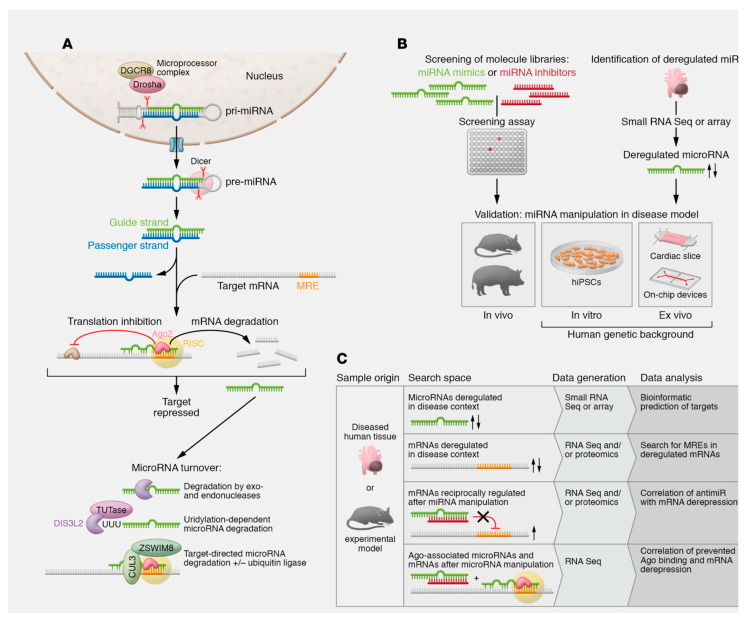
Figure 2.
The biogenesis and function of microRNA (miRNA) are depicted. (A) The mechanism of action and degradation pathways of microRNAs are also significant factors to consider. Canonical microRNA biogenesis involves larger hairpin RNA molecules called pri-miRNAs. These pri-miRNAs arise from RNA Pol II transcription of microRNA genes or clusters or are found within introns. A microprocessor complex, consisting of the endonuclease Drosha, the DiGeorge critical region 8 protein (DGCR8), and other factors, cleaves these pri-miRNAs, activating their functions. The pre-miRNA produced is transported to the cytoplasm and fragmented by the enzyme Dicer. It is tailored to a length of 21 to 22 nucleotides. Occasionally, certain non-canonical mechanisms of microRNA formation bypass the Dicer or microprocessor complex. Afterwards, one of the resulting duplexes, made up of 21–22 nucleotides, is incorporated into the RNA-induced silencing complex (RISC) as the guide strand. Meanwhile, the redundant passenger strand or *-strand undergoes prompt degradation. If both strands are maintained, they can take on individual functions, as shown for cardiovascular miR-21 and miR-126. There are also exceptions where microRNA strands localize to the nucleus and function in unconventional ways. The degradation of microRNAs involves exonucleases XRN-1, PNPase old-35, and RRP41 or the endonuclease Tudor-SN. The nuclease DIS3L2 degrades a subset of microRNAs after modification by terminal uridyltransferases (TUTases). Target-directed microRNA degradation mechanisms have been established, which includes the participation of ubiquitin ligases. (B) Strategies for determining and validating cardiovascular microRNAs relevant to diseases. (C) Methods for identifying microRNA targets. Refs. [34,35,36,37,38,39]. From Laggerbauer B et al. J Clin Invest. 2022 Jun 1;132(11): e159179. ∗ Arrow; increase or decrease.