Abstract
We recently characterized a transposon-induced NaCl-sensitive mutant of Staphylococcus aureus (U. Vijaranakul, M. J. Nadakavukaren, D. O. Bayles, B. J. Wilkinson, and R. K. Jayaswal, Appl. Environ. Microbiol. 63:1889–1897, 1997). To further characterize this mutant, we determined the nucleotide sequence at the insertion site of the transposon on the S. aureus chromosome. Nucleotide sequencing revealed a 1,326-bp open reading frame (ORF442) encoding a hydrophobic 442-amino-acid polypeptide with a calculated molecular mass of 49,058 Da. The hydrophilicity profile of the gene product revealed the existence of 12 hydrophobic domains predicted to form membrane-associated α-helices. Comparison of the amino acid sequence of ORF442 with amino acid sequences in the GenBank database showed extensive homology with the branched-chain-amino-acid transport genes of gram-positive and gram-negative bacteria. This is the first brnQ gene in staphylococci to be described.
Staphylococcus aureus is one of the most halotolerant, nonhalophilic bacteria. It causes a variety of diseases, ranging from simple skin infections to life-threatening diseases such as endocarditis and food poisoning. One of the distinguishing characteristics of S. aureus is its ability to grow in the presence of up to 3.5 M NaCl. It has been reported that high concentrations of NaCl inhibit growth (1), decrease toxin synthesis (22), stimulate synthesis of degradative enzymes (17), increase cell size, and reduce the length of the interpeptide bridge of peptidoglycan (26). However, the mechanisms by which NaCl causes the above physiological and molecular changes in S. aureus are not known.
Osmoregulation in gram-negative bacteria, mainly Escherichia coli and Salmonella typhimurium, has been studied extensively. A number of osmotically regulated genes, such as envZ and ompR, the kdpABC and proU operons, the mal and bet regulons, proQ, phoE, otsB, treA, osmB, rpoS, and algD, have been reported (5, 8, 12). In contrast, there are very few reports of osmotically regulated genes in gram-positive bacteria. In Bacillus subtilis, DegS-DegU (a two-component system) is involved in sensing salt stress (17). Another regulatory gene, clpC (15), that acts downstream from DegS-DegU and ComP-ComA in the regulatory cascade is induced by multiple stresses, including heat shock, ethanol, salt stress, oxygen limitation, and nutrient deprivation (11). Mutations in these regulatory genes lead to increased levels of expression of the alternative sigma factor ςB. The expression of genes controlled by ςB, such as the ect gene and the sigB operon, which codes for ςB and its associated regulatory proteins, was shown to be dramatically induced by salt stress (4). Thus, this sigma factor plays an important role in the increased synthesis of general stress proteins and some salt-specific stress proteins (10). Recently, the sigB operon encoding an alternative sigma factor of S. aureus was cloned and sequenced (16, 28). Whether the sigB operon is involved in salt tolerance in S. aureus remains to be determined.
In an effort to investigate the molecular mechanisms and genes involved in the NaCl tolerance of S. aureus, we isolated NaCl-sensitive mutants by transposon mutagenesis. Recently, one of the NaCl-sensitive mutants was physiologically characterized (27). This NaCl-sensitive mutant showed a pleiotropic phenotype in high salt concentrations. It exhibited normal growth rate and cell division in medium containing a low concentration of NaCl. However, mutant cells grown in medium containing a high concentration of NaCl showed a very long lag phase and increased cell size with the presence of multiple septa. To further characterize this mutant, we cloned and sequenced the mutated gene with the flanking sequences of the transposon (Tn) at the insertion site of the mutant. In this study, we report the cloning, sequencing, and analysis of a gene, brnQ, encoding a branched-chain-amino-acid transport protein of S. aureus. The mutation in brnQ caused the NaCl-sensitive phenotype of S. aureus.
Cloning and nucleotide sequence determination of the region flanking the mutated gene.
A genomic library of the NaCl-sensitive mutant was constructed in cosmid pCP13 (6). The mutant library was screened by colony hybridization to obtain clones containing Tn sequences. Four of 2,000 cosmid mutant clones showed strong hybridization with the radiolabeled Tn917 probe. Southern blot analysis showed that a 6-kb EcoRI fragment of the positive clones hybridized with the Tn probe. This fragment was subcloned into the EcoRI site of plasmid pTZ18R and designated pTSS7.7. Sequencing was performed by either a radioactive protocol with [α-32P]dCTP (ICN Biomedicals, Inc., Costa Mesa, Calif.) and Taq polymerase (U.S. Biochemical Corp., Cleveland, Ohio) or a nonradioactive dye terminator cycle sequencing protocol with AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.) and an automated sequencer, the ABI Prism 310 genetic analyzer (Perkin-Elmer). Nucleotide sequencing of the 6-kb EcoRI fragment with M13 primers and internal primers of the Tn revealed that the cloned fragment had a 2,678-bp region of the Tn and about 3.3-kb of the S. aureus genome.
Cloning and nucleotide sequence determination of the wild-type allele.
To obtain the wild-type allele of the mutated gene, a 400-bp region of DNA flanking Tn917 was used as a probe to screen the cosmid library of RN450, which was the parent strain. Six positive clones obtained by colony hybridization were further subjected to Southern blot hybridization. A 4.8-kb fragment from one of the cosmid clones, CRN4, which showed strong hybridization with the probe was subcloned into pTZ18R and designated pTRN5.
The nucleotide sequence analysis of 1.9 kb (the mutation was localized within this fragment) of the 4.8-kb fragment revealed one complete open reading frame (ORF) of 1,326 bp. This ORF was designated ORF442. As shown in Fig. 1, the putative promoter sequences (−10 and −35) were identified upstream of the initiation site and a termination sequence was located downstream of ORF442. Comparison of the nucleotide sequences of clones pTSS7.7, pTRN5, and CRN4 showed the Tn917 insertion site at 377 nucleotides downstream from the initiation codon of ORF442.
FIG. 1.
Nucleotide sequence of the 1.9-kb chromosomal DNA fragment containing brnQ. The predicted amino acid sequence of BrnQ is shown in single-letter code below the nucleotide sequence. The putative promoter elements (−10 and −35) are underlined. The transcription start site is indicated by +1.
Analysis of the protein product(s) encoded by ORF442.
The deduced translation product of ORF442 is a protein with a calculated molecular mass of 49,058 Da and a pI of 9.96. As shown in Table 1, among 442 amino acids, there are 258 nonpolar amino acids (58.37%), 133 polar amino acids (30.1%), 13 acidic amino acids (2.94%), and 38 basic amino acids (8.6%).
TABLE 1.
Branched-chain-amino-acid carrier proteins of gram-positive and gram-negative bacteria
| Protein | Organism | Amino acid characteristica
|
||||||
|---|---|---|---|---|---|---|---|---|
| n | Nonpolar (%) | Polar (%) | Acidic (%) | Basic (%) | Size (kDa) | pI | ||
| BrnQ | S. aureus | 442 | 58 | 30 | 3 | 9 | 49.058 | 9.96 |
| B. subtilis | 439 | 58 | 30 | 3 | 7 | 46.875 | 9.64 | |
| L. delbrueckii | 446 | 59 | 30 | 4 | 7 | 47.864 | 9.44 | |
| S. typhimurium | 438 | 60 | 30 | 4 | 6 | 46.418 | 9.29 | |
| E. coli | 438 | 59 | 31 | 4 | 6 | 46.092 | 9.14 | |
| BraB | P. aeruginosa | 437 | 60 | 30 | 4 | 5 | 45.28 | 8.83 |
| C. perfringens | 338 | 54 | 33 | 5 | 8 | 35.861 | 9.47 | |
| BraZ | P. aeruginosa | 437 | 61 | 29 | 3 | 6 | 45.271 | 9.66 |
Characteristics are predicted theoretically.
Analysis of the ORF442 transcript.
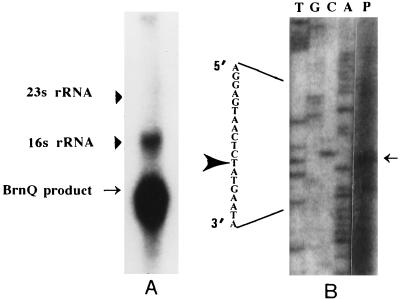
Total RNA from the salt-sensitive mutant and parent strains was isolated as described by Gustafson et al. (9). Ten micrograms of total RNA was electrophoresed on formaldehyde agarose gels (1.0%) and transferred to nitrocellulose membranes. The blot was probed with a radiolabeled 2.5-kb DNA fragment encompassing ORF442. Northern blot analysis revealed that ORF442 codes for a transcript of about 1.3 kb (Fig. 2A). To define the transcription unit more precisely, the 5′ end of the transcript was mapped with the avian myeloblastosis virus reverse transcriptase system (Promega). An 18-base oligonucleotide (5′-GACCCTATCCAATCCGAG-3′) specific to the coding region was annealed with total RNA and extended in a primer extension assay. A sequence ladder was generated with the same primer on a 4.8-kb fragment and was coelectrophoresed to determine the position of the transcription start site. The primer extension showed that the first nucleotide of the mRNA was a T residue corresponding to position 354 in the DNA sequence (Fig. 2B). The transcript initiates from 1 nucleotide upstream of the predicted translation initiation site. Thus, the transcript does not contain the Shine-Dalgarno sequences usually necessary for translation initiation. Although it is an unusual transcript, similar types of transcripts have been reported earlier (23). The mechanism by which ribosomes bind to the mRNA is not known.
FIG. 2.
Transcript analysis of the brnQ gene. (A) Northern blot analysis of ORF442. Ten micrograms of an RNA sample was separated electrophoretically and transferred by Northern blotting onto a membrane. The blot was probed with a radiolabeled 2.5-kb DNA fragment encompassing brnQ. The sizes of the ribosomal RNAs are marked with arrowheads, and the BrnQ product is indicated by an arrow. (B) Mapping of the 5′ end of the brnQ transcript by primer extension analysis. Total RNA from the parent strain was hybridized with an oligonucleotide complementary to the mRNA of the brnQ locus and extended by avian myeloblastosis virus reverse transcriptase (lane P). Lanes T, G, C, and A correspond to a dideoxy sequencing reaction performed with the same primer. The sequence encompassing the initiation start site (marked by an arrowhead) is enlarged.
Sequence homology.
When the nucleotide sequence of ORF442 was compared to known sequences by using National Center for Biotechnology Information BLAST searches, 57% identity to braB of Clostridium perfringens and 72% identity to brnQ of S. typhimurium were found.
The predicted amino acid sequence of ORF442 was used to conduct a homology search. Significant homology of the deduced amino acid sequence of ORF442 to the branched-chain-amino-acid carrier gene (brnQ) of gram-positive and gram-negative bacteria was observed. The deduced amino acid sequence of the translation product of ORF442 revealed identity to BraB of Pseudomonas aeruginosa (33.9%) (13), BrnQ of B. subtilis (33.5%) (3), BraB of C. perfringens (31.6%) (20), BrnQ of Lactobacillus delbrueckii (30.9%) (24), BrnQ of S. typhimurium (27.6%) (21), BrnQ of E. coli (28%), and BraZ of P. aeruginosa (27.7%) (14). The amino acid sequences are extensively conserved over the entire region (Fig. 3). The identity at the N terminus (first 100 amino acids) is particularly prominent, at 45, 45, 42, 44, 41, and 38% between S. aureus and BraB of P. aeruginosa, BrnQ of B. subtilis, BraB of C. perfringens, BrnQ of L. delbrueckii, BrnQ of E. coli, and BrnQ of S. typhimurium, respectively. Therefore, the gene contained by ORF442 was designated brnQ of S. aureus.
FIG. 3.
Alignment of the predicted amino acid sequences of BrnQ of S. aureus (Sa), BrnQ of B. subtilis (Bs), BraB of C. perfringens (Cp), BrnQ of L. delbrueckii (Ld), BrnQ of S. typhimurium (St), BraB of P. aeruginosa (Pa), BrnQ of E. coli (Ec), and BraZ of P. aeruginosa (Pa) by the Clustal W alignment program described by Thompson et al. (25). Identical amino acid residues are shown in boldface type, and similar amino acid residues are indicated by plus signs. The protein sequences were aligned by the insertion of gaps (—) to obtain maximum sequence identity. The following amino acids are similar: A, S, and T; D and E; N and Q; R and K; and I, L, M, V, F, Y, and W.
Further comparisons of the BrnQ protein of S. aureus with other branched-chain-amino-acid carrier proteins showed striking similarities with respect to molecular weight, pI, and percent composition of various amino acids. Hydrophilicity profile comparisons of BrnQ of S. aureus with those of the branched-chain-amino-acid transport carriers also showed strong structural similarities (data not shown). The hydrophilicity profile of BrnQ of S. aureus is highly similar to those reported for other gram-positive and gram-negative bacteria. As shown in Fig. 4, BrnQ of S. aureus contains approximately 12 membrane-spanning segments flanked by short hydrophilic stretches. Thus, BrnQ is extremely hydrophobic.
FIG. 4.
Hydrophilicity profile of the predicted amino acid sequence of BrnQ of S. aureus. The x axis represents amino acid residues measured from the N terminus (hydrophilicity window size, 7). The y axis is an arbitrary scale of hydropathy described previously (18), as modified to represent hydrophilicity (7).
Complementation analysis.
To complement the mutation of a salt-sensitive mutant, a 4.8-kb fragment containing brnQ was subcloned into the shuttle vector pCU1 (2). The resultant plasmid, designated pCUT5, was electroporated into the salt-sensitive mutant. The transformants were tested for the ability to grow in defined medium containing 2.5 M NaCl. All of the transformants were able to grow on solid media containing 2.5 M NaCl. When the transformants were grown in liquid culture, they showed a reduced lag phase (∼30 h) compared to the mutant strain (>60 h). However, the lag phase was still longer in the transformants than that observed in the wild-type strain (∼12 h) grown under identical conditions. Thus, the cloned gene partially complemented the mutation in the trans position. The reasons that only partial complementation was observed are unknown.
ORF442 encodes a highly hydrophobic polypeptide of a calculated molecular mass of 49 kDa and a pI of 9.96. The polypeptide has high homology to the Na+-dependent branched-chain-amino-acid carriers in P. aeruginosa (13, 29) and S. typhimurium (19). In addition, the polypeptide has high homology to the branched-chain-amino-acid carriers in B. subtilis, C. perfringens (20), L. delbrueckii (24), and E. coli. The striking identity of the deduced amino acid sequences throughout the entire length suggests that the branched-chain-amino-acid carriers in these organisms are highly conserved. Based on these analyses, we propose that the cloned gene is a branched-chain-amino-acid carrier gene (brnQ) of S. aureus. BrnQ of S. aureus, BrnB of P. aeruginosa and C. perfringens, and BrnQ of S. typhimurium, B. subtilis, L. delbrueckii, and E. coli are similar in size, pH (basic), and hydropathy profiles. BrnQ of S. aureus, which is an extremely hydrophobic protein, contains 12 membrane-spanning segments flanked by short hydrophilic stretches, as reported for other bacteria. These properties are typical of integral membrane transport proteins. The most abundant amino acid of the BrnQ protein of S. aureus is isoleucine (67 of 442), followed by leucine (62 of 442). Although it has been suggested that in S. aureus, leucine, isoleucine, and valine use the same transporter, there is no report on the branched-chain-amino-acid transport systems in S. aureus. Therefore, this is the first report of a branched-chain-amino-acid carrier (BrnQ) in S. aureus.
To explain the pleiotropic phenotype of the NaCl-sensitive mutant reported earlier (27), we propose the following model for the transport of branched-chain amino acids in S. aureus. There are at least two independent branched-chain-amino-acid transport systems with respect to substrate specificity and affinity. The two systems are a sodium-coupled, branched-chain-amino-acid transport system and a sodium-independent transport system, which is sensitive to environmental stress conditions such as osmolarity, pH, and temperature. Under nonstress conditions, branched-chain amino acids are transported through both systems, whereas under stress conditions, only the sodium-coupled transport system functions. Under low osmotic conditions (low sodium ions), the mutant shows normal growth and cell division, like the parent strain. In the presence of high sodium ion concentrations, the mutant shows a very long lag phase with multiple septa. This may be due to the mutation in the sodium-dependent system and the inability of the sodium-dependent system to transport branched-chain amino acids from the medium. Similarly, at low pH and high temperature, the growth of the mutant is retarded. This also may be due to the sensitivity of the sodium-independent system to transport branched-chain amino acids. To test this hypothesis, we are currently characterizing the gene and gene product with respect to substrate specificity and the kinetics of the uptake of [14C]leucine, [14C]isoleucine, and [14C]valine.
Nucleotide sequence accession number.
The 4.8-kb DNA fragment of which ORF442 was a part was sequenced, and the nucleotide data was deposited in the GenBank database under accession no. U87144.
Acknowledgments
We express our gratitude to Brian J. Wilkinson for helpful discussion and Sean Arkins and David Williams for critical reading of the manuscript.
This study was supported by a student stipend from the American Heart Association Illinois Affiliate to U.V.
REFERENCES
- 1.Armstrong-Buisseret L, Cole M B, Stewart G S A B. A homologue to the Escherichia coli alkyl hydroperoxide reductase AhpC is induced by osmotic upshock in Staphylococcus aureus. Microbiology. 1995;141:1655–1661. doi: 10.1099/13500872-141-7-1655. [DOI] [PubMed] [Google Scholar]
- 2.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, Entian K-D, Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 3.Belitsky B R, Sonenshein A L. Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J Bacteriol. 1997;179:1035–1043. doi: 10.1128/jb.179.4.1035-1043.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csonka L N, Hanson A D. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 6.Darzins A, Chakrabarty A M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galinski E A. Osmoadaptation in bacteria. Adv Microb Physiol. 1995;37:273–328. [PubMed] [Google Scholar]
- 9.Gustafson J, Strassle A, Hachler H, Kayser F H, Bachi B B. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamate synthetase operon. J Bacteriol. 1994;176:1460–1467. doi: 10.1128/jb.176.5.1460-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecker M, Volker U. General stress proteins in Bacillus subtilis. FEMS Microbiol Ecol. 1990;74:197–214. [Google Scholar]
- 12.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino T, Kose K, Uratani Y. Cloning and nucleotide sequence of the gene braB coding for the sodium-coupled branched chain amino acid carrier in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1990;220:461–467. doi: 10.1007/BF00391754. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino T, Kose-Terai K, Uratani Y. Isolation of the braZ gene encoding the carrier for a novel branched-chain amino acid transport system in Pseudomonas aeruginosa PAO. J Bacteriol. 1991;173:1855–1861. doi: 10.1128/jb.173.6.1855-1861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruger E, Volker U, Hecker M. Stress induction of ClpC in Bacillus subtilis and its involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kullik I, Giachino P. The alternate sigma factor sigma B in Staphylococcus aureus: regulation of the SigB operon in response to growth phase and heat-shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 17.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Matsubara K, Ohnishi K, Kiritani K. The third general transport system for branched-chain amino acids in Salmonella typhimurium. J Gen Appl Microbiol. 1988;34:183–189. [Google Scholar]
- 20.Nakamura K, Hashizume E, Shibata T, Nakamura Y, Mala S, Yamani K. Small cytoplasmic RNA (scRNA) gene from Clostridium perfringens can replace the gene for the Bacillus subtilis scRNA in both growth and sporulation. Microbiology. 1995;141:2965–2975. doi: 10.1099/13500872-141-11-2965. [DOI] [PubMed] [Google Scholar]
- 21.Ohnishi K, Hasegawa A, Matsubara K, Date T, Okada T, Kiritani K. Cloning and nucleotide sequence of the brnQ gene, the structural gene for a membrane-associated component of the LIV-II transport system for branched-chain amino acids in Salmonella typhimurium. Jpn J Genet. 1988;63:343–357. doi: 10.1266/jjg.63.343. [DOI] [PubMed] [Google Scholar]
- 22.Regassa L B, Betley M J. High sodium chloride concentrations inhibit staphylococcal enterotoxin C gene (sec) expression at the level of sec mRNA. Infect Immun. 1993;61:1581–1585. doi: 10.1128/iai.61.4.1581-1585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruepp A, Soppa J. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J Bacteriol. 1996;178:4942–4947. doi: 10.1128/jb.178.16.4942-4947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stucky K, Hagting A, Klein J R, Matern H, Henrich B, Konings W N, Plapp R. Cloning and characterization of brnQ, a gene encoding a low-affinity, branched-chain amino acid carrier in Lactobacillus delbrückii subsp. lactis DSM7290. Mol Gen Genet. 1995;249:682–690. doi: 10.1007/BF00418038. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijaranakul U, Nadakavukaren M J, de Jonge B L M, Wilkinson B J, Jayaswal R K. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. J Bacteriol. 1995;177:5116–5121. doi: 10.1128/jb.177.17.5116-5121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vijaranakul U, Nadakavukaren M J, Bayles D O, Wilkinson B J, Jayaswal R K. Characterization of an NaCl-sensitive Staphylococcus aureus mutant and rescue of the NaCl-sensitive phenotype by glycine betaine but not by other compatible solutes. Appl Environ Microbiol. 1997;63:1889–1897. doi: 10.1128/aem.63.5.1889-1897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamato I, Ohki M, Anraku Y. Genetic and biochemical studies of transport systems for branched-chain amino acids in Escherichia coli. J Bacteriol. 1979;138:24–32. doi: 10.1128/jb.138.1.24-32.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]