Abstract
A family of genes encoding laccases has recently been described for the basidiomycete I-62 (CECT 20197). Transcript levels of genes lcc1, lcc2, and lcc3 were analyzed under four different culture conditions to study their expression patterns. Two of the laccase genes were clearly inducible by veratryl alcohol: the lcc1 gene is inducible in early stages of growth, and the lcc2 gene is also inducible but only when the organism reaches the stationary phase. Transcript levels for the third gene, lcc3, were uninduced by veratryl alcohol and repressed by glucose.
Lignin is a complex aromatic biopolymer degradable by a few organisms, like white rot basidiomycetes (7, 22). Industry has an increasing interest in extracellular enzymes from white rot fungi, such as lignin and manganese peroxidases and laccases, due to their potential to degrade both highly toxic phenolic compounds and lignin (1, 8, 14, 29, 31). Elucidation of the catalytic mechanisms exerted by these enzymes, characterization of the proteins, and cloning of the genes encoding them have increased our understanding of the biochemistry and genetics of this quite complex and unique extracellular oxidative system (2, 19, 22, 35–38). The regulation of the expression of genes belonging to families encoding lignin-degrading enzymes, such as lignin peroxidase and manganese peroxidase isozymes produced by Phanerochaete chrysosporium, has been reviewed by Broda et al. (3, 4).
Originally, the numerous laccase isozymes were thought to be posttranslational variants of the same gene product, but several groups have been able to isolate and characterize several laccase genes and cDNA copies (17, 18, 20, 23, 30, 32, 36–38). A total of four different laccase cDNA sequences have been described for Rhizoctonia solani (36), up to five laccase genes have been described for Trametes villosa (37, 38), and three genomic DNA sequences coding for laccases have been described for the basidiomycete I-62 (28), suggesting that at least a part of the biochemical diversity of laccase isozymes must be due to the genomic multiplicity of the laccase gene sequences. Unfortunately, only a few reports up to now have studied the expression of multiple genes encoding different laccase isozymes (10, 37). The basidiomycete I-62 efficiently degrades natural lignin from beech wood, sugar cane bagasse, and wheat straw when cultured under a variety of different physiological conditions (28a). In this report, we demonstrate, by using Northern blot analysis, that the expression of the three laccase genes previously identified and cloned from the basidiomycete I-62 (28) is differentially regulated. Two of these genes are sensitive to induction by veratryl alcohol but at different stages of growth, while the third one clearly appears to be under catabolic repression.
Northern analysis of lcc genes.
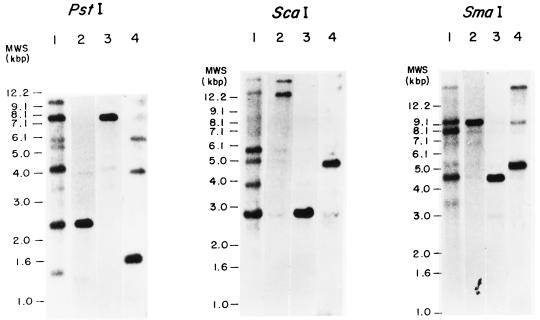
Three genomic sequences encoding laccases from the basidiomycete I-62 corresponding to laccase genes lcc1, lcc2, and lcc3 have been cloned and sequenced (28). To study the expression pattern of each lcc gene, we chose a DNA fragment for each gene that was able to hybridize exclusively with itself. The DNA fragments used to specifically detect the transcripts of the genes encoding the different laccases in the basidiomycete I-62 were a 0.25-kb ScaI-PstI DNA fragment from the lcc1 gene, a 0.5-kb PstI-HindIII fragment from the lcc2 gene, and a 0.5-kb KpnI-PstI fragment from the lcc3 gene. The DNA fragments corresponded mainly to the 5′ coding region, where the different laccase genes are less conserved. A Southern blot containing total DNA from the basidiomycete I-62 completely digested with three restriction enzymes was hybridized, under low-stringency conditions (28), with the internal 0.3-kb PstI-XhoI DNA fragment from the basidiomycete PM1 lac1 gene (9) (Fig. 1, lanes 1). Hybridizations of the same blot with the three lcc-specific probes under highly stringent conditions (33) gave a different pattern of bands for each gene (Fig. 1, lanes 2 to 4). We could thus confirm that the chosen probes were specific for each laccase gene and adequate to perform transcription analysis. Cross hybridization among the selected probes was below 5%. As can be observed, some bands revealed with the heterologous probe (Fig. 1, lanes 1) do not correspond to any of the three cloned genes (Fig. 1, lanes 2 to 4), suggesting the existence of more than three laccase genes in the basidiomycete I-62 genome.
FIG. 1.
Southern analyses of basidiomycete I-62 genomic DNA digested with three restriction enzymes (PstI, ScaI, and SmaI) and hybridized with a heterologous probe from the basidiomycete PM1 lac1 laccase gene under low-stringency hybridization conditions (lanes 1), an lcc1-specific probe (lanes 2), an lcc2-specific probe (lanes 3), and an lcc3-specific probe (lanes 4). MWS, molecular sizes.
To study the regulation of each lcc gene, I-62 was grown under four different culture conditions. A defined culture medium previously described for I-62 (28) was used in this work; it includes 1% glucose as the carbon source, 1 mM ammonium tartrate as the nitrogen source, and 4 mM veratryl alcohol as the inducer of ligninolytic activities. Besides this inducing medium, the basidiomycete I-62 was grown in noninducing medium (1% glucose, 1 mM ammonium tartrate), in fructose medium (1% fructose, 1 mM ammonium tartrate, and 4 mM veratryl alcohol), and under nonlimiting nitrogen conditions (1% glucose, 10 mM ammonium tartrate). All liquid cultures were performed under agitation (100 rpm) at 28°C over 24 days. The inoculum for the cultures was prepared as previously described (28). The extracellular laccase activity has been measured as described elsewhere (28) in the supernatants of basidiomycete I-62 cultures grown in these media and reached a maximal level (3 U/ml) at day 8, followed by a decrease (up to day 12) and a progressive increase in the late stationary phase, under induced conditions (28).
Total RNA was extracted from mycelia harvested on different days during the incubation period (27). Two independent nitrocellulose filters, with total RNA samples from day 4 of growth up to day 24, were prepared for each culture. Four RNA samples were loaded in all gels and used as internal controls for quantitation. Northern analyses were performed with lcc1-, lcc2-, and lcc3-specific probes under high-stringency hybridization conditions (33). Hybridization signals were quantified by using IMAGEQUANT software (Molecular Dynamics) and by following the recommendations of the supplier to get accurate results. We used, as a loading control, hybridization with a 0.83-kb NcoI-KpnI DNA fragment internal to the actin gene of Aspergillus nidulans (13, 15). The values for lcc1, lcc2, and lcc3 were normalized to the actin hybridization signals. We show that the actin heterologous probe is suitable for use in Northern analysis of basidiomycete RNA preparations, indicating the high level of identity among the actin genes from different groups of fungi. The use of actin as the loading control, coupled with the presence of samples common to all Northern analysis membranes, allowed us to rigorously quantitate the laccase transcripts, although we are aware that the carbon source might affect actin transcription (12) and comparisons should be taken with some caution.
Effect of veratryl alcohol on lcc1 and lcc2 transcription.
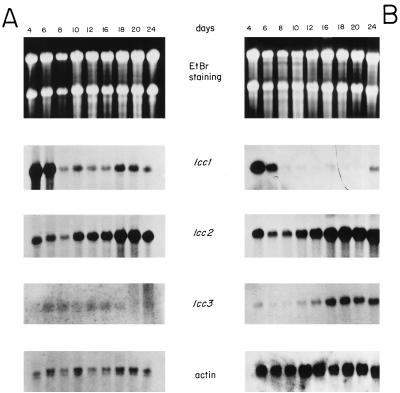
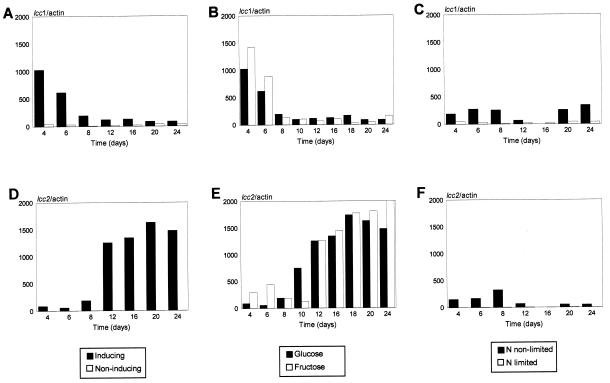
In the media without inducer, the transcripts for all three lcc genes were almost nondetectable (data not shown). Very intense hybridization signals were detected for lcc1 and lcc2 laccase genes under induced conditions, in both inducing and fructose media (Fig. 2). Interestingly, the presence of 4 mM veratryl alcohol in the media significantly increased the transcript level for two genes, lcc1 and lcc2, but in a different way. For lcc1, the increase appeared at the early exponential phase of growth (days 4 and 6), while the lcc2 transcript showed a peak at the end of the incubation period (days 20 and 24), in both inducing and fructose media (Fig. 2). Quantitation of both transcripts by using the actin signal as a loading control showed a clear induction effect (around 1,000-fold) for the lcc1 transcript at the beginning of growth in inducing medium (Fig. 3A), and a similar increase could be observed for the lcc2 transcript during the stationary phase (Fig. 3D). These results clearly prove the induction effect on lcc transcription produced by the addition of veratryl alcohol. Several compounds have been shown to work as inducers for laccase activity, like 2,5-xylidine and p-anisidine in T. villosa and R. solani, respectively (36, 38). In contrast, the expression of lcc3 does not seem to be induced by veratryl alcohol, but it could be sensitive to another inducer. Thus, none of the three laccase genes described up to now for the basidiomycete I-62 appeared to be constitutive, and indeed, almost no laccase activity is detected in noninduced cultures (28).
FIG. 2.
Northern analyses of lcc1, lcc2, and lcc3 transcription patterns. (A) Total RNA samples from mycelium growing on inducing medium; (B) total RNA samples from mycelium growing on fructose medium. The actin hybridization shown in panel B corresponds to a longer exposure time. EtBr, ethidium bromide.
FIG. 3.
Quantitation of lcc1 (A, B, and C) and lcc2 (D, E, and F) transcript levels. Quantitations were performed with a PhosphorImager screen and IMAGEQUANT software (Molecular Dynamics), and values are expressed in arbitrary units. Actin values were used as a loading control for each measurement. (A and D) Inducing and noninducing media; (B and E) glucose and fructose media; (C and F) nonlimiting and limiting nitrogen media. The standard error of quantifications was less than 5%.
Effect of nitrogen and carbon sources on lcc transcription.
Under nonlimiting nitrogen culture conditions, lcc1 and lcc2 transcript levels increased 100-fold compared to the transcript levels under limiting nitrogen conditions, although with both media being uninduced, the transcription levels are lower than those in induced media (Fig. 3C and F). Change in laccase activity responding to nitrogen source availability is a controversial subject. Some authors found that the ligninolytic enzyme activity increased under limiting conditions, and others described the opposite result (4, 6, 19, 25). Our results indicate that the lcc1 and lcc2 laccase genes of I-62 are slightly regulated by nitrogen at the mRNA level. Under non-nitrogen- limiting conditions, the transcript levels of lcc1 and lcc2 subtly increased (Fig. 3C and F), in agreement with the laccase activity levels (28).
The lcc3 transcript was hardly detectable in the inducing medium with glucose as the carbon source, but it appeared after 12 days of growth in the fructose medium. Thus, the most important fact revealed in the study of the regulation of lcc3 gene expression was that it seems to be subjected to carbon catabolite repression (Fig. 2). Interestingly, numerous CreA consensus sequences (24, 34) were found in the 5′ noncoding region of the lcc3 gene (data not shown), suggesting the existence of a carbon catabolite regulatory protein similar to CreA in A. nidulans (11). Nevertheless, the transcription patterns of the lcc1 and lcc2 genes did not change, within our resolution limits, when either glucose or fructose was supplied as the carbon source (Fig. 3B and E). The promoters of the three genes are now under study. Both lcc1 and lcc3 promoters (data not shown) contain multiple putative consensus metal response elements and xenobiotic response elements identical to those described by Brown et al. (5) and Fujisawa-Sehara et al. (16), respectively; these promoters also contain putative binding sites for a NIT2-like protein which could mediate some nitrogen metabolite regulation (21).
The transcription data are in agreement with the laccase activity measured in the supernatants being higher under inducing conditions (28). However, we cannot assign the laccase activity detected to any of the three cloned laccase genes. It is important to remember that there might be more laccase genes which have not yet been cloned or that these laccases might need some posttranslational modification to be active.
Our results show unequivocally that the lcc genes from the basidiomycete I-62 are differentially regulated, like the lcc genes from T. villosa (37) and the genes coding for lignin and manganese peroxidases in other white rot fungi (3, 4, 25, 26).
Acknowledgments
We thank M. Espinosa, G. del Solar, and A. D. W. Dobson for their critical reading of the manuscript.
M. Mansur acknowledges support from fellowships granted by the Instituto de Cooperación Iberoamericano and Dirección de Relaciones Internacionales del Consejo Superior de Investigaciones Cientificas, Madrid, Spain. This work was supported by grant BIO 93-0662-CO4-01 (CICYT, Madrid, Spain).
REFERENCES
- 1.Barr D P, Aust S D. Mechanisms white rot fungi use to degrade pollutants. Environ Sci Technol. 1994;28:78–86. doi: 10.1021/es00051a724. [DOI] [PubMed] [Google Scholar]
- 2.Bourbonnais R, Paice M G, Reid I D, Lanthier P, Yaguchi M. Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol. 1995;61:1876–1880. doi: 10.1128/aem.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broda P, Birch P R J, Brooks P R, Sims P F G. PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl Environ Microbiol. 1995;61:2358–2364. doi: 10.1128/aem.61.6.2358-2364.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broda P, Birch P R J, Brooks P R, Sims P F G. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol Microbiol. 1996;19:923–932. doi: 10.1046/j.1365-2958.1996.474966.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown J A, Alic M, Gold M H. Manganese peroxidase gene transcription in Phanerochaete chrysosporium: activation by manganese. J Bacteriol. 1991;173:4101–4106. doi: 10.1128/jb.173.13.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buswell J A, Cai Y J, Chang S. Effect of nutrient nitrogen and manganese on manganese peroxidase and laccase production by Lentinula (Lentinus) edodes. FEMS Microbiol Lett. 1995;128:81–88. [Google Scholar]
- 7.Buswell J A, Odier E. Lignin biodegradation. Crit Rev Biotechnol. 1987;6:1–60. [Google Scholar]
- 8.Call H P, Mücke I. History, overview and applications of mediated lignolytic systems, especially laccase-mediator-systems (Lignozym®-process) J Biotechnol. 1997;53:163–202. [Google Scholar]
- 9.Coll P M, Tabernero C, Santamaria R, Perez P. Characterization and structural analysis of the laccase I gene from the newly isolated lignolytic basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:4129–4135. doi: 10.1128/aem.59.12.4129-4135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P J, Dobson A D W. Regulation of laccase gene transcription in Trametes versicolor. Appl Environ Microbiol. 1997;63:3444–3450. doi: 10.1128/aem.63.9.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espeso E A, Peñalva M A. Carbon catabolite repression can account for the temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol Microbiol. 1992;6:1457–1465. doi: 10.1111/j.1365-2958.1992.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 13.Fidel S, Doonan J H, Morris N R. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a γ-actin gene. Gene. 1988;70:283–293. doi: 10.1016/0378-1119(88)90200-4. [DOI] [PubMed] [Google Scholar]
- 14.Field J A, de Jong E, Feijoo-Costa G, de Bont J A M. Screening for ligninolytic fungi applicable to the biodegradation of xenobiotics. Trends Biotechnol. 1993;11:44–49. [Google Scholar]
- 15.Fillinger S, Felenbok B. A newly identified gene cluster in Aspergillus nidulans comprises five novel genes localized in the alc region that are controlled both by the specific transactivator AlcR and the general carbon-catabolite repressor CreA. Mol Microbiol. 1996;20:475–488. doi: 10.1046/j.1365-2958.1996.5301061.x. [DOI] [PubMed] [Google Scholar]
- 16.Fujisawa-Sehara A, Yamame M, Fujii-Kuriyama Y. A DNA-binding factor specific for xenobiotic responsive element of P450c gene exists as a cryptic form in cytoplasm: its possible translocation to the nucleus. Proc Natl Acad Sci USA. 1988;263:885–896. doi: 10.1073/pnas.85.16.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germann U A, Müller G, Hunziker P E, Lerch K. Characterization of two allelic forms of Neurospora crassa laccase. J Biol Chem. 1988;263:885–896. [PubMed] [Google Scholar]
- 18.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold M H, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang K, Fujii I, Ebizuka Y, Gomi K, Sankawa U. Molecular cloning and heterologous expression of the gene encoding dihydrogeodin oxidase, a multicopper blue enzyme from Aspergillus terreus. J Biol Chem. 1995;270:21495–21502. doi: 10.1074/jbc.270.37.21495. [DOI] [PubMed] [Google Scholar]
- 21.Jarai G, Truong H N, Daniel-Vedele F, Marzluf G A. NIT2, the nitrogen regulatory protein of Neurospora crassa, binds upstream of nia, the tomato nitrate reductase gene, in vitro. Curr Genet. 1992;21:37–41. doi: 10.1007/BF00318652. [DOI] [PubMed] [Google Scholar]
- 22.Kirk T K, Farrell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 23.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 24.Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Alic M, Gold M H. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Alic M, Brown J A, Gold M H. Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Appl Environ Microbiol. 1995;61:341–345. doi: 10.1128/aem.61.1.341-345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockington R A, Sealy-Lewis H M, Scazzocchio C, Davies R W. Cloning and characterization of the ethanol utilisation regulon in Aspergillus nidulans. Gene. 1985;33:288–294. doi: 10.1016/0378-1119(85)90088-5. [DOI] [PubMed] [Google Scholar]
- 28.Mansur M, Suárez T, Fernández-Larrea J B, Brizuela M A, González A E. Identification of a laccase gene family in the new lignin-degrading basiodiomycete CECT 20197. Appl Environ Microbiol. 1997;63:2637–2646. doi: 10.1128/aem.63.7.2637-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Mansur, M., et al. Unpublished data.
- 29.Messner K, Srebotnik E. Biopulping: an overview of developments in an environmentally safe paper-making technology. FEMS Microbiol Rev. 1994;13:351–364. [Google Scholar]
- 30.Perry C R, Smith M, Britnell C H, Wood D A, Thurston C F. Identification of two laccase genes in the cultivated mushroom Agaricus bisporus. J Gen Microbiol. 1993;139:1209–1218. doi: 10.1099/00221287-139-6-1209. [DOI] [PubMed] [Google Scholar]
- 31.Reid I D, Paice M C. Biological bleaching of kraft pulp by white-rot fungi and their enzymes. FEMS Microbiol Rev. 1994;13:369–376. [Google Scholar]
- 32.Saloheimo M, Niku-Paavola M-L, Knowle J K C. Isolation and structural analysis of the laccase gene from the lignin-degrading fungus Phlebia radiata. J Gen Microbiol. 1991;137:1537–1544. doi: 10.1099/00221287-137-7-1537. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sophianopoulou V, Suárez T, Diallinas G, Scazzocchio C. Operator derepressed mutations in the proline utilisation gene cluster of Aspergillus nidulans. Mol Gen Genet. 1993;236:209–213. doi: 10.1007/BF00277114. [DOI] [PubMed] [Google Scholar]
- 35.Thurston C F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 36.Wahleithner J A, Xu F, Brown K M, Brown S H, Golightly E J, Halkier T, Kauppinen S, Pederson A, Schneider P. The identification and characterisation of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr Genet. 1996;29:395–403. doi: 10.1007/BF02208621. [DOI] [PubMed] [Google Scholar]
- 37.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalbøge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaver D S, Golightly E J. Cloning and characterization of three laccase genes from the white-rot basidiomycete Trametes villosa: genomic organization of the laccase gene family. Gene. 1996;181:95–102. doi: 10.1016/s0378-1119(96)00480-5. [DOI] [PubMed] [Google Scholar]