Abstract
Bacillus pumilus PS213 was found to be able to release acetate from acetylated xylan. The enzyme catalyzing this reaction has been purified to homogeneity and characterized. The enzyme was secreted, and its production was induced by corncob powder and xylan. Its molecular mass, as determined by gel filtration, is 190 kDa, while sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed a single band of 40 kDa. The isoelectric point was found to be 4.8, and the enzyme activity was optimal at 55°C and pH 8.0. The activity was inhibited by most of the metal ions, while no enhancement was observed. The Michaelis constant (Km) and Vmax for α-naphthyl acetate were 1.54 mM and 360 μmol min−1 mg of protein−1, respectively.
Xylan is an important constituent of hemicelluloses, and next to cellulose it is the most abundant renewable polysaccharide in nature. It is a β-1,4-linked d-xylose polymer with arabinofuranose, glucuronic acid, methylglucuronic acid, and acetyl side groups (22). Efficient and complete degradation of xylan requires the cooperation of xylanases and β-xylosidases with the following accessory enzymes: α-arabinofuranosidase, α-methylglucuronidase, acetyl xylan esterase (AXE) (1), and ferulic acid esterase (12). The AXE which liberates acetyl groups from the backbone of xylan has recently been studied in several fungi, including Aspergillus niger (11), Schizophyllum commune (9), Trichoderma reesei (13) and Penicillium purpurogenum (7), and also in bacteria such as Fibrobacter succinogenes (14), Pseudomonas fluorescens (8), Streptomyces lividans (6), and a Thermoanaerobacterium sp. (21).
Bacillus pumilus was reported to degrade xylan via xylanase and β-xylosidase enzymes (17, 18). Both genes coding for these proteins have been isolated (16); however, no accessory enzymes have yet been reported. In this report we describe the purification and biochemical characterization of the AXE from B. pumilus PS213, a strain which also has efficient xylanase activity.
Enzyme production.
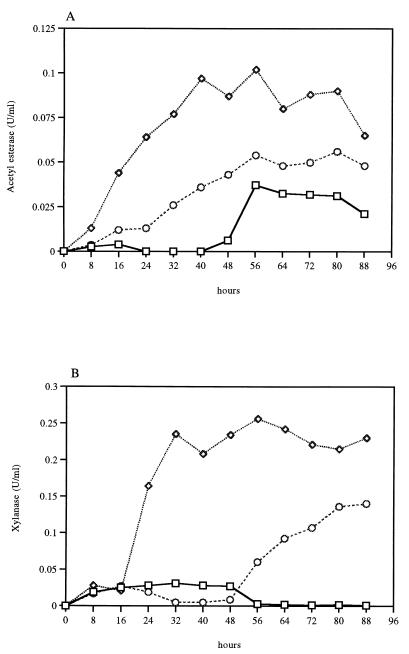
For monitoring the dynamics of xylanase and AXE production, B. pumilus was grown in M9CA medium plus 0.5% corncob powder and in M9CA medium plus 0.5% oat spelt xylan. Samples of the culture supernatant taken every 8 h were tested for both acetylesterase activity and xylanase activity. B. pumilus PS213 produced AXE and xylanase simultaneously when grown in the presence of corncob powder (Fig. 1). This enzyme released acetyl groups from acetylated xylan, as demonstrated by treating the substrate with the purified enzyme. The highest level of activity was found during the stationary phase, between 40 and 72 h of growth in the medium supplemented with corncob powder. In fungi maximal production of these enzymes can take up to several days (4, 9). A lower level of acetylesterase activity was detected in the medium containing 0.5% xylan, and a much lower level was detected in the same medium devoid of corncob powder. The majority (>60%) of the esterase activity was found in the culture supernatant, while the remaining activity was cell associated and was detected in the crude extract prepared as described previously (5). Xylanase activity is also induced by corncob powder and to a lesser extent by xylan. Both xylanase and AXE activities were induced by xylan which is not acetylated, indicating that the two enzymes may be coregulated. The production patterns of both enzymes are strongly related, suggesting cooperativity (Fig. 1).
FIG. 1.
Induction of acetylesterase (A) and xylanase (B) from B. pumilus PS213 grown on M9CA medium either alone (□) or with corncob powder (◊) or xylan (○).
Enzyme purification.
B. pumilus PS213 was grown in 800 ml of M9CA medium (20) supplemented with 0.5% (wt/vol) corncob powder. Purification was performed at room temperature with a low-pressure liquid chromatography system (GradiFrac; Pharmacia Biotech, Uppsala, Sweden). The culture supernatant was subjected to (NH4)2SO4 fractionation (30 and 70% saturation). The 70% pellet was resuspended in 100 mM sodium phosphate (pH 7)–1.7 M (NH4)2SO4, filtered through 0.45-μm-pore-size membrane, and fractionated by hydrophobic interaction chromatography (phenyl Sepharose HP 16/10; Pharmacia Biotech), as described previously (5). Active fractions were pooled and dialyzed against 20 mM bis-Tris buffer (pH 7), concentrated by ultrafiltration with a YM30 membrane (Amicon Inc., Beverly, Mass.), applied to a Q Sepharose fast-flow column, and fractionated (5). Active fractions were pooled, concentrated to 1 ml, and loaded onto a gel filtration column (Sephacryl HR200, column XK16; Pharmacia Biotech), previously equilibrated with 50 mM sodium phosphate–150 mM NaCl (pH 7). Proteins were eluted at a flow rate of 0.5 ml/min, and fractions of 2.5 ml were collected. The column was calibrated with an MW-GF-200 kit (Sigma Chemical Co., St. Louis, Mo.) for molecular weight estimation.
Enzyme purification is summarized in Table 1. After gel filtration, the enzyme was purified to electrophoretic homogeneity (Fig. 2). Only one band was obtained when the purified protein was loaded onto a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel. Purification steps indicated the presence of a single AXE in the supernatant of the B. pumilus culture; multiple AXEs have been found in the culture filtrate of some fungi such as T. reesei (3) and P. purpurogenum (7). Also, in a Thermoanaerobacterium sp., two distinct AXEs have been purified and characterized (21), but they were cell associated, with less than 25% in the culture supernatant, whereas for B. pumilus over 60% of the AXE is secreted.
TABLE 1.
Purification of B. pumilus AXE
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Purifi- cation factor | Yield (%) |
|---|---|---|---|---|---|
| Culture supernatant | 103.4 | 68.7 | 0.66 | 1 | 100 |
| (NH4)2SO4 fractionation | 56 | 51.4 | 0.91 | 1.37 | 74.8 |
| Q Sepharose fast flow | 22.5 | 47.2 | 2.1 | 3.18 | 68.7 |
| Phenyl Sepharose HR | 1.65 | 20.2 | 12.2 | 18.5 | 29.4 |
| Q Sepharose fast flow | 0.35 | 12.5 | 35.9 | 54.4 | 18.2 |
| Sephacryl HR200 | 0.065 | 7.72 | 118.8 | 179.2 | 11.2 |
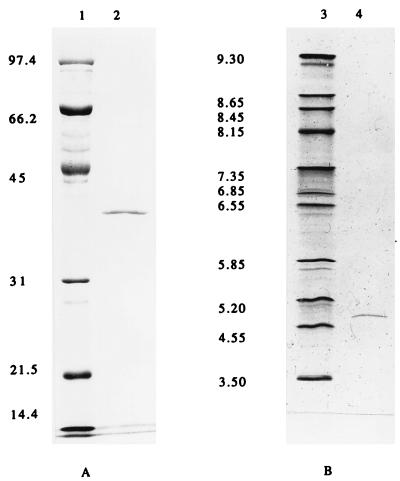
FIG. 2.
SDS-polyacrylamide gel electrophoresis (A) and analytical isoelectric focusing (B) of the purified acetylesterase. Lanes: 1, molecular mass standard; 2, 5 μg of acetylesterase; 3, pI markers, consisting of trypsinogen (9.30), lentil lectin (8.65, 8.45, and 8.15), myoglobin (7.35 and 6.85), human carbonic anhydrase B (6.55), bovine carbonic anhydrase (5.85), β-lactoglobulin A (5.2), soybean trypsin inhibitor (4.55), and amyloglucosidase (3.5); 4, 2.5 μg of acetylesterase.
Enzyme assays.
Acetylesterase activity was measured by using 2 mM α-naphthyl acetate as the substrate (19). AXE activity was measured with acetylated oat spelt xylan, prepared in accordance with the method of Johnson et al. (10). The reaction mixture comprised 500 μl of acetylated xylan (5% suspension in 50 mM sodium phosphate buffer; pH 7.0), 450 μl of 50 mM sodium phosphate buffer (pH 7.0), and 50 μl of purified enzyme (0.79 μg of protein). The incubation was at 37°C with orbital shaking (150 rpm) for 1 h. The deacetylation of xylose tetra-acetate and glucose penta-acetate was determined as described for acetylated oat spelt xylan except that 500 μl of each substrate (1.25 mM; in 50 mM sodium phosphate buffer [pH 7.0]) was used for the reaction. The activity with p-nitrophenyl acetate and 4-methylumbelliferyl acetate was determined by monitoring photometrically the release of p-nitrophenol at a wavelength of 410 nm (λ410) (2) and of 4-methylumbelliferone at λ354 (21). Xylanase activity was tested by measuring the release of reducing sugar from oat spelt xylan by the dinitrosalicilic acid method (15). The liberated acetic acid from acetylated substrates was quantified with an enzymatic analysis kit from Boehringer Mannheim (catalog no. 148261) according to the manufacturer’s instructions. One unit of enzyme released 1 μmol of product min−1 under the assay conditions. The protein concentration was estimated with Bio-Rad protein assay kit I, with bovine serum albumin as the reference.
The enzyme demonstrated a broad spectrum of activity on a variety of substrates, including both aryl and carbohydrate acetyl esters. The activities against acetylated xylan and sugars are presented in Table 2. The enzyme showed a high level of specific activity on acetyl xylan, 40 U/mg, which is comparable to the value of 23 U/mg reported for Schizophyllum commune (9). On the other hand, lower levels of specific activity were reported for the AXEs of Thermomonospora fusca, F. succinogenes, and Thermoanaerobacterium spp. (esterases I and II): 0.6, 8.63, and 5.2 and 12.4 U/mg, respectively. The purified enzyme hydrolyzed p-nitrophenyl acetate and 4-methylumbelliferyl acetate, releasing p-nitrophenol and 4-methylumbelliferone, respectively, with acetic acid.
TABLE 2.
Substrate specificity of purified esterase
| Substrate | Sp act (U/mg)a |
|---|---|
| Acetylated xylan | 40.97 ± 8.00 |
| Xylose tetra-acetate | 37.16 ± 6.25 |
| Glucose penta-acetate | 31.37 ± 5.79 |
| Methylumbelliferyl acetate | 34.00 ± 2.00 |
| p-Nitrophenyl acetate | 32.00 ± 3.00 |
| α-Naphthyl acetate | 118.80 ± 20.28 |
Values are means of triplicate determinations ± standard deviations.
Enzyme characterization.
SDS-polyacrylamide gel electrophoresis (5% stacking gel, 12% resolving gel) was performed by the method of Sambrook et al. (20). Protein bands were stained with Coomassie blue R-250 after electrophoresis. The molecular mass of the purified enzyme was estimated to be 190 kDa by Sephacryl HR200 gel filtration with gel filtration molecular weight markers MW-GF-200 (Sigma). The enzyme consisted of one type of subunit with a molecular mass of 40 kDa on SDS-polyacrylamide gel (Fig. 2A). These data suggested that AXE could be a homotetramer or a homopentamer.
The pI of the AXE was determined by using an Ampholine PAGplate precast polyacrylamide gel (Pharmacia Biotech), with pH values ranging from 3 to 10, and by using the broad-pI calibration kit (Pharmacia Biotech) as the pI marker, in accordance with the instructions of the supplier. The pI value was estimated to be 4.8 (Fig. 2B) from a plot of migration distance versus the pI values of the standards, with the help of an UltroScan XL laser densitometer (Pharmacia Biotech).
The purified protein was subjected to N-terminal amino acid sequence determination by automated Edman degradation on a pulsed liquid-phase protein sequencer (model 470A; Applied Biosystems, Foster City, Calif.) equipped with an on-line phenylthiohydantoin amino acid analyzer (model 120A; Applied Biosystems). The amino acid sequence of an internal fragment of the purified protein was also determined after trypsin digestion. The band of 40 kDa which belongs to the purified AXE revealed the following N-terminal amino acid sequence: MQLFD LFLEE LG. The internal amino acid sequence determined is the following: ALEVI QSFPE VDEHR. The N-terminal sequence, when subjected to a FastA homology search, showed 70% identity with those of two xylanase precursors from Clostridium thermocellum (XynX) and Thermoanaerobacterium saccharolyticum (XynA) (data not shown), while the internal amino acid sequence is 80% identical to that of a cephalosporin-C deacetylase from Bacillus subtilis (data not shown). At this stage we do not know the significance of this homology.
The optimal pH and temperature were determined in the range from pH 3 to 9.5 (50 mM sodium acetate, pH 3 to 5.5; sodium phosphate, pH 6.0 to 7.0; Tris-HCl, pH 7.5 to 9.5) and 4 to 80°C, respectively. For the pH stability determination, samples were incubated in buffers from pH 3.0 to 9.0 and at 37°C for 60 min. For the determination of thermal stability, temperatures of 37, 50, 60, 65, and 70°C were used at pH 7.0 for 135 min, with measurement of residual activity at 15-min intervals. The remaining activity was assayed under standard conditions as described above. The optimal temperature and pH were about 55°C and 8.0, respectively. The optimal pH of 8.0 is similar to the pH values of 7.7 reported for Schizophyllum commune (9) and 7.5 for Streptomyces lividans (6). The optimal temperature and pH were about 55°C and 8.0, respectively. The optimal pH of 8.0 is similar to the pH values of 7.7 reported for Schizophyllum commune (9) and 7.5 for Streptomyces lividans (6). The optimal temperature of 55°C is, however, lower than those reported for other microorganisms such as Streptomyces lividans and a Thermoanaerobacterium sp. (70°C and 80°C, respectively). The AXE was stable at 50°C and was rapidly inactivated at temperatures higher than 60°C, with a half-life of about 1 h at this temperature. The pH stability results showed that AXE was stable in the alkaline pH range, exhibiting almost 100% of its total activity between pH 8.0 and 9.5 (data not shown).
The initial velocity of AXE was determined in 20 mM sodium phosphate buffer (pH 7.0) at 37°C over the substrate concentration range of 0.08 to 4 mM α-naphthyl acetate. A Lineweaver-Burk plot showed a linear response over this concentration range. The Michaelis constant (Km) was 1.54 mM α-naphthyl acetate, and the maximal velocity (Vmax) was 360 μmol min−1 mg−1. The Km value of 1.54 mM determined for the purified esterase is lower than the value of 2.7 reported for F. succinogenes (14) and higher than the Kms of 0.45 and 0.52 of the two esterases from a Thermoanaerobacterium sp. (21), which were determined by using 4-methylumbelliferyl acetate as the substrate. It is much lower than the Km of 23 reported for the acetylesterase from A. niger with p-nitrophenyl acetate as the substrate (11).
Little information has been produced to date on the effect of cations on the AXEs. We found that the activity was not significantly affected by any of the cations tested at a 2 mM concentration, while at 10 mM many of the chemicals affected enzyme activity. B. pumilus AXE is markedly inhibited by Fe3+, Cu2+, Zn2+, Mn2+, Co2+, Ca2+, and Ag+ at a concentration of 10 mM, which is in accordance with the behavior of AXE from Schizophyllum commune (9). The greatest inhibitory effect was recorded with Fe3+; Cu2+ and Zn2+ were the divalent cations with the greatest inhibitory effect. No stimulatory effect was observed.
In conclusion, our results show that B. pumilus PS213 has potent AXE and xylanase activities. Moreover, the AXE purified and characterized here has properties potentially useful for pulp biobleaching by xylanases.
REFERENCES
- 1.Biely P, MacKenzie C R, Puls J, Schneider H. Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Bio/Technology. 1986;4:731–733. [Google Scholar]
- 2.Biely P, Puls J, Schneider H. Acetyl xylan esterases in fungal cellulolytic systems. FEBS Lett. 1985;186:80–84. [Google Scholar]
- 3.Biely P, MacKenzie C R, Schneider H, Puls J. The role of fungal acetyl xylan esterases in the degradation of acetyl xylan by fungal xylanases. In: Kennedy J F, Phillips G O, Williams P A, editors. Wood and cellulosics. New York, N.Y: Ellis Horwood, Ltd.; 1987. pp. 283–289. [Google Scholar]
- 4.Christov L P, Prior B A. Esterases of xylan-degrading microorganisms: production, properties, and significance. Enzyme Microb Technol. 1993;15:460–475. doi: 10.1016/0141-0229(93)90078-g. [DOI] [PubMed] [Google Scholar]
- 5.Degrassi G, Polverino de Laureto P, Bruschi C V. Purification and characterization of ferulate and p-coumarate decarboxylase from Bacillus pumilus. Appl Environ Microbiol. 1995;61:326–332. doi: 10.1128/aem.61.1.326-332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupont C, Daigneault N, Shareck F, Morosoli R, Kluepfel D. Purification and characterization of an acetyl xylan esterase produced by Streptomyces lividans. Biochem J. 1996;319:881–886. doi: 10.1042/bj3190881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egana L, Gutierrez R, Caputo V, Peirano A, Steiner J, Eyzaguirre J. Purification and characterization of two acetyl xylan esterases from Penicillium purpurogenum. Biotechnol Appl Biochem. 1996;24:33–39. [PubMed] [Google Scholar]
- 8.Ferreira L M, Wood T M, Williamson G, Faulds C, Hazlewood G P, Black G W, Gilbert H J. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993;294:349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halgasova N, Kutejova E, Timko J. Purification and some characteristics of the acetyl xylan esterase from Schizophyllum commune. Biochem J. 1994;298:751–755. doi: 10.1042/bj2980751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson K G, Fontana J D, MacKenzie C R. Measurement of acetyl xylan esterase in Streptomyces. Methods Enzymol. 1988;160:551–560. [Google Scholar]
- 11.Linden J, Samara M, Decker S, Johnson E, Boyer M, Pecs M, Adney W, Himmel M. Purification and characterization of an acetyl esterase from Aspergillus niger. Appl Biochem Biotechnol. 1994;45–46:383–393. doi: 10.1007/BF02941813. [DOI] [PubMed] [Google Scholar]
- 12.MacKenzie C R, Bilous D. Ferulic acid esterase from Schizophyllum commune. Appl Environ Microbiol. 1988;54:1170–1173. doi: 10.1128/aem.54.5.1170-1173.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margolles-Clarke E, Tenkanen M, Soderlund H, Penttila M. Acetyl xylan esterase from Trichoderma reesei contains an active-site serine residue and a cellulose binding domain. Eur J Biochem. 1996;237:553–560. doi: 10.1111/j.1432-1033.1996.0553p.x. [DOI] [PubMed] [Google Scholar]
- 14.McDermid K P, Forsberg C W, MacKenzie C R. Purification and properties of an acetylxylan esterase from Fibrobacter succinogenes S85. Appl Environ Microbiol. 1990;56:3805–3810. doi: 10.1128/aem.56.12.3805-3810.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller G L. Use of dinitrosalicylic reagent for determination of reducing groups. Anal Chem. 1959;31:426–428. [Google Scholar]
- 16.Moriyama H, Fukusaki E, Cabrera Crespo J, Shinmyo A, Okada H. Structure and expression of genes coding for xylan-degrading enzymes of Bacillus pumilus. Eur J Biochem. 1987;166:539–545. doi: 10.1111/j.1432-1033.1987.tb13547.x. [DOI] [PubMed] [Google Scholar]
- 17.Panbangred W, Kondo T, Negoro S, Shinmyo A, Okada H. Molecular cloning of the genes for xylan degradation of Bacillus pumilus and their expression in Escherichia coli. Mol Gen Genet. 1983;192:335–341. doi: 10.1007/BF00392172. [DOI] [PubMed] [Google Scholar]
- 18.Panbangred W, Kawaguchi O, Tomita T, Shinmyo A, Okada H. Isolation of two β-xylosidase genes of Bacillus pumilus and comparison of their gene products. Eur J Biochem. 1984;138:267–273. doi: 10.1111/j.1432-1033.1984.tb07911.x. [DOI] [PubMed] [Google Scholar]
- 19.Poutanen K, Sundberg M. An acetyl esterase of Trichoderma reesei and its role in the hydrolysis of acetyl xylans. Appl Microbiol Biotechnol. 1988;28:419–424. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Shao W, Wiegel J. Purification and characterization of two acetyl xylan esterases from Thermoanaerobacterium sp. strain JW/SL-YS485. Appl Environ Microbiol. 1995;61:729–733. doi: 10.1128/aem.61.2.729-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whistler R L, Richards E L. Hemicelluloses. In: Pigman W, Horton D, editors. The carbohydrates—chemistry and biochemistry. New York, N.Y: Academic Press; 1970. pp. 447–469. [Google Scholar]