Abstract
Whipple’s disease is a systemic disorder in which a gram-positive rod-shaped bacterium is constantly present in infected tissues. After numerous unsuccessful attempts to culture this bacterium, it was eventually characterized by 16S rRNA gene analysis to be a member of the actinomycetes. The name Tropheryma whippelii was proposed. Until now, the bacterium has only been found in infected human tissues, but there is no evidence for human-to-human transmission. Here we report the detection of DNA specific for the Whipple’s disease bacterium in 25 of 38 wastewater samples from five different sewage treatment plants in the area of Heidelberg, Germany. These findings provide the first evidence that T. whippelii occurs in the environment, within a polymicrobial community. This is in accordance with the phylogenetic relationship of this bacterium as well as with known epidemiological aspects of Whipple’s disease. Our data argue for an environmental source for infection with the Whipple’s disease bacterium.
Whipple’s disease, which was first described in 1907 as intestinal lipodystrophy (18), is a multisystem disorder characterized by the presence of gram-positive, rod-shaped bacteria in infected human tissues (2). Numerous attempts to culture the bacterium associated with Whipple’s disease have failed (2), and eventually its phylogenetic position within the actinomycetes was established by 16S rRNA gene analysis (11, 19). The name Tropheryma whippelii was proposed (11).
The clinical presentation of Whipple’s disease is heterogeneous. Frequently, patients suffer for years from arthralgias, chronic diarrhea, and weight loss, and less often from dementia or cardiac insufficiency. If untreated, the disease is usually fatal, but with appropriate antibiotic therapy the prognosis is favorable (2). However, the pathogenesis of Whipple’s disease appears to involve more than just an infection with T. whippelii. Immunological abnormalities are presumed to play a necessary role as predisposing factors (2, 7, 8), a view which is strengthened by the detection of T. whippelii in association with AIDS (5).
Two outstanding questions in the epidemiology of Whipple’s disease are the bacterium’s natural habitat and the route of infection. Until now, T. whippelii has never been found outside of infected human hosts, and although an oral route of infection has often been suspected (10), it has not been proven. There is no evidence for human-to-human transmission.
A reassessment of its phylogeny revealed a close relationship of the Whipple’s disease bacterium to typical environmental bacteria, such as the cellulomonads and the rare group B peptidoglycan organisms (6). This would support the hypothesis that T. whippelii may be a soil or water inhabitant. This may also explain the difficulties involved with its culture, as an estimated 80 to 99% of bacteria occurring in such natural environments are not culturable on artificial media (14, 17) and uncultured members of the actinomycete line of descent have been found in different environments and geographical locations (12). Soil and water bacteria tend to concentrate in sewage to form communities in which a large variety of different species are found. Therefore, the search for the Whipple’s disease bacterium was concentrated at sewage treatment plants.
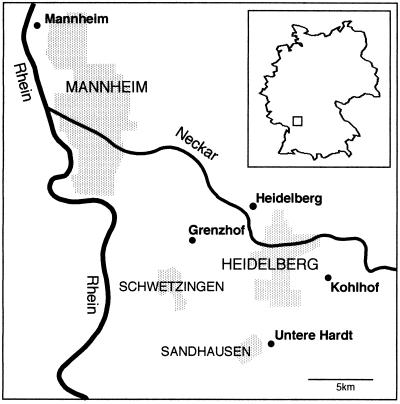
From September 1995 to July 1996, a total of 38 effluent samples from the sedimentation ponds of five different sewage treatment plants were examined by PCR for the presence of the Whipple’s disease bacterium. The sewage treatment plants are located within a distance of 5 to 25 km from Heidelberg, Germany (Fig. 1). Effluent was sampled in 1,000-ml single-use plastic flasks and filtered as described previously (4) in portions of 25 ml through cellulose acetate prefilters (25-mm diameter, 5-μm pore size; Sartorius, Göttingen, Germany). It was then passed through polyvinylidene fluoride filters (25-mm diameter, 0.45-μm pore size; Millipore, Bedford, Mass.). The DNA from the 0.45-μm-pore-size filters was extracted with the EnviroAmp Legionella sample preparation kit (Perkin-Elmer, Norwalk, Conn.), with the addition of 0.2% bovine serum albumin to the water in the final extraction step. Distilled water for negative controls was sampled in the same flasks and treated in the same way as the sewage.
FIG. 1.
Map of the area around Heidelberg, Germany, displaying the rivers Rhine (Rhein) and Neckar, the relevant townships (shaded areas with names in capital letters), and the five sewage treatment plants (dots) from which samples were taken. The inset in the figure shows the localization of the area within Germany.
PCR amplification and detection were performed as described in detail elsewhere (15). Primers whip1 (5′-AGAGATACGCCCCCCGCAA) and whip2 (5′-ATTCGCTCCACCTTGCGA), which amplify a fragment of 267 bp from the 16S rRNA gene of the bacterium, were used. In addition to hybridization with oligonucleotide whip3 (5′-TGGTACAGAGGGTTGCAATA), a second hybridization in a separate reaction was performed with oligonucleotide whip4 (5′-GTAATGGCGGGGACTCACAG). The specificities of primers whip1 and whip2 and of probe whip3 have been tested previously (15), and testing of probe whip4 with the same set of 37 control bacteria gave the same results as those reported for whip3 (15).
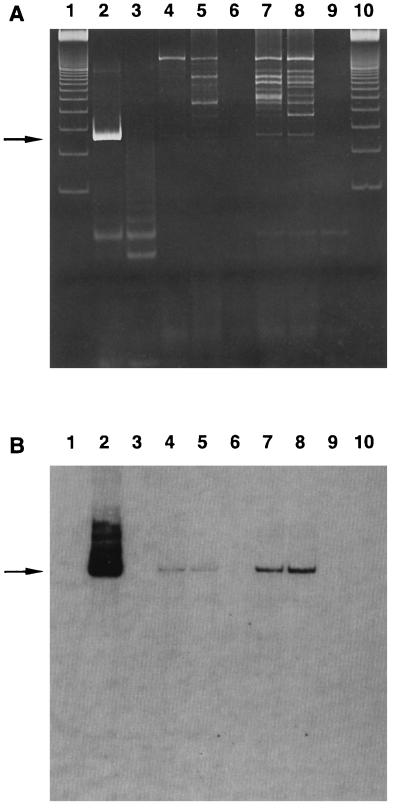
Of the 38 sewage samples tested, 25 were found to be positive by PCR and hybridization with both oligonucleotides (Table 1). Positive samples were found at each of the five sewage treatment plants. Most of the positive PCR products displayed relatively weak bands at the expected size of 267 bp. They also contained some extraneous bands, which were not of the expected size, probably due to the large amount of DNA from other organisms present in sewage. On hybridization, however, the PCR products displayed distinct bands, which were consistent between the two oligonucleotides (Fig. 2).
TABLE 1.
Results of PCR for the Whipple’s disease bacterium from sewage samples
| Sewage treatment plant | No. of samples:
|
|
|---|---|---|
| Tested | Positive | |
| Heidelberg | 14 | 9 |
| Untere Hardt | 7 | 7 |
| Mannheim | 13 | 7 |
| Kohlhof | 2 | 1 |
| Grenzhof | 2 | 1 |
| Total | 38 | 25 |
FIG. 2.
Detection of DNA from the Whipple’s disease bacterium in sewage samples. Shown are the results of analyses of PCR products by polyacrylamide gel electrophoresis (A) and Southern blot hybridization with oligonucleotide whip4 (B). Lanes: 1 and 10, 100-bp DNA marker; 2, positive control consisting of a tissue digest (diluted 10−4) from the intestinal biopsy of a patient with Whipple’s disease (9); 3, negative control consisting of 10 μl of distilled water in the PCR mixture; 4 and 5, sewage samples from Heidelberg; 6 and 9, negative controls consisting of distilled water which was filtered and treated in the same way as the sewage; 7 and 8, sewage samples from Untere Hardt.
To further compare the sequences of the PCR products with the known sequence of the Whipple’s disease bacterium, nine PCR products selected from all five sewage treatment plants were cloned and sequenced. The PCR products were reamplified over 5 cycles by using specific primers with BamHI and EcoRI restriction endonuclease recognition sites (11), ligated into vector pDS1-NOC (3) and transformed by electroporation into Escherichia coli JM101. To screen for positive clones, a series of PCRs (primer whip2 and vector-specific primer 5′-TTGCTTTGTGAGCGGATAACAATTAT) in which pools of 10 E. coli colonies were amplified in each of the reactions was performed. The PCR products were tested by gel electrophoresis, Southern blotting, and hybridization with oligonucleotide whip4. Pools which produced hybridizing bands of the appropriate size were retested to find individual colonies. Plasmids from positive clones were extracted according to standard methods (13), and both strands of the inserts were sequenced manually with the AmpliCycle sequencing kit (Perkin-Elmer), with incorporation of [α-33P]dATP. For each of the nine sewage samples, the sequence of the PCR product was identical to the known sequence determined previously for the Whipple’s disease bacterium (5, 9).
This is the first documented encounter with the Whipple’s disease bacterium outside of the human body. The specific sequence was found in all of the sewage treatment plants from which samples were taken, indicating that the bacterium is a regular member of, and most likely is able to multiply in, such polymicrobial communities. This finding is in agreement with one previous report which found sequences highly similar to, but not identical with, those of the Whipple’s disease bacterium in the eutrophic water of a Mediterranean coastal lagoon (1).
The environmental occurrence of the Whipple’s disease bacterium at the sewage treatment plants is in agreement with the evolutionary relationships of the bacterium (6, 11). It is also consistent with the lack of geographical preference of reported cases of Whipple’s disease in Germany and with the relatively constant incidence of new cases per year (16). In addition, it is consistent with a predominance of outdoor professions among patients with the disease (2). These features and the absence of the Whipple’s disease bacterium in normal human tissues (11, 15) make it highly unlikely that this bacterium is a common commensal which is transferred between warm-blooded hosts in the same way as a variety of other pathogens. However, the complete range of occurrences in the environment and the exact localization of the habitats of the Whipple’s disease bacterium still remain to be determined.
Nucleotide sequence accession numbers.
The GenBank/EMBL accession numbers of the published 16S rRNA gene sequences of the Whipple’s disease bacterium are M77832 (19), M87484 (11), and X99636 (6).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant Ma 1663/2-1).
We thank T. N. Petney (Heidelberg, Germany) for helpful comments on the manuscript and E. Fuchs (Heidelberg, Germany) for providing vector pDS1-NOC.
REFERENCES
- 1.Benlloch S, Rodríguez-Valera F, Martinez-Murcia A J. Bacterial diversity in two coastal lagoons deduced from 16S rDNA amplification and partial sequencing. FEMS Microbiol Ecol. 1995;18:267–280. [Google Scholar]
- 2.Dobbins W O., III . Whipple’s disease. Springfield, Ill: Charles C. Thomas, Publisher; 1987. [Google Scholar]
- 3.Helke A, Geisen R M, Vollmer M, Sprengart M L, Fuchs E. An unstructured mRNA region and a 5′ hairpin represent important elements of the E. coli translation initiation signal determined by using the bacteriophage T7 gene 1 translation start site. Nucleic Acids Res. 1993;21:5705–5711. doi: 10.1093/nar/21.24.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maiwald M, Kissel K, Srimuang S, von Knebel Doeberitz M, Sonntag H-G. Comparison of polymerase chain reaction and conventional culture for the detection of legionellas in hospital water samples. J Appl Bacteriol. 1994;76:216–225. doi: 10.1111/j.1365-2672.1994.tb01619.x. [DOI] [PubMed] [Google Scholar]
- 5.Maiwald M, Meier-Willersen H J, Hartmann M, von Herbay A. Detection of Tropheryma whippelii DNA in a patient with AIDS. J Clin Microbiol. 1995;33:1354–1356. doi: 10.1128/jcm.33.5.1354-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiwald M, Ditton H-J, von Herbay A, Rainey F A, Stackebrandt E. Reassessment of the phylogenetic position of the bacterium associated with Whipple’s disease and determination of the 16S-23S ribosomal intergenic spacer sequence. Int J Syst Bacteriol. 1996;46:1078–1082. doi: 10.1099/00207713-46-4-1078. [DOI] [PubMed] [Google Scholar]
- 7.Marth T, Roux M, von Herbay A, Meuer S C, Feurle G E. Persistent reduction of complement receptor 3 α-chain expressing mononuclear blood cells and transient inhibitory serum factors in Whipple’s disease. Clin Immunol Immunopathol. 1994;72:217–226. doi: 10.1006/clin.1994.1134. [DOI] [PubMed] [Google Scholar]
- 8.Marth T, Neurath M, Cuccherini B A, Strober W. Defects of monocyte interleukin 12 production and humoral immunity in Whipple’s disease. Gastroenterology. 1997;113:442–448. doi: 10.1053/gast.1997.v113.pm9247462. [DOI] [PubMed] [Google Scholar]
- 9.Meier-Willersen H J, Maiwald M, von Herbay A. Morbus Whipple in Assoziation mit opportunistischen Infektionen. Dtsch Med Wochenschr. 1993;118:854–860. doi: 10.1055/s-2008-1059397. [DOI] [PubMed] [Google Scholar]
- 10.Relman D A. Whipple’s disease. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 919–936. [Google Scholar]
- 11.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 12.Rheims H, Spröer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 15.von Herbay A, Ditton H-J, Maiwald M. Diagnostic application of a polymerase chain reaction assay for the Whipple’s disease bacterium to intestinal biopsies. Gastroenterology. 1996;110:1735–1743. doi: 10.1053/gast.1996.v110.pm8964398. [DOI] [PubMed] [Google Scholar]
- 16.von Herbay A, Otto H F, Stolte M, Borchard F, Kirchner T, Ditton H-J, Maiwald M. Epidemiology of Whipple’s disease in Germany. Analysis of 110 patients diagnosed in 1965–95. Scand J Gastroenterol. 1997;32:52–57. doi: 10.3109/00365529709025063. [DOI] [PubMed] [Google Scholar]
- 17.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 18.Whipple G H. A hitherto undescribed disease characterized anatomically by deposits of fat and fatty acids in the intestinal and mesenteric lymphatic tissues. Johns Hopkins Hosp Bull. 1907;18:382–391. [Google Scholar]
- 19.Wilson K H, Blitchington R, Frothingham R, Wilson J A P. Phylogeny of the Whipple’s disease-associated bacterium. Lancet. 1991;338:474–475. doi: 10.1016/0140-6736(91)90545-z. [DOI] [PubMed] [Google Scholar]