Abstract
In meiotic homologous recombination (HR), BRCA2 facilitates loading of the recombinases RAD51 and DMC1 at the sites of double-strand breaks (DSBs). The HSF2BP-BRME1 complex interacts with BRCA2. Its absence causes a severe reduction in recombinase loading at meiotic DSB. We previously showed that, in somatic cancer cells ectopically producing HSF2BP, DNA damage can trigger HSF2BP-dependent degradation of BRCA2, which prevents HR. Here, we report that, upon binding to BRCA2, HSF2BP forms octameric rings that are able to interlock into a large ring-shaped 24-nucleotide oligomer. Addition of BRME1 leads to dissociation of both of these ring structures and cancels the disruptive effect of HSF2BP on cancer cell resistance to DNA damage. It also prevents BRCA2 degradation during interstrand DNA crosslink repair in Xenopus egg extracts. We propose that, during meiosis, the control of HSF2BP-BRCA2 oligomerization by BRME1 ensures timely assembly of the ring complex that concentrates BRCA2 and controls its turnover, thus promoting HR.
Cryo-EM reveals that meiotic HSF2BP concentrates recombination mediator BRCA2 into ring complexes that are disrupted by BRME1.
INTRODUCTION
In vertebrates, both somatic and meiotic homologous recombination (HR) require the BRCA2 protein (1, 2). Its orthologs in fungi (3), plants (4), and invertebrates (5–8) are also essential for meiotic HR, so this role is likely ancestral. Most mechanistic data on BRCA2, however, come from studies in somatic cells due to the strong association of BRCA2 with breast, ovarian, pancreatic, and a number of other types of cancer (9). These studies showed that BRCA2 interacts with and controls RAD51, a recombinase that performs the central HR reactions: homology recognition and strand exchange. Meiotic studies are hindered by the embryonic lethality of the Brca2 knockout in mice. A hypomorphic rescue transgene in a Brca2 knockout mouse strain confirmed the critical role of BRCA2 in mouse meiotic HR (10). It was also demonstrated that BRCA2 interacts with the meiotic recombinase DMC1 (11–14), a paralog of RAD51. This suggested that BRCA2 may contribute to the correct balance in RAD51 and DMC1 loading that is essential in meiotic HR.
New tools to study the role of BRCA2 in meiosis were recently provided by the discovery that, in mouse meiocytes and embryonic stem cells, BRCA2 functions in complex with two previously uncharacterized germline proteins, HSF2BP (also called MEILB2) and BRME1 (15–18). The reported phenotypes of three independent Hsf2bp (17–19) and five independent Brme1 knockout mouse models (16–22) are nearly identical. Loss of these proteins does not lead to embryonic lethality but causes complete spermatogenesis failure due to severe reduction in RAD51 and DMC1 accumulation at meiotic DNA double-strand breaks (DSBs), which prevents crossover formation (17, 18). Loss of BRME1 decreases the number of HSF2BP foci. As HSF2BP directly binds to both BRCA2 and BRME1 (Fig. 1A), it has been proposed that loss of HSF2BP or BRME1 compromises BRCA2 meiotic function, although the mechanism may be different (23) from the suggested “meiotic localizer of BRCA2” model (18).
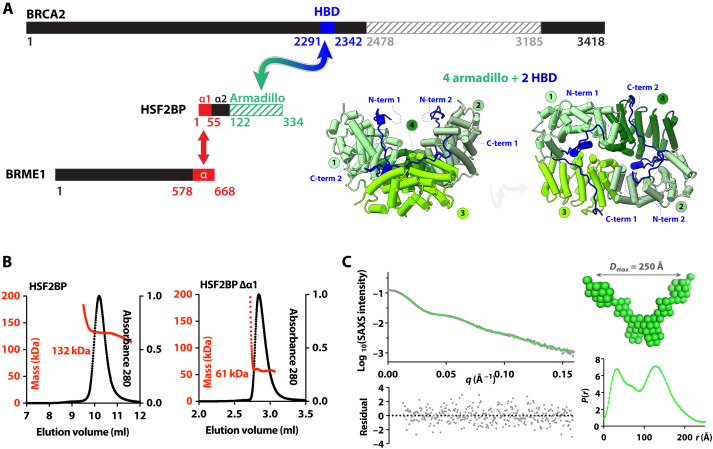
Fig. 1. HSF2BP is a tetramer formed by two dimers interacting through the N-terminal helix α1.
(A) BRCA2, HSF2BP, and BRME1 domains and interactions. Folded domains of known three-dimensional (3D) structure are marked with stripes. Cocrystallized regions of BRCA2 (HBD) and HSF2BP (armadillo) are colored in blue and green, respectively; their 3D structure cartoon is shown in two different orientations [Protein Data Bank (PDB) code: 7BDX]. Regions of HSF2BP and BRME1 that interact (16) are colored in red. (B) SEC-MALS analysis of full-length HSF2BP [monomer weight-average molecular weight (Mw) 37.6 kDa; column: Superdex 200 10/300 GL] and HSF2BP deleted from helix α1 (monomer Mw 32.2 kDa; column: BIOSEC 3). See fig. S1A for replicates. (C) SEC-SAXS curve and resulting distance distribution, obtained on full-length HSF2BP. The SAXS curve is plotted as a function of the scattering angle [experimental curve in black dots; P(r) Fourier Transform in green). The distance distribution P(r) is plotted as a function of the distance. The deduced HSF2BP mass is 156 ± 21 kDa. Calculation of an ab initio model [average model in green spheres; more models in fig. S1 (B and C)] from the SAXS data suggests that HSF2BP has a V shape.
Paradoxically, when HSF2BP is produced ectopically in somatic cancer cells, it suppresses HR instead of supporting it as it does in meiocytes (15, 24). We demonstrated genetically and biochemically that this is due to HSF2BP interaction with BRCA2: In cancer cells, formation of a complex between HSF2BP and BRCA2 impedes BRCA2 function during the repair of lesions induced by DNA interstrand crosslinking agents and poly(adenosine 5′-diphosphate–ribose) polymerase (PARP) inhibitors (but not by ionizing radiation or the I-SceI nuclease) (15, 24). Evolutionary conservation of the interaction between BRCA2 and HSF2BP allowed us to establish, using Xenopus egg extract interstrand crosslink repair assays, that the presence of HSF2BP leads to DNA crosslink-dependent proteasomal degradation of BRCA2 mediated by the p97 segregase (24), which disassembles ubiquitinated protein aggregates (25, 26).
To study the molecular events triggered by the interaction between BRCA2 and HSF2BP, we previously solved the crystal structure of a 51–amino acid BRCA2 peptide (BRCA2-HBD) in complex with the C-terminal armadillo domain of HSF2BP (Fig. 1A). We identified a cryptic motif repeated twice in BRCA2-HBD and encoded by exons 12 and 13, respectively. Each motif binds to one armadillo domain (23, 27). In the complex, two BRCA2-HBD peptides “staple together” two armadillo dimers, resulting in a high-affinity interaction [Kd (dissociation constant) ~ 1 nM]. In addition to the crystallized armadillo domain, HSF2BP contains N-terminal helices α1 and α2 (Fig. 1A) that assemble into oligomers in vitro. Helix α1 homotetramerizes when free in solution and heterotetramerizes when bound to the C-terminal α-helical region of BRME1 (16, 19, 21). The presence of these three HSF2BP oligomerization mechanisms—dimerization via armadillo and α2, tetramerization via α1, and hetero-oligomerization via BRCA2-HBD binding—led us to propose that HSF2BP is a polymerization agent for BRCA2, which could be behind its physiological function in meiotic HR and mediate its pathological effect in somatic HR (23). However, because the N-terminal helices of HSF2BP were missing from previous structural analyses, their effect on the HSF2BP-BRCA2 structure and function was uncharacterized (23, 27). It was also not known how the interaction between HSF2BP helix α1 and BRME1 affects the oligomeric state of the BRCA2-HSF2BP complex and BRCA2 function in cells.
In this work, we obtained a cryo–electron microscopy (cryo-EM) derived model of the complex between full-length HSF2BP and BRCA2-HBD and determined how BRME1 affects the structure of this complex. We revealed that, upon binding to BRCA2, HSF2BP forms an octameric ring and that three of these rings can assemble into a large (⌀ ~ 200 Å) topologically interlocked 24-nucleotide oligomer, with BRCA2 displayed on the outer surface of the complex. We also demonstrated that BRME1 disrupts these ring structures and acts as a protective disaggregation agent in specific cellular conditions. From our structural and cell biology data, we propose a model explaining the opposite effects of HSF2BP on somatic and meiotic HR.
RESULTS
HSF2BP is mainly a V-shaped tetramer in solution
To understand how the N-terminal α-helical region of HSF2BP contributes to the assembly of the HSF2BP-BRCA2 complex, we first characterized full-length HSF2BP (37.6 kDa) and HSF2BP lacking helix α1 (G48-V334; 32.2 kDa) by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). At the concentration of the experiment (~10 to 30 μM), the apparent molar masses of the two samples are 136 ± 7 kDa (n = 2) and 60 ± 2 kDa (n = 2), respectively (Fig. 1B and fig. S1A). This fits with tetrameric and dimeric states for HSF2BP with and without helix α1, respectively. Further characterization of HSF2BP by SEC coupled to small-angle x-ray scattering (SEC-SAXS) showed that, in the conditions of this experiment (protein concentration ~ 50 to 60 μM), the protein has a molar mass of 156 ± 21 kDa (Fig. 1C and table S1). Thus, we confirmed that full-length HSF2BP is mainly tetrameric at concentrations above 10 μM. The SAXS-derived atomic distance distribution curve of HSF2BP is bimodal, with a maximal distance at 250 Å (Fig. 1C). Consistently, ab initio molecular envelopes calculated from this curve using a twofold symmetry hypothesis have a V shape (Fig. 1C and fig. S1, B and C).
HSF2BP and BRCA2 form a ring-shaped complex with BRCA2 on its outer surface
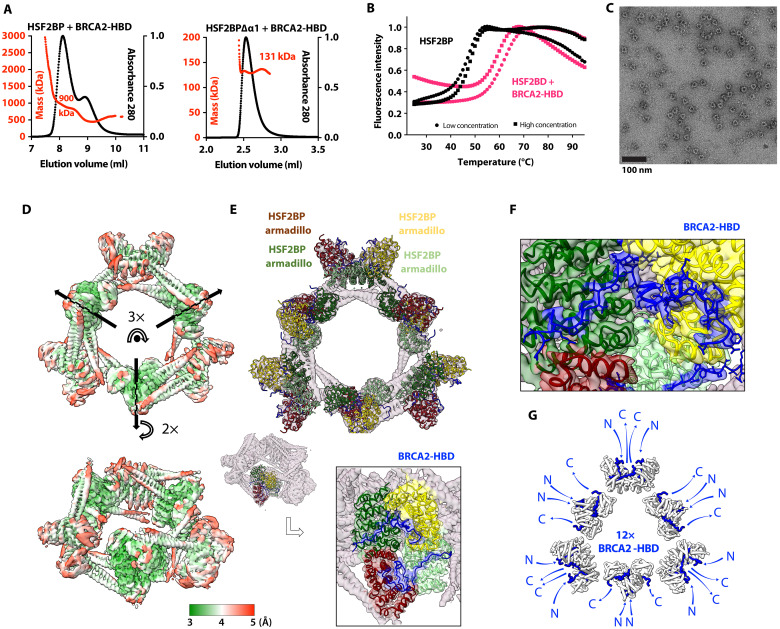
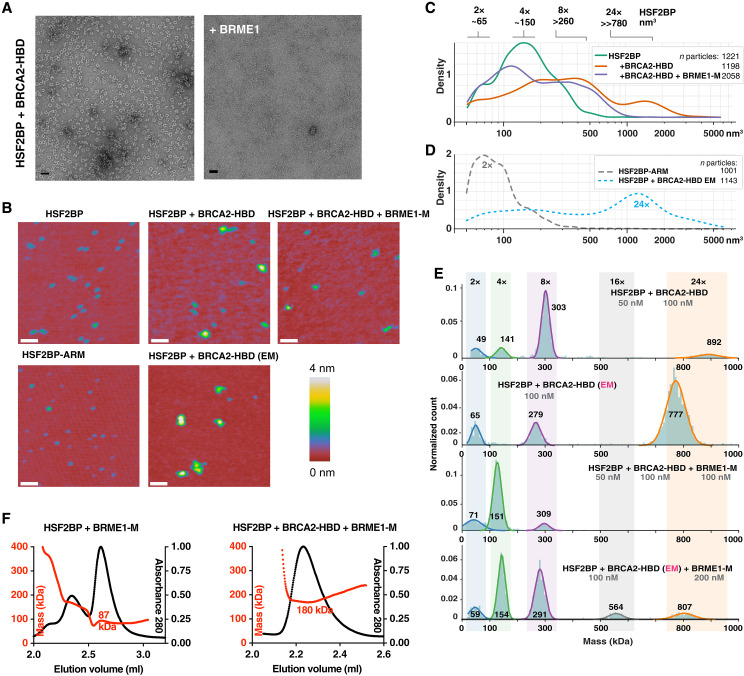
Having characterized the solution structure of free full-length HSF2BP, we measured the change in its oligomeric state induced by BRCA2. We observed that tetrameric HSF2BP mainly forms an 880 ± 30 kDa (n = 2) complex when bound to BRCA2-HBD (N2291-Q2342), as measured by SEC-MALS (Fig. 2A and fig. S1A). Its thermal stability also increases substantially, shifting from 45.9° to 46.7°C to 59° to 60.7°C (Fig. 2B). Negative-staining EM revealed a ring-shaped complex with an outer diameter of about 200 Å (Fig. 2C) and a threefold symmetry (fig. S2A). Further characterization by cryo-EM (fig. S2, B and C) of this complex revealed a D3 symmetry, defined as a threefold symmetry around the axis going through the center of the ring, as well as three twofold symmetries around perpendicular axes going through the center of the globular subvolumes (Fig. 2D). The most resolved cryo-EM map was calculated from 398,000 particles and refined using this symmetry. Resulting local resolutions yield 3 to 4.5 Å and 3.5 to 5 Å in the three inner and outer globular subvolumes, respectively. The tubular-shaped volumes extending out of the globular subvolumes (two tubes on both sides of each subvolume) are less resolved (4 to 8 Å). Further three-dimensional (3D) classification of the cryo-EM particles resulted in the calculation of two cryo-EM maps. Comparison of these maps revealed that the diameter of the ring is slightly variable, with the larger ring being generally better resolved than the tighter ring, except for the inner globular subvolumes that are better resolved in the tighter ring (fig. S2D). The ring flexibility might explain the heterogeneous resolution of the calculated cryo-EM maps. We fitted the published crystal structure of the C-terminal armadillo domain of HSF2BP in complex with BRCA2-HBD (23, 27) into each of the six globular subvolumes of the large ring (Fig. 2E). The shape of the structure as shown in Fig. 1A (3D view on the left) matches the subvolumes as shown in Fig. 2E. Not only the α helices of the armadillo domains but also the BRCA2-HBD peptides can be traced through the cryo-EM map (Fig. 2F and fig. S2C); several side chains can even be identified. The BRCA2 peptides are located on the outer side of the map, their N- and C termini being positioned around the ring-shaped complex (Fig. 2G).
Fig. 2. HSF2BP oligomerizes into a ring-shaped complex upon binding to BRCA2-HBD.
(A) SEC-MALS analysis of HSF2BP bound to BRCA2-HBD (column: Superdex 200 10/300 GL) and HSF2BP deleted from helix α1 bound to BRCA2-HBD (column: BIOSEC 3). See fig. S1A for replicates. (B) Thermal stability of HSF2BP, either free or bound to BRCA2-HBD. The denaturation temperature of HSF2BP shifted from 45.9° to 46.7°C to 59° to 60.7°C upon binding to BRCA2-HBD. (C) Negative-staining EM image obtained on a sample of HSF2BP bound to BRCA2-HBD. (D) Cryo-EM map of HSF2BP bound to BRCA2-HBD. Top and side views are displayed with an electron density threshold of 0.05. The map shows a D3 symmetry. It is colored as a function of the local resolution: from green (3 Å) to red (5 Å). (E) Docking of the crystal structure of the complex between HSF2BP armadillo domain and BRCA2-HBD (PDB: 7BDX) into the cryo-EM map. In the top view, a crystal structure is positioned in each of the six globular subvolumes of the map, whereas in the side view, only one crystal structure is displayed. Each crystal structure contains four armadillo domains (in light green, yellow, green, and maroon) bound to two BRCA2-HBD peptides (in blue). In the boxed side view, the map is colored as the docked chains of the crystal structure. (F) Zoom view on one of the BRCA2 peptides docked into the cryo-EM map. The position of this peptide, represented in blue sticks, readily fits into the cryo-EM density map of the complex between HSF2BP and BRCA2-HBD. The map is colored as the docked chains of the crystal structure. (G) Orientations of the N and C termini of the 12 BRCA2 peptides in the complex. All the BRCA2 extremities are located on the outer surface of the ring shape.
The large HSF2BP-BRCA2 complex is assembled from three interlocked BRCA2-bound HSF2BP diamond-shaped octamers
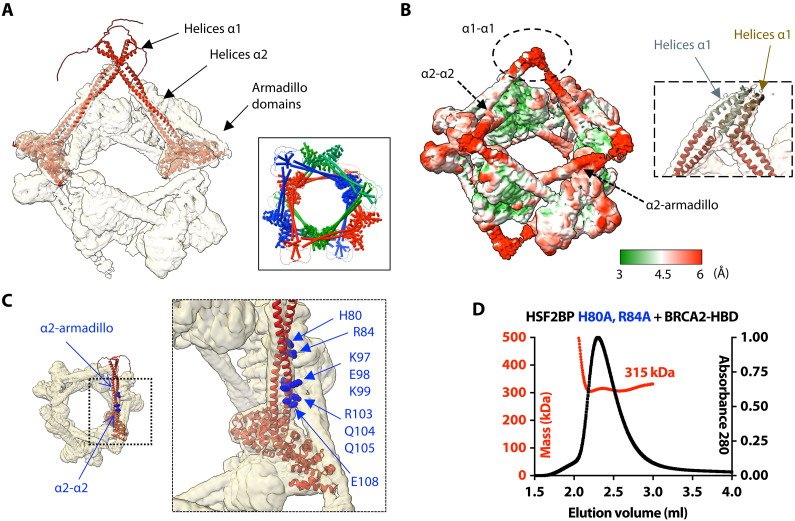
We further analyzed our cryo-EM map of the complex between HSF2BP and BRCA2-HBD to describe the 3D structure of HSF2BP N-terminal α-helical region in this complex. First, we modeled a dimer of HSF2BP using AlphaFold (28). This prediction algorithm proposed five similar models for dimeric HSF2BP with high confidence (fig. S3A). In these models, each HSF2BP monomer is composed of strand β1 (F20-R24), helix α1 (K25-I45), and helix α2 (G48-S136) that overlaps with the armadillo domain (E122-V334). As a validation, we checked that the 3D model of the dimeric armadillo domain is superimposable with the 3D structure observed in the crystal [Protein Data Bank (PDB) code: 7BDX (23) and fig. S3B]. We then docked two dimeric HSF2BP models into the cryo-EM map of the complex between full-length HSF2BP and BRCA2-HBD (Fig. 3A). Helices α2 readily fitted into the electron density tubes extending out of the globular subvolumes corresponding to the armadillo domains bound to BRCA2-HBD. Helices α1, on the other hand, fell into a poorly resolved zone connecting electron densities of two HSF2BP dimers (Fig. 3, A and B). In the resulting model of the complex, four of these helices interact to connect two HSF2BP dimers, so that the HSF2P tetramer exhibits a V-shape structure. Two tetramers interact through the BRCA2-HBD peptides to form an octamer. Three interlocked octamers (displayed in red, blue, and green in Fig. 3A, inset) form the final ring-shaped 24-nucleotide oligomer protein assembly. The N-terminal region of HSF2BP contributes to the assembly of this large complex through interactions between helices α1 within each tetramer and contacts between helices α2 and armadillo domains from different octamers (Fig. 3B).
Fig. 3. The ring-shaped complex is assembled through a large set of interfaces, involving not only armadillo-BRCA2 but also α1-α1, α2-α2, and α2-armadillo contacts.
(A) Docking of two (similar) AlphaFold models of the HSF2BP dimer into the cryo-EM map. The map is displayed with an electron density threshold of 0.035. The cartoon views of the two AlphaFold models are colored in red. Each model consists of a disordered region, a short helix α1, a large helix α2, and an armadillo domain. In the boxed panel, three pairs of HSF2BP tetramers docked into the cryo-EM map are displayed in three different colors. (B) Cryo-EM map displayed with a lower electron density threshold (0.025) and colored as a function of the local resolution. The colors are from green (3 Å) to red (6 Å). A dashed oval identifies a map region corresponding to the four helices α1, whereas arrows indicate regions of α2-α2 and α2-armadillo contacts. In the inset, four helices α2 are docked as in (A) (in red), whereas four parallel helices α1 (in gray and brown) are manually positioned into the cryo-EM density displayed with an electron density threshold (0.010). (C) Position of the α2 residues mutated to test their role in the assembly and/or function of the ring-shaped complex. An AlphaFold model of HSF2BP dimer (in red) is docked into the cryo-EM map. Residues at the α2-α2 and α2-armadillo interfaces are marked with standard and bold labels, respectively. (D) SEC-MALS analysis of the mutant HSF2BP H80A-R84A bound to BRCA2-HBD (column: BIOSEC 3; see also fig. S4).
HSF2BP helices α1 and α2 form the interfaces between the HSF2BP dimers
To further understand how the N-terminal region of HSF2BP contributes to the assembly of this large complex, we modeled the α1 tetramer using AlphaFold (fig. S3C). The best scored model is composed of two antiparallel α1 dimers that cannot be fitted in the density map. However, several parallel tetramers were also proposed by AlphaFold (models 3 to 5). They are similar to each other and closer to what can be obtained by fitting four α1 helices into the cryo-EM density map (fig. S3D). The low resolution of this part of the map is probably due to the variable position of the α1 tetramer relatively to the ring; it prevents a more detailed description of the 3D structure of this tetramer.
We also analyzed how HSF2BP-BRCA2 octamers assemble into the large ring-shaped complex. In our cryo-EM structure, octamers form diamond-shaped rings that interlock to form a 24-nucleotide oligomer complex (Fig. 3A, inset). This suggests that a network of contacts between octamers favors the formation and high thermal stability of the 24-nucleotide oligomer complex and that a dynamic mechanism exists for octamer opening allowing interlocking. Two types of contacts are observed between octamers, involving either coiled coils of helices α2 from both octamers or one of these coiled coils and an armadillo domain (indicated by arrows in Fig. 3, B and C). In each octamer, two α2 coiled coils interact with a second octamer (specifically, with its helices α2 and armadillo domains), whereas the two other α2 coiled coils interact with the third octamer (also with its helices α2 and armadillo domains). We engineered mutations in HSF2BP helix α2 to specifically disrupt these contacts. We mutated a set of selected residues into alanine (Fig. 3C) and measured the oligomeric state of the resulting free and BRCA2-bound HSF2BP variants by SEC-MALS (Fig. 3D and fig. S4). The H80A-R84A variant only assembled into octamers upon binding to BRCA2-HBD; it does not form 24-nucleotide oligomers (Fig. 3D). Thus, not only the α1-α1 interaction but also the α2-armadillo interaction contribute to the formation of the large HSF2BP-BRCA2 oligomer.
HSF2BP helix α1 forms a heterotetramer with an α-helical peptide from the C-terminal region of BRME1
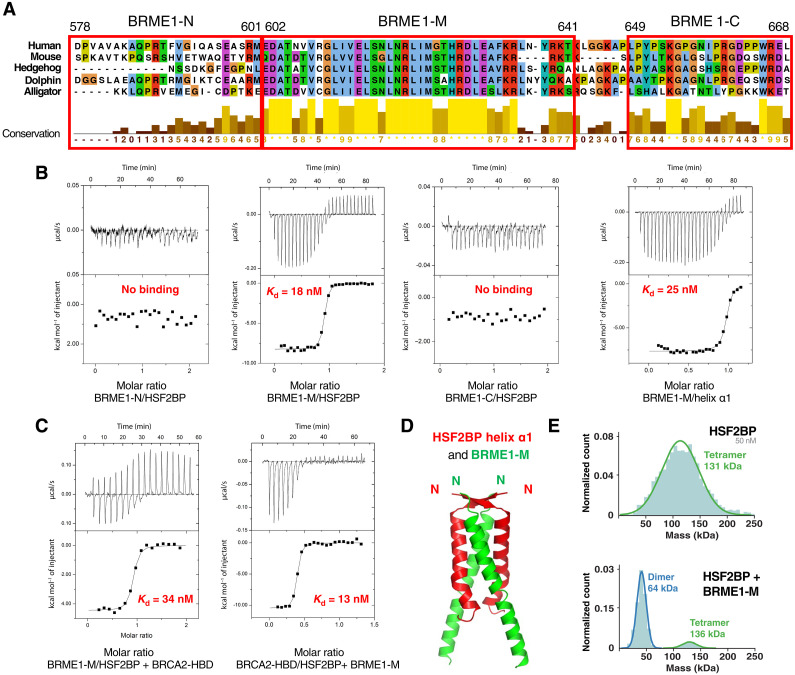
Mouse HSF2BP binds directly to the meiotic protein BRME1 (16). The N-terminal helix α1 of HSF2BP interacts with the C-terminal region of BRME1 (16, 20). To further delineate the binding fragment in human BRME1, we divided its C-terminal region (D578-L668) into three peptides: BRME1-M, corresponding to the central and well-conserved fragment, and BRME1-N and BRME1-C, corresponding to the less conserved flanking regions (Fig. 4A). Isothermal titration calorimetry (ITC) experiments revealed that only BRME1-M detectably bound to HSF2BP and that its affinity was similar when measured against full-length HSF2BP (18 ± 3 nM) or HSF2BP helix α1 (25 ± 5 nM) (Fig. 4B, fig. S5A, and Table 1). Thus, the C-terminal region BRME1-M (E602-K641) directly interacts with the HSF2BP N-terminal helix α1. This interaction is not expected to affect the binding of BRCA2 to the HSF2BP C-terminal armadillo domain (23). Consistently, we found by ITC that BRME1-M is able to interact with HSF2BP bound to BRCA2-HBD (affinity: 34 ± 8 nM) and that BRCA2-HBD is able to interact with HSF2BP bound to BRME1-M (affinity: 13 ± 2 nM) (Fig. 4C). We concluded that HSF2BP simultaneously interacts with BRCA2-HBD and BRME1-M.
Fig. 4. A BRME1 peptide binds to helix α1 and disrupts the HSF2BP tetramer.
(A) Sequence alignment of the C-terminal region of a set of five BRME1 proteins, showing a representative sequence diversity. The sequences corresponding to the three human peptides BRME1-N, BRME1-M, and BRME1-C are boxed. (B) ITC curves identifying the BRME1 sequence binding to HSF2BP. These experiments were all performed at 30°C. Additional experiments performed at 20°C are detailed in fig. S5A and Table 1. (C) ITC curve showing that BRME1-M and BRCA2-HBD do not compete for binding to HSF2BP. These experiments were performed at 20°C. (D) Crystal structure of the HSF2BP peptide E19-V50 (helix α1, in red) bound to the BRME1 peptide E602-K641 (BRME1-M, in green). Each asymmetric unit contained a HSF2BP-BRME1 dimer. The heterotetramer was calculated by application of a twofold crystallography symmetry (table S2). (E) Mass photometry experiment performed on either free HSF2BP (top) or HSF2BP bound to BRME1-M (bottom). The distribution of masses is displayed for samples at a concentration of 50 nM for HSF2BP and 100 nM for BRME1-M. HSF2BP analyzed at 25 nM is shown in fig. S6C.
Table 1. Binding parameters deduced from the ITC analyses.
| Kd (M) (± error) | n | ΔH (kcal/mol) (± error) | ΔG (kcal/mol) | ─TΔS (kcal/mol) | T (K) | |||
|---|---|---|---|---|---|---|---|---|
| HSF2BP versus BRME1-M | 1.1 × 10−08 | (0.4 × 10−08) | 0.77 | −5.7 | (0.07) | −10.6 | −4.90 | 293 |
| HSF2BP versus BRME1-M | 1.8 × 10−08 | (0.3 × 10−08) | 0.88 | −8.2 | (0.06) | −10.7 | −2.50 | 303 |
| HSF2BP versus BRME1-N | Undetectable | 293 | ||||||
| HSF2BP versus BRME1-N | Undetectable | 303 | ||||||
| HSF2BP versus BRME1-C | Undetectable | 293 | ||||||
| HSF2BP versus BRME1-C | Undetectable | 303 | ||||||
| HSF2BP α1 versus BRME1-M | 2.5 × 10−08 | (0.3 × 10−08) | 0.96 | −7.6 | (0.06) | −10.5 | −2.90 | 303 |
| HSF2BP α1 versus BRME1-M | 2.1 × 10−08 | (0.4 × 10−08) | 0.93 | −1.2 | (0.05) | −10.2 | −9.00 | 293 |
| HSF2BP + BRME1-M versus BRCA2-HBD | 1.3 × 10−08 | (0.2 × 10−08) | 0.37 | −10.5 | (0.13) | −10.4 | −0.10 | 293 |
| HSF2BP + BRME1-M versus BRCA22213–2342 | 7.8 × 10−09 | (1.8 × 10−09) | 0.35 | −10.9 | (0.12) | −12.6 | −1.70 | 293 |
| HSF2BP + BRCA2-HBD versus BRME1-M | 3.4 × 10−08 | (0.8 × 10−08) | 0.87 | −4.4 | (0.06) | −10 | −5.60 | 293 |
| HSF2BP + BRCA2-HBD versus BRME1-M | 2.0 × 10−08 | (1.9 × 10−08) | 0.8 | −5.8 | (0.22) | −10.2 | −4.40 | 293 |
To elucidate how HSF2BP helix α1 interacts with BRME1-M, we crystallized HSF2BP helix α1 both alone and bound to BRME1-M. We solved the crystal structure of the human HSF2BP fragment from E19 to G48. Each monomer consists of a β strand (residues F20-R24) and an α-helix (residues K25-L46) (fig. S5B). The crystal asymmetric unit contains two peptides interacting through antiparallel β strands and parallel α helices that assemble into a tetramer by application of a twofold crystallography symmetry (table S2). This tetrameric structure is similar to the best model proposed by AlphaFold (fig. S3D). However, we have no experimental evidence supporting the presence of such an antiparallel conformation in the free full-length HSF2BP tetramer, and we reported that this conformation is not consistent with the cryo-EM map of HSF2BP bound to BRCA2-HBD. We also solved the crystal structure of helix α1 bound to BRME1-M. Each BRME1-M monomer contains a large α helix (residues T605 to R638) (Fig. 4D). The asymmetric unit contains a HSF2BP helix α1 and a BRME1-M peptide that forms a parallel heterodimer. Two of these dimers assemble into a parallel tetramer by application of a twofold crystallographic symmetry (table S2). The β strands from the two HSF2BP peptides form an antiparallel β sheet. Together, our x-ray crystallography analyses provided an antiparallel structure for the α1 tetramer and a parallel arrangement for the helices of the α1-BRME1 heterotetramer. Our cryo-EM data and AlphaFold modeling suggested that homotetramerization of α1 in a parallel configuration is responsible for the formation of the ring. On the basis of the x-ray structure of α1 bound to BRME1-M, we further hypothesized that BRME1-M α helix competes with HSF2BP helix α1, thus disassembling α1 tetramers. To experimentally validate this hypothesis, we performed mass photometry analyses on HSF2BP in the absence and presence of BRME1-M. We observed that, indeed, the HSF2BP tetramer is dissociated into dimers after addition of BRME1-M (Fig. 4E).
BRME1 binding to HSF2BP dissociates HSF2BP-BRCA2 ring complexes
We then proceeded to determine the effect of BRME1 on the structure of the HSF2BP-BRCA2 complex. We observed using negative-staining EM that addition of BRME1-M to a 2:1 mix of HSF2BP and BRCA2-HBD resulted in a near-complete disappearance of the large ring-shape complexes (Fig. 5A). To replicate this marked effect under different experimental conditions and to quantify it, we performed single-molecule analysis using scanning force microscopy (SFM; Fig. 5, B to D). HSF2BP alone appeared on SFM scans as spherical objects with a volume distribution centered around 95 and 190 nm3. We surmised that under the low protein concentrations used for single-molecule SFM experiments, HSF2BP oligomer equilibrium may be shifted toward dimers and interpreted the two populations of objects as a mix of dimers and tetramers (Fig. 5, B and C, and fig. S6, A and B). This interpretation is consistent with the mass photometry data recorded on HSF2BP at different concentrations, which revealed that, whereas at 50 nM HSF2BP is a tetramer (Fig. 4E), at 25 nM, it is a mix of dimers and tetramers (fig. S6C). We also observed by SFM that the armadillo domain alone appeared as smaller spherical objects with a volume distribution centered on 35 and 90 nm3, which we assigned to monomers and dimers, respectively (Fig. 5D). By contrast, the volume distribution of the complexes formed by HSF2BP and BRCA2-HBD revealed much larger objects, with peaks at ~550 and ~2000 nm3 (Fig. 5, B and C, and fig. S6, A and B). Taking the 95 nm3 volume of HSF2BP dimers as the smallest unit and correcting for the increase in measured volume due to the ringed shape and scanning precision, we assigned the minor subpopulation of the largest objects (~2000 nm3) to the 24-nucleotide oligomers. This was confirmed by SFM analysis of the sample of HSF2BP-BRCA2-HBD that was purified by SEC and used in the EM analyses (Fig. 5D). Addition of BRME1-M to the HSF2BP-BRCA2-HBD sample used for the first SFM measurements resulted in a substantial reduction in apparent object size, which, when quantified, corresponded to a complete disappearance of the 24-nucleotide oligomer population (Fig. 5, B and C, and fig. S6, A and B). Octameric species were still observed, and the number of tetramer- and dimer-sized objects was increased.
Fig. 5. A BRME1 peptide disrupts the ring-shaped complex.
(A) Negative-staining EM images recorded on the complex formed by HSF2BP and BRCA2-HBD in the absence (left) and presence (right) of the peptide BRME1-M. The conditions are the same as in Fig. 2C. (B) Representative SFM images of HSF2BP, its armadillo domain (HSF2BP-ARM), HSF2BP + BRCA2-HBD, and HSF2BP + BRCA2-HBD + BRME1-M. The complexes were assembled just before deposition on mica for SMF analysis. Only the sample prepared for EM was purified by gel filtration. Scale bars, 100 nm. (C) Kernel density plots of the particle volume distributions for HSF2BP and its complexes with BRCA2-HBD and BRCA2-HBD + BRME1-M. The number of particles is indicated in the legend, and the replicate of the experiment is shown in fig. S6 (A and B). (D) Kernel density plots of particle volume distributions of the HSF2BP-ARM fragments and HSF2BP + BRCA2-HBD complexes prepared following the protocol used for EM. These were used to determine the volumes of the dimers and the ring-shaped 24-nucleotide oligomer, respectively. The experiment was done once, and the number of analyzed particles is indicated. (E) Mass photometry experiments performed on HSF2BP in the presence of BRCA2-HBD (freshly assembled or cryo-EM samples) and/or BRME1-M. The distributions of masses are displayed at the indicated concentrations of proteins. The cryo-EM sample was diluted down to 100 nM, and a twofold excess of BRME1-M was used to observe the impact of BRME1-M on the 24-nucleotide oligomer complex. Replicates are presented in fig. S6D. (F) SEC-MALS analysis of the complex between HSF2BP and BRME1-M in the absence (left) or presence (right) of BRCA2-HBD (column: BIOSEC 3). Replicates are presented in fig. S6E.
To support these SFM results, we performed additional mass photometry analyses of both freshly prepared samples of BRCA2-HBD bound HSF2BP and the sample used for cryo-EM studies. We observed that, at the concentrations used, addition of BRCA2-HBD to HSF2BP triggers the formation of octamers and, in smaller amounts, 24-nucleotide oligomers (Fig. 5E and fig. S6D). Further addition of BRME1-M disassembles the 24-nucleotide oligomer complexes, yielding to a mix between octamers, tetramers, and dimers. Similarly, addition of BRME1-M to the cryo-EM sample caused disassembly of the ring-shaped complex and enrichment in double-octamers, octamers, and tetramers (Fig. 5E). Together, our EM, SFM, and mass photometry results consistently demonstrated that BRME1 can act as a dissociation factor for HSF2BP-BRCA2 oligomers.
Last, we characterized the complex among HSF2BP, BRCA2-HBD, and BRME1-M using SEC-MALS. We first purified HSF2BP bound to BRME1-M and verified using SEC-MALS that it is a dimer (Figs. 5F and 6E). We then prepared a sample of the ternary complex at the highest possible concentration (1 mg/ml; the solubility limit of this complex in our conditions) in the presence of an excess of BRCA2-HBD and BRME1-M peptides and performed a SEC-MALS analysis on this sample. We observed that the ternary complex has a molecular mass of 181 ± 1 kDa (n = 2) (Fig. 5F and fig. S6E). The theoretical mass of four HSF2BP bound to four BRME1-M and two BRCA2-HBD being 182 kDa, we concluded that this complex contains a tetrameric form of HSF2BP. Thus, the HSF2BP-BRCA2 complex dissociates into a tetramer of BRCA2-bound HSF2BP in the presence of BRME1.
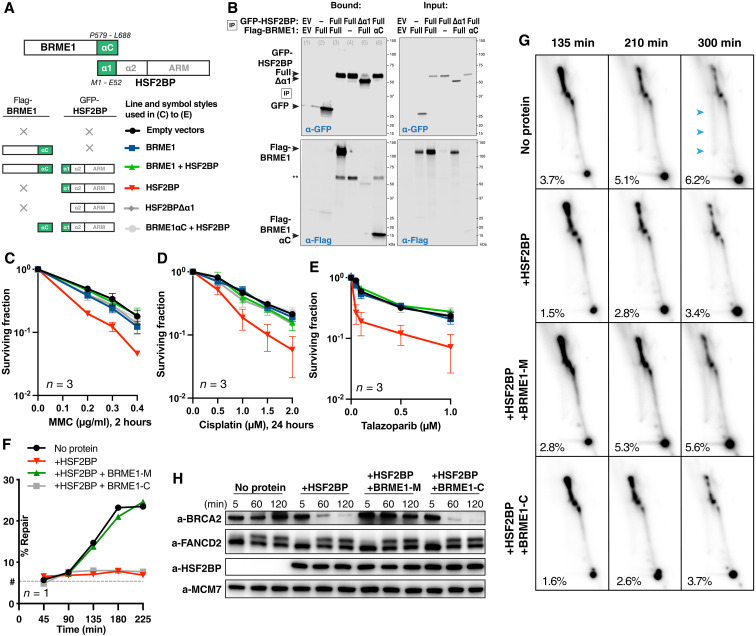
Fig. 6. BRME1 protects cancer cells from HSF2BP and prevents BRCA2 degradation during interstrand crosslink repair.
(A) Schematic of the six HSF2BP and BRME1 fragment combinations used in (B) to (E), next to the corresponding line styles; × — protein not present. Interacting α helices are shown as green blocks. (B) Co-immunoprecipitation of HSF2BP and BRME1 variants used in clonogenic survivals. Proteins eluted from the anti–green fluorescent protein (GFP) beads were detected by immunoblotting with the indicated antibodies. (C to E) Clonogenic survival of HeLa cells stably producing HSF2BP and BRME1 variants, as indicated in (A), and treated with mitomycin C (MMC), cisplatin, or talazoparib. The experiments were repeated three times, and means and SEM are plotted. (F) Efficiency of synthetic cisplatin interstrand crosslink repair in Xenopus egg extract (24) in the presence or absence of HSF2BP, the BRME1 peptides BRME1-M (binding HSF2BP), and BRME1-C (not binding HSF2BP; Fig. 4, A and B) (24). Replicate is shown in fig. S7 (C and D). (G) HR intermediate formation during the repair of synthetic cisplatin DNA interstrand crosslink in Xenopus egg extract monitored by 2D agarose gel electrophoresis in the presence or absence of HSF2BP, the BRME1 peptides BRME1-M, and BRME1-C (see Fig. 4, A and B). The X-arc that contains HR intermediates is indicated by blue arrows in the top right panel, the percentage of the signal localizing to it is indicated, as detailed previously (24), and replicate is shown in fig. S7E. (H) Effect of HSF2BP and BRME1-M or BRME1-C peptides on the endogenous Xenopus BRCA2 protein during the time course (5 to 120 min) of the interstrand crosslink repair reaction. Antibodies used for immunoblotting are indicated (24), and replicate is shown in fig. S7F.
BRME1 protects cancer cells from BRCA2 inhibition by HSF2BP
In our previous work, we demonstrated that ectopic HSF2BP compromises BRCA2 function and causes BRCA2 proteasomal degradation. This degradation involved the p97 segregase (24), an hexameric protein that unfolds and disassembles ubiquitylated substrates, to pull proteins out of membranes, segregate proteins from partners for downstream activity, or unfold proteins for proteasomal degradation (29). Given the dissociative effect of BRME1 on HSF2BP-BRCA2 multimers, we hypothesized that deaggregation by BRME1 will cancel the effect of HSF2BP. To test this hypothesis, we stably produced in HeLa cells HSF2BP alone and together with BRME1 (Fig. 6, A and B) and measured their effect on resistance to DNA interstrand crosslinking agents and PARP inhibitors (Fig. 6, C to E). BRME1 on its own had no effect on the cell sensitivity to DNA damage, but it completely abolished the sensitization induced by HSF2BP, consistent with our hypothesis. The C-terminal HSF2BP-binding part of BRME1 had the same protective effect as the full-length protein. Moreover, while full-length HSF2BP sensitized cells to DNA damage, its variant lacking helix α1 required for homotetramerization and the formation of a diamond-shaped HSF2BP-BRCA2 ring did not. These results strongly suggest that disrupting HSF2BP-BRCA2 multimerization is sufficient to protect BRCA2 function. Together, we confirmed that formation of HSF2BP-BRCA2 octameric rings compromises BRCA2 role in interstrand crosslink repair, and this effect is prevented when HSF2BP cannot tetramerize or BRME1 prevents the formation of HSF2BP-BRCA2 rings.
To further distinguish the function of octamer versus 24-nucleotide oligomer ring-shaped complexes, we tested the HSF2BP variant H80A-R84A that exclusively forms octamers when bound to BRCA2-HBD (Fig. 3D). Measuring the impact of overexpressing this variant in HeLa cells showed that, upon DNA damage, HSF2BP H80A-R84A compromised cell survival as HSF2BP wild type (fig. S7, A and B). We propose that the HSF2BP capacity to induce BRCA2 degradation necessitates the formation of HSF2BP-BRCA2 octameric rings, but not the assembly of the 24-nucleotide oligomer ring-shaped complex.
BRME1 prevents HSF2BP-induced BRCA2 degradation
To establish the mechanism by which HSF2BP attenuates BRCA2 function, we previously studied the repair of a single chemically defined and site-specific cisplatin DNA interstrand crosslink in a plasmid replicating in Xenopus egg extract (24). These biochemical experiments showed that both Xenopus and human HSF2BP inhibit replication-dependent restoration of the genetic information at the crosslink as measured by the regeneration of a restriction enzyme site. We demonstrated using 2D agarose gel electrophoresis that the HR step and not the preceding steps (recognition, signaling, crosslink unhooking by nucleases, or translesion synthesis) of the reaction was inhibited and that the immediate reason for the inhibition was HSF2BP-induced BRCA2 degradation. To determine whether the protective effect of BRME1 we observed in cancer cells ectopically producing HSF2BP would also manifest biochemically, we performed the same Xenopus egg extract assays (Fig. 6, F to H, and fig. S7, C to F) in the presence of either the BRME1-M peptide, which binds HSF2BP α1, or the adjacent BRME1-C peptide, which does not bind HSF2BP (Fig. 4, A and B). Consistent with the cancer cell data, BRME1-M reverted the inhibitory effect of HSF2BP on the interstrand crosslink repair reaction (Fig. 6F) and specifically suppressed the reduction in the formation of HR repair intermediates monitored by 2D agarose DNA gel electrophoresis (Fig. 6G); the nonbinding BRME1-C peptide had no such effect. Moreover, HSF2BP-induced BRCA2 degradation was completely abolished by the BRME1-M, but not the BRME1-C peptide (Fig. 6H). Combined with the structural and cancer cell data, this suggests that BRME1 acts as a disaggregation factor for HSF2BP-BRCA2 complexes, preventing their recognition by the p97/VCP segregase and degradation by the proteasome.
DISCUSSION
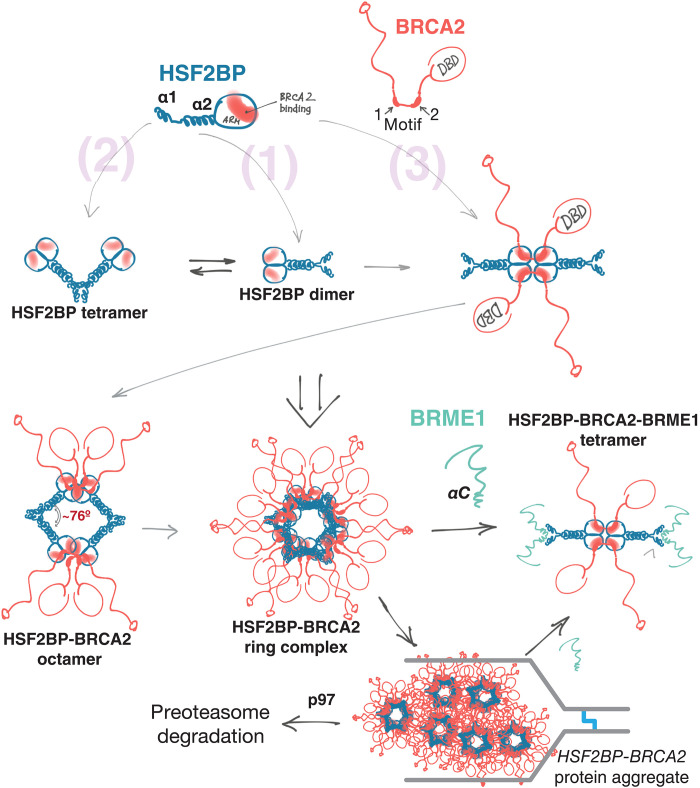
The HR mediator BRCA2 and the recently described meiotic proteins HSF2BP and BRME1 participate in meiotic HR. They are essential for male fertility. Recent studies revealed that, in germline cells, BRCA2 functions in a tight, likely constitutive, complex with HSF2BP and BRME1. BRCA2 directly binds to HSF2BP via a unique repeat-mediated oligomerization-inducing mechanism. BRME1 also directly binds to HSF2BP and phenocopies it. Here, we extended our structural characterization of this germline BRCA2 complex (23) by studying the full-length HSF2BP, rather than only its armadillo domain, in complex with several BRCA2 and BRME1 peptides. Our findings support our previous proposal that HSF2BP can act as a polymerization factor for BRCA2 due to three oligomerization mechanisms: two intrinsic to HSF2BP (homo-oligomerization via α1 and dimerization via α2 + armadillo) and one extrinsic, mediated by the repeats encoded by exons 12 and 13 of BRCA2 (Fig. 7). In addition, our study reveals several properties that we did not anticipate from the previous analyses: Tetramerization of HSF2BP into a V-shaped structure revealed by SAXS and formation of ring-shaped HSF2BP/BRCA2-HBD complexes, including a diamond-shaped octameric ring consisting of eight HSF2BP and four BRCA2-HBD molecules, and a 880-kDa hetero-oligomer consisting of three interlocked octameric rings revealed by cryo-EM and supported by SFM, mass photometry, SEC-MALS, AlphaFold modeling, and x-ray crystallography (Figs. 1 to 3). We also delineated a BRME1 α-helical peptide that can bind and displace helix α1 of HSF2BP, thus disrupting the tetramer formed by HSF2BP and the ring-shaped complexes formed by HSF2BP bound to BRCA2-HBD (Figs. 4 and 5). Together, our results support an aggregator-disaggregator model, in which HSF2BP induces BRCA2 oligomerization, whereas BRME1 disrupts the ring-shaped HSF2BP-BRCA2 oligomers.
Fig. 7. Oligomeric states of HSF2BP and the “aggregator-disaggregator” model proposed to explain the effects of HSF2BP and BRME1 on BRCA2 in somatic and germline cells.
Oligomers observed experimentally are indicated in bold. Two intrinsic [(1) and (2)] and one BRCA2-mediated (3) oligomerization mechanisms result in the formation of a constitutive homodimer, a V-shaped tetramer, and a large ring complex. This concentrates and organizes BRCA2, but at replication fork stalled at crosslink, it results in aggregation and proteasomal degradation of BRCA2. BRME1 competes with one of the oligomerization mechanisms (2), resulting in the formation of a 4× HSF2BP:2× BRCA2:4× BRME1 complex and thus preventing or reversing BRCA2 aggregation.
The remarkable ring structure of the HSF2BP-BRCA2 24-nucleotide oligomer complex revealed by our cryo-EM analysis comes from three properties of HSF2BP we identified: (i) HSF2BP is a tetrameric protein with a V shape, the angle at the bottom of the V being defined by the α1-mediated tetramerization constraint; (ii) the relatively constrained V shape favors the formation of an HSF2BP octamer upon binding to the BRCA2 peptide. In this octamer, the α2-coiled coils lie in the same plane. They outline a diamond shape, the angle at the vertex formed by α1 helices (~76°) being favored by the HSF2BP tetramerization mode revealed in this study and the supplementary angle at the vertex formed by the armadillo domains (~104°) being defined by the BRCA2-dependent tetrameric structure described previously (23). The structural origin of the angle value at the α1 vertex is not clear, and it may have a degree of flexibility in solution, but we postulate that this angle value must be constrained to explain why HSF2BP bound to BRCA2-HBD organizes into closed diamond-shaped octamers rather than linear polymers; (iii) at high concentration (μM), the octamers associate to form a 24-nucleotide oligomer, in which the α2-coiled coils and armadillo domains interact through interfaces that are partially evolutionary conserved (fig. S8, A and B). While the details of these interfaces are missing, we have identified a double mutant, H80A-R84A, which only forms octamers upon binding to BRCA2-HBD (Fig. 3D). H80 and R84 are conserved through evolution (fig. S9A). They are not essential for the disruptive function of HSF2BP in cancer cells. However, their role might be important in meiosis or other functional contexts.
We also noted the presence of three cysteine pairs located at the HSF2BP dimerization interface and were curious whether they are required for HSF2BP function and could be a point of redox regulation for HSF2BP oligomerization (fig. S9, A to D). The triple C78A-C120S-C128A substitution variant sensitized HeLa cells less than the wild-type protein. The C128R variant and the L186P variant were found in families with premature ovarian insufficiency (30). L186 is also located at the HSF2BP dimerization interface. We suggest that all these mutations destabilize the HSF2BP oligomerization interface. Together, we surmise that (i) conservation of the interfaces (armadillo and α2 regions involved in dimerization, α1 regions involved in tetramerization, and the groove binding to BRCA2; fig. S8A) reflects the functional importance of the ring-shaped complexes, (ii) dimerization of HSF2BP is essential for most of its functions, and (iii) formation of the octamers upon binding to BRCA2-HBD is important for the disruptive function of HSF2BP in cancer cells; these octamers can trigger unloading of BRCA2 from the DNA damage site and further degradation of BRCA2 by the proteasome.
The distinct structure of the HSF2BP-BRCA2 ring-shaped 24-nucleotide oligomer complex makes it tempting to speculate how this spatial organization might be functionally relevant. In our mass photometry experiments, we observed single, double, and triple octamers at 10 to 100 nM, close to nucleoplasmic concentrations. The micromolar concentrations we used for cryo-EM are higher but may well be reached within the repair foci, the relevant functional context for HSF2BP-BRCA2 (31). Despite the poor definition of the interfaces between octamers in our cryo-EM model, we identified among the candidate residues two evolutionarily conserved positions that are specifically required for the assembly of the 24-nucleotide oligomer from the octamers, which further suggests that this is a positively selected feature rather than an experimental artefact. There are multiple precedents for the specific role of ring-shaped complexes in DNA metabolism and HR specifically, from sliding clamps and hexameric helicases to the octameric ring formed by DMC1. However, the ring we characterized here is larger than the ring complexes that encompass DNA: It has an inner diameter of 100 Å compared to 25 Å for hexameric helicases or 35 Å for proliferating cell nuclear antigen (PCNA) (32, 33). Also, the inner surface of HSF2BP-BRCA2 is negatively charged (fig. S8C), and HSF2BP has no detectable affinity for DNA (23), whereas the inner surfaces of the DNA-encompassing rings are positively charged. It is notable that the opening of the rings we characterized is sufficiently large to accommodate protein-coated single-stranded DNA (ssDNA) [~40 to 80 Å; (34, 35)] and that positioning of BRCA2 on the outer surface would not hinder this. The ring structures could thus facilitate BRCA2 retention by clamping around coated DNA structure. HSF2BP is proposed to facilitate BRCA2 recruitment or retention of BRCA2 at resected meiotic DSBs by interacting with the ssDNA-binding proteins (RPA, SPATA22, or MEIOB) that coat them. However, efforts to demonstrate these direct high-affinity interactions have so far been unsuccessful. Multiple low-affinity interactions could ensure that the rings lock in around the correct substrate during meiotic HR. Another role for the ring in meiosis can be hypothesized on the basis of the proximity between the HBD and the DMC1-binding PhePP motif of BRCA2. The motifs are separated by a flexible linker, and it is conceivable that HSF2BP-BRCA2 ring could form multiple connections with the octameric ring formed by DMC1 (36, 37), positioning it to facilitate meiotic HR. New separation of function mouse models are required to test these hypotheses (34, 35).
A major finding here is the disruptive role of BRME1 on the ring structures. However, a strong phenotypic and cytological similarity was observed between BRME1 and HSF2BP mutants, which suggests that the two proteins function as a complex in meiosis. In this context, we propose that, in the presence of BRME1, an alternative HSF2BP tetrameric conformation is formed, linked by two BRCA2 molecules and blocked by four BRME1 molecules from forming higher-order assemblies. This is supported by the appearance of particles with close to tetrameric volumes, as observed in our SFM and mass photometry experiments, in the presence of both BRCA2-HBD and BRME1-M (Fig. 5, C and E). It is also supported by SEC-MALS experiments performed at higher concentration and in the presence of excess of peptides, in which only a tetramer is observed (Fig. 5F). This tetramer may carry out some or all of the known HSF2BP functions in meiosis. It has a twofold symmetry and could bridge two other entities as has been shown for monopolin, a V-shaped dimeric protein bridging kinetochores during yeast meiosis (38). BRME1 could also serve as a regulatory factor that ensures that the HSF2BP-BRCA2 ring is assembled at the right time and place, preventing ectopic BRCA2 polymerization that can lead to BRCA2 degradation, as revealed by our study of cancer cells discussed below. The release of BRME1 from HSF2BP could be controlled by posttranslational modification or other mechanisms, such as alternative splicing or competition with partners.
While the relevance of the ring-shaped BRCA2-HSF2BP complex in meiosis is speculative at the moment, we suggest that our findings resolve the arguably most puzzling observation about HSF2BP, namely, its apparently opposite effects on somatic versus meiotic HR (Figs. 6 and 7). We previously found that ectopic HSF2BP in human cancer cells or Xenopus egg extracts suppresses HR during interstrand crosslink repair by inducing BRCA2 degradation. This degradation is dependent on the p97 segregase, which processes protein aggregates (23). We hypothesized that the polymerizing effect of HSF2BP on BRCA2 turns into aggregation in ectopic settings, thus inducing p97-dependent BRCA2 degradation. It was however not clear what restricts this polymerization both spatially, stopping the formation of endless HSF2BP-BRCA2 chains via α1- and BRCA2-linked armadillo connections, and temporally, restraining it to specific ectopic contexts. Circularization into octamers and ring complexes allows HSF2BP-BRCA2 to diffuse through the cytoplasm without forming linear chain aggregates. The disaggregating effect of BRME1 explains why aggregation does not happen in testis and embryonic stem cells. Aggregation followed by proteasomal degradation happens only at the stalled replication fork. It may be triggered by high local concentration of the proteins in foci, additional oligomerizing interactions in these foci with other proteins, or local posttranslational modifications. Alternatively, the aggregation-sensing machinery may be more active at the crosslink-blocked fork, which is known to recruit ubiquitin ligases and the VCP/p97 segregase itself. BRME1 was not present in the cancer cells that we used during our first analyses, explaining why we initially observed a disruptive effect for HSF2BP. Differential expression of HSF2BP and BRME1 as well as alternative splicing of BRCA2 exon 12 (17, 23, 39–41) provide cancer cell with an evolutionary platform to drift between genomic instability and drug resistance.
In conclusion, HSF2BP organizes BRCA2 into ring-shaped structures, consisting of BRCA2-bound HSF2BP octamers that can assemble into 24-nucleotide oligomers. The rings prevent extensive polymerization of BRCA2-bound HSF2BP, display BRCA2 on their outer surface, and have openings that can accommodate objects the size of protein-coated ssDNA. Under specific DNA damage conditions, assembly of HSF2BP-BRCA2 octamers can trigger BRCA2 aggregation and degradation, resulting in genomic instability. When HSF2BP is expressed physiologically, the formation of the ring-shaped complexes is regulated by BRME1. In the presence of both BRCA2 and BRME1, HSF2BP is a tetramer that could bridge two entities during HR. We propose that HSF2BP and BRME1 timely concentrate and organize BRCA2 and other bound proteins to facilitate BRCA2-mediated DSB repair in meiocytes.
MATERIALS AND METHODS
Protein expression and purification
Human HSF2BP—full-length wild type, its substitution variants (H80A-R84A; Q104A-E108A), or the truncation variant lacking helix α1 (from G48 to V334)—was expressed using a pETM11 (6×His-TEVsite) expression vector in Escherichia coli BL21 (DE3) Rosetta2 pLysS or Star strains. A starter culture [LB + kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml)] was grown overnight at 37°C and used to inoculate 1 liter of LB, which was grown at 37°C until the optical density at 600 nm (OD600) reached 0.6. Expression was induced by addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside and continued at 20°C overnight. Harvested cells were resuspended in 25 mM tris-HCl (pH 8), 500 mM NaCl, 5 mM β-mercaptoethanol, and EDTA-free Protease Inhibitor Cocktail (Roche) and disrupted by sonication. Lysates were supplemented with 1 mM MgCl2, treated by benzonase nuclease at 4°C for 30 min, and then centrifuged at 15,000g at 4°C for 30 min. After filtration (0.4 μm), the supernatant was loaded on a chromatography HisTrap FF crude 5 ml column (GE Healthcare) equilibrated with 25 mM tris-HCl (pH 8), 500 mM NaCl, and 5 mM β-mercaptoethanol. HSF2BP (variant) was eluted with a linear gradient of imidazole. The tag was cleaved by the TEV protease (at a ratio of 2% w/w) during an ON dialysis at 4°C against 25 mM tris-HCl (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. The protein solution was loaded on a HisTrap column, and the tag-free HSF2BP was collected in the flow through. Last, a SEC was performed on HiLoad Superdex 10/300 200 pg equilibrated in 25 mM tris (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. The quality of the purified protein was analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and the protein concentration was determined by spectrophotometry from the absorbance at 280 nm.
The gene coding for BRCA2-HBD (BRCA2 residues 2291 to 2342, including mutation C2332T to avoid oxidation problems) was optimized for expression in bacteria and synthesized by GenScript. It was cloned in a pET-22b vector. This vector was used to express a fusion protein comprising BRCA2-HBD, a TEV site, GB1, and a 6xHis tag in BL21 DE3 Star cells. BRCA2-HBD was purified as previously reported (23). Peptides coding for HSF2BP helix α1 (residues 19 to 50) and BRME1 C-terminal region (BRME1-N: residues 578 to 601; BRME1-M: 602 to 641; BRME1-C: 649 to 668) were purchased from GeneCust.
The complex between HSF2BP and BRCA2-HBD was prepared by mixing the two proteins at a molar ratio of 1:1.5. The mix was concentrated by centrifugation at 4500g using a 3-kDa cutoff membrane at 4°C and then loaded on a size exclusion column Superdex 200 increase 10-300 GL (GE Healthcare) equilibrated with 25 mM tris-HCl (pH 7.5), 150 or 250 mM NaCl, and 5 mM β-mercaptoethanol. The peak fractions were collected and further analyzed by EM.
The thermal stability of HSF2BP either free or bound to BRCA2-HBD was evaluated using the simplified ThermoFluor Assay available on the High Throughput Crystallization Laboratory (HTX Lab) of the EMBL Grenoble (42). Samples were analyzed at different concentrations: HSF2BP at 7 and 14 mg/ml and HSF2BP bound to BRCA2-HBD at 1 and 12 mg/ml.
Mass photometry
Mass photometry experiments were recorded on a Refeyn TwoMP system. Samples were prepared at 1 μM and diluted in 25 mM tris-HCl buffer (pH 7.5), 250 mM (or 150 mM when indicated) NaCl, and 5 mM β-mercaptoethanol before the experiments to reach concentrations comprised between 25 and 100 nM. A twofold excess of peptides (either BRCA2-HBD or BRME1-M) was used to measure the impact of peptide binding on HSF2BP oligomerization. Calibration of the instrument was performed in the same buffer with carbonic anhydrase, bovine serum albumin (BSA), and urease covering a range of molecular weights from 29 to 816 kDa. Data were analyzed using the provided Discover analysis software. Masses were estimated by multi-Gaussian fitting using the online PhotoMol software (https://spc.embl-hamburg.de/app/photoMol) (43).
SEC coupled to multi-angle light scattering
SEC-MALS was used to measure the molecular masses of the complexes in solution. Therefore, HSF2BP (full-length wild type or its substitution and truncation variants) was loaded in the presence or absence of BRCA2-HBD and BRME1-M on a Superdex 200 10/300 GL (GE Healthcare; flow rate of 500 μl/min; Figs. 1B and 2A) or a BIOSEC 3 column (Agilent; flow rate of 200 μl/min) using a HPLC Agilent system coupled to MALS/QELS/UV/RI (Wyatt Technology). The chromatography buffer was 25 mM tris-HCl buffer (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. The proteins were injected at 0.8 to 7 mg/ml in 20 to 100 μl. Data were analyzed using the ASTRA software; a calibration was performed with BSA as a standard. To represent the data, the normalized absorbance at 280 nm was overlaid with the molar mass (Da), and both parameters were plotted as a function of the elution volume.
SEC coupled to small angle x-ray scattering
SEC-SAXS is available on the SWING beamline at synchrotron SOLEIL to obtain a distance distribution corresponding to each sample in solution. The free HSF2BP protein was analyzed using a BIOSEC 3 column (Agilent) equilibrated in 25 mM tris-HCl (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. The protein was loaded at a concentration of 6 mg/ml to observe an elution peak at an OD280nm of 1. The SAXS intensity was measured every second during the elution as a function of the scattering angle. The final SAXS curve is the sum of the curves recorded on the elution peak. A distance distribution curve P(r) was calculated, whose Fourier transform fits to the SAXS curve. The residual values are the experimental minus the fitted SAXS intensity values and divided by the experimental errors. The distance distribution P(r) was plotted in arbitrary units as a function of the distance, the maximal distance being 250 Å.
Sample preparation for EM
The quality of the EM sample was checked by negative-staining EM. Therefore, the complex between HSF2BP and BRCA2-HBD was diluted to approximately 0.05 mg/ml in 25 mM tris-HCl (pH 7.5) and 250 mM NaCl. Three microliters was deposited on an airglow-discharged carbon-coated grid. Excess liquid was blotted, and the grid rinsed with 2% (w/v) aqueous uranyl acetate. The grids were visualized at 100 kV with a TECNAI Spirit (FEI) transmission electron microscope (Thermo Fisher Scientific, New York, NY, USA) equipped with a K2 Base 4k × 4k camera (Gatan, Pleasanton CA, USA). A nondiluted sample (2 to 4 mg/ml) concentrated in 25 mM tris-HCl (pH 7.5), 150 mM NaCl, and 5 mM β-mercaptoethanol was further prepared for cryo-EM. An aliquot of 2.5 μl was deposited onto freshly glow-discharged, Quantifoil R2/2 300-mesh gold grids. These grids were frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 100% humidity, 4°C, using a blotting force of 1 and a blotting time of 5 s.
Cryo-EM data acquisition and image processing
For cryo-EM analysis, 9531 movies were collected using the EPU software on a Titan Krios electron microscope (Thermo Fisher Scientific) operating at 300 kV, equipped with a Selectris X filter and slit open at 10 eV, and a Falcon 4i direct electron detection camera (table S3). The defocus range was between −0.4 and − 1.8 μm, with a pixel size of 0.73 Å. The total dose was 40 e/Å2 distributed on 40 frames. Movie frames were aligned using MotionCor2 (44), with applied dose compensation. Motion-corrected micrographs were imported into CryoSPARC (45). Contrast transfer function (CTF) parameters were estimated using CTFFIND4 (46).
Micrographs were examined, and those with a CTF fit resolution higher than 5.5 Å or a relative ice thickness larger than 1.2 were excluded. The remaining 8962 micrographs were used for blob picking as described in Fig. 2B. After several rounds of particle picking and classification, first three volumes were calculated and refined. These volumes corresponded to open and closed rings; particles used to build closed rings were selected for further classification and template picking. After several iterations including template picking, 2D classification, ab initio volume calculation, and heterogeneous refinement, seven 2D classes were selected to train the Topaz tool (47), which was then able to pick a larger set of particles. These particles were enriched in side views of the complex. A minor family of particles showing two interlocked rings was observed and excluded. The remaining particles were used to calculate three new volumes, which corresponded again to open and closed rings. A set of particles consistent with closed rings were further used for another Topaz training. Using this tool, a new set of particles was picked and added to the first set of particles picked by Topaz. Duplicates were removed, and three volumes were calculated and refined. One of these volumes had the shape of an open ring, and the two other volumes were well-resolved rings. The 398,000 particles corresponding to these two volumes were used to calculate the final cryo-EM map of the complex (fig. S2C). Then, the 186,000 and 212,000 particles were separately used to calculate two new cryo-EM maps (fig. S2D). Whereas the Gold-Standard Fourier Shell Correlation (GSFSC) resolution of the first map is 3.3 Å, the resolutions of the two other maps are slightly lower (3.5 to 3.6 Å). However, the local resolution distribution of these two maps is different, with the map built from 212,000 particles showing overall a better resolution, and the map built from 186,000 particles showing better resolved inner armadillo tetramers but poorly resolved outer tetramers and coiled coils.
Building and refinement of the cryo-EM model
The density map of the complex was of sufficient quality (between 3 and 8 Å) to fit the crystal structure of the armadillo domain of HSF2BP in complex with BRCA2-HBD (PDB codes: 7BDX) in ChimeraX (48, 49). The map regions in which the armadillo domains were fitted showed a local resolution of 3 to 5 Å. In the peripheral region of the complex, corresponding to the N-terminal helices of HSF2BP, the local resolution map was in the range 5 to 8 Å. In these regions, AlphaFold guided model building. The crystal structure of the HSF2BP armadillo tetramer bound to BRCA2-HBD was fitted in the high-resolution cryo-EM map at a level contour of 0.035 (Fig. 2A) using the standard fit routine from ChimeraX. In these conditions, 79% of the atoms were in the density (5855 within 7403 atoms). The AlphaFold model of a dimer of full-length HSF2BP was similarly fitted in the high-resolution cryo-EM map. When it was fitted by positioning the armadillo domain in one of the three inner subvolumes at a level contour of 0.025 (Fig. 2B), 68% of the atoms were in the density (3602 within 5272 atoms). When it was fitted by positioning the armadillo domain in one of the three outer subvolumes at a level contour of 0.025 (Fig. 2B), 71% of the atoms were in the density (3761 within 5272 atoms). All figures were prepared using UCSF Chimera and UCSF ChimeraX. Conservation scores were calculated using Consurf [https://consurf.tau.ac.il/ (50)].
Crystallization and structure determination
Synthetic peptides corresponding to helix α1 of HSF2BP (E19-V50) and BRME1-M (E602-K641) were dissolved in 25 mM tris (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. The concentration of the helix α1 peptide was 10 mg/ml. This peptide was mixed with the BRME1-M peptide to reach a concentration of 5 mg/ml. Crystallization assays were carried out at 277 K in the HTX Lab (EMBL Grenoble). Several crystallization conditions were identified within 1 day, and the crystals were grown within 2 weeks. They were prepared for x-ray diffraction experiments using the CrystalDirect harvesting and processing robot (51). The best crystals diffracted at 1.47-Å (α1) and 1.98-Å (α1/BRME1-M) resolution. They were obtained by sitting drop vapor diffusion against a reservoir containing 0.2 M LiCl; 0.1 M sodium acetate trihydrate (pH 5); 20% polyethylene glycol, molecular weight 800 (PEG-6000) for α1; 0.1 M Hepes sodium salt (pH 7.5); and 40% PEG-200 for α1/BRME1-M. Diffraction data were collected on the MASSIF-1 beamline (EMBL Grenoble) of the ESRF synchrotron. Datasets were indexed, integrated, and scaled using the X-ray Detector Software (XDS) (52). The 3D structures were solved by molecular replacement using the PHENIX Phaser software (53, 54) and an input coordinate file calculated by AlphaFold2 (28), were iteratively improved by manual reconstruction in COOT (55), and were refined using the PHENIX Refine and BUSTER (Global Phasing Limited) (56) software. A summary of crystallographic statistics is shown in table S2.
ITC experiments
Using a VP-ITC Calorimeter (GE Healthcare), interactions between the HSF2BP protein (either full length or the peptide corresponding to helix α1) and the BRME1 peptides (BRME1-N, BRME1-M, and BRME1-C) were characterized. The experiments were performed at two temperatures, 293 and 303 K, and duplicated. The buffer was 25 mM tris buffer (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. A total of 8 to 13 μM of HSF2BP (either full length or helix α1) in the cell was titrated with 80 to 130 μM BRME1 peptide (BRME1-N, BRME1-M, or BRME1-C) in the injection syringe. We also tested the interactions of HSFB2P bound to BRCA2-HBD with BRME1-M and HSF2BP bound to BRME1-M with BRCA2 fragments. The experimental conditions were similar, with 10 μM complex in the cell and either 200 μM BRME1-M or 55 to 60 μM BRCA2 fragments in the injection syringe. In all cases, 10 μl of the syringe volume was injected every 210 s, except for the first injection which was of 2 μl and which was ignored in the final data analysis. This analysis was performed using the Origin 7.0 software provided by the manufacturer to obtain the stoichiometries, equilibrium constants, and thermodynamic parameters of the binding reactions. A summary of the ITC data is shown in Table 1.
Scanning force microscopy
Images were obtained on a NanoScope IV SFM (Digital Instruments; Santa Barbara, CA) operating in tapping mode in air with a type J scanner using silicon probes, ACT-W, with a tip radius < 10 nm, and a resonance frequency range of 200 to 400 kHz (AppNano, Santa Clara, CA). Images were processed using NanoScope Analysis (Bruker) for background flattening. The protein volumetric analyses were done using Image SXM 1.89 (National Institutes of Health IMAGE version modified by Steve Barrett, Surface Science Research Centre, University of Liverpool, Liverpool, UK). Kernel density analysis and plotting were done using the R software.
Sample preparation for SFM
All dilutions, reactions and depositions were done in a buffer containing 25 mM tris-HCl (pH 7.5), 250 mM NaCl, and 5 mM β-mercaptoethanol. HSF2BP was diluted 10,000 times to final concentration of 12 nM and deposited onto a freshly cleaved mica (Muscovite mica, V5 quality, EMS). After 30 s, the mica was washed with H2O and dried with a stream of filtered air. HSF2BP and BRCA-HBD2288–2337 complex was prepared by mixing 80 μM HSF2BP with 120 μM BRCA-HBD2288–2337, followed by incubation on ice for 15 min. One half of the reaction was diluted 10,000 times, deposited onto a freshly cleaved mica, washed with H2O, and dried with a stream of filtered air. Another half of the reaction was further incubated with BRME1 peptide (molar ratio HSF2BP:BRME1-M = 1:1) on ice for 10 min. Sample was diluted 10,000 times, deposited on mica, washed with H2O, and dried with a stream of filtered air.
Cell culture, constructs, and stable cell line generation
HeLa (human cervical adenocarcinoma, female origin) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin (200 U/ml), and streptomycin (200 μg/ml). Expression constructs for stable production of green fluorescent protein (GFP)–HSF2BP, Flag-BRME1, and their truncation variants were engineered as described before (17) in the PiggyBac vectors by Gibson assembly and verified by sequencing. The constructs were cotransfected in combinations shown in Fig. 6A and together with the PiggyBac transposase expression plasmid [hyPBase (57)] using Lipofectamine 3000 (Thermo Fisher Scientific) in six-well plates seeded with 400,000 HeLa cells the day before. Selection with puromycin (1.5 μg/ml) for GFP-HSF2BP constructs and G418 (800 μg/ml) for Flag-BRME1 constructs started 2 days after transfection and was maintained for 8 days. The resulting resistant mixed cell population was used for clonogenic survival assays.
Clonogenic survival assays
Clonogenic survival assays were performed in six-well plates in technical duplicates. Untreated control wells were seeded with 100 cells per well in 2 ml of media. Higher seeding densities of 400 and 1000 cells per well were used in the wells treated with higher drug concentrations. One day after seeding, the following drugs were added: mitomycin C (MMC; Sigma-Aldrich, M4287-2MG), cisplatin, and talazoparib (BMN-673, Axon Medchem, #2502). After 2-hour (MMC) or overnight (cisplatin and talazoparib) incubation, drug-containing media was removed, wells were rinsed with phosphate-buffered saline (PBS) and refilled with 2 ml of fresh media. Colonies were stained [0.25% Coomassie brilliant blue (CBB), 40% methanol, and 10% acetic acid] on day 10 after seeding. Plates were photographed using a digital camera, and images were analyzed using OpenCFU software to quantify the colonies.
Co-immunoprecipitation and immunoblotting
Cells were grown in 10-cm dish to near confluence, washed twice with PBS, and lysed in situ in 1 ml of NETT buffer [100 mM NaCl, 50 mM tris (pH 7.5), 5 mM EDTA (pH 8.0), and 0.5% Triton X-100) supplemented immediately before use with protease inhibitor cocktail (Roche) and Pefabloc (0.4 mg/ml; Roche) (NETT++). After 5 to 10 min, cells were scraped off and collected in 1.5-ml microcentrifuge tubes; lysis was continued for additional 15 to 20 min on ice, then mixtures were centrifuged (15 min, 4°C, 14,000 rcf), and 70 μl of the supernatant was collected as input sample, mixed with 2× Laemmli sample buffer, and denatured for 5 min at 95°C. The rest of the supernatant was mixed with 8 μl of anti-GFP beads (ChromoTek, gta-20). The mixture was incubated for 2 hours at 4°C while rotating and washed three times in NETT++ buffer, and bound proteins were eluted with 70 μl of 2× sample buffer and incubated for 5 min at 95°C. A total of 15 μl of bound and 7 μl of input sample were separated on 4 to 15% TGX SDS-PAGE gel (Bio-Rad, #456-1086) and transferred to polyvinylidene difluoride membrane (Immobilon-FL, Millipore, IPFL00010). Gels were stained with colloidal CBB after transfer and scanned to ascertain equal loading. Immunoblotting was performed following standard procedures with anti-GFP rabbit polyclonal antibody (Invitrogen, A11122) and anti-Flag mouse mAb (M2 antibody, Sigma-Aldrich, F3165) antibodies, followed by detection with fluorescently labeled secondary antibodies were used: anti-mouse CF680 (Sigma-Aldrich, #SAB460199) and anti-rabbit CF770 (Sigma-Aldrich, #SAB460215). Membranes were scanned using Odyssey CLx imaging system (LI-COR).
Interstrand crosslink repair assay
Interstrand crosslink repair assays were performed as described (58, 59). Xenopus egg extracts [high-speed supernatant (HSS) and nucleoplasmic egg extract (NPE)] and a plasmid containing a site-specific cisplatin interstrand crosslink (pICL) were prepared as described previously (60, 61). pICL (9 ng/μl) and pQuant (0.45 ng/μl) were first incubated in a HSS of egg cytoplasm for 20 min at room temperature, which promotes the assembly of prereplication complexes on the DNA. Addition of two volumes NPE supplemented with 32P-α-dCTP (deoxycytidine triphosphate) triggers a single round of DNA replication. Where indicated, His-tagged human HSF2BP (0.45 μM), BRME1-M peptide (1.35 μM), or BRME1-C peptide (1.35 μM) was added to NPE before mixing with HSS. Aliquots of replication reaction (4 μl) were stopped at various times with 45 μl of Stop solution II [50 mM tris (pH 7.5), 0.5% SDS, and 10 mM EDTA]. Samples were incubated with ribonuclease (0.13 μg/μl) for 30 min at 37°C followed by proteinase K (0.5 μg/μl) overnight at room temperature. DNA was extracted using phenol/chloroform, ethanol-precipitated in the presence of glycogen (20 μg), and resuspended in 4 μl of TE [10 mM tris (pH 7.5) and 1 mM EDTA]. Crosslink repair was analyzed by digesting 1 μl of extracted DNA with Hinc II or Hinc II and Sap I, separation on a 0.8% agarose gel in 1x TBE buffer, and quantification using Typhoon TRIO+ (GE Healthcare) and ImageQuant TL software (GE Healthcare). Repair efficiency was calculated as described (62).
2D gel electrophoresis
2D gel electrophoresis was performed as described previously (63). Replication intermediates of pICL at various times were extracted and digested with Hinc II. Fragments were then separated on a 0.4% agarose gel in 0.5× TBE buffer at 0.86 V/cm for 24 hours at room temperature. The lanes of interest were cut out, cast across the top of the second-dimension gel consisting of 1% agarose with ethidium bromide (0.3 μg/ml), and run in 0.5× TBE containing ethidium bromide (0.3 μg/ml) with buffer circulation at 3.5 V/cm for 14.5 hours at room temperature. The gel was dried on Amersham Hybond-XL membrane and exposed to a phosphor screen. DNA was visualized using a Typhoon TRIO+.
Acknowledgments
We warmly thank C. Velours (I2BC) and A. Collange (Synchrotron SOLEIL) for the SEC-MALS experiments, A. Thureau (Synchrotron SOLEIL) for discussions about the SAXS data analysis, F. Dupeux (IBS) and all the HTX Lab staff for the crystallogenesis and crystallography experiments, M.-H. Le Du (I2BC) for helpful advice in crystallogenesis, M. Vos and the NanoImaging Core at Institut Pasteur (Paris) for the support with sample preparation and image acquisition, S. Bressanelli (I2BC) for the support during the cryo-EM data analyses, B. Raynal and the molecular biophysics platform at Institut Pasteur (Paris) for the help during the mass photometry analyses, and the staff of the I2BC computing facility (I2BC/SICS) for facilitating data management and accessibility. We acknowledge SOLEIL (SWING beamline) and ESRF (MASSIF-1 beamline) for provision of synchrotron radiation and beamlines staff for assistance in collecting data.
Funding: The research leading to these results has received funding from the European Community’s Seventh Framework Program H2020 under the project iNEXT discovery (grant agreement no. 871037). It has also benefited from the cryo-EM platforms of Institut Pasteur supported by the French Government’s Investissement d’Avenir program EQUIPEX Centre d’analyse de systèmes complexes dans les environnements complexes (CACSICE, ANR-11-EQPX-008) and of I2BC supported by the French Infrastructure for Integrated Structural Biology (FRISBI, ANR-10-INSB-05) and by the CEA. This study was supported by the Oncode Institute, which is partly financed by the Dutch Cancer Society (KWF). We thank the Josephine Nefkens Cancer Program for infrastructure support. P.K. was supported by the European Research Council (ERC) through an ERC Consolidator Grant (ERCCOG 101003210-XlinkRepair) and by the Oncode Institute, which is partly financed by the Dutch Cancer Society (KWF). K.S. was supported by the Kanae Foundation for the Promotion of Medical Science and the Japanese Biochemical Society. S.M. was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM).
Author contributions: Conceptualization; S.M., R.K., A.N.Z., and S.Z.-J. Methodology: R.G., S.M., K.S., M.O., P.L., V.R., A.-A.A., and A.N.Z. Formal analysis: R.G., S.M., K.S., P.L., C.W., P.K., A.N.Z., and S.Z.-J. Investigation: R.G., S.M., K.S., P.L., M.O., A.-A.A., J.M.W., G.D., and A.N.Z. Writing—original draft preparation: A.N.Z. and S.Z.-J. Writing—review and editing: R.G., S.M., A.N.Z., and S.Z.-J. Visualization: A.N.Z. and S.Z.-J. Supervision: S.M., C.W., P.K., R.K., A.N.Z., and S.Z.-J. Project administration: A.N.Z. and S.Z.-J. Funding acquisition: R.K. and S.Z.-J. All authors have read and agreed to the published version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The coordinates and structure factors file corresponding to the crystal structures of HSF2BP helix α1 and the complex between HSF2BP helix α1 and BRME1-M described in the study were deposited in the PDB (wwPDB) under the entry codes 8A50 and 8A51, respectively. The SAXS data reported in this study were deposited in the Small Angle Scattering Biological Data Bank (SASBDB) under the entry code SASDSM7. The cryo-EM map generated in this study was deposited in the wwPDB database under accession code EMD-16432.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S3
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Holloman W. K., Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 18, 748–754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zelensky A. N., Kanaar R., Wyman C., Mediators of homologous DNA pairing. Cold Spring Harb. Perspect Biol. 6, a016451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kojic M., Kostrub C. F., Buchman A. R., Holloman W. K., BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 10, 683–691 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Siaud N., Dray E., Gy I., Gérard E., Takvorian N., Doutriaux M.-P., Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23, 1392–1401 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg-Shukron A., Rachmiel M., Renbaum P., Gulsuner S., Walsh T., Lobel O., Dreifuss A., Ben-Moshe A., Zeligson S., Segel R., Shore T., Kalifa R., Goldberg M., King M.-C., Gerlitz O., Levy-Lahad E., Zangen D., Essential role of BRCA2 in ovarian development and function. N. Engl. J. Med. 379, 1042–1049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klovstad M., Abdu U., Schüpbach T., Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLOS Genet. 4, e31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko E., Lee J., Lee H., Essential role of brc-2 in chromosome integrity of germ cells in C. elegans. Mol. Cells 26, 590–594 (2008). [PubMed] [Google Scholar]
- 8.Martin J. S., Winkelmann N., Petalcorin M. I. R., Mcilwraith M. J., Boulton S. J., RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 25, 3127–3139 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Silvestri V., Leslie G., Rebbeck T. R., Neuhausen S. L., Hopper J. L., Nielsen H. R., Lee A., Yang X., McGuffog L., Parsons M. T., Andrulis I. L., Arnold N., Belotti M., Borg Å., Buecher B., Buys S. S., Caputo S. M., Chung W. K., Colas C., Colonna S. V., Cook J., Daly M. B., de la Hoya M., de Pauw A., Delhomelle H., Eason J., Engel C., Evans D. G., Faust U., Fehm T. N., Fostira F., Fountzilas G., Frone M., Garcia-Barberan V., Garre P., Gauthier-Villars M., Gehrig A., Glendon G., Goldgar D. E., Golmard L., Greene M. H., Hahnen E., Hamann U., Hanson H., Hassan T., Hentschel J., Horvath J., Izatt L., Janavicius R., Jiao Y., John E. M., Karlan B. Y., Kim S.-W., Konstantopoulou I., Kwong A., Laugé A., Lee J. W., Lesueur F., Mebirouk N., Meindl A., Mouret-Fourme E., Musgrave H., Yie J. N. Y., Niederacher D., Park S. K., Pedersen I. S., Ramser J., Ramus S. J., Rantala J., Rashid M. U., Reichl F., Ritter J., Rump A., Santamariña M., Saule C., Schmidt G., Schmutzler R. K., Senter L., Shariff S., Singer C. F., Southey M. C., Stoppa-Lyonnet D., Sutter C., Tan Y., Teo S. H., Terry M. B., Thomassen M., Tischkowitz M., Toland A. E., Torres D., Vega A., Wagner S. A., Wang-Gohrke S., Wappenschmidt B., Weber B. H. F., Yannoukakos D., Spurdle A. B., Easton D. F., Chenevix-Trench G., Ottini L., Antoniou A. C., Cancer risks associated with BRCA1 and BRCA2 pathogenic variants. J. Clin. Oncol. 40, 1529–1541 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharan S. K., Pyle A., Coppola V., Babus J., Swaminathan S., Benedict J., Swing D., Martin B. K., Tessarollo L., Evans J. P., Flaws J. A., Handel M. A., BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131, 131–142 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Martinez J. S., von Nicolai C., Kim T., Ehlen A., Mazin A. V., Kowalczykowski S. C., Carreira A., BRCA2 regulates DMC1-mediated recombination through the BRC repeats. Proc. Natl. Acad. Sci. U.S.A. 113, 3515–3520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeliger K., Dukowic-Schulze S., Wurz-Wildersinn R., Pacher M., Puchta H., BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 193, 364–375 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Thorslund T., Esashi F., West S. C., Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 26, 2915–2922 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dray E., Siaud N., Dubois E., Doutriaux M.-P., Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 140, 1059–1069 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.I. Brandsma, H. Odijk, A. Oostra, D. Dekkers, K. Bezstarosti, N. van den Tempel, J. Demmers, J. Dorsman, D. van Gent, R. Kanaar, A. N. Zelensky, Overexpression of HSF2BP, a new BRCA2 interactor, results in a Fanconi anemia-like phenotype, in Balancing Pathways in DNA Double Strand Break Repair (2016), pp. 98–140; https://repub.eur.nl/pub/80038.
- 16.Zhang J., Gurusaran M., Fujiwara Y., Zhang K., Echbarthi M., Vorontsov E., Guo R., Pendlebury D. F., Alam I., Livera G., Emmanuelle M., Wang P. J., Nandakumar J., Davies O. R., Shibuya H., The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells. Nat. Commun. 11, 2055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandsma I., Sato K., van Rossum-Fikkert S. E., van Vliet N., Sleddens E., Reuter M., Odijk H., van den Tempel N., Dekkers D. H. W., Bezstarosti K., Demmers J. A. A., Maas A., Lebbink J., Wyman C., Essers J., van Gent D. C., Baarends W. M., Knipscheer P., Kanaar R., Zelensky A. N., HSF2BP interacts with a conserved domain of BRCA2 and is required for mouse spermatogenesis. Cell Rep. 27, 3790–3798.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., Fujiwara Y., Yamamoto S., Shibuya H., A meiosis-specific BRCA2 binding protein recruits recombinases to DNA double-strand breaks to ensure homologous recombination. Nat. Commun. 10, 722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felipe-Medina N., Caburet S., Sánchez-Sáez F., Condezo Y. B., de Rooij D. G., Gómez-H L., Garcia-Valiente R., Todeschini A. L., Duque P., Sánchez-Martin M. A., Shalev S. A., Llano E., Veitia R. A., Pendás A. M., A missense in HSF2BP causing Primary Ovarian Insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1. eLife 9, e56996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang Y., Huang T., Liu H., Liu Y., Liang H., Yu X., Li M., Zhai B., Yang X., Wei Y., Wang G., Chen Z., Wang S., Zhang L., MEIOK21: A new component of meiotic recombination bridges required for spermatogenesis. Nucleic Acids Res. 48, gkaa406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takemoto K., Tani N., Takada-Horisawa Y., Fujimura S., Tanno N., Yamane M., Okamura K., Sugimoto M., Araki K., Ishiguro K.-I., Meiosis-specific C19orf57/4930432K21Rik/BRME1 modulates localization of RAD51 and DMC1 to DSBs in mouse meiotic recombination. Cell Rep. 31, 107686 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Li M., Feng H., Lin Z., Zheng J., Liu D., Guo R., Li J., Li R. H. W., Ng E. H. Y., Huen M. S. Y., Wang P. J., Yeung W. S. B., Liu K., The novel male meiosis recombination regulator coordinates the progression of meiosis prophase I. J. Genet. Genomics 47, 451–465 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ghouil R., Miron S., Koornneef L., Veerman J., Paul M. W., Du M.-H. L., Sleddens-Linkels E., van Rossum-Fikkert S. E., van Loon Y., Felipe-Medina N., Pendas A. M., Maas A., Essers J., Legrand P., Baarends W. M., Kanaar R., Zinn-Justin S., Zelensky A. N., BRCA2 binding through a cryptic repeated motif to HSF2BP oligomers does not impact meiotic recombination. Nat. Commun. 12, 4605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato K., Brandsma I., van Rossum-Fikkert S. E., Verkaik N., Oostra A. B., Dorsman J. C., van Gent D. C., Knipscheer P., Kanaar R., Zelensky A. N., HSF2BP negatively regulates homologous recombination in DNA interstrand crosslink repair. Nucleic Acids Res. 48, 2442–2456 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramadan K., p97/VCP- and Lys48-linked polyubiquitination form a new signaling pathway in DNA damage response. Cell Cycle 11, 1062–1069 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Ju J.-S., Miller S. E., Hanson P. I., Weihl C. C., Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease*. J. Biol. Chem. 283, 30289–30299 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pendlebury D. F., Zhang J., Agrawal R., Shibuya H., Nandakumar J., Structure of a meiosis-specific complex central to BRCA2 localization at recombination sites. Nat. Struct. Mol. Biol. 28, 671–680 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franz A., Ackermann L., Hoppe T., Ring of change: CDC48/p97 drives protein dynamics at chromatin. Front. Genet. 7, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S., Xu W., Xu B., Gao S., Zhang Q., Qin Y., Guo T., Pathogenic variations of homologous recombination Gene HSF2BP identified in sporadic patients with premature ovarian insufficiency. Front. Cell Dev. Biol. 9, 768123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miné-Hattab J., Liu S., Taddei A., Repair foci as liquid phase separation: Evidence and limitations. Genes 13, 1846 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishna T. S. R., Kong X.-P., Gary S., Burgers P. M., Kuriyan J., Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Dieckman L. M., Freudenthal B. D., Washington M. T., PCNA structure and function: Insights from structures of PCNA complexes and post-translationally modified PCNA. Subcell. Biochem. 62, 281–299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribeiro J., Dupaigne P., Petrillo C., Ducrot C., Duquenne C., Veaute X., Saintomé C., Busso D., Guerois R., Martini E., Livera G., The meiosis-specific MEIOB–SPATA22 complex cooperates with RPA to form a compacted mixed MEIOB/SPATA22/RPA/ssDNA complex. DNA Repair 102, 103097 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Yates L. A., Aramayo R. J., Pokhrel N., Caldwell C. C., Kaplan J. A., Perera R. L., Spies M., Antony E., Zhang X., A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Nat. Commun. 9, 5447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinebuchi T., Kagawa W., Enomoto R., Tanaka K., Miyagawa K., Shibata T., Kurumizaka H., Yokoyama S., Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol. Cell 14, 363–374 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Passy S. I., Yu X., Li Z., Radding C. M., Masson J.-Y., West S. C., Egelman E. H., Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl. Acad. Sci. U.S.A. 96, 10684–10688 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corbett K. D., Yip C. K., Ee L.-S., Walz T., Amon A., Harrison S. C., The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142, 556–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meulemans L., Mesman R. L. S., Caputo S. M., Krieger S., Guillaud-Bataille M., Caux-Moncoutier V., Léone M., Boutry-Kryza N., Sokolowska J., Révillion F., Delnatte C., Tubeuf H., Soukarieh O., Bonnet-Dorion F., Guibert V., Bronner M., Bourdon V., Lizard S., Vilquin P., Privat M., Drouet A., Grout C., Calléja F. M. G. R., Golmard L., Vrieling H., Stoppa-Lyonnet D., Houdayer C., Frebourg T., Vreeswijk M. P. G., Martins A., Gaildrat P., Skipping nonsense to maintain function: The paradigm of BRCA2 Exon 12. Cancer Res. 80, 1374–1386 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Li L., Biswas K., Habib L. A., Kuznetsov S. G., Hamel N., Kirchhoff T., Wong N., Armel S., Chong G., Narod S. A., Claes K., Offit K., Robson M. E., Stauffer S., Sharan S. K., Foulkes W. D., Functional redundancy of exon 12 of BRCA2 revealed by a comprehensive analysis of the c.6853A>G (p.I2285V) variant. Hum. Mutat. 30, 1543–1550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bièche I., Lidereau R., Increased level of exon 12 alternatively spliced BRCA2 transcripts in tumor breast tissue compared with normal tissue. Cancer Res. 59, 2546–2550 (1999). [PubMed] [Google Scholar]
- 42.Dupeux F., Röwer M., Seroul G., Blot D., Márquez J. A., A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr. D Biol. Crystallogr. 67, 915–919 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Niebling S., Veith K., Vollmer B., Lizarrondo J., Burastero O., Schiller J., García A. S., Lewe P., Seuring C., Witt S., García-Alai M., Biophysical screening pipeline for cryo-EM grid preparation of membrane proteins. Front. Mol. Biosci. 9, 882288 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A. J., Berger B., Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N., ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zander U., Hoffmann G., Cornaciu I., Marquette J.-P., Papp G., Landret C., Seroul G., Sinoir J., Röwer M., Felisaz F., Rodriguez-Puente S., Mariaule V., Murphy P., Mathieu M., Cipriani F., Márquez J. A., Automated harvesting and processing of protein crystals through laser photoablation. Acta Crystallogr. D Struct. Biol. 72, 454–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L.-W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.G. Bricogne, E. Blanc, M. Brandl, C. Flensburg, P. Keller, W. Paciorek, P. Roversi, A. Sharff, O. S. Smart, C. Vonrhein, T. O. Womack, BUSTER version 2.10.3 (Global Phasing Ltd., 2020). [Google Scholar]
- 57.Yusa K., Zhou L., Li M. A., Bradley A., Craig N. L., A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 108, 1531–1536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Räschle M., Knipscheer P., Knipsheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., Walter J. C., Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knipscheer P., Räschle M., Smogorzewska A., Enoiu M., Ho T. V., Schärer O. D., Elledge S. J., Walter J. C., The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sparks J., Walter J. C., Extracts for analysis of DNA replication in a nucleus-free system. Cold Spring Harb. Protoc. 2019, 10.1101/pdb.prot097154, (2019). [DOI] [PubMed] [Google Scholar]
- 61.Enoiu M., Ho T. V., Long D. T., Walter J. C., Schärer O. D., Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol. Biol. 920, 203–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knipscheer P., Räschle M., Schärer O. D., Walter J. C., Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol. Biol. 920, 221–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Long D. T., Raschle M., Joukov V., Walter J. C., Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S3
Legend for data S1
Data S1