Abstract
To construct a bacterial catalyst for bioconversion of toluene and several alkyl and chloro- and nitro-substituted derivatives into the corresponding benzoates, the upper TOL operon of plasmid pWW0 of Pseudomonas putida was fully reassembled as a single gene cassette along with its cognate regulatory gene, xylR. The corresponding DNA segment was then targeted to the chromosome of a P. putida strain by using a genetic technique that allows deletion of all recombinant tags inherited from previous cloning steps and leaves the otherwise natural strain bearing exclusively the DNA segment encoding the phenotype of interest. The resulting strains grew on toluene as the only carbon source through a two-step process: conversion of toluene into benzoate, mediated by the upper TOL enzymes, and further metabolism of benzoate through the housekeeping ortho-ring cleavage pathway of the catechol intermediate.
Many Pseudomonas strains are able to employ as their sole carbon source a variety of unusual chemicals, including a wide range or aromatic hydrocarbons and their derivatives (25, 30). The catabolic pathways involved (often encoded by plasmids) are composed of distinct steps, each of which can be split from the others with the tools of recombinant DNA and expressed independently (28). Not infrequently, the enzymes involved in one or more steps in a catabolic pathway have activity on structural analogs of the natural substrates and can thereby give rise to derivatives of interest for industry (29). This is the case for certain enzymes included in the catabolic pathway for toluene, m-xylene, p-xylene, and ethyl benzene, encoded by the TOL plasmid pWW0 of Pseudomonas putida mt-2 (14, 21). Degradation of toluene through the TOL pathway has two major steps, namely, bioconversion of this substrate into benzoate and subsequent catabolism of its aromatic ring down to Krebs cycle metabolites via a catechol intermediate that is cleaved in meta by one of the pathway enzymes. The enzymes that determine conversion of toluene to benzoate are encoded by the so-called upper pathway (Fig. 1), and they happen to have activity not only on the hydrocarbon but also on a variety of chloro and nitro derivatives (1–3). Therefore, the expression of the collective enzymes of the upper pathway away from the remaining TOL genes is predicted to yield a collection of nitro- and halo-substituted benzoates. However, the development of a bacterial catalyst for these biotransformations has been impossible because of the difficulty in reassembling the entire upper TOL operon along with its cognate regulatory gene (xylR; 21), away from the meta-cleavage activity.
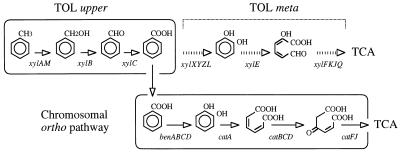
FIG. 1.
Biodegradation of toluene by P. putida through alternative meta or ortho lower pathways. P. putida strains carrying the TOL plasmid pWW0 perform sequential oxidation of the CH3 group of toluene to alcohol, aldehyde, and acid through the action of the enzymes xylene monooxygenase (encoded by xylAM), benzyl alcohol dehydrogenase (encoded by xylB), and benzaldehyde dehydrogenase (encoded by xylC), respectively (14, 21). The resulting benzoate is then channeled into a lower route involving meta-ring cleavage of the catechol intermediate through the action of the products of the TOL genes xylXYZL, xylE, and xylFKJQ, all the way down to Krebs cycle intermediates (TCA, tricarboxylic acid cycle). In the absence of the TOL meta pathway, benzoate can be channeled through an alternative, chromosomally encoded route (involving the ben genes) for formation of catechol (30). This is then subjected to ortho-ring cleavage by the product of the catA gene (catechol 1,2-dioxygenase) and further converted to metabolic intermediates. The meta-cleavage pathway is generally incompatible with the degradation of chloroaromatics (2, 30).
A more general problem of using recombinant strains in biotransformations or bioremediation of aromatic hydrocarbons is the instability of the cloned genes (frequently borne by plasmids) and the inheritance of marker genes (antibiotic or not antibiotic) used for selection (4, 5). The presence of antibiotic resistance genes in strains destined for environmental release is particularly undesirable under U.S. and European Union regulations (5). While the problem of stability has been alleviated by the use since 1990 of Tn5-derived transposon vectors for stable chromosomal insertion of cloned genes (6, 7, 16), the resulting strains still inherit DNA segments unrelated to the desired phenotype (i.e., those of the selection markers included in the transposon vector). As a consequence, the resulting biocatalysts are still quite different from those which could arise during natural evolution, by processes involving exclusively shuffling of DNA segments through transposition (27, 30) or DNA slippage (15).
In this work, we have employed transposon vectors with excisable selection markers (18) for insertion of the reconstructed and fully functional upper TOL pathway into the chromosome of P. putida KT2442 (7). The resulting strains convert toluene into benzoate and further metabolize this compound through the housekeeping ortho-ring cleavage pathway of the catechol intermediate. Due to the loss of recombinant markers during the construction process, these genetically engineered strains differ minimally from their nonrecombinant counterparts.
Reconstruction of the upper TOL operon as a single catabolic segment.
A gene cassette bearing the complete upper operon along with its native toluene-responsive regulator gene, xylR, was constructed by using the strategy shown in Fig. 2. The origin of the upper TOL sequences was plasmids pED3306 and pRL4, which span overlapping segments of the operon (Fig. 2). pED3306 (22) consists of pBR322 containing a 10-kb HindIII fragment (D fragment) with xylUWCMA (31) and the adjacent upstream region. pRL4 (kindly provided by S. Harayama, Kamaishi City, Japan) contains a 7.1-kb BamHI fragment generated upon insertion of Tn5 into the TOL plasmid downstream of the upper operon. This fragment includes the xylMABN sequences along with an unrelated segment of Tn5. During early attempts to splice different portions of the upper operon, it was noticed that subcloning of a 5.8-kb XbaI-ClaI segment from pRL4 spanning xylMABN was impossible in a multicopy plasmid. To overcome the problem, we used a monocopy plasmid vector with an Ω element (9) in front of the cloned DNA to prevent any expression of the inserted sequences. The specialized vector pCK02 is a derivative of the low-copy-number, chloramphenicol-resistant vector pCK01 (10), in which a 2.0-kb Ω-EcoRI element from pHP45Ω (9) has been inserted into the EcoRI site to stop any transcription arising from the lac promoter present in the plasmid. pCK02 was cleaved with XbaI and AccI and ligated to the 5.8-kb XbaI-ClaI fragment of pRL4 containing xylMABN. The resulting plasmid (pCK03; Fig. 2) was then cleaved with XmaI and XbaI (thereby releasing the Ω element) to receive a 3.3-kb XmaI-XbaI fragment of pED3306 containing the Pu promoter and the downstream genes xylUWC (31). This reconstructed the entire upper TOL pathway downstream of its native promoter, Pu, in plasmid pCK04. Two additional restriction sites, KpnI and AvrII, were then created at the 5′ end of the operon sequence by insertion of a linker at the single XmaI site of the plasmid, thus producing plasmid pCK04AK. In addition, a 2.4-kb DNA segment spanning the regulatory gene xylR was prepared as a KpnI fragment by adding KpnI linkers to the ends of the HpaI fragment released upon digestion of plasmid pTK19 (11) with this enzyme. The KpnI segment was then inserted at the single KpnI site of pCK04AK, giving rise to pCK04AxylR. This plasmid carries a ca. 12-kb NotI insert (named hereafter the upp TOL catabolic segment; Fig. 2) with the entire TOL upper pathway, along with its cognate regulatory gene, xylR.
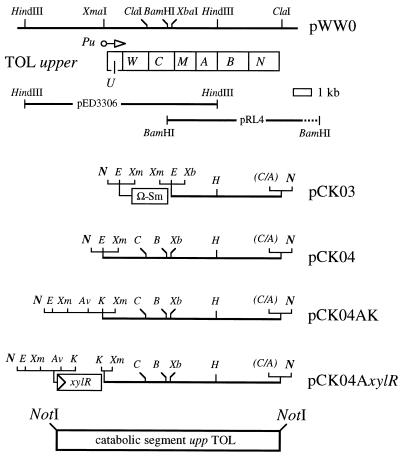
FIG. 2.
Reconstruction of the upper TOL gene cluster as a single catabolic segment. The organization of the xyl genes of the upper TOL operon is shown at the top, aligned with the restriction map of the portion of the pWW0 plasmid involved (14, 31). Genes xylUW and xylN are not required for degradation of toluene, but they form part of the same transcriptional unit. The location and orientation of the toluene-responsive promoter of the system, Pu, are also indicated. Pu becomes activated by the XylR protein (which maps at a distant position in the pWW0 plasmid; 21) when cells encounter toluene or xylenes in the medium (1, 21). The DNA inserts of plasmids pED3306 and pRL4 used as the source of the xyl genes are indicated. The organization of the constructions that preceded the reassembly of the operon along with the xylR gene in pCK04AxylR is shown (see the text for an explanation). The various segments were cloned between the NotI sites of vector pCK01. This is a Cmr plasmid with a pSC101 replicon (i.e., monocopy) and an α-lac fragment in which the polylinker of pUC18 is flanked by NotI sites. This allows the excision of the cloned DNA segments as NotI fragments for further insertion in the single NotI site of the transposon vector of pJMS11 (see Fig. 3). Restriction sites: N, NotI; E, EcoRI; Xm, XmaI; Xb, XbaI; H, HindIII; C, ClaI; A, AccI; Av, AvrII.
The functionality of every enzymatic step of the pathway was verified in vivo as follows. Presentation of indole to Escherichia coli cells bearing pCK04AxylR (Fig. 2) in the presence of 3-methylbenzyl alcohol resulted in accumulation of a dark-blue precipitate (indigo), which revealed the activity of the enzyme xylene monooxygenase (22), which is encoded by xylMA (Fig. 1). When the same strain was exposed to vapors of 3-methylbenzyl alcohol, we observed the transient formation of a product which migrated the same as 3-methylbenzaldehyde in a high-performance liquid chromatography system (data not shown), thus indicating the functionality of the second enzyme of the pathway, benzyl alcohol dehydrogenase (encoded by xylB). Finally, pCK04AxylR was cotransformed along with the compatible xylS+ plasmid pKT570 (20, 21) in the specialized strain E. coli CC118Pm-lacZ, which bears a chromosomal lacZ fusion to the m-toluate/XylS-responsive promoter Pm (17). In the presence of 3-methylbenzyl alcohol, this strain accumulated high levels of β-galactosidase, i.e., ≥5,000 Miller units (23), which is equivalent to 15-fold induction. This was evidence of the conversion of the aromatic substrate into m-toluate by the enzyme benzaldehyde dehydrogenase (encoded by xylC) because m-toluate activates XylS, which in turn activates the chromosomal Pm-lacZ fusion of the host E. coli strain (21). These observations indicated that the 12-kb upp TOL catabolic segment contains all of the activities of the TOL upper pathway required for the bioconversions of interest (31). Although the products of the xylUW (31) and xylN (14) genes do not have a known role in the degradation of toluene, they were kept in their original configuration within the upper TOL operon to ensure reliable performance of the system.
Insertion of the hybrid mini-Tn5 [upp TOL] transposon into P. putida and excision of the selection marker.
Cloning of the NotI insert of pCK04AxylR spanning the upper TOL operon plus xylR (Fig. 2) into the unique NotI site of pJMS11 (Fig. 3) gave rise to plasmid pCK05, the organization of which is sketched in Fig. 4. The minitransposon assembled in pJMS11 contains a DNA segment encoding a selection marker (npt, which encodes Kmr) and a visual marker (xylE, encoding catechol 2,3-dioxygenase). The latter causes colonies to become yellow upon spraying with catechol (C23O+ phenotype). This xylE/npt marker segment is flanked by two tandem res sequences recruited from the multimer resolution system (mrs) of plasmid RP4 (8, 12, 13, 18). The mrs system is a site-specific recombination mechanism involving parA, a site-specific resolvase that promotes the deletion of any supercoiled DNA placed between two directly oriented res sites. The result of this process is excision of the intervening nucleotide sequence when cells express the cognate resolvase encoded by gene parA (8, 18).
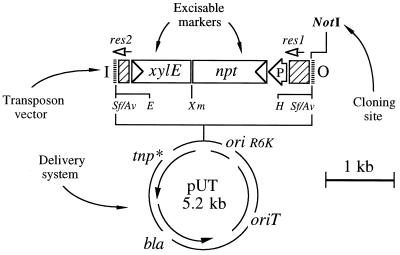
FIG. 3.
Organization of pJMS11. This plasmid is the delivery vector for the minitransposon shown at the top and was constructed by deleting the NotI insert of pJMS10 (18) and further religation to recover a unique NotI cloning site. The suicide donation system (bottom) is that of pUT (7) and includes the Tn5 transposase gene devoid of NotI sites (tnp∗), and Apr selection marker (bla), an origin of transfer for RP4-mediated mobilization (oriT), and the origin of replication of plasmid R6K, which is dependent on the π protein encoded by the pir gene carried by specialized λpir E. coli hosts. At the top are the elements included in the mini-Tn5 transposon vector portion of the plasmid. Besides the single NotI site used for cloning of heterologous DNA segments, the predominant feature of this plasmid is the presence of a DNA segment carrying the xylE (C23O) and npt (Kmr) genes flanked by tandem res sites. The arrow-shaped box in front of the npt gene indicates the orientation of its promoter (P). Similarly, the triangles in front of the xylE and npt boxes indicate the orientation of their structural genes. The res sequences are represented by the shaded boxes. Cloning of a DNA segment at the single NotI site of the plasmid gives rise to a hybrid mobile element flanked by the 19-bp I and O ends of Tn5. Restriction sites: E, EcoRI; Xm, XmaI; H, HindIII; Sf, SfiI; Av, AvrII.
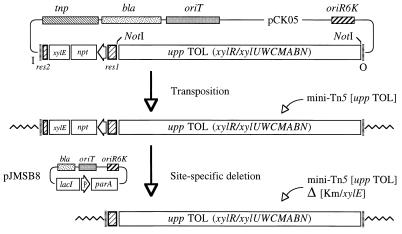
FIG. 4.
Insertion of the upp TOL catabolic segment into the chromosome of P. putida KT2442 and deletion of selection markers. The organization of pCK05, the delivery plasmid for the mobile element mini-Tn5 [upp TOL], is shown at the top (the vector part is not to scale). It includes all of the features of pJMS11 (Fig. 3) plus the NotI insert of pCK04AxylR (upp TOL segment) within the boundaries of the I and O ends of Tn5. This plasmid was mobilized to P. putida KT2442, and the insertion of the mini-Tn5 [upp TOL] transposon was selected with kanamycin and verified upon spraying of the exconjugants with catechol. In a subsequent step, the Kmr/xylE markers were deleted from P. putida KT2442::mini-Tn5 [upp TOL] by mobilization of the suicide plasmid pJMSB8 (which bears the parA sequence downstream of lacI/Plac). The final result is stable inheritance of the remainder of the hybrid transposon, i.e., mini-Tn5 [upp TOL] Δ[Kmr/xylE].
On this basis, pCK05 became the delivery plasmid for the mobile element mini-Tn5 [upp TOL]. Insertion of this minitransposon into the chromosome of P. putida KT2442 was carried out by following published protocols (7, 18). Insertions were selected through resistance to kanamycin and verified upon spraying of the exconjugants with catechol. The insertion frequency of mini-Tn5 [upp TOL], defined as the ratio of the number of inserts to the number of recipients (7), was 10−5, a value within the range of other mini-Tn5 transposons carrying smaller DNA segments (16). Of 50 Kmr and piperacillin-sensitive yellow colonies tested, 30 were able to grow on toluene as the only carbon source (Fig. 5).
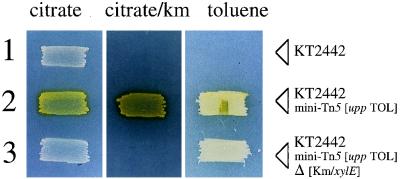
FIG. 5.
Phenotypes of P. putida KT2442 inserted with the upp TOL catabolic segment. Shown is the result of patching three related P. putida KT2442 strains in minimal medium with the carbon source and the antibiotic indicated in each case and spraying them with 1% catechol after growth. Strain 1 is P. putida KT2442 without any insertion. It can grow only in the medium with citrate and remains white. Strain 2 is P. putida KT2442 inserted with the complex transposon mini-Tn5 [upp TOL]. The presence of the xylE and npt markers in the transposon (Fig. 4) causes the patch to become yellow (C23O+) and to grow in the presence of kanamycin (Km), as well as on toluene, as the only carbon source. Strain 3 is a derivative of strain 2 in which the xylE/npt portion of mini-Tn5 [upp TOL] has been deleted in vivo. Where indicated, toluene was provided as a saturating vapor phase.
Sensitivity to piperacillin ensured the occurrence of authentic transposition events (7), since it rules out the integration of the whole delivery plasmid into the P. putida chromosome. The 30 clones with the phenotype caused by insertions of mini-Tn5 [upp TOL] were separately subjected to triparental mating (7) with E. coli CC118(pJMSB8) and the helper strain E. coli HB101(RK600). The pJMSB8 plasmid is used for transient expression of the RP4 resolvase gene parA (18), whereas RK600 assists in the conjugal transfer of pJMSB8 from E. coli to Pseudomonas (7). To achieve transient expression of parA, overnight cultures of E. coli(pJMSB8), P. putida KT2442:: mini-Tn5 [upp TOL], and E. coli HB101(RK600) were separately washed, mixed at 10:1:10, respectively, and then spotted onto the surface of a Luria broth plate. After 4 to 8 h of incubation at 30°C, spots were streaked onto the surface of M9 minimal medium plates with 10 mM citrate as the carbon source (23) but without antibiotics. This allowed growth of Pseudomonas but not of the E. coli strains present in the mating, which cannot use citrate as a carbon source. The plates were then sprayed with catechol to reveal the loss of the xylE marker (along with Kmr). With this procedure, >80% of the colonies had the phenotype anticipated for those losing the xylE/npt marker segment of the transposon while retaining the ability to grow on toluene as the only carbon source (Fig. 5). Further analysis of the strains by Southern blotting (26) and PCR verified the presence of the entire upp TOL element in the chromosome of P. putida KT2442 and the loss of the xylE/npt cassette (data not shown).
Characteristics of P. putida derivatives bearing the mini-Tn5 [upp TOL] Δ[Kmr/xylE] insert.
The strains resulting from the insertion of mini-Tn5 [upp TOL] and subsequent removal of the Kmr/xylE segment inherited a new phenotype (growth on toluene due to the acquisition of the upp TOL segment) with no other additional markers. Although the concepts for the genetic manipulations were laid out before (18), this work demonstrates their practical utility and its extension in the design of novel bacterial biocatalysts. The recruitment of a DNA fragment to the chromosome of P. putida through this procedure resembles the natural mechanisms of insertion and deletion of DNA that occur during natural adaptation processes (15, 27) to such an extent that the constructed strains can be considered quasi-natural. In fact, the final product of the manipulations described here is somewhat reminiscent of that of the naturally occurring genetic event designated “transposition without transposase” by Rappleye and Roth (24). Although in our case, the transposase of Tn5 is used to effect the insertion of the DNA segment, the tnp gene is not inherited by the resulting strain (7, 16). This makes it extremely unlikely that the insertions can be moved again. In this respect, the new DNA segment(s) added to the genome of the recipient strain (in our case, the upper TOL pathway) can be even more stable and predictable than many catabolic genes of natural Pseudomonas isolates, the DNA sequences of which are systematically subjected to rearrangements (30). In contrast, no loss of the hybrid mini-Tn5 insertion was detected after 1,000 generations (data not shown).
The strains bearing mini-Tn5 [upp TOL] Δ[Kmr/xylE] grew on toluene as the only carbon source (Fig. 5). The rationale of such a phenotype is the conversion of toluene to benzoate, mediated by the upper TOL enzymes encoded and expressed by the insertion and the further metabolism of benzoate through the housekeeping ortho-ring cleavage pathway of the catechol intermediate (Fig. 1). This pathway for degradation of toluene, which avoids a meta-cleavage step, is infrequent in natural isolates and will permit us to tackle the biodegradation of difficult chemical species such as chlorotoluenes (1, 2, 19). In a different context, the separate expression of the entire upper TOL pathway away from the meta operon should allow bioconversion of an array of substituted toluenes to the corresponding acids (2, 3).
Acknowledgments
We are indebted to S. Molin, C. Sternberg, and S. Harayama for facilitating some of the materials and ideas exploited in this work. K. N. Timmis and D. Pieper are also gratefully acknowledged for their support.
This work was funded by contract ENV4-CT95-0141 (ENVIRONMENT) with the European Union and by grant AMB94-1038-CO2-02 from the Spanish Comisión Interministerial de Ciencia y Tecnología.
S.P. and J.M.S.-R. contributed equally to this work.
REFERENCES
- 1.Abril M-A, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkmann U, Reineke W. Degradation of chlorotoluenes by in vivo constructed hybrid strains: problems of enzyme specificity, induction and prevention of meta-pathway. FEMS Microbiol Lett. 1992;96:81–88. doi: 10.1016/0378-1097(92)90460-6. [DOI] [PubMed] [Google Scholar]
- 3.Delgado A, Wubbolts M G, Abril M-A, Ramos J L. Nitroaromatics are substrates for the TOL plasmid upper-pathway enzymes. Appl Environ Microbiol. 1992;58:415–417. doi: 10.1128/aem.58.1.415-417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lorenzo V. Genetic engineering strategies for environmental applications. Curr Opin Biotechnol. 1992;3:227–231. doi: 10.1016/0958-1669(92)90097-3. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V. Designing microbial systems for gene expression in the field. Trends Biotechnol. 1994;12:365–371. doi: 10.1016/0167-7799(94)90037-X. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Herrero M, Jakubzik V, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived mini-transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 8.Eberl L, Kristensen C, Givskov M, Grohmann E, Gerlitz M, Schwab H. Analysis of the multimer resolution system encoded by the parCBA operon of the broad-host-range plasmid RP4. Mol Microbiol. 1994;12:131–141. doi: 10.1111/j.1365-2958.1994.tb01002.x. [DOI] [PubMed] [Google Scholar]
- 9.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 10.Fernández S, de Lorenzo V, Pérez-Martín J P. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 11.Fernández S, Shingler V, de Lorenzo V. Cross-regulation by XylR and DmpR activators of Pseudomonas putida suggests that transcriptional control of biodegradative pathways evolves independently of catabolic genes. J Bacteriol. 1994;176:5052–5058. doi: 10.1128/jb.176.16.5052-5058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlitz M, Hrabak O, Schwab H. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. J Bacteriol. 1990;172:6194–6203. doi: 10.1128/jb.172.11.6194-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grinter N J, Brewster G, Barth P T. Two mechanisms necessary for the stable inheritance of plasmid RP4. Plasmid. 1989;22:203–214. doi: 10.1016/0147-619x(89)90003-6. [DOI] [PubMed] [Google Scholar]
- 14.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartnett C S, Neidle E L, Ngai K L, Ornston L N. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate-3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990;172:956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessler B, Timmis K N, de Lorenzo V. The organization of the Pm promoter of the TOL plasmid reflects the structure of its cognate activator protein XylS. Mol Gen Genet. 1994;244:596–605. doi: 10.1007/BF00282749. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen C, Eberl L, Sánchez-Romero J M, Giskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehning A, Happe B, Timmis K N, Pieper D. Metabolism of chlorotoluenes by Burkholderia sp. strain PS12 and toluene dioxygenase of Pseudomonas putida F1: evidence for monooxygenation by toluene and chlorobenzene dioxygenases. Appl Environ Microbiol. 1997;63:1974–1979. doi: 10.1128/aem.63.5.1974-1979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrbach P R, Zeyer J, Reineke W, Knackmuss H-J, Timmis K N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984;158:1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marqués S, Ramos J L. Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways. Mol Microbiol. 1993;9:923–929. doi: 10.1111/j.1365-2958.1993.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 22.Mermod N, Harayama S, Timmis K N. New route to bacterial production of indigo. Bio/Technology. 1986;4:321–324. [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 24.Rappleye C A, Roth J R. Transposition without transposase: a spontaneous mutation in bacteria. J Bacteriol. 1997;179:2047–2052. doi: 10.1128/jb.179.6.2047-2052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reineke W, Knackmuss H J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1989;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Shapiro J A. Natural genetic engineering of the bacterial genome. Curr Opin Genet Dev. 1993;3:845–848. doi: 10.1016/0959-437x(93)90003-8. [DOI] [PubMed] [Google Scholar]
- 28.Timmis K, Steffan R, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 29.van Beilen J B, Wubbolts M, Chen Q, Nieboer M, Witholt B. Molecular biology of pseudomonads. T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.) Washington, D.C: ASM Press; 1996. Effects of two-liquid-phase systems and expression of alk genes on the physiology of alkane-oxidizing strains; pp. 35–47. [Google Scholar]
- 30.van der Meer J R, de Vos W M, Harayama S, Zehnder A. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams P A, Shaw L M, Pitt C W, Vrecl M. xylUW, two genes at the start of the upper pathway operon of TOL plasmid pWW0, appear to play no essential part in determining its catabolic phenotype. Microbiology. 1997;143:101–107. doi: 10.1099/00221287-143-1-101. [DOI] [PubMed] [Google Scholar]