transcriptome profile in Muc2+ cells isolated from GWTM2, GS4M2, and GA3M2 mice by RNA sequencing (RNA-seq). Gene ontology (GO) analysis of up-regulated genes in Muc2+ cells from GS4M2 and GA3M2 mice showed functional associations with inflammatory response, epithelial cell differentiation, and biosynthetic process (Fig. 3A), but down-regulated genes were linked to ion transport and metabolic process (Fig. 3B). Heatmap also showed that mucin-related genes were among the up-regulated genes, while transport activity-related genes were down-regulated (Fig. 3C). These changes mainly took place in the proximal colon (Fig. 3C and fig. S4A), which was in agreement with the more obvious tumefaction phenotype in the proximal colon.
Fig. 3. Disruption of BMP signaling in Gli1+ stromal cells enhances mucin production and suppresses the ion transport activity in colon goblet cells.
(A) GO analysis of the up-regulated genes in Muc2-mCherry+ cells from GS4M2 and GA3M2 mice compared with GWTM2 mice. (B) GO analysis of the down-regulated genes in Muc2-mCherry+ cells from GS4M2 and GA3M2 mice compared with GWTM2 mice. (C) Heatmap of the up-regulated mucin genes (red) and the down-regulated ion transport genes (blue) in Muc2-mCherry+ cells from indicated mice. FC, fold change. (D) Representative images of Muc2-mCherry-labeled distal colon organoids cocultured with Gli1-tdTomato+ cells from indicated mice for 3 days. Three hundred organoids cocultured with 10,000 Gli1-tdTomato+ cells. Scale bars, 100 μm. (E) Fluorescence-activated cell sorting (FACS) analysis and quantitation of Muc2-mCherry+ cells from distal colon organoids cocultured with Gli1+ cells from indicated mice for 3 days. SSC, side scatter. (F) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of mucin and ion transport genes in distal colon organoids cocultured with Gli1+ cells from indicated mice for 3 days. (G) Representative images of wild-type (WT) distal colon organoids cocultured with Gli1-tdTomato+ cells from indicated mice for 3 days and the spheroid ratio quantification. Scale bars, 100 μm. (H) Representative images of H&E-stained organoid sections from the coculture system with Gli1-tdTomato+ cells from indicated mice for 3 days. Scale bars, 500 μm. *P < 0.05, **P < 0.01, and ***P < 0.001 by one-way ANOVA test in (E) and (G) and two-way ANOVA followed by Tukey’s multiple comparisons test in (F).
To determine the effect of Gli1+ stromal cells on Muc2+ goblet cells in vitro, we cocultured primary intestinal Gli1+ cells with the organoids derived from distal colon crypts of Muc2-mCherry mice. The Gli1-CreERT2;Rosa26-tdTomato (GWTT), Gli1-CreERT2;Smad4fl/fl;Rosa26-tdTomato (GS4T), and Gli1-CreERT2;Alk3fl/fl;Rosa26-tdTomato (GA3T) mice were generated to isolate Gli1+ cells. Coculture with Gli1+ cells from GS4T and GA3T mice apparently elevated Muc2-mCherry cells (Fig. 3, D and E). In addition, the expression level of Alpi, Muc2, and E-Cad were increased, but the expression of Chgb, Ki67, and Lgr5 was not (fig. S4B), which is consistent with the above observation in the GS4 or GA3 intestinal epithelium (Fig. 1H and fig. S1, E to G). Furthermore, the mucin family members were up-regulated, while the ion transport-related genes were down-regulated (Fig. 3F). In addition, the spheroid ratio was increased after coculture with Gli1+ cells from GS4T and GA3T mice (Fig. 3G), the squamous epithelium can also be observed in the cocultured spheroids (Fig. 3H). These results together suggest that BMP signaling blockade in Gli1+ stromal cells enhances mucin production and suppresses ion transport in goblet cells, which contributes to the intestinal mucosa tumefaction in the large intestine.
BMP inhibits cell proliferation and increases IL production of stromal cells
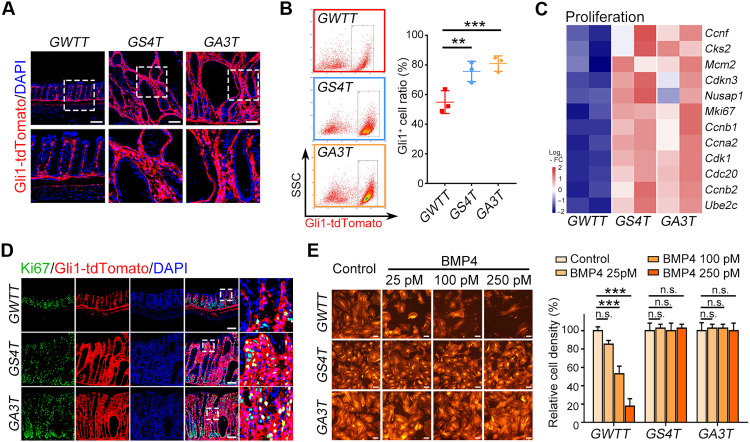
To better understand the mechanism in which BMP acts on Gli1+ stromal cells to influence epithelium homeostasis, we assessed the change of Gli1+ stromal cells. Our results showed that the Gli1+ stromal cell number was notably increased in the colon after ablation of Alk3 or Smad4 (Fig. 4, A and B, and fig. S5A), while no notable changes were observed in the small intestine (fig. S5B). Consistently, Ki67+ stromal cells and proliferation genes were increased after Alk3 or Smad4 KO (Fig. 4, C and D, and fig. S5C). In addition, BMP treatment inhibited the proliferation of wild-type Gli1+ cells but not that of Alk3 or Smad4 KO cells (Fig. 4E). These results suggest that BMP inhibits cell proliferation to balance the stromal cell pool, while loss of BMP expands stromal cells and leads to microenvironment turbulence.
Fig. 4. Inactivation of BMP signaling enhances the proliferation of Gli1+ stromal cells.
(A) Representative images of Gli1-tdTomato signals in distal colon sections at 30 dpt. Scale bars, 100 μm. (B) FACS analysis and quantitation of Gli1-tdTomato+ cells in distal colon from indicated mice at 30 dpt. (C) Heatmap of proliferation-related genes in Gli1-tdTomato+ cells from the indicated mice at 30 dpt. (D) Representative images of Ki67 immunofluorescence staining in distal colon sections of indicated mice at 30 dpt. Scale bars, 100 μm. (E) Representative images of WT, Smad4-KO, or Alk3-KO Gli1-tdTomato+ cells and the quantification of relative cell density at day 3 after BMP4 treatment. Scale bars, 100 μm. **P < 0.01 and ***P < 0.001 by one-way ANOVA test in (B) and (E).
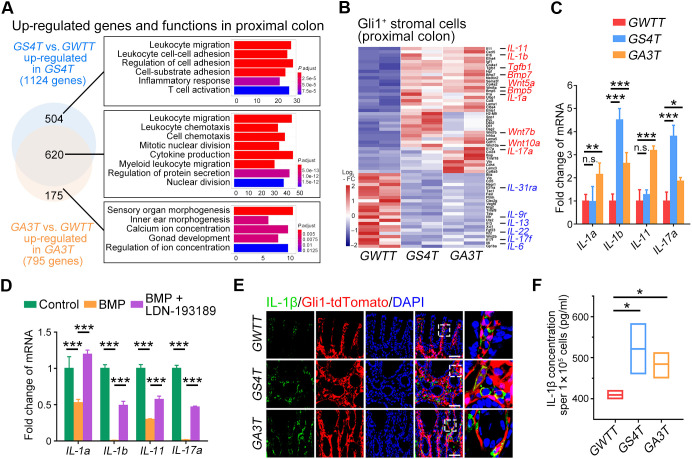
To assess how Gli1+ stromal cells affect the epithelium after BMP signaling disruption, we performed RNA-seq and GO analysis of Gli1-tdTomato+ cells. The results showed that commonly up-regulated genes upon Alk3 or Smad4 KO were involved in leukocyte migration, cell division, and cytokine production (Fig. 5A and fig. S6A). By analysis of cytokines, we found that IL genes, such as IL-1, IL-17, and IL-11, were up-regulated upon Alk3 or Smad4 KO, while IL-6, IL-13, and IL-22 were down-regulated (Fig. 5B and fig. S6B). Increased mRNA expression of IL-1a, IL-1b, IL-11, and IL-17a was confirmed in cultured Alk3 or Smad4 KO Gli1+ stromal cells (Fig. 5C), and BMP specifically suppressed their expression (Fig. 5D). Higher protein levels of IL-1α and IL-1β were detected in Gli1+ stromal cells of intestinal tissues (Fig. 5E and fig. S6C). Increased skipped exon of IL-1b was found in Gli1+ stromal cell after Alk3 or Smad4 KO (fig. S6D). An increased IL-1β protein level was found in the conditional medium of Alk3 KO or Smad4 KO Gli1+ cells (Fig. 5F). These results indicate that BMP stimulates expression of IL-1a, IL-1b, IL-17a, and IL-11 in Gli1+ stromal cells.
Fig. 5. Inactivation of BMP signaling increases IL secretion in Gli1+ stromal cells.
(A) GO analysis of the up-regulated genes in Gli1-tdTomato+ cells from GS4T and GA3T mice compared with GWTT mice. (B) Heatmap of the up-regulated (red) and down-regulated genes (blue) in Gli1-tdTomato+ cells from indicated mice. (C) qRT-PCR analysis of IL genes detected in Gli1-tdTomato+ cells from indicated mice. (D) qRT-PCR analysis of IL genes detected in WT Gli1-tdTomato+ cells at day 3 after the treatment with 25 pM BMP4 or 1 μM LDN-193189. (E) Representative images of IL-1β immunofluorescence in distal colon sections of indicated mice at 30 dpt. Scale bars, 100 μm. (F) Elisa quantitation of IL-1β levels from in vitro cultured Gli1-tdTomato+ cells. *P < 0.05, **P < 0.01, and ***P < 0.001 by two-way ANOVA followed by Tukey’s multiple comparisons test in (C) and (D) and one-way ANOVA test in (F).
IL-1 and IL-17 promote goblet cell differentiation and enhance mucin secretion through NF-κB signaling
To investigate the effect of ILs on goblet cells, we treated colonic organoids derived from the crypts of Muc2-mCherry mice with IL-1, IL-11, and IL-17. Inhibition of Wnt and Notch signaling with IWP-2/DAPT has been shown to promote goblet cell differentiation (35). IWP-2/DAPT greatly enhanced Muc2-mCherry+ cells (Fig. 6, A and B). Notably, IL-17 or IL-1, but not IL-11, also increased Muc2-mCherry+ cells (Fig. 6, A and B). Consistently, the goblet cell marker genes were increased upon IL-1 or IL-17 treatment (Fig. 6C and fig. S7A). Conversely, IL-1 receptor antagonist (IL-1Ra) suppressed mucin mRNA levels in colonic organoids cocultured with Alk3 or Smad4 KO Gli1-tdTomato+ cells (Fig. 6D). IL-1 and IL-17 also enhanced mucin expression in small intestinal organoids (fig. S7, B to D). Mucin accumulation or cavity structures were only found in the large intestine but not in the small intestine of Alk3 or Smad4 KO mice, perhaps due to more Gli1+ cells in the colon (4).
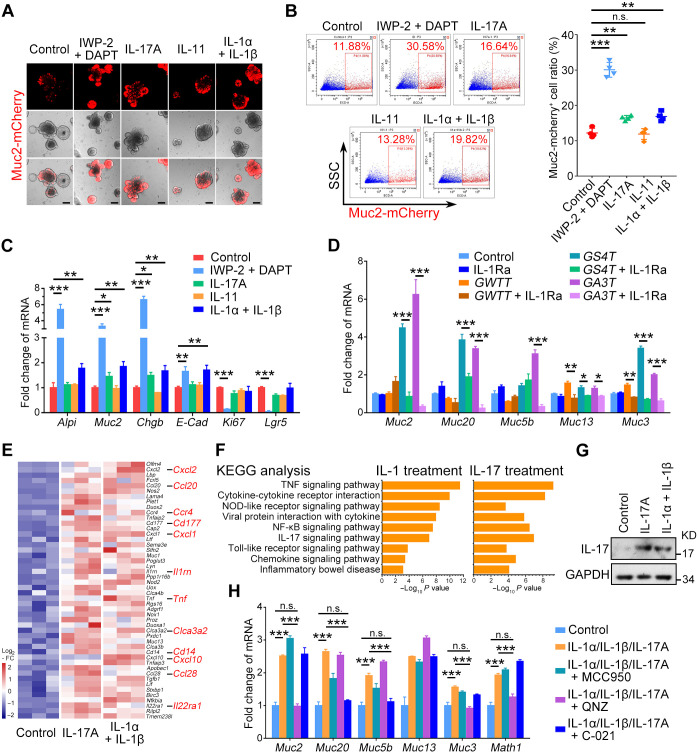
Fig. 6. IL-1 and IL-17 promote goblet cell differentiation and enhance mucin production in mouse colonic organoids through NF-κB signaling.
(A) Representative images of Muc2-mCherry-labeled distal colon organoids at day 3 after the treatments with 2 μM IWP-2, 10 μM DAPT, or 10 nM ILs. Scale bars, 100 μm. (B) FACS analysis and quantitation of Muc2-mCherry+ cells from distal colon organoids at day 3 after the treatments with 2 μM IWP-2, 10 μM DAPT, or 10 nM ILs. (C) qRT-PCR analysis of epithelial marker genes in distal colon organoids at day 3 after the treatments with 2 μM IWP-2, 10 μM DAPT, or 10 nM ILs. (D) qRT-PCR analysis of mucin genes in distal colon organoids cocultured with Gli1-tdTomato+ cells from indicated mice and treated with 50 nM IL-1Ra for 3 days. (E) Heatmap of the up-regulated genes in distal colon organoids after the treatments with 10 nM ILs for 12 hours. (F) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis for the up-regulated signaling pathways after the indicated treatments for 12 hours. (G) Immunoblot analysis of the expression level of IL-17 in distal colon organoids treated with 10 nM ILs for 3 days. (H) qRT-PCR analysis of mucin genes in distal colon organoids after the indicated treatments for 3 days. *P < 0.05, **P < 0.01, and ***P < 0.001 by one-way ANOVA test in (B) and two-way ANOVA followed by Tukey’s multiple comparisons test in (C), (D), and (H).
Gli1+ stromal cells also secrete BMP ligands, such as BMP5 and BMP7 (Fig. 5B and fig. S6B), which may contribute to goblet differentiation (9, 10). To investigate the influence of BMP on goblet cells, we added the BMP receptor inhibitor LDN-193189 in the colonic organoids and Gli1+ stromal cell coculture system. As shown in fig. S7E, LDN-193189 increased Lgr5 and Alpi expression but did not suppress the increased expression of Muc2. These results together suggest that BMP suppresses IL expression in Gli1+ stromal cells to control goblet cell differentiation.
To explore how IL-1/17 affect the intestinal epithelium, RNA-seq was performed after the organoids were treated for 12 hours. Chemokines and cytokines were up-regulated by IL-1 or IL-17 (Fig. 6E and fig. S7F). These increased genes involved in tumor necrosis factor (TNF) signaling, nonobese diabetic (NOD)–like signaling, NF-κB signaling, and cytokine-cytokine receptor interaction, as revealed by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Fig. 6F). IL-1/17 treatment could increase IL-17 expression in colon organoids (Fig. 6G and fig. S7G). To define the pathways involved in IL-1/17 signaling, three inhibitors were used: QNZ (EVP4593) for NF-κB signaling and TNF-α production, MCC950 for NOD-like signaling, and C-021 for CCR4 signaling. We found that QNZ inhibited the increased expression of Muc2 and Math1 (a transcript factor for goblet differentiation), while C-021 partly impaired the increased expression of Muc20 and Muc5b induced by IL-1 or IL-17 (Fig. 6H). These results suggest that IL-1 and IL-17 promote goblet cell differentiation and enhance mucin secretion through NF-κB signaling and CCR4 signaling.
To further explore the role of IL-1/17, RNA-seq and gene expression analysis were performed after the organoids were treated with IL-1 or IL-17 for 3 days. Mucins and solute carrier (SLC) family members were regulated by IL-1 and IL-17, including the increased expression of Muc1 and Muc13 and the decreased expression of Slc16a10 and Chp2 (fig. S7H). The decreased expression of ion transport-related genes Slc16a10 (MOT10) and Chp2 (CHP2) were further confirmed at mRNA and protein levels (fig. S7, I and J), which was consistent with the above results (Fig. 3F). GO analysis also revealed that IL-1 and IL-17 up-regulated the genes related to regulation of cell morphogenesis, cell shape, and mucus secretion while decreasing the ones related to regulation of mitotic cell cycle, cellular amino acid metabolic process, and cilium organization (fig. S7K).
IL-1 signals are increased in human colitis and enhance mucin secretion in human colonic organoids
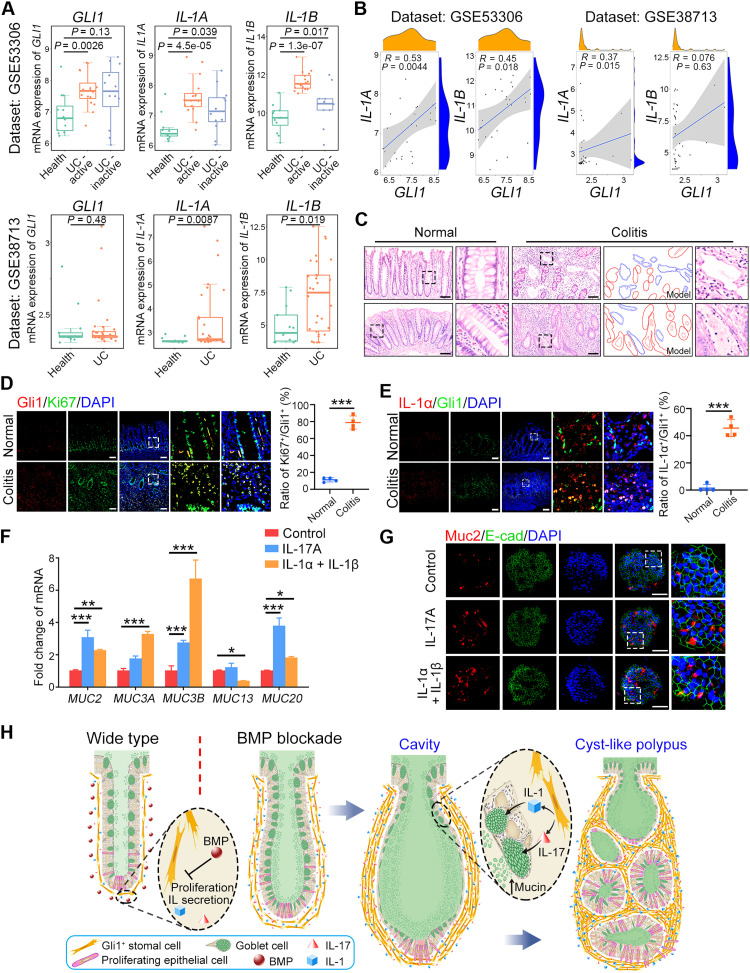
It has been reported that GLI1 is associated with colitis and promotes colitis-associated tumorigenesis (36). Our KEGG analysis also indicated that IL-1 and IL-17 might function in IBD pathogenesis (Fig. 6F). We then asked whether the regulation of goblet cells by IL from stromal cells contributes to human colitis development. We first analyzed the expression levels of GLI1, IL-1A, and IL-1B with the published dataset (37, 38) and found that they were increased in ulcerative colitis compared to health tissues (Fig. 7A). GLI1 expression was positively correlated with IL-1A and IL-1B in these datasets (Fig. 7B). Similar to the Alk3 KO and Smad4 KO mice (Fig. 2C), cavity structures filled with mucins were also observed in human colitis samples (Fig. 7C and fig. S8A). Ki67 signals in GLI1+ cells was increased in human colitis (Fig. 7D), so as the secretion of IL-1α and IL-1β (Fig. 7E and fig. S8B). IL-1β expression was also increased in GLI1+ cells in human colorectal cancer (fig. S8C). In addition, cytokines and inflammatory response were also up-regulated in the dextran sodium sulfate (DSS)–induced mouse colitis model, such as the CXC subfamily, CC subfamily and IL-1 signaling (fig. S8D). The increased protein level and positive cell ratio of IL-1α or IL-1β were observed in DSS-induced colitis (fig. S8, E and F). The increased colocalizations of GLI1 with IL-1β or Ki67 were also detected (fig. S8, G and H). Decreased p-Smad1/5 level was observed in human colitis and DSS-induced colitis (fig. S8, I and J) (39), indicating that the inactivation of BMP signaling may account for high proliferation capacity of GLI1+ cells shown by up-regulated Ki67 signals. Furthermore, IL-17 and IL-1 also up-regulated mucin gene expression and Muc2 protein level in human colonic organoids (Fig. 7, F and G). These data indicate that high IL levels may contribute to pathogenesis of human colitis.
Fig. 7. Increased IL-1 signals enhance the mucin secretion in human colitis samples.
(A) Box plots of mRNA levels of GLI1 and ILs in healthy and colitis specimens (using datasets GSE53306 and GSE38713). In the box plots, the middle line depicts the median and the whiskers in the min-to-max range. UC, ulcerative colitis. (B) The positive correlation between GLI1 and IL-1A or IL-1B in human datasets GSE53306 and GSE38713. (C) Representative images of H&E staining in human large intestine and colitis sections. The red lines show the column epithelial cell structure and the blue lines show the squamous epithelial cell structure in colitis model. Scale bars, 100 μm. (D) Gli1 and Ki67 immunostaining and quantification of human colon derived from healthy and ulcerative colitis specimens. Scale bars, 100 μm. (E) IL-1α and Gli1 immunostaining and quantification of human colon derived from healthy and ulcerative colitis specimens. Scale bars, 100 μm. (F) qRT-PCR analysis of mucin genes in human large intestinal organoids after treated with 10 nM IL-1 or IL-17 for 3 days. (G) Representative images of Muc2 and E-cadherin immunofluorescence costaining of human colon organoids after treated with IL-1 or IL-17 for 3 days. Scale bars, 100 μm. (H) A working model shows that disruption of BMP signaling in Gli1+ stromal cells increases goblet cell number and promotes mucin production through IL-1 and IL-17. *P < 0.05, **P < 0.01, and ***P < 0.001 by one-way ANOVA test in (D) and (E) and two-way ANOVA followed by Tukey’s multiple comparisons test in (F).
DISCUSSION
The interplay between intestinal epithelium and stromal cells is critical for intestinal functionality and homeostasis (40), and dysfunction of any of them can drive pathological development (40, 41). However, most studies in the past focus on polyposis initiation and tumorigenesis due to dysfunction of BMP signaling in the intestinal epithelium (5, 42). It is unclear whether BMP regulates stromal cell activity and whether stromal BMP signaling plays a role in intestinal epithelium homeostasis. Here, we demonstrate that Alk3 or Smad4 KO in Gli1+ cells results in tumefaction, cavity formation, and polyposis development in the large intestine. We further show that interruption of BMP signaling provokes a high production of IL-1 and IL-17, which stimulate abnormal mucin secretion in goblet cells (Fig. 7H).
Goblet cells secrete mucins to form mucous layers, which stabilize the bacterial biofilm and prevent the epithelium from pathogen invasion (34). The destruction of mucosal barrier and imbalance of mucin dynamics may lead to enteritis development (43). For example, reduction in goblet cell number, impairment of mucin secretion and shrinkage of mucus layer are associated with IBD in human and colitis in mouse model (44, 45). However, consequences initiated by the abnormal accumulation of mucins are obscure. Our results show that excessive mucin production accounts for the formation of cavity structure, which further results in epithelium tumefaction and changes the epithelial morphology from column to squamous structure. Goblet cell emptying can be found in normal mucus secretion, and the column structure can be recovered from membrane recombination and replenishment of mucin granules (46, 47). We hypothesize that persistent mechanical force produced by the cavity structure prevents the column-structure recovery and maintains the squamous structure. That abnormal epithelial morphology and barrier are associated with human ulcerative colitis and DSS-induced mouse colitis may induce the regenerative and proliferative progress of the intestinal epithelium and result in the colonic polyposis development (48–51).
Gli1, a transcription factor acting downstream of Hedgehog signaling (52), can label a subgroup of mesenchymal cells that produce Wnt ligands and contribute to the maintenance of colon stem cells (4). In addition to growth factors, cytokines and chemokines produced from stromal cells are also pivotal intestinal niche factors to regulate epithelium activity (53). However, regulation on these cytokine or chemokine-secreted stromal cells is not fully understood. Our results revealed that BMP signaling suppresses the expansion of Gli1+ stromal cells and IL production, which is critical for appropriate homeostatic control of goblet cells in their amount and their mucin-secreting activity. However, how BMP signaling regulates Gli1+ stromal cells requires further exploration.
The mucosa tumefaction, abnormal cavity structures, and mesenchymal cell overexpansion have been reported in the large intestine after the KO of Alk3 in Foxl1- or Col1a2-positive cells, which may be due to intestinal immune and extracellular microenvironment and CXCL12 signaling (54, 55). In addition to these phenotypes, we also observed that the abnormal mucin accumulation was associated with cavity formation and the epithelial squamous shape transformation in the mice with Alk3 or Smad4 KO in Gli1+ cells. We further found that IL-1/17 from Gli1+ cells may account for enhanced goblet differentiation and mucin secretion. These results suggest that disruption of BMP signaling in intestinal stromal cells can remodel the intestinal microenvironment and regulate the epithelial cell fate by regulating cytokine production.
IL-1 can induce gastrointestinal inflammation (56), promote colitis and tumor development (57), and associate with therapy nonresponse and unfavorable tumor prognosis (57, 58). Most of these functions of IL-1 are associated with immune cells. It is not known whether IL-1 can directly regulate epithelial cells. We found that IL-1 promotes mucin secretion in goblet cells and leads to the excessive mucin accumulation. NF-κB signaling and CCR4 signaling may contribute to these processes. Nonetheless, our results uncover a previously unknown link of subepithelial stromal cells to control goblet cell function in the intestinal epithelium and provide insights into development of therapeutic treatment for intestinal colitis.
MATERIALS AND METHODS
Mice and ethics statement
Gli1-CreERT2 mice were from L. Baojie (59); Alk3fl/fl mice were provided by Y. Mishina (60); Smad4fl/fl mice were provided by Y. Xiao (61); Rosa26-Loxp-STOP-Loxp-tdTomato mice were obtained from the Jackson Laboratory; Muc2-mCherry mice and Apoa1-mCherry mice were commercially generated by Gem Pharmatech (Nanjing). Both male and female mice ranging from 2 to 4 months old in age were used. No statistical method was used to predetermine sample size. In general, we used at least three mice per genotype in each experiment. For Cre induction, mice were intraperitoneally injected with 100 μl of tamoxifen dissolved in oil at 20 mg/ml for five consecutive days. All animal studies were performed in accordance with the guidelines and under the approval of the Institutional Animal Care and Use Committee of Tsinghua University (YGC-19).
Isolation of mouse intestinal crypts and organoid culture
Mouse intestine was split longitudinally and washed three times with cold phosphate-buffered saline (PBS). Villi were carefully scraped away, and small pieces (5 mm) of intestine were incubated in 0.5 mM EDTA in PBS for 30 min at 4°C. These pieces were then vigorously suspended in cold PBS, and the mixture was passed through 70-μm cell strainer (BD Biosciences). Large intestine was directly scraped after incubated in 0.5 mM EDTA in PBS for 30 min at 4°C. The crypts were enriched through centrifugation (3 min at 1000 rpm). Then, the crypts were embedded in Matrigel (BD Biosciences) and seeded on 24-well plate. Small intestinal organoids were cultured as previously described (7), and additional CHIR-99021 (2.5 μM; Selleck) was added into large intestinal organoids. For chemical treatments, IWP-2 (2 μM; Selleck), DAPT (10 μM; Selleck), or indicated ILs (10 nM; Novoprotein) were added to the cultured medium. After 3 or 6 days, RNA or single-cell suspension was selected for quantitative reverse transcription polymerase chain reaction (qRT-PCR) or fluorescence-activated cell sorting (FACS) analysis, respectively.
Patient-derived colon tissues and organoid culture
The human materials used in this study have been previously described (62). Briefly, intestine tissues were surgically resected and sampled from patients who had been diagnosed with intestine tumors at the Peking University Third Hospital, Beijing, China. All tissue samples were obtained with informed consent, and the study was approved by the Peking University Third Hospital Medical Science Research Ethics Committee (M2018083). The human colon organoids were cultured as previously described (62).
Isolation and culture of mesenchymal cells
Mouse large intestine was split longitudinally and trimmed into small pieces. After shocked in cold PBS vigorously, tissues were transferred into cold PBS with 1 mM DTT and incubated in 37°C shaker (10 min at 250 rpm). Then, tissues were transferred into cold PBS with 0.5 M EDTA and 1 M Hepes and incubated in 37°C shaker for two times (10 min at 250 rpm), which was followed by digestion with collagenase IV (10,000 U/ml; Gibco) in RPMI 1640 medium (Gibco) and incubated in 37°C incubator for 45 min. Last, tissue pieces were then vigorously suspended in culture medium containing 10% fetal bovine serum (ExCell) and 1% penicillin-streptomycin (HyClone) in MEM alpha basic medium (Gibco), and the mixture was passed through 70-μm cell strainer (BD Biosciences). The cell suspension was added into culture medium and cultured in 10-cm dishes.
Immunohistochemistry and immunofluorescence staining
Intestines isolated from the indicated mice were fixed with 4% formaldehyde solution for overnight and dehydrated with gradient concentration ethanol. Then, tissues were embedded in paraffin with paraffin embedding machine (Arcadia H+C; Leica), and sections were prepared with paraffin slicing machine (RM2255; Leica). For frozen sections, intestine tissues were fixed with 4% formaldehyde solution for 2 hours at 4°C, followed by dehydrating in 30% sucrose solution at 4°C overnight. Next, the tissues were embedded in optimal cutting temperature compound (Sakura) and stored at −80°C. Sections were prepared with freezing slicing machine (CM1950; Leica). For staining, the sections were permeabilized with PBST solution (3% bovine serum albumin and 0.1% Triton X-100 in PBS) and then incubated overnight with the primary antibody at 4°C. The fluorescein-labeled secondary antibodies (1:800; the Jackson Laboratory) for immunofluorescence or secondary horseradish peroxidase–conjugated goat anti-rabbit antibody (ZSGB-BIO) for immunohistochemistry were added for 2 hours at room temperature. Confocal laser scanning (FV3000; Olympus) or 3,3′-diaminobenzidine chromogenic fluid (ZSGB-BIO) was used to detect the staining signals.
Chemical staining
Intestine tissue sections were rehydrated in gradient concentration ethanol and PBS. For hematoxylin and eosin staining, hematoxylin (ZSGB-BIO) was added onto slides and covered tissue region for 2 min, followed by washing in a large amount of water. Then, section was covered by eosin (Beyotime) for 2 min and washed by water flow. For mucin staining, Alcian Blue & Nuclear Fast Red Staining Kit (BASO) was carried out according to the manufacturer’s instructions. For PAS staining, the AB-PAS Staining Kit (Solarbio) was used. In addition, the BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime) was performed for alkaline phosphatase staining. The sections were dehydrated and sealed by neutral gum (ZSGB-BIO) for 48 hours, and sections were scanned by a pathological section scanner (KF-PRO-120; KFBIO).
Antibodies
Rabbit anti-Ki67 (1:300; ab15580, Abcam), rabbit anti-Mucin 2 (1:300; ab272692, Abcam), rabbit anti-lysozyme (1:300; ab108508, Abcam), rabbit anti-chromogranin A (1:300; ab15160, Abcam), mouse anti–E-cadherin (1:1000; 610182, BD Biosciences), mouse anti–α-catenin (1:500; 610194, Biosciences), mouse anti–β-catenin (1:500; 610154, Biosciences), rabbit anti-BMPR1A (1:1000; sc-20736, Santa Cruz Biotechnology), rabbit anti-Smad4 (1:1000; sc7154, Santa Cruz Biotechnology), rabbit anti-Olfm4 [1:300; 39141, Cell Signaling Technology (CST)], mouse anti-vimentin (1:500; 550513, BD Biosciences), rabbit anti–IL-1α (1:50; ab300499, Abcam), rabbit anti–IL-1α (1:50; 16765-1-AP, Proteintech), mouse anti–IL-1β (1:200; sc-52012, Santa Cruz Biotechnology), rabbit anti-MOT10 (1:1000; YT7177, ImmunoWay), rabbit anti-CHP2 (1:1000; YT0915, ImmunoWay), rabbit anti–p-Smad1/5 (1:100; 9516, CST), mouse anti-Ki67 (1:200; 9449, CST), and rabbit anti-Gli1 (1:80; 3538, CST).
FACS analysis
For stromal cells isolated from mice, dissociated cells were passed through 40-μm cell strainer (BD Biosciences), and single tdTomato positive cells were sorted by flow cytometry (MoFlo Astrios EQ; Beckman). For cultured organoids derived from Muc2-mCherry mice, organoids embedded in Matrigel were suspended in cold PBS after medium removal and pelleted by centrifugation (3 min at 1000 rpm). The organoids were dissociated in TrypLE (Invitrogen) for 15 min at 37°C and passed through 40-μm cell strainer, and single Muc2-mCherry positive cells were sorted by flow cytometry (CytoFLEX S; Beckman).
qRT-PCR analysis
The cultured organoids were precipitated, and RNA was purified using the RNeasy Kit (mf167-01, Mei5 Biotech) and converted into cDNA using the NovoScript One-Step RT-PCR kit (Novoprotein). Real-time PCR reactions were performed in triplicates on a LightCycler 480 (Roche). Primers used were listed in table S1.
Bulk RNA-seq and GO and KEGG analysis
RNA-seq was performed with the Illumina sequencing platform, and the quality assessment was done by FastQC (version 0.11.9). Then, the data were uniquely mapped to the mm10 genome by STAR (version 2.5.3), and the normalized gene counts were performed by DESeq2 (version 1.28.1) (63, 64). The analysis of differentially expressed genes (DEGs) and expression values was performed by DESeq2 (version 1.28.1) software (64). Genes with absolute log2-transformed fold changes ≥ 1 were regarded as DEGs, and a threshold of P value < 0.05 was used. The primary analysis data of Muc2-mCherry+ cells and Gli1-tdTomato+ cells are presented in tables S2 and S3. The normalized gene expression data of distal colon organoids treated with IL-1 or IL-17 for 12 or 72 hours are presented in table S4. The heatmap of gene fold changes was generated by Pheatmap (version 1.0.12). The intersection analysis of differential genes of each component was displayed in the VennDiagram (version 1.7.3). DEGs’ GO enrichment and KEGG analysis were carried out by ClusterProfiler (version 3.16.1) (65).
ELISA analysis
For in vitro cultured Gli1+ cells, 1 × 105 cells were cultured for 24 hours. After the medium was discarded, the cells were washed twice with preheated PBS, and MEM-α medium without serum or phenol red (Gibco) was used for further culture. One day later, the culture medium was collected for IL-1β enzyme-linked immunosorbent assay (ELISA) detection according to the manufacturer’s instructions (RK00006, ABclonal).
Gene expression analysis from colitis datasets
Microarray data were derived from Gene Expression Omnibus (GEO) dataset [GSE38713 (37) and GSE53306 (38)]. The GEO datasets were directly accessed through the R (v4.2.1) package GEOquery to retrieve normalized expression data (66). Then, the R (v4.2.1) package ggpubr (https://cran.r-project.org/web/packages/ggpubr/index.html) was used to generate box plots and calculate respective P value, and Wilcoxon rank sum tests were used to compare two conditions.
Statistics
All experiments were performed independently at least three times with three replicates. Data shown in column and dot graphs represent means ± SD. Statistical differences were analyzed using one-way analysis of variance (ANOVA) test or two-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, and ***P < 0.001. All statistical analysis was performed with GraphPad Prism 8 (Windows 10).
Acknowledgments
We thank X. Yang for Smad4fl/fl mice and Y. Mishina for Alk3fl/fl mice. We thank M. Zhang, Y. Li, and Y. Jin for technical assistance. We thank H. Liu for reading manuscript and other colleagues in the Chen laboratory for stimulating discussion.
Funding: This work was supported by grants from the National Natural Science Foundation of China (31988101 to Y.-G.C., 32300586 to Y.W., and 31900550 to Y.L.), the Guangdong Basic and Applied Basic Research Foundation (2021A1515111215 to Y.W.), and the China Postdoctoral Science Foundation (2021 M703230 and 2022 T150653 to Y.W). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: Y.W., R.L., and Y.-G.C. designed the study. Y.W. and R.L. carried out most of the experiments. Z.Z. and S.Y. performed the bioinformatic analysis. C.X. and S.W. helped with the functional experiments. W.F. provided clinical samples. B.L. provided Gli1-CreERT2 mice and instructed the experiments. Y.W., Y.L., and Y.-G.C. wrote the manuscript. All authors contributed to intellectual inputs.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All RNA-seq data have been deposited in the GEO under accession no. GSE234340.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Legends for tables S1 to S4
Legends for movies S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
Movies S1 to S3
REFERENCE AND NOTES
- 1.Y. Wang, W. Song, S. Yu, Y. Liu, Y. G. Chen, Intestinal cellular heterogeneity and disease development revealed by single-cell technology. Cell Regen. 11, 26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.G. Zhu, J. Hu, R. Xi, The cellular niche for intestinal stem cells: A team effort. Cell Regen. 10, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Shoshkes-Carmel, Y. J. Wang, K. J. Wangensteen, B. Tóth, A. Kondo, E. E. Massasa, S. Itzkovitz, K. H. Kaestner, Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.B. Degirmenci, T. Valenta, S. Dimitrieva, G. Hausmann, K. Basler, GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453 (2018). [DOI] [PubMed] [Google Scholar]
- 5.N. McCarthy, E. Manieri, E. E. Storm, A. Saadatpour, A. M. Luoma, V. N. Kapoor, S. Madha, L. T. Gaynor, C. Cox, S. Keerthivasan, K. Wucherpfennig, G. C. Yuan, F. J. de Sauvage, S. J. Turley, R. A. Shivdasani, Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell 26, 391–402.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K. Miyazono, S. Maeda, T. Imamura, BMP receptor signaling: Transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 16, 251–263 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Z. Qi, Y. Li, B. Zhao, C. Xu, Y. Liu, H. Li, B. Zhang, X. Wang, X. Yang, W. Xie, B. Li, J. D. J. Han, Y. G. Chen, BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. Beumer, B. Artegiani, Y. Post, F. Reimann, F. Gribble, T. N. Nguyen, H. Zeng, M. van den Born, J. H. van Es, H. Clevers, Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20, 909–916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. Beumer, J. Puschhof, F. Y. Yengej, L. Zhao, A. Martinez-Silgado, M. Blotenburg, H. Begthel, C. Boot, A. van Oudenaarden, Y. G. Chen, H. Clevers, BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38, 110438 (2022). [DOI] [PubMed] [Google Scholar]
- 10.B. A. Auclair, Y. D. Benoit, N. Rivard, Y. Mishina, N. Perreault, Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology 133, 887–896 (2007). [DOI] [PubMed] [Google Scholar]
- 11.J. R. Howe, J. L. Bair, M. G. Sayed, M. E. Anderson, F. A. Mitros, G. M. Petersen, V. E. Velculescu, G. Traverso, B. Vogelstein, Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat. Genet. 28, 184–187 (2001). [DOI] [PubMed] [Google Scholar]
- 12.A. P. Haramis, H. Begthel, M. van den Born, J. van Es, S. Jonkheer, G. J. A. Offerhaus, H. Clevers, De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686 (2004). [DOI] [PubMed] [Google Scholar]
- 13.S. Ouahoud, R. J. Jacobs, M. P. Peppelenbosch, G. M. Fühler, J. Heijmans, S. Diks, M. E. Wildenberg, L. J. A. C. Hawinkels, L. L. Kodach, P. W. Voorneveld, J. C. H. Hardwick, Kinome-wide analysis of the effect of statins in colorectal cancer. Br. J. Cancer 124, 1978–1987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. Nowarski, R. Jackson, R. A. Flavell, The stromal intervention: Regulation of immunity and inflammation at the epithelial-mesenchymal barrier. Cell 168, 362–375 (2017). [DOI] [PubMed] [Google Scholar]
- 15.G. Zhu, R. Xi, MRISCs protect colonic stem cells from inflammatory damage. Cell Regen. 10, 24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Biton, A. L. Haber, N. Rogel, G. Burgin, S. Beyaz, A. Schnell, O. Ashenberg, C. W. Su, C. Smillie, K. Shekhar, Z. Chen, C. Wu, J. Ordovas-Montanes, D. Alvarez, R. H. Herbst, M. Zhang, I. Tirosh, D. Dionne, L. T. Nguyen, M. E. Xifaras, A. K. Shalek, U. H. von Andrian, D. B. Graham, O. Rozenblatt-Rosen, H. N. Shi, V. Kuchroo, O. H. Yilmaz, A. Regev, R. J. Xavier, T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. R. Howitt, S. Lavoie, M. Michaud, A. M. Blum, S. V. Tran, J. V. Weinstock, C. A. Gallini, K. Redding, R. F. Margolskee, L. C. Osborne, D. Artis, W. S. Garrett, Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. Waddell, J. E. Vallance, A. Hummel, T. Alenghat, M. J. Rosen, IL-33 induces murine intestinal goblet cell differentiation indirectly via innate lymphoid cell IL-13 secretion. J. Immunol. 202, 598–607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G. W. He, L. Lin, J. DeMartino, X. Zheng, N. Staliarova, T. Dayton, H. Begthel, W. J. van de Wetering, E. Bodewes, J. van Zon, S. Tans, C. Lopez-Iglesias, P. J. Peters, W. Wu, D. Kotlarz, C. Klein, T. Margaritis, F. Holstege, H. Clevers, Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation. Cell Stem Cell 29, 1333–1345.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. A. Lindemans, M. Calafiore, A. M. Mertelsmann, M. H. O’Connor, J. A. Dudakov, R. R. Jenq, E. Velardi, L. F. Young, O. M. Smith, G. Lawrence, J. A. Ivanov, Y. Y. Fu, S. Takashima, G. Hua, M. L. Martin, K. P. O’Rourke, Y. H. Lo, M. Mokry, M. Romera-Hernandez, T. Cupedo, L. E. Dow, E. E. Nieuwenhuis, N. F. Shroyer, C. Liu, R. Kolesnick, M. R. M. van den Brink, A. M. Hanash, Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. Gronke, P. P. Hernández, J. Zimmermann, C. S. N. Klose, M. Kofoed-Branzk, F. Guendel, M. Witkowski, C. Tizian, L. Amann, F. Schumacher, H. Glatt, A. Triantafyllopoulou, A. Diefenbach, Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.W. Xia, J. Bai, X. Wu, Y. Wei, S. Feng, L. Li, J. Zhang, G. Xiong, Y. Fan, J. Shi, H. Li, Interleukin-17A promotes MUC5AC expression and goblet cell hyperplasia in nasal polyps via the Act1-mediated pathway. PLOS ONE 9, e98915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C. Schiering, T. Krausgruber, A. Chomka, A. Fröhlich, K. Adelmann, E. A. Wohlfert, J. Pott, T. Griseri, J. Bollrath, A. N. Hegazy, O. J. Harrison, B. M. J. Owens, M. Löhning, Y. Belkaid, P. G. Fallon, F. Powrie, The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Z. Hodzic, E. M. Schill, A. M. Bolock, M. Good, IL-33 and the intestine: The good, the bad, and the inflammatory. Cytokine 100, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. H. Duerr, K. D. Taylor, S. R. Brant, J. D. Rioux, M. S. Silverberg, M. J. Daly, A. H. Steinhart, C. Abraham, M. Regueiro, A. Griffiths, T. Dassopoulos, A. Bitton, H. Yang, S. Targan, L. W. Datta, E. O. Kistner, L. P. Schumm, A. T. Lee, P. K. Gregersen, M. M. Barmada, J. I. Rotter, D. L. Nicolae, J. H. Cho, A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.B. G. Feagan, W. J. Sandborn, G. D'Haens, J. Panés, A. Kaser, M. Ferrante, E. Louis, D. Franchimont, O. Dewit, U. Seidler, K. J. Kim, M. F. Neurath, S. Schreiber, P. Scholl, C. Pamulapati, B. Lalovic, S. Visvanathan, S. J. Padula, I. Herichova, A. Soaita, D. B. Hall, W. O. Böcher, Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn's disease: A randomised, double-blind, placebo-controlled phase 2 study. Lancet 389, 1699–1709 (2017). [DOI] [PubMed] [Google Scholar]
- 27.T. Nishina, Y. Deguchi, D. Ohshima, W. Takeda, M. Ohtsuka, S. Shichino, S. Ueha, S. Yamazaki, M. Kawauchi, E. Nakamura, C. Nishiyama, Y. Kojima, S. Adachi-Akahane, M. Hasegawa, M. Nakayama, M. Oshima, H. Yagita, K. Shibuya, T. Mikami, N. Inohara, K. Matsushima, N. Tada, H. Nakano, Interleukin-11-expressing fibroblasts have a unique gene signature correlated with poor prognosis of colorectal cancer. Nat. Commun. 12, 2281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. M. Nicolas, M. Pesic, E. Engel, P. K. Ziegler, M. Diefenhardt, K. B. Kennel, F. Buettner, C. Conche, V. Petrocelli, E. Elwakeel, A. Weigert, A. Zinoveva, M. Fleischmann, B. Häupl, C. Karakütük, H. Bohnenberger, M. H. Mosa, L. Kaderali, J. Gaedcke, M. Ghadimi, F. Rödel, M. C. Arkan, T. Oellerich, C. Rödel, E. Fokas, F. R. Greten, Inflammatory fibroblasts mediate resistance to neoadjuvant therapy in rectal cancer. Cancer Cell 40, 168–184.e13 (2022). [DOI] [PubMed] [Google Scholar]
- 29.S. Fu, H. Zhu, S. Li, Y. Wang, Y. Bei, J. Xiao, Telocytes in cardiac protection. Curr. Stem Cell Res. Ther. 11, 390–394 (2016). [DOI] [PubMed] [Google Scholar]
- 30.O. N. Karpus, B. F. Westendorp, J. L. M. Vermeulen, S. Meisner, J. Koster, V. Muncan, M. E. Wildenberg, G. R. van den Brink, Colonic CD90+ crypt fibroblasts secrete semaphorins to support epithelial growth. Cell Rep. 26, 3698–3708.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 31.J. E. Kim, L. Fei, W. C. Yin, S. Coquenlorge, A. Rao-Bhatia, X. Zhang, S. S. W. Shi, J. H. Lee, N. A. Hahn, W. Rizvi, K. H. Kim, H. K. Sung, C. C. Hui, G. Guo, T. H. Kim, Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat. Commun. 11, 334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Gao, Q. Yu, H. Zhang, Z. Wang, T. Zhang, J. Xiang, S. Yu, S. Zhang, H. Wu, Y. Xu, Z. Wang, L. Shen, G. Shu, Y. G. Chen, H. Liu, L. Shen, B. Li, A resident stromal cell population actively restrains innate immune response in the propagation phase of colitis pathogenesis in mice. Sci. Transl. Med. 13, eabb5071 (2021). [DOI] [PubMed] [Google Scholar]
- 33.S. M. van Neerven, N. E. de Groot, L. E. Nijman, B. P. Scicluna, M. S. van Driel, M. C. Lecca, D. O. Warmerdam, V. Kakkar, L. F. Moreno, F. A. Vieira Braga, D. R. Sanches, P. Ramesh, S. ten Hoorn, A. S. Aelvoet, M. F. van Boxel, L. Koens, P. M. Krawczyk, J. Koster, E. Dekker, J. P. Medema, D. J. Winton, M. F. Bijlsma, E. Morrissey, N. Léveillé, L. Vermeulen, Apc-mutant cells act as supercompetitors in intestinal tumour initiation. Nature 594, 436–441 (2021). [DOI] [PubMed] [Google Scholar]
- 34.E. E. L. Nystrom, B. Martinez-Abad, L. Arike, G. M. H. Birchenough, E. B. Nonnecke, P. A. Castillo, F. Svensson, C. L. Bevins, G. C. Hansson, M. E. V. Johansson, An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 372, eabb1590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.X. Yin, H. F. Farin, J. H. van Es, H. Clevers, R. Langer, J. M. Karp, Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Liu, B. Qian, L. Zhou, G. Shen, Y. Tan, S. Liu, Z. Zhao, J. Shi, W. Qi, T. Zhou, X. Yang, G. Gao, Z. Yang, IL25 enhanced colitis-associated tumorigenesis in mice by upregulating transcription factor GLI1. Front. Immunol. 13, 837262 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.N. Planell, J. J. Lozano, R. Mora-Buch, M. C. Masamunt, M. Jimeno, I. Ordás, M. Esteller, E. Ricart, J. M. Piqué, J. Panés, A. Salas, Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut 62, 967–976 (2013). [DOI] [PubMed] [Google Scholar]
- 38.X. Zhao, J. Fan, F. Zhi, A. Li, C. Li, A. E. Berger, M. P. Boorgula, S. Barkataki, J. P. Courneya, Y. Chen, K. C. Barnes, C. Cheadle, Mobilization of epithelial mesenchymal transition genes distinguishes active from inactive lesional tissue in patients with ulcerative colitis. Hum. Mol. Genet. 24, 4615–4624 (2015). [DOI] [PubMed] [Google Scholar]
- 39.M. A. J. Koppens, H. Davis, G. N. Valbuena, E. J. Mulholland, N. Nasreddin, M. Colombe, A. Antanaviciute, S. Biswas, M. Friedrich, L. Lee; Oxford IBD Cohort Investigators, L. M. Wang, V. H. Koelzer, J. E. East, A. Simmons, D. J. Winton, S. J. Leedham, Bone morphogenetic protein pathway antagonism by Grem1 regulates epithelial cell fate in intestinal regeneration. Gastroenterology 161, 239–254.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. M. Jacob, S. E. di Carlo, I. Stzepourginski, A. Lepelletier, P. D. Ndiaye, H. Varet, R. Legendre, E. Kornobis, A. Benabid, G. Nigro, L. Peduto, PDGFRα-induced stromal maturation is required to restrain postnatal intestinal epithelial stemness and promote defense mechanisms. Cell Stem Cell 29, 856–868.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 41.M. Quante, S. P. Tu, H. Tomita, T. Gonda, S. S. W. Wang, S. Takashi, G. H. Baik, W. Shibata, B. DiPrete, K. S. Betz, R. Friedman, A. Varro, B. Tycko, T. C. Wang, Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 19, 257–272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. Kobayashi, K. A. Gieniec, J. A. Wright, T. Wang, N. Asai, Y. Mizutani, T. Lida, R. Ando, N. Suzuki, T. R. M. Lannagan, J. Q. Ng, A. Hara, Y. Shiraki, S. Mii, M. Ichinose, L. Vrbanac, M. J. Lawrence, T. Sammour, K. Uehara, G. Davies, L. Lisowski, I. E. Alexander, Y. Hayakawa, L. M. Butler, A. C. W. Zannettino, M. O. Din, J. Hasty, A. D. Burt, S. J. Leedham, A. K. Rustgi, S. Mukherjee, T. C. Wang, A. Enomoto, M. Takahashi, D. L. Worthley, S. L. Woods, The balance of stromal BMP signaling mediated by GREM1 and ISLR drives colorectal carcinogenesis. Gastroenterology 160, 1224–1239.e30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.G. M. Birchenough, E. E. Nystrom, M. E. Johansson, G. C. Hansson, A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.K. Fyderek, M. Strus, K. Kowalska-Duplaga, T. Gosiewski, A. Wędrychowicz, U. Jedynak-Wąsowicz, M. Sładek, S. Pieczarkowski, P. Adamski, P. Kochan, P. B. Heczko, Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J. Gastroenterol. 15, 5287–5294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. Van der Sluis, B. A. De Koning, A. C. De Bruijn, A. Velcich, J. P. Meijerink, J. B. Van Goudoever, H. A. Büller, J. Dekker, I. Van Seuningen, I. B. Renes, A. W. Einerhand, Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129 (2006). [DOI] [PubMed] [Google Scholar]
- 46.B. Dolan, A. Ermund, B. Martinez-Abad, M. E. V. Johansson, G. C. Hansson, Clearance of small intestinal crypts involves goblet cell mucus secretion by intracellular granule rupture and enterocyte ion transport. Sci. Signal. 15, eabl5848 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D. Yang, A. Jacobson, K. A. Meerschaert, J. J. Sifakis, M. Wu, X. Chen, T. Yang, Y. Zhou, P. V. Anekal, R. A. Rucker, D. Sharma, A. Sontheimer-Phelps, G. S. Wu, L. Deng, M. D. Anderson, S. Choi, D. Neel, N. Lee, D. L. Kasper, B. Jabri, J. R. Huh, M. Johansson, J. R. Thiagarajah, S. J. Riesenfeld, I. M. Chiu, Nociceptor neurons direct goblet cells via a CGRP-RAMP1 axis to drive mucus production and gut barrier protection. Cell 185, 4190–4205.e25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. Yu, K. Tong, Y. Zhao, I. Balasubramanian, G. S. Yap, R. P. Ferraris, E. M. Bonder, M. P. Verzi, N. Gao, Paneth cell multipotency induced by notch activation following Injury. Cell Stem Cell 23, 46–59.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. Ayyaz, S. Kumar, B. Sangiorgi, B. Ghoshal, J. Gosio, S. Ouladan, M. Fink, S. Barutcu, D. Trcka, J. Shen, K. Chan, J. L. Wrana, A. Gregorieff, Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125 (2019). [DOI] [PubMed] [Google Scholar]
- 50.K. Murata, U. Jadhav, S. Madha, J. van Es, J. Dean, A. Cavazza, K. Wucherpfennig, F. Michor, H. Clevers, R. A. Shivdasani, Ascl2-dependent cell dedifferentiation drives regeneration of ablated intestinal stem cells. Cell Stem Cell 26, 377–390.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.P. Cheung, J. Xiol, M. T. Dill, W. C. Yuan, R. Panero, J. Roper, F. G. Osorio, D. Maglic, Q. Li, B. Gurung, R. A. Calogero, Ö. H. Yilmaz, J. Mao, F. D. Camargo, Regenerative reprogramming of the intestinal stem cell state via hippo signaling suppresses metastatic colorectal cancer. Cell Stem Cell 27, 590–604.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Q. Ding, S. I. Fukami, X. Meng, Y. Nishizaki, X. Zhang, H. Sasaki, A. Dlugosz, M. Nakafuku, C. C. Hui, Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 9, 1119–11S1 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Q. Hou, J. Huang, H. Ayansola, H. Masatoshi, B. Zhang, Intestinal stem cells and immune cell relationships: Potential therapeutic targets for inflammatory bowel diseases. Front. Immunol. 11, 623691 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. M. Allaire, S. A. B. Roy, C. Ouellet, É. Lemieux, C. Jones, M. Paquet, F. Boudreau, N. Perreault, Bmp signaling in colonic mesenchyme regulates stromal microenvironment and protects from polyposis initiation. Int. J. Cancer 138, 2700–2712 (2016). [DOI] [PubMed] [Google Scholar]
- 55.S. Ouahoud, B. F. Westendorp, P. W. Voorneveld, S. Abudukelimu, P. J. Koelink, E. Pascual Garcia, J. F. I. Buuren, T. J. Harryvan, K. J. Lenos, T. van Wezel, J. A. Offerhaus, A. Fariña-Sarasqueta, S. Crobach, M. Slingerland, J. C. H. Hardwick, L. J. A. C. Hawinkels, Loss of bone morphogenetic protein signaling in fibroblasts results in CXCL12-driven serrated polyp development. J. Gastroenterol. 58, 25–43 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Tu, G. Bhagat, G. Cui, S. Takaishi, E. A. Kurt-Jones, B. Rickman, K. S. Betz, M. Penz-Oesterreicher, O. Bjorkdahl, J. G. Fox, T. C. Wang, Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell 14, 408–419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. Garlanda, A. Mantovani, Interleukin-1 in tumor progression, therapy, and prevention. Cancer Cell 39, 1023–1027 (2021). [DOI] [PubMed] [Google Scholar]
- 58.M. Friedrich, M. Pohin, M. A. Jackson, I. Korsunsky, S. J. Bullers, K. Rue-Albrecht, Z. Christoforidou, D. Sathananthan, T. Thomas, R. Ravindran, R. Tandon, R. S. Peres, H. Sharpe, K. Wei, G. F. M. Watts, E. H. Mann, A. Geremia, M. Attar; Oxford IBD Cohort Investigators; Roche Fibroblast Network Consortium, F. Barone, M. Brenner, C. D. Buckley, M. Coles, A. P. Frei, K. G. Lassen, F. M. Powrie, S. McCuaig, L. Thomas, E. Collantes, H. H. Uhlig, S. N. Sansom, A. Easton, S. Raychaudhuri, S. P. Travis, F. M. Powrie, IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat. Med. 27, 1970–1981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. Ahn, A. L. Joyner, Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Y. Mishina, M. C. Hanks, S. Miura, M. D. Tallquist, R. R. Behringer, Generation of Bmpr/Alk3conditional knockout mice. Genesis 32, 69–72 (2002). [DOI] [PubMed] [Google Scholar]
- 61.X. Yang, C. Li, P. L. Herrera, C. X. Deng, Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–81 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Y. Wang, W. Song, J. Wang, T. Wang, X. Xiong, Z. Qi, W. Fu, X. Yang, Y. G. Chen, Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 217, e20191130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.G. Yu, L. G. Wang, Y. Han, Q. Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. Davis, P. S. Meltzer, GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 to S4
Legends for movies S1 to S3
Tables S1 to S4
Movies S1 to S3