Abstract
Understanding natural and traditional medicine can lead to world-changing drug discoveries. Despite the therapeutic effectiveness of individual herbs, traditional Chinese medicine (TCM) lacks a scientific foundation and is often considered a myth. In this study, we establish a network medicine framework and reveal the general TCM treatment principle as the topological relationship between disease symptoms and TCM herb targets on the human protein interactome. We find that proteins associated with a symptom form a network module, and the network proximity of an herb’s targets to a symptom module is predictive of the herb’s effectiveness in treating the symptom. These findings are validated using patient data from a hospital. We highlight the translational value of our framework by predicting herb-symptom treatments with therapeutic potential. Our network medicine framework reveals the scientific foundation of TCM and establishes a paradigm for understanding the molecular basis of natural medicine and predicting disease treatments.
The general principles of traditional Chinese medicine are rooted in the proximity of proteins in the protein interaction network.
INTRODUCTION
Understanding the therapeutic effects of traditional and natural medicine can lead to drug discoveries that reshape world welfare. For example, aspirin (acetylsalicylic acid) is extracted from willow bark, a traditional medicine practice since thousands of years ago (1). More recently, the 2015 Nobel Prize was given to the discovery of the malaria-treating artemisinin, extracted from qinghao (Artemisia annua), an herb used in traditional Chinese medicine (TCM) (2). As a famous practice of natural medicine, TCM is a personalized and holistic approach to treating diseases using natural medical products tailored to a patient’s symptoms, offering a rich pool of therapeutic candidates (3–5). However, although clinical data and studies of single herbs/prescriptions (6, 7) showed that certain TCM herbal treatments are effective, the general mechanistic principle of how TCM selects herbs to treat diseases remains unknown. Two major challenges exist in understanding the mechanistic root of TCM: (i) The lack of scientific foundation in classic TCM theory obstructs the understanding of TCM from a modern biomedical perspective; (ii) the complexity of herbs’ chemical composition and the often-unknown therapeutic protein targets of the chemicals makes conventional brute-force herb/chemical screening infeasible. Therefore, to understand and exploit the therapeutic mechanisms of TCM, it is necessary to establish a framework that can connect TCM knowledge to modern biomedical science and can handle the complexity of herb composition and target data.
An in silico strategy to understand the therapeutic effect of a natural product is to leverage the multiple protein targets of its composing chemicals via network pharmacology (8) and network medicine (9–12). Network pharmacology emphasizes the “network target, multi-components” paradigm that complements conventional research’s focus on single targets. This approach has helped researchers identify herbal chemicals with therapeutic potentials, better understand mechanisms of action, and discover drugs (13–15). However, existing TCM network pharmacology studies are limited to single herbs or single prescriptions, unable to explain the totality of TCM herb-disease relations. Moreover, many network pharmacology approaches only consider herbs/drugs that target disease genes directly, unable to account for network effects, e.g., when the impact of perturbing a target emerges further downstream and is mediated by protein interactions. Here, we propose avenues to overcome these limitations and improve our understanding of the therapeutic effects of natural products.
Network medicine leverages the human protein-protein interactome (PPI) to reveal disease and drug patterns (9). The PPI is a network consisting of nodes that are proteins that link to each other by physical (binding) interactions. Network medicine showed that disease-associated proteins tend to form locally clustered modules in the PPI, and shorter network distance between two disease modules is indicative of their comorbidity (16); moreover, drug efficacy can be predicted by leveraging the network relation between drug targets and disease modules (17, 18), leading to the development of drug-repurposing methodologies (19). These methods have been successful in identifying drug-repurposing candidates for coronavirus disease 2019 (COVID-19) and in understanding the network patterns of effective drugs (20). Furthermore, some of these tools have already affected clinical practice, like the network-based diagnostic tool available for patients with rheumatoid arthritis (21). Unlike earlier network pharmacology approaches, network medicine characterizes drug-disease relations by capturing the network effects based on protein interactions from the PPI, enabling more accurate predictions.
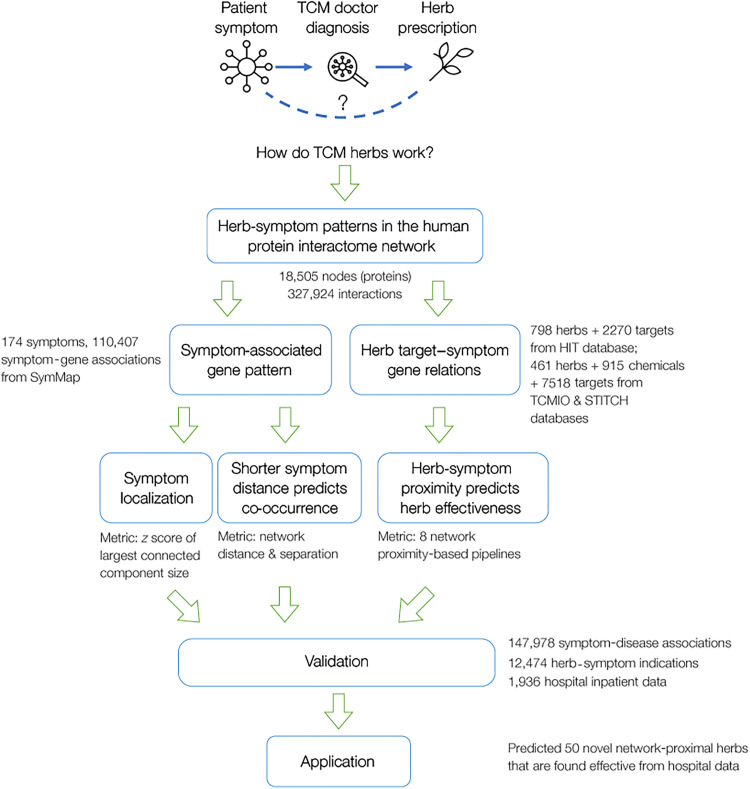
In this study, we develop a network medicine framework that theorizes the scientific basis of TCM as the topological relationship between symptom-associated proteins and herb targets on the protein interactome. By focusing on symptoms rather than diseases, our approach aligns with the TCM practice of diagnosing and treating patients based on their symptom phenotypes. We discover that proteins associated with a symptom tend to cluster into a local PPI module, and the network proximity between an herb’s targets and a symptom module is indicative of the herb’s effectiveness in treating the symptom. We validate our network medicine framework with empirical data and hospital patient data and highlight its potential in identifying herb discovery/repurposing opportunities. The design of our study is presented in Fig. 1.
Fig. 1. Study design.
To explore the mechanisms of how TCM treats disease/symptoms, we develop a generic framework that characterizes TCM mechanisms as the network-based relation between symptom-associated proteins and herb targets in the human PPI. After collecting the symptom-associated proteins and herb-target data, we designed multiple network-based metrics to unveil the network patterns connecting them, including symptom localization, symptom-symptom relation, and herb-symptom proximity. We validated these relations by showing that our network-based framework captures symptom-disease relations and herb-symptom effectiveness, leveraging online public databases and a hospital inpatient dataset. We highlight the potential application of our work in predicting herb-symptom treatments.
RESULTS
Symptom-associated proteins form modules in the protein interactome
Connecting TCM to the modern biomedical literature is challenging, due to the absence of the concept of “disease” in TCM. As a result, previous findings based on diseases, e.g., disease modules (16, 22, 23), are not directly applicable to TCM. To bridge this gap, we propose the use of symptom phenotypes to characterize the indications and effects of TCM and study the PPI pattern of symptoms. This approach is based on the fact that TCM clinical diagnosis and treatments are based on symptom phenotypes (24, 25), and is further supported by the availability of disease taxonomy and protein/gene association data in symptom phenotypes (26, 27).
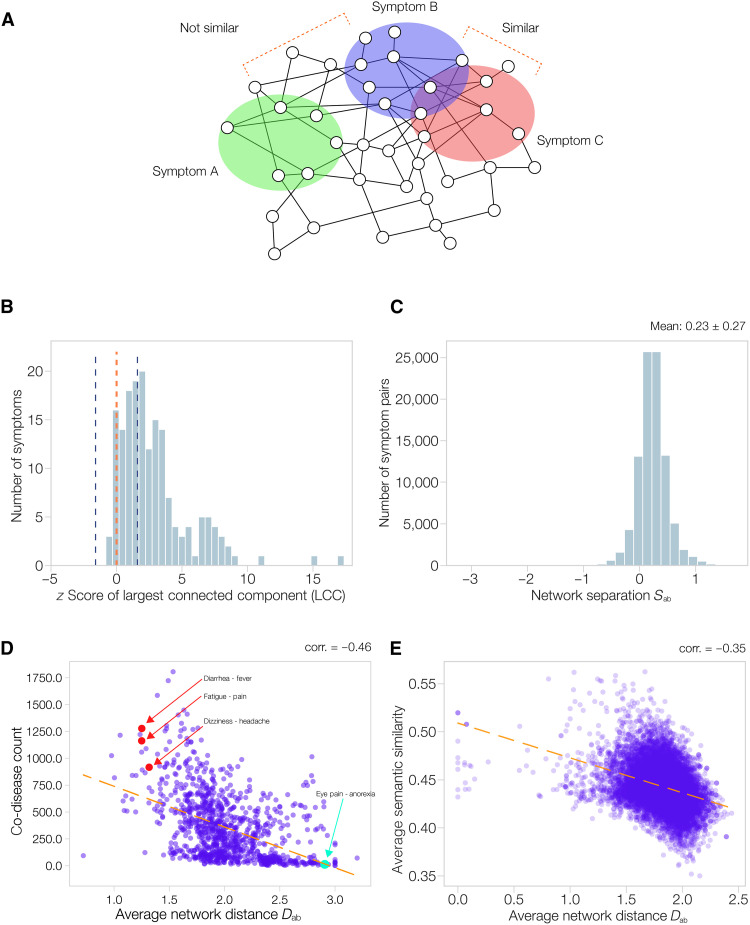
We rely on a curated symptom-gene association dataset (28) (see Materials and Methods and data S1) to identify genes associated with each symptom, and then map these genes onto their corresponding proteins in the PPI (see Materials and Methods and data S2). We focus on 174 symptoms with at least 20 associated proteins. We find that for 108 of these 174 symptoms, their associated proteins form a connected component significantly larger than random expectation (z > 1.6; Fig. 2A and data S3). This suggests that the symptom-associated proteins agglomerate into a localized module in the PPI. In addition, we found that proteins associated with different symptoms are distant from each other (Fig. 2B), characterized by the average network separation metric (see Materials and Methods) Sab = 0.23, larger than the random expectation of zero. This suggests that different symptoms perturb different regions of the PPI.
Fig. 2. Symptom pattern in the human PPI.
(A) Schematic illustrating that proteins associated with a symptom form localized modules on the human protein interactome, and the inter-module network distance is indicative of symptom similarity. (B) Distribution of largest-connected-component z score formed by symptom-associated proteins, for 174 symptoms. One hundred eight out of 174 symptoms form significantly clustered local modules (z > 1.6). The blue dotted lines indicate z = ±1.6, and the red dotted line indicates z = 0. (C) Distribution of network separation (Sab) of all symptom pairs. The average ⟨Sab⟩ is larger than zero, the random expectation. This suggests that different symptoms perturb different/specific regions in the PPI, by forming modules distant from each other. (D) The average interactome network distance (Dab) of a symptom pair negatively correlates with the symptoms’ co-occurrence in diseases (co-disease count), with Pearson’s correlation −0.46. Each dot represents a symptom pair. We highlight in red examples of similar and co-occurring symptoms, such as fever-diarrhea (Dab = 1.25, co-disease count = 1278), fatigue-pain (Dab = 1.25, co-disease count = 1163), and dizziness-headache (Dab = 1.32, co-disease count = 917). We also highlight in green an example symptom pair with high network distance and less co-occurrence, eye pain and anorexia (Dab = 2.91, co-disease count: 13). (E) The interactome network distance of a symptom pair negatively correlates with the biological similarity of the genes associated with the symptoms.
We also ask if the network distance between symptom modules on the interactome can reveal clinically relevant relations between the symptoms. To do this, we calculated the average network distances between the PPI modules of two symptoms (Dab; see Materials and Methods). Then, we leverage 147,978 symptom-disease associations (29) to compute the number of shared diseases of symptoms. We found the number of shared diseases of two symptoms to negatively correlate with their PPI modules’ distance Dab (Fig. 2D, Pearson’s correlation = −0.46, P = 3.1 × 10−55), indicating that a closer network distance between symptom modules predicts their co-occurrence in diseases. We also investigate if the network distance between symptoms can predict their biological similarity. To do so, we leverage the Gene Ontology (GO) semantic similarity of genes (see Materials and Methods) (30), which characterizes the similarity of two genes based on their similarity in GO annotations. We found a symptom pair’s overall GO semantic similarity to negatively correlate with their average network distance Dab (Fig. 2E, Pearson’s correlation = −0.35, P < 1 ×10−100). Furthermore, we observed significant negative correlations between network distance and GO semantic similarity, for each of the three branches of GO ontology, cellular component, biological process, and molecular function (text S1 and fig. S2). Together, these findings indicate that two symptom modules with closer distance in the PPI are more likely to co-occur in the same disease, and to be more biological similar. We provide symptom pairwise network distance Dab & Sab along with co-disease count and GO semantic similarity in data S3.
For example, the symptom pair fever and diarrhea has a network distance Dab = 1.25, much shorter than the average symptom distance <Dab> = 2.01 ± 0.37 (z = −2.1, P = 0.018), and a co-disease count of 1278, much higher than the average co-disease count 236 ± 264 (z = 3.9, P = 4.8 × 10−5). Diarrhea and fever co-occur in many diseases such as inflammatory diseases (e.g., inflammatory gastroenteropathy) (31) and virus-induced infectious diseases [e.g., severe acute respiratory syndrome coronavirus 2 (32)]. These co-occurrences may be also rooted in the two symptoms’ 27 shared genes, including inflammatory biomarkers [e.g., PIK3R1 and TNF (33)] and the cytokines [e.g., IL1A and IL7R (34)]. Their associated pathways tend to be related to the inflammatory immune processes, such as the Janus kinase/signal transducers and activators of transcription pathway (35) and cytokine-mediated signaling pathway (36). Other frequently co-occurring symptom pairs (highlighted in red in Fig. 2D) include fatigue-pain (Dab = 1.25, co-disease count = 1163) and dizziness-headache (Dab = 1.32, co-disease count = 917). In contrast, symptoms with higher network distance have less co-occurrence in diseases and are not considered similar, such as eye pain and anorexia (highlighted in green in Fig. 2D), which has a large Dab = 2.91 and a low co-disease count of 13.
Herb-symptom network proximity indicates therapeutic effectiveness
We investigate TCM herbs’ therapeutic effects by analyzing their protein targets in the PPI. One challenge of this approach is the complexity of assessing the effects of targets for each herb, as each herb contains numerous chemicals, and each chemical can bind to multiple protein targets (37, 38). To overcome this challenge, we refined herb-chemical-target data and designed a multimodal network-based approach to characterizing the PPI relationship between herb targets and symptom modules. We rely on two groups of datasets: (i) We directly use herb-target data from the recently updated HIT 2.0 database (39), compiled by text-mining literature abstracts for compound-target relations, followed by manual review. After name mapping, this dataset yields 798 herbs and 2270 protein targets, with an average of 162.9 ± 185.5 targets per herb. (ii) We compile herb-target data by integrating herb chemical composition data from the TCMIO database with chemical-target data from STITCH. The TCMIO database (40) contains a recent and comprehensive compilation of the TCMSP, TCMID, and TCM-ID databases (41–43), which focus on chemicals with potential therapeutic effects. We then use STITCH (44) for chemical-target data, keeping only targets with experimental evidence. In the end, we obtained 461 herbs with target data, consisting of 915 chemicals with therapeutic potential, and together targeting 7518 unique proteins. On average, each herb has 61.9 ± 61.5 (potentially therapeutic) chemicals, and each chemical has 69.7 ± 311.4 targets (see data S4; see also text S1 and fig. S3 for distribution plots).
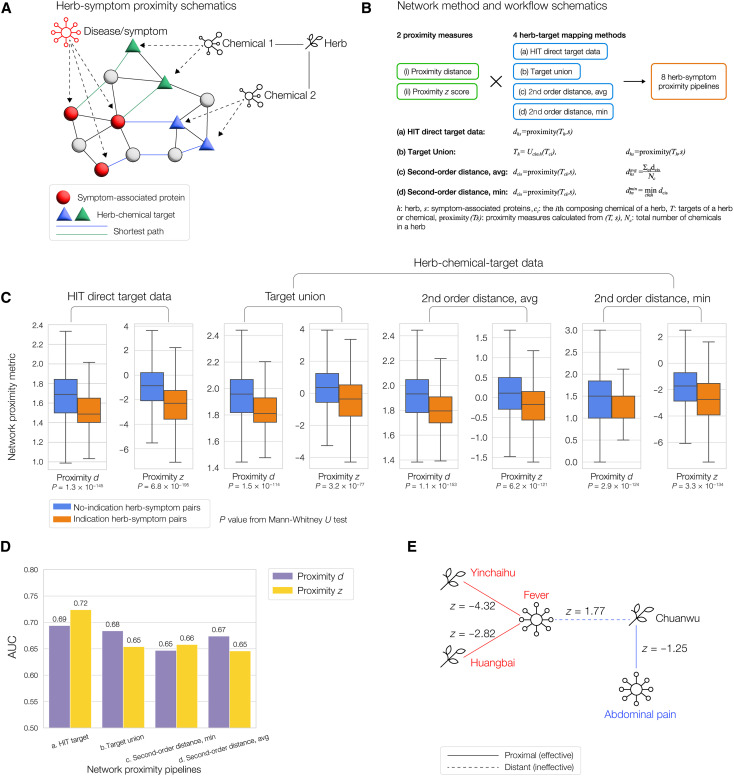
To characterize the network-based relations between herbs and symptoms, we develop a multimodal approach comprising eight pipelines, each of which produces a network-based metric for each herb-symptom pair (see schematics and workflow in Fig. 3, A and B). Our pipelines are driven by the hypothesis that herbs effective for treating a symptom must target proteins proximal to the symptom-associated proteins in the PPI, similar to the network pattern observed in drug-disease relations (17). To quantify the network-based relationship between a set of targets and a set of symptom-associated proteins, we use two metrics: (i) the proximity distance, which is the average distance between herb targets to their closest symptom-associated protein(s), and (ii) the proximity z score, which measures how the proximity distance differs from random expectation (see Materials and Methods). For both metrics, lower values indicate a closer network relation between the target set and the symptom module. For the proximity z score, z < 0 means more proximal than random, z > 0 means more distant than random, and z = 0 means neutral. We also designed four herb-target mapping methods from (a) HIT target data and (b to d) integrated herb-chemical-target data (see Materials and Methods). Together, the combination of the two distance/proximity metrics (proximity d and z) and the four herb-target mapping methods (a to d) resulted in eight pipelines for herb-symptom network metrics (Fig. 3B), for each herb-symptom pair. We compute the network metrics for all herb-symptom pairs using the eight pipelines (see data S5).
Fig. 3. Herb-symptom network proximity predicts effectiveness.
(A) Schematics of the herb-symptom network proximity metric, based on shortest paths between herb-chemical targets and symptom-associated proteins in the protein interactome. (B) Workflow of the multimodal approach for eight herb-symptom proximity pipelines, with definitions of the metrics. (C) Results of the eight pipelines of network metrics for herb-symptom pairs categorized as indicated or non-indicated. Indicated herb-symptom pairs (orange bars) show lower proximity metrics (shorter network distance) than the non-indicated herb-symptom pairs (blue bars), consistently over all eight pipelines. (D) AUC (area under the receiver operating characteristic curve) performance evaluation of the eight herb-symptom proximity pipelines, using the known herb-symptom indications as positive cases. (E) Example demonstrating herb-symptom proximity: Herbs Yinchaihu and Huangbai are proximal (having highly negative network proximity z score) to the fever symptom and are used to treat fever in practice, whereas the Chuanwu herb is distant (having positive z score) to fever but proximal to abdominal pain, thus it is not used to treat fever but to treat abdominal pain.
To evaluate the performance of the proposed network metrics, we leverage as ground truth a dataset of expert-curated herb-symptom indications from SymMap (45), where the herb is recognized by Chinese Pharmacopoeia (CHPH; the authoritative TCM data, 2015 edition) to be effective against the symptom. We map these herb-symptom indication pairs into our eight pipelines, resulting in 1480 indications in pipelines 1 and 2, and 1325 indications in pipelines 3 to 8. To evaluate our network proximity hypothesis, first, we make box-and-whiskers plots of the network metrics for all eight pipelines (Fig. 3C), comparing indicated herb-symptom pairs (orange bars) against non-indicated herb-symptom pairs (blue bars). We found that the orange bars were consistently lower than the blue bars across all pipelines, indicating that known effective herb-symptom pairs are more proximal, compared to other herb-symptom pairs. In addition, we calculate the AUC (area under the receiver operating characteristic curve, or AUROC) as an accuracy metric for all pipelines, using the herb-symptom pairs with indications as positive cases, and the herb-symptom pairs without indications as negative cases (Fig. 3D). The obtained AUC values between 0.65 and 0.72 indicate that all pipelines are highly predictive compared to the random expectation of AUC = 0.5. Both proximity distance and the proximity statistical z score were found to be predictive, with neither consistently outperforming the other. The best-performing pipeline is the HIT target dataset with proximity z score, with the highest AUC = 0.72. To our knowledge, there is no other generic method that predicts an herb’s effectiveness against a symptom/disease from the PPI, so we compare this result with previous predictions on drug-disease relations. The observed best AUC value of 0.72 in this work is higher than the best AUC of 0.66 observed in generic drug-disease effectiveness (17), and the best AUC of 0.63 in drug-COVID effectiveness (20). Overall, the (relatively) high AUCs and their consistency across all pipelines indicate that network proximity has predictive power regarding TCM herb-symptom effectiveness. This result is especially remarkable considering the high noise in such large-scale and multi-faceted data, and the diversity of symptom/disease-herb relationships.
To illustrate the role of herb-symptom proximity (Fig. 3E), we consider the “fever” symptom and herbs effective in treating it. We use HIT proximity z score (pipeline P2) here and in the rest of the paper when demonstrating examples because it is the best-performing pipeline according to the AUC score. Herbs with highly negative network proximity z scores to the fever symptom-associated protein module include Yinchaihu (Radix Stellariae, Starwort root, z score: −4.32), which is recognized by the CHPH to treat fever and is prescribed by TCM doctors in practice to treat, e.g., asthenic fever in the late stage of febrile diseases (46). This herb treats fever by regulating a series of inflammatory processes, such as nuclear factor κB and mitogen-activated protein kinase (47). Another well-recognized herb used by TCM doctors to treat fever, Huangbai (Phellodendri Chinensis Cortex, Phellodendron Bark, z score: −2.82), is used to treat pneumonia and tuberculosis (48). Berberine, one of the main active chemical components of Huangbai, has anti-inflammatory and antipyretic effects (49). On the other hand, an herb distant from the fever symptom module in the PPI, such as Chuanwu (Radix Aconitum, aconite root, z score: 1.77), is unlikely to be effective against fever, consistent with expert knowledge. Chuanwu is network-proximal to abdominal pain (z score: −1.25) and is recognized by the CHPH and prescribed in practice for pain relief due to its anti-inflammatory, analgesic, and antitumor effects (50).
Validation of herb-symptom relation with hospital inpatient data
In this section, we validate the effectiveness of our network medicine framework in predicting symptom relations and herb-symptom proximity using real-world patient data. We collected the electronic medical record (EMR) data of 1936 liver cirrhosis inpatient cases treated at Hubei Provincial Hospital of Traditional Chinese Medicine in Wuhan. Data on patient symptoms and their changes (before and after treatment) were extracted from the admission and discharge records using a clinical information extraction tool (Human-machine Cooperative Phenotypic Spectrum Annotation System, HCPSAS; www.tcmai.org) (51). We manually mapped the symptoms in the patient data from Chinese to Unified Medical Language System (UMLS) terms to enable symptom-gene associations (see Materials and Methods). Similarly, we map herbs from their Chinese names in the data to herb IDs to obtain their chemical composition and target data. The resulting patient dataset contains a total of 114 symptoms, 218 herbs, and 23,413 herb-symptom pairs (data S6).
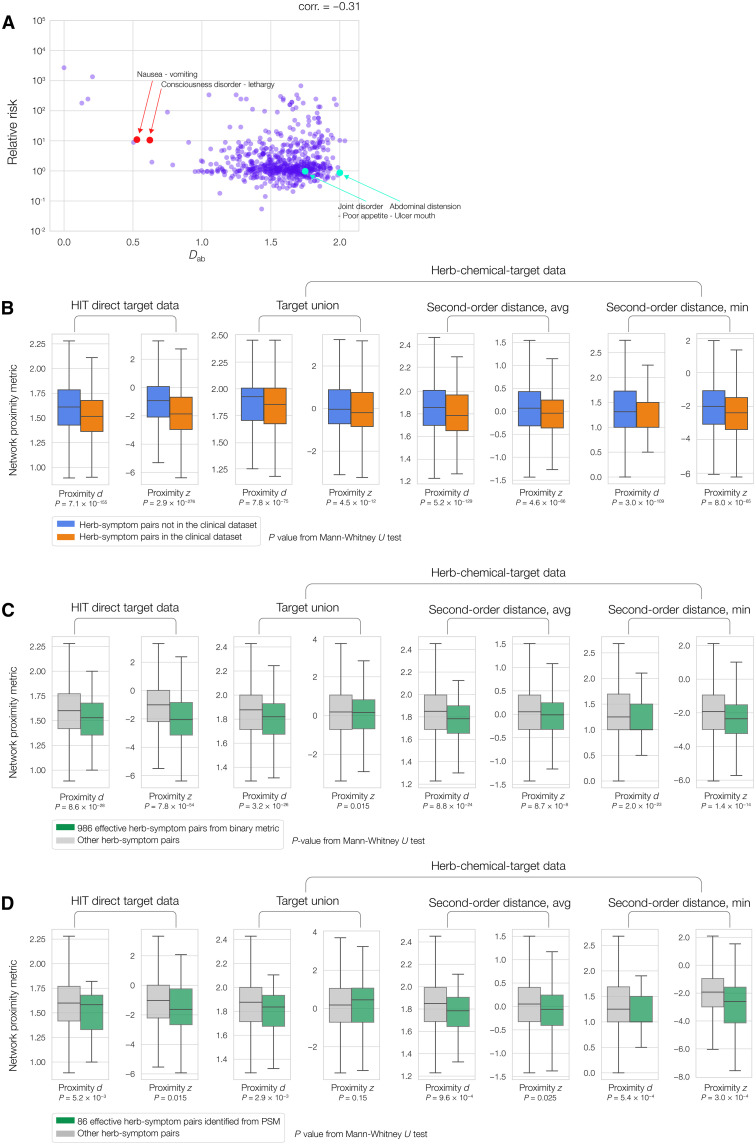
To validate the relation between a symptom pair’s network distance and their co-occurrence, we computed the relative risk (RR) between each symptom pair. RR is a standard statistic that measures the strength of an association (in this case, the co-occurrence of two symptoms), defined as the ratio of the probabilities of the exposed and unexposed groups. Then, we computed the PPI network distance Dab between each pair of symptoms (see data S7 and found a negative Pearson’s correlation of −0.31 (P = 1.4 × 10−15) between symptom pairs’ RR and Dab (Fig. 4A). This negative correlation indicates that symptoms with shorter network distance in the PPI are more likely to co-occur. Examples of co-occurring symptom pairs include nausea and vomiting (Dab = 0.53, RR = 11.00) as well as consciousness disorder and lethargy (Dab = 0.62, RR = 10.6). Conversely, symptom pairs with longer network distance do not have high RR, for example, joint disorder and poor appetite (Dab = 1.75, RR = 0.98) as well as abdomen distention and ulcer mouth (Dab: 2.00, RR:0.87). These results validate our hypothesis that shorter network distances in the PPI can indicate co-occurrence of symptoms. We do not observe high negative correlations on the high network distance (right-hand) side in Fig. 4A, suggesting that while symptoms with short PPI distance tend to co-occur more frequently, symptoms with long PPI distance co-occur randomly.
Fig. 4. Validation of network medicine framework with hospital inpatient data.
(A) Patient symptom data show a negative Pearson’s correlation between symptom pair relative risk (in log scale) and network distance Dab, validating that shorter network distance between symptoms is predictive of their co-occurrence. (B) Herbs used by doctors in patient data (orange boxes) are significantly more proximal to symptoms than herbs not used in patient data (blue boxes), consistently observed over all eight pipelines, indicating that network proximity captures doctors’ knowledge. (C) The 986 effective herb-symptom pairs identified from the binary effectiveness metric (green boxes) have lower network metrics than other herb-symptom pairs (gray boxes) in all eight pipelines, i.e., network proximity metrics can predict the effective herb-symptom pairs. (D) The 86 effective herb-symptom pairs identified from propensity score matching (green boxes) have lower network metrics than other herb-symptom pairs (gray boxes) in seven of all eight pipelines, i.e., network proximity metrics can predict the significantly effective herb-symptom pairs.
Next, we used three different methods to validate the hypothesis that the network proximity of an herb-symptom pair can predict the herb’s effectiveness in treating the symptom. First, we show that network proximity captures doctors’ knowledge in prescribing herbs against symptoms (see data S8), by comparing the network proximity of herb-symptom pairs in the patient dataset (representing herbs prescribed by doctors) against that of herb-symptom pairs absent from the clinical dataset (representing herbs not prescribed by doctors). We observe in Fig. 4B that, for all eight proximity pipelines, the herb-symptom pairs in the patient dataset (orange boxes) have significantly lower network proximity metrics than the herb-symptom pairs not observed in the patient dataset (blue boxes), with P values ranging from 3.6 × 10−07 to 9.0 × 10−39. In other words, TCM doctors tend to prescribe herbs whose therapeutic targets are proximal to the disease/symptom module in the PPI. This supports our hypothesis that proximal herbs are more likely to be effective and aligns with doctors’ expert knowledge (data S8).
Second, we explore if network proximity predicts effective herb-symptom pairs in the patient dataset. As the patient data do not contain any metric of herb effectiveness, we need to define one. The ideal metric would be the statistical significance of effectiveness for each herb-symptom pair, but that would require more data than currently available. We therefore define a binary (i.e., true or false) herb effectiveness measure by comparing patients’ symptom recovery rate after treatment versus the symptom’s baseline recovery rate (see Materials and Methods). In short, an herb is considered effective if the patient receiving it recovers better than not receiving it. Using this definition, we identified 986 effective herb-symptom pairs with a frequency of at least 10 (i.e., at least 10 patients with the symptom and treated with the herb). We observe in Fig. 4C that for all eight pipelines, the effective herb-symptom pairs (green bars) consistently have significantly lower network proximity metrics than other herb-symptom pairs (gray bars), with P values ranging from 8.0 × 10−57 to 1.5 × 10−2 (see Materials and Methods and data S8). Therefore, these 986 effective herb-symptom pairs support our hypothesis that effective herb-symptom pairs tend to be network-proximal.
Third, we focus on a subset of the herb-symptom pairs with sufficient frequency to perform bioinformatics analysis with statistical significance. We apply a propensity score matching (PSM) method to a data subset of 888 herb-symptom pairs with at least 30 cases where the patient with this symptom is treated with the herb. PSM means that for each herb-symptom pair, we matched the patients with this symptom and treated with this herb (i.e., the case group) to a control group where the patients have the same symptom but are not treated with this herb, and we adjusted for potential confounders (e.g., age, gender, history of hypertension, diabetes, coronary artery disease, and chronic kidney disease) of the patients in the control group (see Materials and Methods). After PSM, we identified 86 herb-symptom pairs where the case group has a significantly higher symptom recovery rate than the control group (P < 0.05, chi-square test), i.e., the herb treatment is effective by statistical significance. We found that these 86 effective herb-symptom pairs are network-proximal, compared to all herb-symptom pairs (data S9). As shown in Fig. 4D, seven of the eight pipelines indicated that the effective pairs have significantly lower network metrics (P value ranging from 3.0 × 10−4 to 2.5 × 10−2), confirming that network proximity is a good predictor of herb effectiveness in patient data. Herb-symptom pairs with proximity metrics from all eight pipelines, together with an indicator of whether it is in patient data, an indicator for presence in the clinical dataset, an indicator for effectiveness identified by the binary metric, and an indicator for effectiveness identified from PSM, are provided in data S8.
As an example, we consider the herb-symptom pair Baizhu (Atractylodis Macrocephalae Rhizoma, Rhizome of Largehead Atractylodes) used for poor appetite. Network proximity shows a negative z score = −2.45 between Baizhu’s protein targets and the poor appetite symptom module, meaning the herb’s target is proximal to the symptom’s associated proteins, suggesting Baizhu’s potential effectiveness in improving poor appetite. To evaluate from patient data the effectiveness of the herb, we match patients with poor appetite who are treated with Baizhu (case group), to patients with poor appetite but not treated with Baizhu (control group), and compare their symptom recovery rate. We observe that in the matched patients, Baizhu significantly improved the recovery rate of poor appetite (79.53% case group recovery rate versus 72.51% control group recovery rate, P = 0.0316), consistent with the network proximity prediction. This aligns with the known use of Baizhu to treat gastrointestinal dysfunction according to the CHPH. Studies showed that atractylenolide I, sourced from Baizhu, regulates gastrointestinal function and promotes the absorption of nutrients (52), supporting the effectiveness of Baizhu in improving a patient’s appetite.
Herb-symptom proximity is robust under chemical filtering
A challenge to a deeper explanation of the therapeutic role of specific herbs is the limited specificity of the herb-chemical-target dataset. A small set of common chemicals tends to appear in many foods (text S1 and fig. S3D). Given their prevalence in food, these chemicals are less likely to be responsible for the specific therapeutic effect of herbs. To ensure that these common chemicals do not undermine the herb-symptom proximity, we filter out common chemicals with high frequency in food as identified based on the FooDB database (www.foodb.ca) (text S1) (53). We find that 704 chemicals out of the total 915 chemicals survived this filter. We then compute herb-symptom network proximity using these 704 chemicals and repeat our accuracy analysis using indication data and inpatient data with effectiveness metrics. We find that the results are consistent with the network proximity hypothesis but did not improve its predictive power: The AUCs from the indication data are above 0.5 but have declined (see text S1 and fig. S4A); on the other hand, all eight pipelines show consistently better network proximity (i.e., lower metrics) for the significantly effective herbs identified by PSM (fig. S4D). Other metrics did not change as much. Since the results after the chemical filtering still show that effective herb-symptom pairs tend to be network-proximal, we conclude that chemical filtering does not affect the herb-symptom proximity hypothesis.
Our network medicine framework reveals herb discovery and repurposing opportunities
We demonstrate the utility of our network medicine framework for predicting herb candidates to treat symptoms. By calculating the network proximity metrics between herb-symptom pairs, we identified several promising candidates for further investigation. For example, we found a negative proximity z score = −2.86 between the herb-symptom pair Chaihu (Bupleuri Radix, Root of Chinese Thorowax)-abdomen distention, predicting the herb as potentially effective against the symptom. Although the CHPH does not explicitly record this herb for the treatment of abdomen distension, it does record that Chaihu is used to treat distension in the chest and ribs, similar to distension in the abdomen. In support of this prediction, our patient dataset showed that (i) Chaihu is frequently prescribed in practice to treat abdomen distention, with herb-symptom co-occurrence frequency at 381, significantly higher than the average frequency of 106.8 ± 106.5 for all herb-symptom pairs and (ii) in the PSM matched patients, Chaihu significantly improved the recovery rate from abdomen distention (88.71% versus 83.73%, P = 0.0458). Chaihu contains chemicals such as saikosaponins, which can relieve abdomen distension caused by dyspepsia and ascites of liver cirrhosis (54), suggesting its potential effectiveness against abdomen distention. Furthermore, we identified effective herb-symptom pairs less frequent in clinical practice, such as the “Cangzhu (Atractylodis Rhizoma, Rhizome of Swordlike Atractylodes)-abdominal pain” pair, which has a negative proximity z score = −3.08 and has a significantly improved recovery rate (93.55% versus 70.97%, P = 0.0461). Studies have shown that the volatile oil component of Cangzhu has an anti-acetylcholine effect, which can relieve abdominal pain symptoms caused by intestinal spasms (55). We also found potentially effective herb-symptom pairs that are rarely reported, such as the “Baiji (Bletillae Rhizoma, Bletilla Striata Rchb.F.)-edema” pair, which has a highly negative proximity z score of −4.12 with improved recovery rate (83.33% versus 67.86%, P = 0.0195). This suggests that Baiji, an astringent hemostatic conventionally used to relieve gastrointestinal bleeding (56), might be effective in relieving edema. These findings of potentially effective herb-symptom treatments highlight the predictive power of the network medicine framework in identifying herb discovery or repurposing candidates. We provide a list of 50 herb-symptom pairs that are both network-proximal and effective after PSM in patient data but are not yet recorded in the CHPH (Table 1). They are promising treatment candidates that may be tested in follow-up studies. The comprehensive herb-symptom pair network proximity result, which may provide more candidate effective herbs against a symptom, can be found in data S5.
Table 1. Fifty herb-symptom pairs with negative network proximity z score (i.e., predicted as potentially effective) and significantly effective in propensity score matched patient data.
They are promising candidates for herb-symptom treatment discovery/repurposing but are not yet recorded in the Chinese Pharmacopoeia. The table is ordered from the most negative proximity z score to the least negative. The third column is the network proximity z score; the fourth column is the number of patients in the case/control group after propensity score matching; the fifth and sixth are the recovery rates of the case group and the control group; the last column is the P value for the recovery rate difference, from a chi-square test. A similar table with more herb name mapping is provided in data S9.
| Herb | Latin name | Symptom | Proximity z score | Number of patients | Case group recovery rate | Control group recovery rate | P value |
|---|---|---|---|---|---|---|---|
| Bei Sha Shen | Glehnia littoralis | Edema | −6.31 | 79 | 77.22% | 58.23% | 1.07 × 10−02 |
| Jin Yin Hua | Flos lonicerae | Edema | −5.58 | 78 | 73.08% | 53.85% | 1.26 × 10−02 |
| Hu Ji Sheng | Viscum coloratum | Edema | −5.31 | 46 | 91.30% | 69.57% | 1.80 × 10−02 |
| Xiang Fu | Cyperus rotundus | Edema | −4.32 | 63 | 73.02% | 53.97% | 2.64 × 10−02 |
| Chi Shao | Paeonia obovata | Edema | −4.3 | 98 | 80.61% | 68.37% | 4.93 × 10−02 |
| Bai Ji | Bletillae rhizoma | Edema | −4.12 | 84 | 83.33% | 67.86% | 1.95 × 10−02 |
| Ku Shen | Sophora flavescens | Abdomen distention | −3.37 | 40 | 92.50% | 72.50% | 3.94 × 10−02 |
| Chen Pi | Citrus aurantium | Body pain | −3.16 | 87 | 93.10% | 81.61% | 2.25 × 10−02 |
| Cang Zhu | Atractylodes lancea | Abdominal pain | −3.08 | 31 | 93.55% | 70.97% | 4.61 × 10−02 |
| Xiang Fu | Cyperus rotundus | Fatigue | −2.96 | 201 | 87.56% | 80.10% | 4.21 × 10−02 |
| Mu Xiang | Radix aucklandiae | Fatigue | −2.9 | 149 | 92.62% | 85.23% | 4.23 × 10−02 |
| Chai Hu | Bupleurum chinense | Abdomen distention | −2.86 | 381 | 88.71% | 83.73% | 4.58 × 10−02 |
| Sha Ren | Amomum villosum | Poor appetite | −2.77 | 112 | 81.25% | 67.86% | 2.14 × 10−02 |
| Zhe Bei | Fritillariae thunbergii Bulbus | Abdomen distention | −2.76 | 64 | 95.31% | 81.25% | 2.79 × 10−02 |
| Chen Pi | Citrus aurantium | Fatigue | −2.67 | 579 | 87.39% | 83.25% | 4.63 × 10−02 |
| Shan Zha | Crataegus cuneata | Abdominal pain | −2.67 | 53 | 81.13% | 62.26% | 3.11 × 10−02 |
| Zhi Zi | Fructus gardeniae | Poor appetite | −2.48 | 73 | 84.93% | 71.23% | 4.54 × 10−02 |
| Fang Ji | Aristolochia fangchi | Abdomen distention | −2.36 | 85 | 95.29% | 84.71% | 4.08 × 10−02 |
| Yi Zhi Ren | Alpinia oxyphylla | Fatigue | −2.34 | 38 | 92.11% | 68.42% | 2.11 × 10−02 |
| Shan Yao | Dioscorea batatas | Edema | −2.29 | 150 | 80.67% | 70.00% | 3.21 × 10−02 |
| Fang Feng | Saposhnikovia divaricata | Abdomen distention | −2.27 | 67 | 91.04% | 76.12% | 1.97 × 10−02 |
| Zhe Bei | Fritillariae thunbergii Bulbus | Poor appetite | −2.21 | 67 | 86.57% | 71.64% | 3.36 × 10−02 |
| Fang Feng | Saposhnikovia divaricata | Abdominal pain | −2.16 | 30 | 83.33% | 60.00% | 4.49 × 10−02 |
| Chai Hu | Bupleurum chinense | Fatigue | −2.15 | 593 | 88.20% | 82.63% | 6.63 × 10−03 |
| Lian Qiao | Forsythia suspensa | Cough | −1.94 | 56 | 89.29% | 75.00% | 4.84 × 10−02 |
| Xuan Shen | Scrophularia ningpoensis | Poor appetite | −1.8 | 64 | 82.81% | 67.19% | 4.12 × 10−02 |
| Zhi Shi | Citrus aurantium | Fatigue | −1.77 | 399 | 88.47% | 79.95% | 9.64E-04 |
| Hong Hua | Carthamus tinctorius | Poor appetite | −1.73 | 37 | 89.19% | 67.57% | 4.81 × 10−02 |
| Tai Zi Shen | Radix pseudostellariae | Insomnia | −1.72 | 234 | 87.18% | 77.35% | 5.38 × 10−03 |
| Ban Bian Lian | Lobelia chinensis | Fatigue | −1.68 | 86 | 86.05% | 72.09% | 2.45 × 10−02 |
| Yu Jin | Curcuma aromatica | Abdomen distention | −1.59 | 351 | 90.31% | 84.90% | 2.95 × 10−02 |
| Fang Feng | Saposhnikovia divaricata | Insomnia | −1.56 | 55 | 90.91% | 72.73% | 1.34 × 10−02 |
| Hui Xiang | Foeniculi fructus | Fatigue | −1.49 | 31 | 93.55% | 70.97% | 4.61 × 10−02 |
| Hu Ji Sheng | Viscum coloratum | Poor appetite | −1.36 | 45 | 86.67% | 64.44% | 1.42 × 10−02 |
| Chuan Xiong | Chuanxiong rhizoma | Poor appetite | −1.35 | 146 | 80.82% | 68.49% | 1.54 × 10−02 |
| Zhi Mu | Anemarrhena asphodeloides | Poor appetite | −1.32 | 102 | 82.35% | 70.59% | 4.76 × 10−02 |
| Hu Ji Sheng | Viscum coloratum | Insomnia | −0.84 | 34 | 94.12% | 67.65% | 1.36 × 10−02 |
| Hou Po | Magnoliae officinalis Cortex | Fatigue | −0.71 | 369 | 86.45% | 80.22% | 2.31 × 10−02 |
| Zhi Mu | Anemarrhena asphodeloides | Insomnia | −0.69 | 85 | 91.76% | 77.65% | 1.06 × 10−02 |
| Xuan Fu Hua | Flos inulae | Fatigue | −0.68 | 93 | 89.25% | 77.42% | 3.04 × 10−02 |
| Xu Zhang Qing | Cynanchi Paniculati Radix Et Rhizoma | Fatigue | −0.54 | 35 | 94.29% | 71.43% | 2.64 × 10−02 |
| Suan Zao Ren | Ziziphus Jujuba var. spinosa | Poor appetite | −0.44 | 187 | 84.49% | 75.94% | 3.78 × 10−02 |
| Huang Lian | Coptis chinensis | Fatigue | −0.41 | 288 | 88.54% | 80.90% | 1.08 × 10−02 |
| Sheng Ma | Cimicifuga foetida | Fatigue | −0.4 | 65 | 92.31% | 73.85% | 5.00 × 10−03 |
| Niu Xi | Achyranthes aspera | Insomnia | −0.36 | 83 | 84.34% | 71.08% | 4.02 × 10−02 |
| Bai Xian Pi | Dictamni cortex | Poor appetite | −0.33 | 32 | 81.25% | 56.25% | 3.10 × 10−02 |
| Zhi Shi | Citrus aurantium | Yellow skin | −0.21 | 124 | 54.84% | 35.48% | 2.20 × 10−03 |
| Zhe Bei | Fritillariae thunbergii Bulbus | Insomnia | −0.12 | 49 | 93.88% | 77.55% | 4.33 × 10−02 |
| Di Ding | Corydalis bungeana | Fatigue | −0.02 | 78 | 91.03% | 76.92% | 1.64 × 10−02 |
| Jing Jie | Schizonepeta tenuifolia | Insomnia | −0.01 | 38 | 89.47% | 60.53% | 8.07 × 10−03 |
DISCUSSION
In this work, we established a network medicine framework to reveal the scientific principle of TCM herbal treatment, by mapping symptom-associated proteins and herb targets onto the human PPI and analyzing their topological relations. We found that proteins associated with a symptom cluster into localized modules and observed that a short network distance between two symptom modules is predictive of the symptoms’ co-occurrence and their GO semantic similarity. We showed that the network proximity between an herb’s targets and a symptom module is predictive of the herb’s effectiveness in treating that symptom, validated with indication data curated from the CHPH. We then comprehensively validated our framework with patient data, showing that higher RR of symptoms in patients correlates with shorter interactome distance, and herb-symptom proximity predicts herb-symptom treatment effectiveness. Last, we identified herb-symptom pairs that are predicted to be effective based on network proximity and proven effective in the patient data but not yet recognized by the TCM community, highlighting the translational value of our framework in prioritizing effective herb treatment against diseases, which can further lead to drug discovery and repurposing opportunities.
To our knowledge, our framework is the first scientific theory that uncovers the generic mechanistic principle of a traditional medicine system, demonstrating the translation of traditional/empirical practice into modern biomedical knowledge. We are also the first to have studied and validated TCM herb effectiveness on a systematic level, given that previous research is limited to single herbs or single prescriptions. Our network medicine framework opens up a paradigm to study the effectiveness and the molecular basis of natural medicine. In contrast to existing network pharmacology approaches which often assume that herb/drug targets must directly target diseases/symptoms, our whole-interactome approach is more general, as we have observed that herbs/drugs can be effective even if they act on the appropriate network neighborhood (17, 20). We designed multiple pipelines to extract the network-based relation between herbs, chemical targets, and symptoms, overcoming the complexity challenge of herb-chemical-target data. Our approach combining computational network science and patient data offers a powerful cross-disciplinary way to prioritize chemicals/herbs with therapeutic potentials and discover herb treatment predictions against specific diseases.
Our work opens up multiple follow-up directions for future work. For example, prioritizing effective herb-symptom pairs can lead to phenotype-based herb/chemical screening, identifying potential treatments against specific disease phenotypes. Furthermore, our framework might be further investigated to reveal interactome patterns that prioritize therapeutic chemicals from an effective herb’s chemical composition. Next, given that the herb-chemical-target data and symptom-gene association data are still incomplete (e.g., we lack the chemical dose dependence in herb-chemical mapping), improving and refining these data can enhance our framework’s ability to capture herb-disease treatment relations. We also used a dataset focusing on patients with liver cirrhosis, with a subset of the data available for statistical significance in bioinformatic analysis. Extending the size of the patient data could offer better support for our framework. Another future direction is to explore alternative methods/metrics to tackle the diversity/complexity challenge in the herb-chemical-target relations, better capturing herb-symptom relations. Here, we used distance-based metrics to define the network relations; developing more complicated methods may increase the predictive power; or consensus algorithms may help balance the results of multiple prediction pipelines (20). For example, we also tested network embedding-based methods, finding that they can also capture herb-symptom proximity (text S1 and fig. S5). Last, the PPI pattern of TCM prescriptions, or herb combinations, is another under-explored direction. Recent works showed that co-prescribed herbs tend to be close in the protein interactome (57, 58). According to the classic TCM concept, each herb in a prescription has a specific effect, which is often complementary to the effects of other co-prescribed herbs. Our established framework to study herb-symptom relations may improve the understanding of how a combination of herbs works against a given symptom or symptom set.
MATERIALS AND METHODS
Symptom-gene association data
We used symptom data from SymMap, which integrates disease-gene association from DisGeNet (59) and MalaCards (60). The data contain 110,407 associations with 11,362 unique diseases represented by UMLS concept codes and 13,271 unique genes. To obtain high-quality symptom-gene associations, we used the concept of “dual phenotypes” (DP) (61), such as obesity, fever, and insomnia, which are regarded as both diseases and symptoms. Thus, the symptom-gene associations are straightforwardly the corresponding disease-gene associations, for diseases with DP properties. To identify these kinds of phenotype terms (e.g., symptom) from databases, we filtered an integrated DP-genotype association dataset by limiting the semantic types of UMLS concepts as symptoms from the disease-gene associations (62). We obtained 16,049 associations between 490 symptoms with concept unified identifiers code and 4193 genes. To ensure the reliability of the symptom-associated gene data, we focus on the 174 symptoms with at least 20 associated genes and discarded the symptoms with fewer gene associations due to potential incompleteness. The compiled symptom-gene association dataset is provided in data S1.
Human protein interactome
We use the human PPI from our previous work on predicting COVID-treating drugs (20). The PPI is assembled using experimentally validated protein interactions including: (i) binary interactions, derived from high-throughput yeast two-hybrid experiments, three-dimensional protein structures; (ii) interactions identified by affinity purification followed by mass spectrometry; (iii) kinase substrate interactions; (iv) signaling interactions; and (v) regulatory interactions. The final PPI used in our study contains 18,505 proteins and 327,924 interactions between them (data S2).
Herb, chemical, and target data
We used herb data from (i) the recently updated HIT 2.0 database (39), and (ii) the TCMIO database (40), a comprehensive collection of TCMSP, TCMID, and TCM-ID databases (41–43). The HIT database has straightforward herb-target data, so we directly map the herbs and targets to our herb name data and protein interactome. For the other TCM databases, we consider an herb as an assembly of chemicals and use their chemical composition data. The TCM databases focus on chemicals with potential therapeutic effects, identified from experiments (e.g., mass spectrometry) with quality control (63, 64). Then, we obtain the protein targets of each chemical from the STITCH database (44), keeping only targets with experimental evidence. The compiled herb-chemical-target association datasets are provided in data S4. In addition to herb-chemical-target data, we also used an herb-symptom indication dataset from SymMap (45), an expert-curated list of herb-symptom pairs recognized by doctors as effective treatments.
Patient data: Symptom-herb associations
We have collected the EMR data of liver cirrhosis inpatient cases from Hubei Provincial Hospital of Traditional Chinese Medicine in Wuhan, which included the full clinical profiles of patients. TCM clinical named entities, such as symptoms and their trajectory (e.g., symptom recovery) were extracted from the admission and discharge records using text-mining methods based on a clinical information extraction tool (HCPSAS, www.tcmai.org). The resulting dataset contains 1936 inpatients with herb prescription records, which usually consisted of 16 to 18 herbs used in combination for treatment. We considered that if a prescription is given to a patient, then all herbs included in this prescription are associated with all symptoms the patient has. For example, if a patient has five symptoms and is given 16 herbs, then we will include all 5 × 16 = 80 herb-symptom pairs. Yet, not every herb-symptom pair is expected to be effective. Last, we obtained 5106 symptom-herb associations which involve 55 symptoms and 218 herbs (see data S10 for herb ID and name mapping). All admission data of these patients were verified and standardized by trained medical researchers to ensure accurate terminological mappings.
Patient data: Symptom terminology mapping and processing
To connect clinical patient data to symptom gene data, we manually mapped the Chinese terms of symptoms and herbs in patient data to the English terms in symptom-gene associations. This was done by trained medical researchers to ensure accurate terminological mappings. A total of 315 English symptom terms with associated genes are mapped to 92 Chinese symptom terms in patient data. Note that multiple UMLS codes could correspond to one TCM symptom. For example, C0277799 and C0015967 were both mapped to 发热 (fever). Symptom-gene association data for patients’ symptoms are provided in data S6.
Metrics
LCC and LCC z score
We characterize the localization of a node set in the network with the z score of the node set’s largest connected component (LCC) (16). We first compute the size of the LCC formed by the node set, and then compare the observed LCC size against the random expectation generated from simulations preserving the degree of the nodes (17). The LCC z score is the difference between the observed LCC size and the mean of randomization μ(random Lcc), divided by the SD of the randomization σ(random Lcc):
An LCC z score larger than 1.6 indicates the observed LCC is significantly larger than random expectation. An implementation of the code for LCC size and its z-score computation can be found in (16).
Network distance Dab and network separation Sab
We measure the network relation between two node sets (e.g., target modules of herbs A and B) using the network distance Dab and network separation Sab metrics. The network distance Dab, also denoted as ⟨dAB⟩, is the average of network distances between all node pairs in two node sets. The network separation metric Sab was designed to characterize disease-disease relation and drug-drug relation (16, 18):
The network separation metric compares the mean shortest distance within the interactome between the nodes of each node set, ⟨dAA⟩ and ⟨dBB⟩, to the mean shortest distance ⟨dAB⟩ between node sets A and B. In ⟨dAB⟩, targets associated with both herbs A and B have a zero distance by definition. The random expectation of sAB is zero. A negative sAB means the two node sets are located in the same network neighborhood, while a positive sAB means the two node sets are topologically separated.
An implementation of network separation computation in Python can be found in (16).
Symptom semantic similarity
To evaluate the biological similarity between a pair of symptoms, we use semantic similarity (30) to characterize the biological similarity of genes associated with the symptoms. We used the Python package pygosemsim (https://github.com/mojaie/pygosemsim) to compute the GO semantic similarity between a pair of genes. The package allows automatic collection of GO .obo file from http://geneontology.org and annotations from http://geneontology.org/gene-associations/goa_human.gaf.gz. It then computes Lin similarity (65) as semantic similarity, combining all three branches of GO ontology (cellular component, biological process, and molecular function). In addition, we also compute the semantic similarity for each of the three GO ontology branches and show a similar correlation trend (text S1 and fig. S2). For the semantic similarity of two symptom modules, we compute the average GO semantic similarity of all pairs of genes between the two symptoms.
Network proximity distance and z score
Given that T, the set of herb targets, and s, the set of symptom-associated proteins, denote dist(t0, s0) as the shortest path length between nodes t0 ∈ T and s0 ∈ s in the network, we define the network proximity distance metric (referred to as “proximity distance d” in the main text) as the average distance over targets to their closest symptom-associated protein (17):
Then, we convert this absolute distance d to a relative proximity z score, by simulating the random expectation of distances between two randomly selected groups of proteins, matching the size and degrees of the original S and T sets. To avoid repeatedly selecting the same high-degree nodes, we use degree-binning (17). Denote the mean of the reference random expectation of distances as μrand(T, s) and the SD as σrand(T, s), we define the network proximity z score as
The proximity z score measures how the proximity distance differs from random expectation, with z = 0 being neutral, z < 0 being more proximal than random, and z > 0 being more distant from random. For both proximity distance d and proximity z score, the lower the metric value, the closer the two node sets are on the network. Note the proximity z score is a stochastic measure because of the randomized simulation. In other words, identical repeated computations do not yield identical z scores. An implementation of network proximity metrics computation can be found in either (17) or (20).
Herb-target mapping methods to obtain herb-symptom distance for each herb-symptom pair
We used four herb-target mapping methods to obtain herb-symptom distance from (a) HIT direct target data and (b to d) herb-chemical-target data:
(a) HIT data directly associate targets to each herb, so we compute the two proximity measures straightforwardly as an herb-symptom metric.
In contrast, the TCM herb-chemical-target dataset does not have direct herb-target associations, so no direct herb-symptom relation metric can be applied. Therefore, we design three herb-target mapping methods (b to d) to obtain herb-symptom network metrics:
(b) Target union: We define an herb’s target set as the union of the targets of all composing chemicals of the herb, and then we interpret herb target-symptom proximity measures as herb-symptom relation metrics.
In (c) and (d), we define a second-order herb-symptom distance from first-order chemical-symptom distances: first, for every chemical-symptom pair, we compute chemical-symptom proximity metrics using the chemical’s targets; then, we define the second-order herb-symptom distance as follows:
(c) The average of all chemical-symptom distances from the herb’s composing chemicals, i.e., assuming the effect of an herb is the average effect of its composing chemicals.
(d) The smallest of all chemical-symptom distances from the herb’s composing chemicals, i.e., assuming the effect of an herb is dominated by the chemical most proximal to a symptom.
We give the mathematical formulae of the metrics below.
Notations: dhs, herb-symptom distance; h, herb; s, symptom-associated proteins; ci, the ith composing chemical of an herb; T, targets of an herb or chemical, proximity(T, s),proximity measures calculated from (T, s), i.e., proximity d(T, s) or z(T, s).
In the cases where herb targets are directly associated, i.e., (a) the HIT database or (b) target union, the herb-symptom distance metric is, straightforwardly, the proximity metric(s) between herb targets and symptom proteins as dhs = proximity(Th, s). In (a), HIT data Th is given directly; in (b), the targets of an herb are defined as the union of targets from all the herb’s composing chemicals: Th = ⋃ci∈h(Tci).
When herb targets are not directly available in (c) and (d), we define second-order herb-symptom distance metrics from first-order chemical-symptom distances. The first-order chemical-symptom distance is the proximity distance or z score for a chemical ci and a symptom s, using targets of this chemical, denoted as dcis = proximity(TCi, s). Then, based on this first-order distance, we define the second-order herb-symptom distance (b) as the (c) average or (d) minimum of all first-order distances:
(c) Average: , with Nc being the total number of chemicals in this herb.
(d) Minimum:.
Together, these four herb-target mapping methods crossing the two proximity measures yield eight herb-symptom proximity pipelines.
Patient data: Binary effective metric for TCM effectiveness in hospital patient dataset
To evaluate the effectiveness of TCM in the patient dataset, we define a binary (i.e., true or false) herb effectiveness measure by comparing patients’ symptom recovery rate after herb treatment versus the symptom’s baseline recovery rate. Specifically, we define for each symptom a baseline “recovery percentage,” meaning the percentage of patients recovered from the symptom by the time they leave the hospital. Then, we define for each herb-symptom pair a “recovery percentage after treatment”, meaning the percentage of patients with a given symptom and treated with a specific herb that has recovered from the symptom. An herb-symptom pair is considered “effective,” if patients’ recovery percentage after treatment is higher than the symptom’s (baseline) recovery percentage without TCM treatment; otherwise, the herb-symptom pair is considered ineffective.
Using the above definition, we obtain 986 effective herb-symptom pairs with a frequency of at least 10 (i.e., at least 10 patients with the symptom and treated with the herb), and with all eight pipeline proximity scores available. As the herb-symptom pairs used in the patient dataset are already more proximal than random (Fig. 4B), the patient dataset overrepresents positive/effective herbs. For this reason, we cannot compare the effective pairs against other pairs within the patient dataset, as they are both positive samples. Instead, in Fig. 4C, we compare the effective pairs against all herb-symptom pairs. The same comparison is used in the PSM-matched herb-symptom pairs in Fig. 4D.
Patient data: Propensity score matching
We used PSM in the patient dataset to remove the biases of patient basic information on herb treatment outcomes. PSM is a statistical matching technique that attempts to estimate the effect of a treatment, policy, or other intervention by accounting for the covariates that predict receiving the treatment (66). For a designated herb-symptom pair, we matched the patients with the herb-symptom pair against other herbs treating the same symptom, to evaluate the effectiveness of the herb-symptom pair. For example, for the Baizhu-fatigue pair, the fatigue patients who received Baizhu therapy at any point during hospitalization were defined as the case group for the Baizhu-fatigue pair. Fatigue patients who did not receive Baizhu treatment from the control group. We adjusted for baseline characteristics (e.g., age and sex) and high-incidence comorbidity characteristics of patients in the two groups. The most common comorbidities selected to control include esophageal and gastric varices, abdominal effusion, hypoproteinemia, hypertension, and diabetes. In the PSM analysis, the nearest-neighbor method was applied to create a 1:1 matched control sample. Mann-Whitney U test (two-tailed) and chi-square test (one-tailed) were used to compare the differences of variables between the two groups to ensure that there was no statistical difference in these variables between the groups after matching.
Of the total 888 herb-symptom pairs, approximately 50% (436 pairs) exhibited higher clinical effectiveness compared to the control groups. Among these pairs, we identified 86 herb-symptom pairs where the case group had a significantly higher symptom recovery rate than the control group (P < 0.05, chi-square test). The other 350 herb-symptom pairs are effective but did not show a statistically significant difference (P > 0.05). This is likely attributed to the limited sample sizes, as ~65% of the herb-symptom pairs consist of sample sizes below 100 patients.
Statistics
We used standard statistics including mean ± SD, standard score , Mann-Whitney U test (one-tailed) for proximity pipeline comparisons, chi-squared test (one-tailed), and Mann-Whitney U test (two-tailed) for PSM result.
Acknowledgments
We thank M. Sebek, J. Cheng, I. do Valle, D. M. Gysi, R. Hua, N. Wang, and S. Matsuoka (RIKEN CSRS) for helpful discussions, as well as D. Koshkina for help in designing the figures.
Funding: This work is supported by funding from the A.-L.B. laboratory, the X.Z. laboratory, and the R.A. laboratory. A.-L.B. is supported by NIH grant 1P01HL132825, American Heart Association grant 151708, and European Research Council grant 810115-DYNASET. X.Z. is supported by the National Natural Science Foundation of China (82174533) and the Natural Science Foundation of Beijing (M21012). R.A. is supported by NSF MCB 1715826. X.G. is supported by NUIST faculty startup fund 1523142301052.
Author contributions: X.G. designed the research, with inputs from Z.S., X.Z., and A.-L.B. X.G., Z.S., X.W., D.Y., J.L., X.L., and B.L. collected and processed the data. X.G., Z.S., and X.W. analyzed the data. X.G., Z.S., R.A., X.Z., and A.-L.B. wrote the paper, with inputs from all authors.
Competing interests: A.-L.B. is a cofounder of Scipher Medicine Inc., which applies network medicine strategies to biomarker development and personalized drug selection, and of Naring Inc., which applies data science to health. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. We used existing code for computation, for which we provided references or links at each corresponding method subsection.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S5
Legends for data S1 to S15
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S15
REFERENCES AND NOTES
- 1.E. Raviña Rubira, The Evolution of Drug Discovery: From Traditional Medicines to Modern Drugs (Wiley-VCH, 2011). [Google Scholar]
- 2.X.-Z. Su, L. H. Miller, The discovery of artemisinin and the Nobel Prize in Physiology or Medicine. Sci. China Life Sci. 58, 1175–1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L. Li, H. Yao, J. Wang, Y. Li, Q. Wang, The role of chinese medicine in health maintenance and disease prevention: Application of constitution theory. Am. J. Chin. Med. 47, 495–506 (2019). [DOI] [PubMed] [Google Scholar]
- 4.X. Zhou, Y. Li, Y. Peng, J. Hu, R. Zhang, L. He, Y. Wang, L. Jiang, S. Yan, P. Li, Q. Xie, B. Liu, Clinical phenotype network: The underlying mechanism for personalized diagnosis and treatment of traditional Chinese medicine. Front. Med. 8, 337–346 (2014). [DOI] [PubMed] [Google Scholar]
- 5.S. Jafari, M. Abdollahi, S. Saeidnia, Personalized medicine: A confluence of traditional and contemporary medicine. Altern. Ther. Health Med. 20, 31–40 (2014). [PubMed] [Google Scholar]
- 6.L. Wang, G.-B. Zhou, P. Liu, J.-H. Song, Y. Liang, X.-J. Yan, F. Xu, B.-S. Wang, J.-H. Mao, Z.-X. Shen, S.-J. Chen, Z. Chen, Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 105, 4826–4831 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Yao, X. Zhang, Z. Wang, C. Zheng, P. Li, C. Huang, W. Tao, W. Xiao, Y. Wang, L. Huang, L. Yang, Deciphering the combination principles of traditional Chinese medicine from a systems pharmacology perspective based on Ma-huang Decoction. J. Ethnopharmacol. 150, 619–638 (2013). [DOI] [PubMed] [Google Scholar]
- 8.S. Li, B. Zhang, Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 11, 110–120 (2013). [DOI] [PubMed] [Google Scholar]
- 9.A. L. Barabási, N. Gulbahce, J. Loscalzo, Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. A. Yildirim, K. I. Goh, M. E. Cusick, A. L. Barabási, M. Vidal, Drug-target network. Nat. Biotechnol. 25, 1119–1126 (2007). [DOI] [PubMed] [Google Scholar]
- 11.A. L. Hopkins, Network pharmacology. Nat. Biotechnol. 25, 1110–1111 (2007). [DOI] [PubMed] [Google Scholar]
- 12.A. L. Barabási, Network medicine — from obesity to the “diseasome”. N. Engl. J. Med. 357, 404–407 (2007). [DOI] [PubMed] [Google Scholar]
- 13.S. Li, T.-P. Fan, W. Jia, A. Lu, W. Zhang, Network pharmacology in traditional Chinese medicine. Evid. Based Complement. Alternat. Med. 2014, 138460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Z. Zhou, B. Chen, S. Chen, M. Lin, Y. Chen, S. Jin, W. Chen, Y. Zhang, Applications of network pharmacology in traditional Chinese medicine research. Evid. Based Complement. Alternat. Med. 2020, 1646905 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.X. Lai, X. Wang, Y. Hu, S. Su, W. Li, S. Li, Editorial: Network pharmacology and traditional medicine. Front. Pharmacol. 11, 1194–1194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. Menche, A. Sharma, M. Kitsak, S. D. Ghiassian, M. Vidal, J. Loscalzo, A.-L. Barabási, Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.E. Guney, J. Menche, M. Vidal, A.-L. L. Barábasi, Network-based in silico drug efficacy screening. Nat. Commun. 7, 10331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.F. Cheng, I. A. Kovács, A. L. Barabási, Network-based prediction of drug combinations. Nat. Commun. 10, 1197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.F. Cheng, R. J. Desai, D. E. Handy, R. Wang, S. Schneeweiss, A. L. Barabási, J. Loscalzo, Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 9, 2691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. M. Gysi, Í. Do Valle, M. Zitnik, A. Ameli, X. Gan, O. Varol, S. D. Ghiassian, J. J. Patten, R. A. Davey, J. Loscalzo, A. L. Barabási, Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. U.S.A. 118, e2025581118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.T. Mellors, J. B. Withers, A. Ameli, A. Jones, M. Wang, L. Zhang, H. N. Sanchez, M. Santolini, I. Do Valle, M. Sebek, F. Cheng, D. A. Pappas, J. M. Kremer, J. R. Curtis, K. J. Johnson, A. Saleh, S. D. Ghiassian, V. R. Akmaev, Clinical validation of a blood-based predictive test for stratification of response to tumor necrosis factor inhibitor therapies in rheumatoid arthritis patients. Netw. Syst. Med. 3, 91–104 (2020). [Google Scholar]
- 22.H. Cui, S. Srinivasan, D. Korkin, Enriching human interactome with functional mutations to detect high-impact network modules underlying complex diseases. Genes 10, 933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Choobdar, M. E. Ahsen, J. Crawford, M. Tomasoni, T. Fang, D. Lamparter, J. Lin, B. Hescott, X. Hu, J. Mercer, T. Natoli, R. Narayan; DREAM Module Identification Challenge Consortium, A. Subramanian, J. D. Zhang, G. Stolovitzky, Z. Kutalik, K. Lage, D. K. Slonim, J. Saez-Rodriguez, L. J. Cowen, S. Bergmann, D. Marbach, Assessment of network module identification across complex diseases. Nat. Methods 16, 843–852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. Cummings, The role of neuropsychiatric symptoms in research diagnostic criteria for neurodegenerative diseases. Am. J. Geriatr. Psychiatry 29, 375–383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. H. Siddiqi, S. F. Taylor, D. Cooke, A. Pascual-Leone, M. S. George, M. D. Fox, Distinct symptom-specific treatment targets for circuit-based neuromodulation. Am. J. Psychiatry 177, 435–446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.X. Zhou, L. Lei, J. Liu, A. Halu, Y. Zhang, B. Li, Z. Guo, G. Liu, C. Sun, J. Loscalzo, A. Sharma, Z. Wang, A systems approach to refine disease taxonomy by integrating phenotypic and molecular networks. EBioMedicine 31, 79–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. A. Aronowitz, When do symptoms become a disease? Ann. Intern. Med. 134, 803–808 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Z. Shu, J. Wang, H. Sun, N. Xu, C. Lu, R. Zhang, X. Li, B. Liu, X. Zhou, Diversity and molecular network patterns of symptom phenotypes. NPJ Syst. Biol. Appl. 7, 41 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.X. Zhou, J. Menche, A. L. Barabási, A. Sharma, Human symptoms-disease network. Nat. Commun. 5, 4212 (2014). [DOI] [PubMed] [Google Scholar]
- 30.G. Yu, F. Li, Y. Qin, X. Bo, Y. Wu, S. Wang, GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010). [DOI] [PubMed] [Google Scholar]
- 31.L. R. Schiller, D. S. Pardi, J. H. Sellin, Chronic diarrhea: Diagnosis and management. Clin. Gastroenterol. Hepatol. 15, 182–193.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 32.F. D’Amico, D. C. Baumgart, S. Danese, L. Peyrin-Biroulet, Diarrhea during COVID-19 infection: Pathogenesis, epidemiology, prevention, and management. Clin. Gastroenterol. Hepatol. 18, 1663–1672 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Kuparinen, S. Marttila, J. Jylhävä, L. Tserel, P. Peterson, M. Jylhä, A. Hervonen, M. Hurme, Cytomegalovirus (CMV)-dependent and -independent changes in the aging of the human immune system: a transcriptomic analysis. Exp. Gerontol. 48, 305–312 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Y. Shao, J. Saredy, K. Xu, Y. Sun, F. Saaoud, C. Drummer, Y. Lu, J. J. Luo, J. Lopez-Pastrana, E. T. Choi, X. Jiang, H. Wang, X. Yang, Endothelial immunity trained by coronavirus infections, DAMP stimulations and regulated by anti-oxidant NRF2 may contribute to inflammations, myelopoiesis, COVID-19 cytokine storms and thromboembolism. Front. Immunol. 12, 653110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. Seago, L. Hilton, E. Reid, V. Doceul, J. Jeyatheesan, K. Moganeradj, J. McCauley, B. Charleston, S. Goodbourn, The Npro product of classical swine fever virus and bovine viral diarrhea virus uses a conserved mechanism to target interferon regulatory factor-3. J. Gen. Virol. 88, 3002–3006 (2007). [DOI] [PubMed] [Google Scholar]
- 36.M. Mahapatro, L. Erkert, C. Becker, Cytokine-mediated crosstalk between immune cells and epithelial cells in the gut. Cell 10, 111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.I. F. do Valle, H. G. Roweth, M. W. Malloy, S. Moco, D. Barron, E. Battinelli, J. Loscalzo, A.-L. Barabási, Network medicine framework shows that proximity of polyphenol targets and disease proteins predicts therapeutic effects of polyphenols. Nat. Food 2, 143–155 (2021). [DOI] [PubMed] [Google Scholar]
- 38.A.-L. Barabási, G. Menichetti, J. Loscalzo, The unmapped chemical complexity of our diet. Nat. Food. 1, 33–37 (2020). [Google Scholar]
- 39.D. Yan, G. Zheng, C. Wang, Z. Chen, T. Mao, J. Gao, Y. Yan, X. Chen, X. Ji, J. Yu, S. Mo, H. Wen, W. Han, M. Zhou, Y. Wang, J. Wang, K. Tang, Z. Cao, HIT 2.0: An enhanced platform for herbal ingredients’ targets. Nucleic Acids Res. 50, D1238–D1243 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Z. Liu, C. Cai, J. Du, B. Liu, L. Cui, X. Fan, Q. Wu, J. Fang, L. Xie, TCMIO: A comprehensive database of traditional chinese medicine on immuno-oncology. Front. Pharmacol. 11, 439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Ru, P. Li, J. Wang, W. Zhou, B. Li, C. Huang, Z. Guo, W. Tao, Y. Yang, X. Xu, Y. Li, Y. Wang, L. Yang, TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Chem. 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. Huang, D. Xie, Y. Yu, H. Liu, Y. Shi, T. Shi, C. Wen, TCMID 2.0: A comprehensive resource for TCM. Nucleic Acids Res. 46, D1117–D1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.X. Chen, H. Zhou, Y. B. Liu, J. F. Wang, H. Li, C. Y. Ung, L. Y. Han, Z. W. Cao, Y. Z. Chen, Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 149, 1092–1103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. Szklarczyk, A. Santos, C. von Mering, L. J. Jensen, P. Bork, M. Kuhn, STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–D384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Y. Wu, F. Zhang, K. Yang, S. Fang, D. Bu, H. Li, L. Sun, H. Hu, K. Gao, W. Wang, X. Zhou, Y. Zhao, J. Chen, SymMap: An integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 47, D1110–D1117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. Dong, X. Zhou, J. Ma, H. Zhou, X. Fu, Chemical constituents of Stellaria dichotoma var. lanceolata and their anti-inflammatory effect on lipopolysaccharide-stimulated RAW 264.7 cells. Chem. Nat. Compd. 57, 158–162 (2021). [Google Scholar]
- 47.S. J. Bae, J. W. Choi, B. J. Park, J. Lee, E. K. Jo, Y. H. Lee, S. B. Kim, J. M. Yuk, Protective effects of a traditional herbal extract from Stellaria dichotoma var. lanceolata against Mycobacterium abscessus infections. PLOS ONE 13, e0207696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J. N. Oliwa, J. M. Karumbi, B. J. Marais, S. A. Madhi, S. M. Graham, Tuberculosis as a cause or comorbidity of childhood pneumonia in tuberculosis-endemic areas: A systematic review. Lancet Respir. Med. 3, 235–243 (2015). [DOI] [PubMed] [Google Scholar]
- 49.M. Chu, R. Ding, Z. Y. Chu, M. B. Zhang, X. Y. Liu, S. H. Xie, Y. J. Zhai, Y. D. Wang, Role of berberine in anti-bacterial as a high-affinity LPS antagonist binding to TLR4/MD-2 receptor. BMC Complement. Altern. Med. 14, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Y. T. Chan, N. Wang, Y. Feng, The toxicology and detoxification of Aconitum: Traditional and modern views. Chin Med. 16, 1–4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Q. Zou, K. Yang, Z. Shu, K. Chang, Q. Zheng, Y. Zheng, K. Lu, N. Xu, H. Tian, X. Li, Y. Yang, Y. Zhou, H. Yu, X. Zhang, J. Xia, Q. Zhu, J. Poon, S. Poon, R. Zhang, X. Zhou, Phenonizer: A fine-grained phenotypic named entity recognizer for chinese clinical texts. Biomed. Res. Int. 2022, 3524090 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.B. Zhu, Q. L. Zhang, J. W. Hua, W. L. Cheng, L. P. Qin, The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review. J. Ethnopharmacol. 226, 143–167 (2018). [DOI] [PubMed] [Google Scholar]
- 53.C. The Metabolomics Innovation (2021); www.foodb.ca.
- 54.F. Yang, X. Dong, X. Yin, W. Wang, L. You, J. Ni, Radix Bupleuri: A review of traditional uses, botany, phytochemistry, pharmacology, and toxicology. Biomed. Res. Int. 2017, 7597596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Gu, L. Li, H. Huang, B. Wang, T. Zhang, Antitumor, antiviral, and anti-inflammatory efficacy of essential oils from Atractylodes macrocephala Koidz. Produced with different processing methods. Molecules 24, 2956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. Chen, J. Zhong, C. J. Zhang, Y. Wang, A. G. He, S. T. Xia, First report of Fusarium fujikuroi causing black rot of Bletilla striata (Baiji) in China. Plant Dis. 103, 377 (2018). [Google Scholar]
- 57.Y. Wang, H. Yang, L. Chen, M. Jafari, J. Tang, Network-based modeling of herb combinations in traditional Chinese medicine. Brief. Bioinform. 22, bbab106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.N. Wang, N. Du, Y. Peng, K. Yang, Z. Shu, K. Chang, D. Wu, J. Yu, C. Jia, Y. Zhou, X. Li, B. Liu, Z. Gao, R. Zhang, X. Zhou, Network patterns of herbal combinations in traditional chinese clinical prescriptions. Front. Pharmacol. 11, 590824 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Piñero, J. M. Ramírez-Anguita, J. Saüch-Pitarch, F. Ronzano, E. Centeno, F. Sanz, L. I. Furlong, The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845–D855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.N. Rappaport, M. Twik, I. Plaschkes, R. Nudel, T. Iny Stein, J. Levitt, M. Gershoni, C. P. Morrey, M. Safran, D. Lancet, MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 45, D877–D887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.K. Yang, N. Wang, G. Liu, R. Wang, J. Yu, R. Zhang, J. Chen, X. Zhou, Heterogeneous network embedding for identifying symptom candidate genes. J. Am. Med. Inform. Assoc. 25, 1452–1459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O. Bodenreider, The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 32, 267D–2270D (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.H. Gao, Z. Wang, Y. Li, Z. Qian, Overview of the quality standard research of traditional Chinese medicine. Front. Med. 5, 195–202 (2011). [DOI] [PubMed] [Google Scholar]
- 64.M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. M. Lean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. Boya P, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O’Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. Van Leer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. O. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein, N. Bandeira, Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D. Lin, An Information-Theoretic Definition of Similarity (ICML, 1998).
- 66.P. R. Rosenbaum, D. B. Rubin, The central role of the propensity score in observational studies for causal effects. Biometrika 70, 41–55 (1983). [Google Scholar]
- 67.B. Perozzi, R. Al-Rfou, S. Skiena, DeepWalk: Online learning of social representations, in Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY (ACM, 24 August 2014), pp. 701–710. [Google Scholar]
- 68.A. Grover, J. Leskovec, node2vec: scalable feature learning for networks, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA (ACM, 13 August 2016), pp. 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. Tang, M. Qu, M. Wang, M. Zhang, J. Yan, Q. Mei, LINE: large-scale information network embedding, in Proceedings of the 24th International Conference on World Wide Web, Florence, Italy (ACM, 18 May 2015), 1067–1077. [Google Scholar]
- 70.D. Wang, P. Cui, W. Zhu, Structural deep network embedding, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA (ACM, 13 August 2016), pp. 1225–1234. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S5
Legends for data S1 to S15
References
Data S1 to S15