Abstract
The NCI-MATCH (Molecular Analysis for Therapy Choice) trial (NCT02465060) was launched in 2015 as a genomically-driven, signal-seeking precision medicine platform trial — largely for patients with treatment-refractory, malignant solid tumors. Having completed in 2023, it remains one of the largest tumor-agnostic, precision oncology trials undertaken to date. Nearly 6000 patients underwent screening and molecular testing, with a total of 1473 (inclusive of continued accrual from standard next-generation sequencing) being assigned to one of 38 substudies. Each substudy was a Phase II trial of a therapy matched to a genomic alteration, with a primary endpoint of objective tumor response by RECIST criteria. Here, we summarize the outcomes of the initial 27 substudies in NCI-MATCH, which met its signal-seeking objective with 7/27 positive substudies (25.9%). In this Perspective, we discuss key aspects of the design and operational conduct of the trial, highlighting important lessons for future precision medicine studies.
Introduction
It has long been known that certain malignancies are primarily driven by ‘driver’ mutations and that inhibition of the affected pathways can lead to a substantial anti-tumor response and survival advantage. A well-established example is chronic myelogenous leukemia, driven by a translocation of chromosome 9 and 22 which results in formation of the BCR/ABL fusion oncogene. The use of tyrosine kinase inhibitors against this driver has reduced the annual mortality by a factor of 10 – from 10–20% annually to 1–2% annually (1). Other notable examples include EGFR mutations in a subset of lung cancers and breast cancers driven by overexpression of HER2, to name but a few (2) — leading to the idea of precision oncology, whereby a treatment is targeted to specific molecular driver.
It has also become clear that some driver mutations can occur across different tumor histologies but crucially, these can confer susceptibility to targeted therapies in some tumor sites but not in others. An early example of this complexity was provided by drugs directed to the V600E mutation in the BRAF gene; while high response rates (40% or more) (3) were observed in melanoma patients with the aberration, colorectal cancer patients were almost uniformly resistant (4). With the development of massively parallel high-throughput genomic sequencing (next-generation sequencing, NGS) more than a decade ago (5), the broader picture of DNA aberrations across cancers began to emerge, along with myriad potential therapeutic opportunities.
With these considerations in mind, the ECOG-ACRIN Cancer Research Group collaborated with the National Cancer Institute (NCI) to design a trial that would systematically evaluate the activity of the many emerging targeted drugs across a range of different cancer diagnoses. Planning began in 2013 and the NCI-MATCH trial launched in 2015 — at that time representing the largest tumor-histology agnostic, genomically-driven clinical trial yet undertaken (6,7). At the time of its planning, numerous studies attested to the variability of results obtained from different next-generation sequencing (NGS) platforms (8,9), prompting implementation of a uniform assay (Oncomine™, Thermo-Fisher) in four credentialed laboratories, to be applied across all samples (10). Updates to the assay platform were made to keep pace with scientific developments in the field, including designated immunohistochemical biomarkers for patient selection (11), all the while maintaining a consistent platform for the screened population. When the screening reached the target accrual of 6000 in July 2017, the trial was continued using commercial and academic laboratory testing, until it closed in January 2023. In this Perspective, we summarize the key features of the trial design and conduct, as well as some key challenges and how we addressed these along the way. We discuss the outcomes and limitations, and what they mean for patients and clinicians. The results provide a perspective on the interpretation of the NCI-MATCH trial, and its implications for the design of successor studies.
Trial Design and Conduct
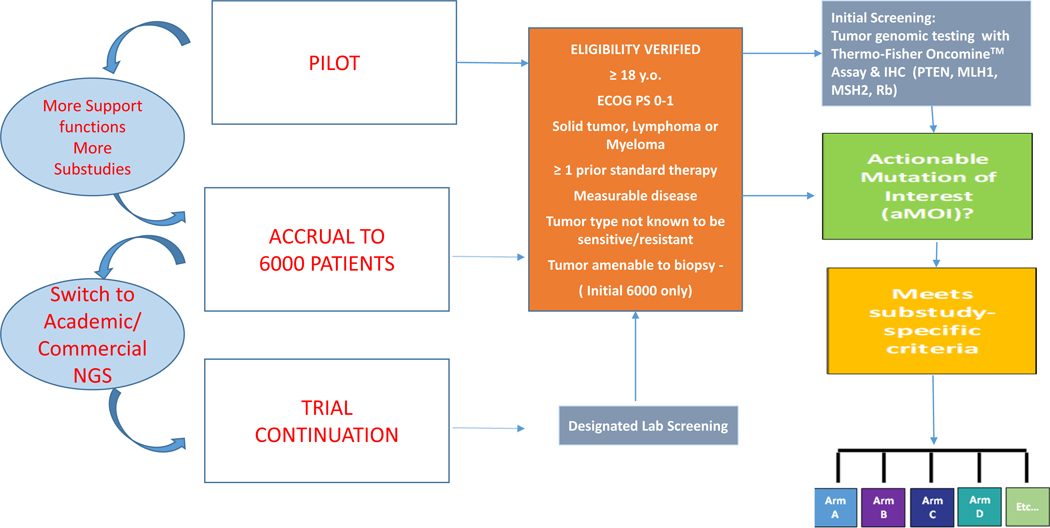
The design of NCI-MATCH (Fig 1) has been outlined in detail (6,7). Each target molecular aberration had to be supported as a driver of tumor growth, and the paired drug demonstrated to inhibit either the driver itself, or the growth pathway activated by it. Each list of qualifying molecular aberrations for a gene was defined through database searches for new experimental or clinical data, and was updated regularly as the trial proceeded.
Figure 1. NCI-MATCH Platform Trial Design.
The trial was implemented in three parts, sequentially: a pilot phase (795 patients) that led to multiple modifications; a screening accrual phase (~6000 patients); and a continuation phase whereby patients were recruited from a network of academic and commercial laboratories. In the initial 6000-patient screening period, eligibility was assessed prior to obtaining a dedicated biopsy, while during the continuation period, tumor samples were assayed as part of standard clinical practice, and eligibility assessed when a candidate mutation was identified. In either case, eligible patients with qualifying tumor molecular aberrations were assigned to a therapeutic substudy, to receive treatment directed to their molecular profile. As a further quality control measure, for patients tested at designated labs, confirmation sequencing was conducted by the NCI-MATCH Central Labs and only patients whose tumor genomic profile was confirmed in this manner were included for efficacy endpoints. Each substudy was a separate Phase II trial constituting a drug-genomic driver pair. Patients were assigned to substudies with the assistance of a decision tool (MATCHBox), overseen for appropriateness by a team of medical oncologists, laboratory scientists, and bioinformaticists from the trial leadership that reviewed every patient. In each substudy, the initial aim was to accrue 35 patients, assuming 31 would be eligible and start protocol treatment (analyzable); there was provision to increase accrual to 70 in selected arms. The primary endpoint for each substudy was objective response rate (ORR), defined as rate of complete or partial response, as assessed by RECIST guidelines (12).
Pilot Phase of Trial Initiation
The NCI-MATCH trial incorporated an initial pilot phase of accrual to assess the distribution of genomic aberrations (given limited sequencing data at the time for metastatic cancers), and to evaluate the performance of all elements needed to provide therapeutic options to patients across a wide geographic area. In the initial three months, the trial had 10 treatment arms, and the central network comprising four laboratories – harmonized to run assays on an identical platform – was responsible for all the sequencing (6). Unanticipated enthusiasm in the research community for this trial revealed a number of practical needs — including higher tissue processing and sequencing throughput to enable acceptable turnaround times, and a support desk and broad educational program for clinical providers. The pilot also revealed the need for a substantially higher total screening accrual, since estimates of prevalence based on The Cancer Genome Atlas data (from less advanced cancers) were incorrect by a factor of two or more, and also revealed the need for a greater number of treatment arms, to provide options for patients with less prevalent actionable mutations or alterations. Similar needs had been identified in the earlier SHIVA trial (13), conducted with a small number of treatment arms across multiple institutions in France — indicating the importance of large numbers of therapeutic options in future precision oncology trials.
As had been hoped at the outset, most of the accrued patients were not from the four most common tumor types (breast, lung, colon and prostate cancers); over 60% were from less common or rare histologic types [Table 1] (6). Furthermore, new biopsy specimens of metastatic disease were successfully acquired (7), with the aim of capturing new genomic changes that may have occurred since earlier collection of primary tumor tissue (6).
Table 1.
Total accrual by Disease to NCI-MATCH.
| Screening Cohort N = 6390 |
Outside Assay N = 762 |
|
|---|---|---|
| Colorectal Cancer | 963 (15.1%) | 76 (10%) |
| Breast | 764 (12%) | 85 (11.2%) |
| Ovarian | 610 (9.5%) | 38 (5.0%) |
| NSCLC | 485 (7.6%) | 53 (7.0%) |
| Pancreas | 413 (6.5%) | 25 (3.3%) |
| Uterine Cancer | 402 (6.3%) | 70 (9.2%) |
| Liver and Hepatobiliary Cancer | 290 (4.5%) | 39 (5.1%) |
| Sarcoma | 288 (4.5%) | 29 (3.8%) |
| Head and Neck | 239 (3.7%) | 34 (4.5%) |
| Neuroendocrine Cancer | 214 (3.3%) | 11 (1.4%) |
| Gastroesophageal Cancer | 211 (3.3%) | 34 (4.5%) |
| Prostate Cancer | 157 (2.5%) | 33 (4.3%) |
| Bladder/Urothelial | 108 (1.7%) | 17 (2.2%) |
| Cervical Cancer | 103 (1.6%) | 9 (1.2%) |
| CNS | 103 (1.6%) | 106 (13.9%) |
| Small Cell Lung Cancer | 90 (1.4%) | 4 (0.5%) |
| Melanoma | 85 (1.3%) | 14 (1.8%) |
| Kidney | 83 (1.3%) | 12 (1.6%) |
| Lymphoma | 55 (0.9%) | 1 (0.1%) |
| Mesothelioma | 55 (0.9%) | 2 (0.3%) |
| Anal Cancer | 52 (0.8%) | 6 (0.8%) |
| Myeloma | 1 (<0.1%) | 1 (0.1%) |
| Other | 619 (9.7%) | 63 (8.3%) |
Screening of 6000 patients with a Dedicated Assay
Following the pilot phase, 24 available treatment arms (subsequently expanded to 38 in total) were approved, and a revised goal for 6000 patients to be screened was set. With the increase in available therapies, the treatment assignment rate increased from ~8% to 17.8%, despite each arm of the trial excluding patients if it was already known (based on phase 2 or 3 data) that the drug was either active or inactive in that patient’s cancer type (7). The proportion of patients with an actionable mutation (one for which any targeted therapy was available within NCI-MATCH or outside the trial) was over twice this rate, at 37.6% — indicating that nearly two of five patients with advanced cancer may have a candidate treatment revealed by next-generation sequencing (NGS) of their tumor. Given the continuing development of targeted drugs and therapeutic protocols, this finding in an unselected population may be a starting point for future trials designed to investigate and establish the efficacy of molecularly targeted therapies and their contribution to patient outcome. The proportion of screened patients who were actually registered for treatment was 12.4%, representing 70% of all patients assigned to a treatment arm; interim disease progression or consent withdrawal are believed to be the major contributors to the fractional registration.
Trial Continuation beyond Central Molecular Screening
At the conclusion of the biopsy screening portion of the trial in July 2017 (two years ahead of schedule), 5961 patients had been enrolled for molecular profiling, and 11 treatment substudies had completed full accrual. By that time, the sequencing landscape had also changed, and greater methodological consistency resulted in high reproducibility of genomic findings across a variety of sequencing platforms (9). Also, there was a desire to speed up the identification of patients with rare targetable tumor mutations. To this end, NCI-MATCH expanded its reach by engaging a network of academic and commercial labs (termed the NCI-MATCH Designated Laboratory Network), that performed NGS assays as routine care at sites participating in the trial. After a careful vetting process, a total of 30 laboratories (12 commercial and 18 academic), were approved to identify patients for this next phase of the trial, resulting in the treatment of an additional 512 patients. In addition, evidence accumulating external to the trial did not show a major difference between molecular abnormalities found at initial diagnosis and those found in metastatic disease (2). Accordingly, the trial was continued using the archived specimens that are usually analyzed in genomic laboratories, without a requirement for a new biopsy. This approach has now been implemented for all current NCI-sponsored solid tumor precision medicine trials.
Outcomes and Evaluation of NCI-MATCH
Was the trial feasible?
Feasibility was a concern at the outset of the trial. Thus, the statistical plan included monitoring for insufficient accrual and lack of activity among the arms, as well as stopping rules built into the individual substudies. Concerns about accrual were swept away promptly: registration of 6000 patients in just 15 months was unmatched in the history of ECOG-ACRIN therapeutic studies. Several other genomically-targeted therapy studies conducted in the same time-frame also support feasibility. One example is the SHIVA trial, conducted in France, with 10 treatment arms (13). Though there were limitations in terms of the availability of therapies in the SHIVA trial, it accrued 741 patients in 21 months. Another trial in France that required re-biopsy (MOSCATO-01) accrued 1035 patients over 51 months (14). Hyman et al reported on a study of neratinib in 141 patients with a variety of cancers whose tumors harbored mutations in ERRB2 and ERRB3 (15). A large trial of genomically-defined maintenance therapy in colorectal cancer (MODUL) has been completed with a total accrual of 824 patients (16). The VIKTORY trial in Korea accrued 772 patients with gastric cancer in a period of 52 months (17). These and other trials attest to the ability of the oncology community, researchers, and patients together, to develop and implement precision oncology trials. Response data are now available for on a substantial proportion (27/38) of the NCI-MATCH treatment arms (discussed below), and eight arms have been closed based on very low prevalence of the targeted aberration.
Did NCI-MATCH achieve its signal-seeking goal?
The goal of NCI-MATCH was to understand the activity of molecularly targeted therapy applied to cancer gene-defined subsets across different tumor histologies, and to document response — regardless of tumor histologic type. The purpose was signal-seeking: a specific target number of positive Phase II studies was not defined, nor was it attempted to define a concept of ‘tumor-agnostic activity’ in which an agent could be defined as broadly active in cancers with the targeted molecular alteration. Sample size and power calculations stipulated that, for a substudy with 31 analyzable patients, five or more responses (partial or complete) would be required, i.e., objective response rate (ORR) ≥ 5/31 (16%). Given the admixture of multiple tumor types to be accrued in each arm, this ORR was considered indicative of activity across tumor types and worthy of further investigation in a tumor-type agnostic fashion.
A summary of the outcomes of the 27 substudies reported to date is shown in Table 2. Seven of the 27 arms (25.9%) met the pre-specified criterion for positivity. Other arms had lower response rates, some of which may support future development with combinations, or with single agents in specific tumor types. In fact, at least one response was observed in 22/27 (81.5%) of the substudies. Across all the treatment arms reported to date, the overall response rate among evaluable patients was 79/765 (10.3%). In terms of breadth of activity, two of the arms – nivolumab in mismatch repair-deficient tumors and dabrafenib plus trametinib in BRAF V600 mutant tumors (37, 21) – identified activity across a broad range of malignancies. These results contributed to tumor type-agnostic registration for nivolumab, and an accelerated approval for the combination of dabrafenib plus trametinib in patients with BRAF V600-mutant tumors. These FDA approvals provide strong support for the impact of genomically-driven trials for patients, especially those with less common diseases. It should be noted that despite the modest impact of early inhibitors, signals were also obtained in select genotypes with PI3K-PTEN-AKT pathway aberrations. The AKT inhibitors capivasertib and ipatasertib had almost identical response rates in AKT E17-mutated cancers, a mutually validating result (34,40). Copanlisib in PIK3CA-mutated tumors also provided a signal of activity, with a response rate of 16% and 38% of patients free of progression at 6 months (38). These results also provide additional impetus for the study of mutation-specific agents directed to these targets.
Table 2.
Outcomes of the initial 27 substudies (of 38 total) in NCI-MATCH.
| Arm | Molecular Aberration | Treatment | N Enrolled | N Evaluable† | Number of Responses (%) | 6-month PFS | Ref | Met Endpoint?* |
|---|---|---|---|---|---|---|---|---|
| A | EGFR activating mutations | afatinib | 19 | 14 | 1 (7.1%) | 8.9% | 18 | No |
| B | HER2 activating mutations | afatinib | 40 | 37 | 1 (2.7%) | 12.0% | 19 | No |
| F | ALK fusions | crizotinib | 5 | 4 | 2 (50.0%) | 25% | 20 | Yes |
| G | ROS1 fusions | crizotinib | 4 | 4 | 1 (25.0%) | 50% | 20 | No |
| H | BRAF V600E or V600K mutations | Dabrafenib/trametinib | 35 | 29 | 11 (37.9%) | 68.4% | 21 | Yes |
| I | PIK3CA mutation without RAS mutation or PTEN loss | taselisib | 70 | 61 | 0.0% | 19.9% | 22 | No |
| J | HER2 amplification | Trastuzumab/pertuzumab | 35 | 25 | 3 (12%) | 25.3% | 23 | No |
| K2 | FGFR mutation/fusion | erdafitinib | 35 | 21 | 3 (14.3%) | 36.8% | 24 | Yes |
| M | TSC1 or TSC2 Mutations | TAK-228 | 49 | 34 | 5 (14.7%) | 28.7% | 25 | No |
| N | PTEN aberration, with + expression on IHC | GSK2636771 | 24 | 22 | 0.0% | 4.8% | 26 | No |
| P | PTEN loss by IHC | GSK2636771 | 35 | 32 | 0.0% | 3.3% | 26 | No |
| Q | HER2 amplification | ado-trastuzumab emtansine | 38 | 36 | 2 (5.6%) | 23.6% | 27 | No |
| R | BRAF fusions/non-V600 mutations | trametinib | 35 | 32 | 1 (3.0%) | 17% | 28 | No |
| S1 | NF1 mutation | trametinib | 50 | 46 | 2 (4.3%) | 20.5% | 29 | No |
| S2 | GNAQ or GNA11 mutation | trametinib | 4 | 4 | 1 (25%) | 50% | 29 | No |
| T | SMO or PTCH1 mutations | vismodegib | 34 | 22 | 2 (9.1%) | 22.4% | 30 | No |
| U | NF2 mutation | Defactinib | 35 | 30 | 1 (3.3%) | 22.8% | 31 | No |
| V | C-kit mutations | Sunitinib | 10 | 8 | 2 (25%) | 25% | 32 | No |
| W | FGFR pathway aberrations | AZD4547 | 52 | 48 | 4 (8.3%) | 15.0% | 33 | No |
| Y | AKT mutations | capivasertib | 35 | 35 | 10 (28.6%) | 50.0% | 34 | Yes |
| Z1A | NRAS mutations | binimetinib | 53 | 47 | 1 (2.1%) | 29.2% | 35 | No |
| Z1B | CCND1/2/3 amp and Rb + | palbociclib | 40 | 32 | 0.0% | 16.0% | 36 | No |
| Z1D | dMMR status | nivolumab | 47 | 42 | 15 (35.7%) | 51.3% | 37 | Yes |
| Z1F | PIK3CA | copanlisib | 35 | 25 | 4 (16.0%) | 38% | 38 | Yes |
| Z1H | PTEN mut without PTEN protein loss | copanlisib | 35 | 23 | 1 (4.3%) | 14.3% | 39 | No |
| Z1K | AKT mutation | Ipatasertib | 35 | 26 | 6 (23.1%) | 52.4% | 40 | Yes |
| Z1L | BRAF Fusions or Non-V600E, Non-V600K BRAF Mutations | Ulixertinib | 35 | 26 | 0.0% | 5% | 41 | No |
Eligible, treated and variant confirmed by central laboratory testing.
A substudy with 31 or more analyzable patients was to be called positive if the null hypothesis of objective response rate (ORR) ≤ 5% could be rejected at the one-sided type 1 error rate of 1.8%; if there were fewer than 31 analyzable patients, a type I error of 5.0% was used. This requires five or more responses (partial or complete) for a substudy with 31 analyzable patients, i.e., ORR > 5/31 (16%).
Therefore, even in a heavily pretreated population, the goal of identifying signals was met, although the majority of the arms did not meet the activity threshold, and none of the positive arms had response rates in the range of highly active targeted single agents, e.g. imatinib in gastrointestinal stromal tumor (GIST) (42). Consequently, NCI-MATCH led to additional questions about what factors influence response to a given agent when the targeted mutation is present. Illuminating such complexity may be possible upon completion of additional sequencing (whole exome, RNAseq, and others) and circulating tumor DNA (ctDNA) analyses of pretreatment and progression samples — and may encourage combination studies at earlier stages of patients’ treatment courses.
Limitations of NCI-MATCH
Design Limitations.
This trial does not provide an evaluation of the efficacy of using genomics to target molecular abnormalities in patients with metastatic cancer. The ‘match rate’ refers only to patients who had the index molecular variant when an appropriate substudy was available. At the time of screening, the appropriate substudy for any given patient might not have been available, and thus the patient could not be accrued. In regard to response, NCI-MATCH was a signal-seeking trial, with response assessed in a tumor-agnostic way — and as such the analysis could not provide response rates for specific tumor types as, by design, it was not powered to accrue enough patients to do so.
Operational Limitations.
Even with a central institutional review board, the time it took to open a substudy was long, sometimes approaching a year. Various scenarios contributed, including issues with drug supply or change in formulation, awaiting phase I results, protocol preparation, education of site staff and individual site review, and stress on the clinical and ethical protocol evaluation system. In some cases, not all patients accrued could be evaluated for the primary endpoint, due to lack of tissue availability for confirmatory sequencing by the NCI-MATCH assay. In future, this confirmatory step should not be necessary, given the high reproducibility of the assay.
Diversity of Accrual.
The racial/ethnic composition of the patients accrued to NCI-MATCH did not mirror distributions within the US population at large. Among all registered patients, 9.3% were Black, 5.6% were Hispanic and 3.9% were Asian. All these frequencies appear to be lower than catchment area population representation, although a formal analysis has not been conducted and comparator data are difficult to collect, due to the large number of sites (n=367) that accrued at least one patient. Proportions of different groups were almost identical in both the initial screening phase and subsequent periods of accrual, consistent with similar profiles of participating oncologists. In the absence of detailed information on social determinants of health for participants in the NCI-MATCH accrual, there remain questions as to how geographically and socioeconomically representative the trial population is. More detailed reporting of social determinants of health, together with pro-active outreach to underserved populations, is needed to provide access for a diverse population to precision oncology trials.
Implications of the NCI-MATCH Trial
This trial was established during a period of controversy over the value of genomically-driven clinical trials, whether or not such trials could be accomplished by the National Clinical Trials Network, and whether any patient benefit would be realized. The results have highlighted specific topics to be addressed, based upon the lessons learned (Box 1). These fall into four main categories: the definition of rare tumors, the relative contributions of tissue of origin and molecular subtype to outcomes, next steps to design genomic cancer clinical trials, and understanding of co-mutations and the tumor microenvironment.
Box 1. Key lessons from NCI-MATCH to guide future precision medicine trials.
Pro-active outreach is needed to assure optimal patient (and provider) diversity.
Rare tumors are an area of unmet need that can be met (at least in part) with genomic trials; but novel trial designs and regulatory approaches are needed.
A clinically-relevant definition of ‘driver mutation’ will be helped by rigorous criteria for evidence to support the matching of therapy to mutation, enabling greater therapeutic activity.
Circumvention of resistance mechanisms will be helped by intervention earlier in the disease course and progress will be accelerated by combination approaches involving targeted therapies and immunotherapies.
Trial design should encompass as many therapeutic options for as many molecular aberrations as possible, so as to have an impact commensurate with the collective effort required.
Defining Rare Tumors
Rare tumors have typically been specified by histology or tissue of origin, and are conventionally considered those with a prevalence of less than 15 per 100,000 population (43). By this definition, all but 11 tumor types are classified as rare. Pediatric tumors also need to be considered by these criteria, and the initiation of the Pediatric-MATCH clinical trial (NCT03155620) recognizes this need (44); a retrospective review of all screened cases applied the WHO International Classification of Diseases for Oncology (ICD-O-3), and this provided greater diagnostic specificity compared to the MedDRA disease coding captured during the study (unpublished). In addition, the advent of broadly-available sequencing suggests two further rare tumor groups: unusual genomic profiles within a particular tumor type, and unusual genomic drivers across tumor types. Certain genomic aberrations are closely associated with some rare tumor subtypes, but may also be found sporadically in other tumor types, e.g. neurotrophic tyrosine receptor kinase (NTRK) fusions (45) (NTRK inhibitors have received disease-agnostic approval).
A question of therapeutic interest has long been whether the histologic tumor type or the genomic driver should be the more important consideration in choosing a treatment. Regardless of how a ‘rare tumor’ is defined, it is evident that a precision medicine approach is required for the 25% of adults who have a tumor type that is below a prevalence of 15/100,000, and for additional genomic and histopathological subsets of cancer. We emphasize the precision approach since it has both immediate and long-term implications for cancer diagnosis and treatment: immediate, in that many therapies are already approved, or are in development for specific genomic aberrations. These rare tumors provide a proof of principle: that outcomes are better when a vulnerability can be identified, even when we may not have all the tools at our disposal to effect a cure. But as importantly, the classification of these rare tumors opens for patients the possibility of clinical research options that would otherwise not be available, permits the design of a series of trials to build upon positive results, and may facilitate regulatory decision-making. Further, the NCI-MATCH trial shows that such treatment can be delivered in community as well as academic settings.
The finding that 38% of the accrual to the NCI-MATCH trial was in rare or uncommon cancers (defined histologically) raises issues relevant to treatment of these patients (46). Overall, the frequency of ‘actionable’ genomic aberrations is similar among rare versus common cancers, as is the degree of benefit from interventions (47). Further, the use of more extensive analyses such as whole-genome sequencing may identify actionable aberrations in as many as 62% of a large sample (including rare and common cancers) (48). Therefore, a strong rationale exists to examine genomic characteristics of all rare tumors, given that they have fewer treatment options to begin with.
How are the successes in this setting to be made widely available (or commercialized, which generally amounts to the same thing) to patients with rare tumors? The FDA Oncology Center of Excellence has provided extensive guidance to address this issue, with a recent focus on the opportunities to bring real-world evidence to bear on trials in rare tumors (). In genomic subgroups in which a response signal has been observed, how will it be feasible to develop studies to render this signal more effective, if not curative? Large-scale deep-sequencing analyses of samples from patients treated within substudies of NCI-MATCH are ongoing, and will be hypothesis-generating as to molecular characteristics that influence response (49). In future trials, such comprehensive analysis methods (that are available at major centers) would need to be incorporated into the clinical trial procedures within a practical time-frame, using tissues or other samples that can be acquired, transported, stored and analyzed uniformly.
Impact of Tissue of Origin vs Molecular Subtype in Response
Though not the primary goal of NCI-MATCH, the trial has made several contributions to this dialogue in precision medicine. One of the most revealing and impactful results emerged from the BRAF substudy (Arm H in Table 2), in which patients harboring a BRAF V600 mutation were offered a combination of dabrafenib (a BRAF inhibitor) and trametinib (a MEK inhibitor) as dual inhibition of the MAP kinase signaling pathway. As noted previously, RAF-directed therapy has proven highly effective in some diseases (such as melanoma) (3), but is almost inactive in colon cancer with a BRAF V600 mutation (4). In NCI-MATCH, tumor types such as these (as well as lung and thyroid cancers), in which the effect of combined therapy was already known, were excluded from the substudy. Patients in the initial 35-patient cohort harboured a broad range of histologies and the response rate (38%) was substantial (21). These results strongly suggest that response to inhibiting this pathway is indeed disease-agnostic. With additional disease-specific sensitivities observed in a parallel Novartis trial (50), the FDA was approached by Novartis and ECOG-ACRIN for a disease-agnostic indication, which was approved under Accelerated Approval provisions in June 2022.
These findings contrast with those of capivasertib and ipatasertib (arms Y and Z1K, respectively) in AKT E17K-mutated cancers, where lower response rates (around 20%) were identified in the substudies for each agent. In both treatment arms, the aberration was identified most frequently in women’s cancers, including breast, endometrial, ovarian, and cervical cancers. Furthermore, responses were largely confined to these tissue types, suggesting that both tissue of origin and mutation are required for response in these circumstances. Future trials will be needed to further address this issue.
Relevance to Genomic Cancer Trials
Beyond demonstration of feasibility, one must ask how these results inform the development of future studies directed to patients with genomically-defined cancers. The characterization of molecular changes in individual cancers has led the field of precision medicine and has greatly changed the outcomes of treatment for specific patients. One need only look at the falling death rates from lung cancer (where EGFR and ALK-directed therapies have had a clear impact) (51), or the improved outcomes for patients with metastatic melanoma (once it became possible to target mutant BRAF) (52), to appreciate that effective molecular medicine has changed standards of care in oncology. It may also be pointed out that the incremental survival benefits observed in patients with metastatic melanoma harboring mutant BRAF, translates to higher cure rates when brought forward to the adjuvant treatment setting (53). In the NCI-MATCH trial, in all but three arms, the intervention tested was a single agent. The limitations of single agent therapies in this trial echo what has been observed for many years with traditional chemotherapy and anti-HER2 therapy in breast cancer, for example. In this context, the overall response rate of 10.3% observed in NCI- MATCH (across all arms) may be viewed as meaningful for future research in the area. The implications of these results are as much strategic for therapeutics as they are specific to trial design. Sequencing of cancer tissues is needed to identify potential vulnerabilities in the tumor, and this trial provides an impetus to direct treatment to these vulnerabilities as early as possible in a patient’s course. It is recognized that single agent treatments will have limited effects, and that rational combinations to overcome resistance should be explored early, especially in potentially curative settings.
This was not a trial in which therapy directed to resistant disease could be explored. It does, however, set the stage for future studies of this nature, and a trial called ComboMATCH (NCT05564377), also to be coordinated by ECOG-ACRIN, will investigate combinations of therapies in similar molecular subtypes of cancer (54). The combinations to be tested in ComboMATCH will require in vivo evidence of efficacy for the combination in well-characterized relevant tumor models and will be restricted to targeted therapies. Given that many of the frequently co-occurring mutations are currently considered undruggable, research on combinations with immune therapies and other modalities should be a high priority.
Co-Mutations and the Tumor Microenvironment
A major finding of the molecular analysis of NCI-MATCH (and other sequencing studies of advanced cancers (47,48)) was that most patients whose tumors had a qualifying mutation also had at least one co-occurring mutation that was known – based on preclinical evidence – to contribute to drug resistance. Some of these have already been outlined (7), and additional sequencing and ctDNA analysis is underway to characterize more completely the tumors of treated patients. Most of these additional mutations (for example in TP53, KRAS, p16, MYC) are currently undruggable, although candidate molecules are in development. Nearly all arms of NCI-MATCH were targeting oncogenic alterations that are known to be truncal. This circumstance facilitated reliance on the Designated Laboratory Network and use of archival tumor specimens as a source for sequencing. However, many of the co-occurring alterations were subclonal, and so possibly acquired in response to prior therapy (55). For trials aiming to overcome resistance mediated by these co-occurring alterations, repeat tumor biopsies or ctDNA analysis of mutation profiles continue to be important considerations.
Alternative approaches to addressing resistance in these subclones should also be explored. Hahn et al have recently characterized cancer targets as either ‘intrinsic’ to the cancer itself (such as oncogenes, as well as epigenetic, metabolic, transcriptional or signaling dysregulation, and DNA damage response aberration) or ‘extrinsic’, involving cellular components of the tumor microenvironment (such as immune cells, cancer-associated fibroblasts, and blood vessels) – all of which contribute to tumor growth and progression, and are therefore plausible targets (56). The combination of targeted therapies with agents directed at the tumor microenvironment is yielding regimens with markedly enhanced activity. The multi-kinase inhibitor cabozantinib, combined with the anti-PD-1 antibody nivolumab in kidney cancer, is a salient recent example (57) — as is vemurafenib (BRAF inhibitor) with rituximab (anti-CD20) in hairy-cell leukemia (58), with many more trials in prospect. Precision cancer studies should include collection of appropriate specimens for research into which molecular or patient characteristics contribute to response or resistance to combined targeted and immunological treatments. A precision medicine trial that would address such combinations directed to genomically-defined subsets is a current need with considerable therapeutic potential.
Conclusion
In addition to its impact in clinical research, the NCI-MATCH trial emphasizes the importance of tumor DNA sequencing as part of the standard evaluation of most patients with cancer, given that more than one-third of patients will be shown to have a cancer for which the outcome can be improved with targeted therapy. Since the disease type alone cannot predict the molecular characteristics of individual tumors, implementing such interventions is not currently possible without genomic sequence information.
There are often questions as to the value of routine sequencing; we emphasize that NCI-MATCH was not set up to address this question. Relating the number of responses to the number screened is not meaningful; while some 17% of those screened could be assigned to a treatment arm, there was a total of 38% with driver mutations for whom a treatment could have been available. Assignment to treatment in NCI-MATCH depended on whether the substudy was open or closed in general, and for the patient’s tumor type in particular — as some tumors were more likely to have a given genomic variant, and the numbers of such tumors in a given substudy were restricted to allow a broader recruitment of tumor types, as required by study design. In addition, patients with tumors for which a treatment was known to be effective were not eligible, since the purpose of the trial was to address what was unknown. For all these reasons, while the value of a genomic screening policy is not represented by any proportion of responders to numbers screened in NCI-MATCH, the results clearly support availability of NGS to patients with advanced cancer.
Nevertheless, it is abundantly clear with emerging data that diseases such as melanoma, lung cancer, colon cancer, and gynecologic malignancies, as well as some rarer cancers, all now have expanded treatment options resulting from available detailed sequence analyses. Furthermore, the increasing technological capacity of sequencing and other emerging genomic characterization tools to identify evolution of sub-populations of tumor cells over time holds the promise of early approaches to target resistant clones. Although not addressed by NCI-MATCH, germline sequencing of cancer patients is also receiving considerable attention, and has been implemented in Pediatric MATCH (44,59).
These results have implications for the critical elements of future platform trial designs: rigorous molecular characterization and objective assignment of patients to treatment arms; tumor sample acquisition to enable retrospective additional sequencing and immune system evaluation to define better who will respond; pathology review of tumor specimens from treated patients as a key quality factor; and sufficient numbers of treatment arms to warrant the efforts both in the cooperative groups that run the trial, and in the community, where the resources for trial activation and management compete with other research priorities. Put another way, to be motivated to open a complex trial, oncologists must have confidence that their patients will benefit through availability of novel therapies that would be otherwise inaccessible.
Footnotes
All authors made substantial contributions to conception and design, or acquisition of data, or analysis and interpretation of data; assisted in drafting the article and/or revising it critically for important intellectual content; approved the final version to be published.
Contributor Information
Peter J. O’Dwyer, University of Pennsylvania, Philadelphia, PA, USA.
Robert J. Gray, Dana-Farber Cancer Institute – ECOG-ACRIN Biostatistics Center, Boston, MA, USA.
Keith T. Flaherty, Massachusetts General Hospital, Boston, MA, USA.
Alice P. Chen, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA.
Shuli Li, Dana-Farber Cancer Institute – ECOG-ACRIN Biostatistics Center, Boston, MA, USA.
Victoria Wang, Dana-Farber Cancer Institute – ECOG-ACRIN Biostatistics Center, Boston, MA, USA.
Lisa M. McShane, Biometric Research Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA
David Patton, Center for Biomedical Informatics & Information Technology, National Cancer Institute, Bethesda, MD, USA.
James V. Tricoli, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA
P. Mickey Williams, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
A. John Iafrate, Massachusetts General Hospital, Boston, MA, USA.
Jeffrey Sklar, Yale University, New Haven, CT, USA.
Edith P. Mitchell, Thomas Jefferson University Hospital, Philadelphia, PA, USA
Naoko Takebe, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA.
David J. Sims, Frederick National Laboratory for Cancer Research, Frederick, MD, USA
Brent Coffey, Center for Biomedical Informatics & Information Technology, National Cancer Institute, Bethesda, MD, USA.
Tony Fu, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Mark Routbort, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Larry V. Rubinstein, Biometric Research Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA
Richard F. Little, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA
Carlos L. Arteaga, UT Southwestern Simmons Comprehensive Cancer Center, Dallas, TX, USA
Donna Marinucci, ECOG-ACRIN Cancer Research Group, Philadelphia, PA, USA.
Stanley R. Hamilton, City of Hope National Medical Center, Duarte, CA, USA.
Barbara A. Conley, Cancer Diagnosis Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA.
Lyndsay N. Harris, Cancer Diagnosis Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA.
James H. Doroshow, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD, USA.
REFERENCES
- 1.Jabbour E and Kantarjian H. Chronic Myelogenous Leukemia: 2020 Update on diagnosis, therapy and monitoring. Am J Hematol. 2020. Jun;95(6):691–709. doi: 10.1002/ajh.25792. [DOI] [PubMed] [Google Scholar]
- 2.Waarts MR, Stonestrom AJ, Park YC and Levine RL. Targeting mutations in cancer. J Clin Invest. 2022. Apr 15;132(8):e154943. doi: 10.1172/JCI154943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010. Aug 26;363(9):809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopetz S, Desai J, Chan E, Hecht JR, O’Dwyer PJ, Maru D, Morris V, Janku F, Dasari A, Chung W, Issa JP, Gibbs P, James B, Powis G, Nolop KB, Bhattacharya S, Saltz L. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J Clin Oncol. 2015. Dec 1;33(34):4032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008. Apr 17;452(7189):872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Gray R, Chen A, Li S, Patton D, Hamilton SR, Williams PM, Mitchell EP, Iafrate AJ, Sklar J, Harris LN, McShane LM, Rubinstein LV, Sims DJ, Routbort M, Coffey B, Fu T, Zwiebel JA, Little RF, Marinucci D, Catalano R, Magnan R, Kibbe W, Weil C, Tricoli JV, Alexander B, Kumar S, Schwartz GK, Meric-Bernstam F, Lih CJ, McCaskill-Stevens W, Caimi P, Takebe N, Datta V, Arteaga CL, Abrams JS, Comis R, O’Dwyer PJ, Conley BA; NCI-MATCH Team. The Molecular Analysis for Therapy Choice (NCI-MATCH) trial: lessons for genomic trial design. J Natl Cancer Inst. 2020. Oct 1;112(10):1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty KT, Gray RJ, Chen AP, Li S, McShane LM, Patton D, Hamilton SR, Williams PM, Iafrate AJ, Sklar J, Mitchell EP, Harris LN, Takebe N, Sims DJ, Coffey B, Fu T, Routbort M, Zwiebel JA, Rubinstein LV, Little RF, Arteaga CL, Comis R, Abrams JS, O’Dwyer PJ, Conley BA; NCI-MATCH team. Molecular Landscape and Actionable Alterations in a Genomically Guided Cancer Clinical Trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J Clin Oncol. 2020. Nov 20;38(33):3883–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E, Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Commitee ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xuan J, Yu Y, Qing T, Guo L, Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340:284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lih CJ, Harrington RD, Sims DJ, Harper KN, Bouk CH, Datta V, Yau J, Singh RR, Routbort MJ, Luthra R, Patel KP, Mantha GS, Krishnamurthy S, Ronski K, Walther Z, Finberg KE, Canosa S, Robinson H, Raymond A, Le LP, McShane LM, Polley EC, Conley BA, Doroshow JH, Iafrate AJ, Sklar JL, Hamilton SR, Williams PM. Analytical Validation of the Next-Generation Sequencing Assay for a Nationwide Signal-Finding Clinical Trial: Molecular Analysis for Therapy Choice Clinical Trial. J Mol Diagn. 2017. Mar;19(2):313–327. doi: 10.1016/j.jmoldx.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoury JD, Wang WL, Prieto VG, Medeiros LJ, Kalhor N, Hameed M, Broaddus R, Hamilton SR. Validation of Immunohistochemical Assays for Integral Biomarkers in the NCI-MATCH EAY131 Clinical Trial. Clin Cancer Res. 2018. ;24:521–531. doi: 10.1158/1078-0432.CCR-17-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009. Jan;45(2):228–47. [DOI] [PubMed] [Google Scholar]
- 13.Le Tourneau C, Delord JP, Gonçalves A, Gavoille C, Dubot C, Isambert N, Campone M, Trédan O, Massiani MA, Mauborgne C, Armanet S, Servant N, Bièche I, Bernard V, Gentien D, Jezequel P, Attignon V, Boyault S, Vincent-Salomon A, Servois V, Sablin MP, Kamal M, Paoletti X; SHIVA investigators. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015. Oct;16(13):1324–34. [DOI] [PubMed] [Google Scholar]
- 14.Massard C, Michiels S, Ferté C, Le Deley MC, Lacroix L, Hollebecque A, Verlingue L, Ileana E, Rosellini S, Ammari S, Ngo-Camus M, Bahleda R, Gazzah A, Varga A, Postel-Vinay S, Loriot Y, Even C, Breuskin I, Auger N, Job B, De Baere T, Deschamps F, Vielh P, Scoazec JY, Lazar V, Richon C, Ribrag V, Deutsch E, Angevin E, Vassal G, Eggermont A, André F, Soria JC. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017. Jun;7(6):586–595. doi: 10.1158/2159-8290. [DOI] [PubMed] [Google Scholar]
- 15.Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, Juric D, Quinn DI, Moreno V, Doger B, Mayer IA, Boni V, Calvo E, Loi S, Lockhart AC, Erinjeri JP, Scaltriti M, Ulaner GA, Patel J, Tang J, Beer H, Selcuklu SD, Hanrahan AJ, Bouvier N, Melcer M, Murali R, Schram AM, Smyth LM, Jhaveri K, Li BT, Drilon A, Harding JJ, Iyer G, Taylor BS, Berger MF, Cutler RE Jr, Xu F, Butturini A, Eli LD, Mann G, Farrell C, Lalani AS, Bryce RP, Arteaga CL, Meric-Bernstam F, Baselga J, Solit DB. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018. Feb 8;554(7691):189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmoll HJ, Arnold D, de Gramont A, Ducreux M, Grothey A, O’Dwyer PJ, Van Cutsem E, Hermann F, Bosanac I, Bendahmane B, Mancao C, Tabernero J. MODUL-a multicenter randomized clinical trial of biomarker-driven maintenance therapy following first-line standard induction treatment of metastatic colorectal cancer: an adaptable signal-seeking approach. J Cancer Res Clin Oncol. 2018. Jun;144(6):1197–1204. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Kim ST, Kim K, Lee H, Kozarewa I, Mortimer PGS, Odegaard JI, Harrington EA, Lee J, Lee T, Oh SY, Kang JH, Kim JH, Kim Y, Ji JH, Kim YS, Lee KE, Kim J, Sohn TS, An JY, Choi MG, Lee JH, Bae JM, Kim S, Kim JJ, Min YW, Min BH, Kim NKD, Luke S, Kim YH, Hong JY, Park SH, Park JO, Park YS, Lim HY, Talasaz A, Hollingsworth SJ, Kim KM, Kang WK. Tumor Genomic Profiling Guides Patients with Metastatic Gastric Cancer to Targeted Treatment: The VIKTORY Umbrella Trial. Cancer Discov. 2019. Oct;9(10):1388–1405. doi: 10.1158/2159-8290. [DOI] [PubMed] [Google Scholar]
- 18.Reckamp KL, Song Z, Gettinger SN, Mitchell EP, Wright JJ, Moscow JA, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II trial of afatinib in patients with EGFR-mutated solid tumors excluding lung cancer: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) Subprotocol A. Presented at: 34th EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; October 26–28, 2022; Barcelona, Spain. Abstract 235 [Google Scholar]
- 19.Bedard PL, Li S, Wisinski KB, Yang ES, Limaye SA, Mitchell EP, Zwiebel JA, Moscow JA, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of Afatinib in patients with tumors with HER2 activating mutations: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) sub-protocol EAY131-B. JCO Precis Oncol. 2022; 6: e2200165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansfield AS, Wei Z, Mehra R, Shaw AT, Lieu CH, Forde PM, Drilon AE, Mitchell EP, Wright JJ, Takebe N, Sharon E, Hovelson D, Tomlins S, Zeng J, Poorman K, Malik N, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams M, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Crizotinib in patients with tumors harboring ALK or ROS1 rearrangements: results from the NCI-MATCH Trial (EAY131) sub-protocols F and G. NPJ Precis Oncol. 2022; 6(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salama AKS, Li S, Macrae ER, Park JI, Mitchell EP, Zwiebel JA, Chen HX, Gray RJ, McShane LM, Rubinstein LV, Patton D, Williams PM, Hamilton SR, Armstrong DK, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Dabrafenib and Trametinib in Patients With Tumors With BRAF V600E Mutations: Results of the NCI-MATCH Trial Subprotocol H. J Clin Oncol. 2020. Nov 20;38(33):3895–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krop IE, Jegede O, Grilley-Olson JE, Lauring JD, Mitchell EP, Zwiebel JA, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Kono SA, Ford JM, Garcia AA, Sui XD, Siegel RD, Slomovitz BM, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of Taselisib in PIK3CA-mutated solid tumors other than breast and squamous lung cancer: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) Sub-protocol I. JCO Precis Oncol. 2022; 6: e2100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connolly RM, Wang X, Hyman DM, Grivas P, Mitchell EP, Wright JJ, Sharon E, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, Arteaga CL, Harris L, O’Dwyer PJ, Chen AP, Flaherty KT. Activity of trastuzumab and pertuzumab in patients with non-breast/gastroesophageal HER2 amplified tumors: Results of the NCI-MATCH trial (EAY131) Subprotocol J. Ann Oncol. 2020; 31(S4): S479–S480. ABSTRACT 553P [Google Scholar]
- 24.Mita AC, Wei Z, Mayer IA, Cheng HH, Mitchell EP, Wright JJ, Ivy SP, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams M, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Erdafitinib in patients with tumors harboring FGFR gene mutations or fusions: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) Sub-protocol K2. Mol. Cancer Ther. 2021; 20(12). ABSTRACT LBA003 [Google Scholar]
- 25.Hays JL, Song Z, Paik PK, Iyer GV, Mitchell EP, Wright JJ, Doyle L, Gray RJ, Wang V, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) - Phase 2 Study of MLN0128 (TAK-228) in Patients with Tumors with TSC1 or TSC2 Mutations: Sub-protocol EAY131-M. Presented at: 34th EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; October 26–28, 2022; Barcelona, Spain. Abstract 73 [Google Scholar]
- 26.Janku F, Jegede O, Puhalla SL, Konstantinopoulos PA, Meric-Bernstam F, Mitchell EP, Zwiebel JA, McShane L, Li S, Rubinstein LV, Doyle LA, Patton DR, Conley BA, O’Dwyer PJ, Harris LN, Arteaga CL, Williams P, Hamilton SR, Chen AP, Flaherty KT. NCI-MATCH Arms N & P; Phase II study of PI3K-beta inhibitor GSK2636771 in patients (pts) with cancers (ca) with PTEN mutation/deletion (mut/del) or PTEN protein loss. Ann Oncol. 2018; 29(8). ABSTRACT 418PD [Google Scholar]
- 27.Jhaveri K, Wang X, Makker V, Luoh S, Mitchell EP, Zwiebel JA, Sharon E, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams P, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Ado-trastuzumab emtansine (T-DM1) in patients with HER2 amplified tumors excluding breast and gastric/gastro-esophageal junction (GEJ) adenocarcinomas: Results from the NCI-MATCH Trial (EAY131) Sub-protocol Q. Ann Oncol. 2019; 30(11): 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson DB, Zhao F, Noel MS, Riely GJ, Mitchell EP, Wright JJ, Chen HX, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams P, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Trametinib activity in patients with solid tumors and lymphomas harboring BRAF Non-V600 mutations or fusions: Results from NCI-MATCH (EAY131). Clin Cancer Res. 2020; 26(8): 1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisinski KB, Flamand Y, Wilson MA, Luke JJ, Tawbi HA, Hong F, Mitchell EP, Zweibel JA, Chen HX, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Behrens RJ, Pennington KP, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Trametinib in Patients with NF1, GNAQ or GNA11 Mutant Tumors: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) Subprotocols S1 and S2. Ann Oncol [DOI] [PMC free article] [PubMed]
- 30.Tsao AS, Song Z, Ho AL, Mehnert JM, Mitchell EP, Wright JJ, Takebe N, Gray RJ, Wang V, McShane LM, Rubinstein LV, Patton DR, Williams M, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of vismodegib in patients with SMO or PTCH1 mutated tumors: Results from NCI-MATCH ECOG-ACRIN Trial (EAY131) Subprotocol T.. J Clin Oncol. 2022; 40(16_suppl). ABSTRACT 3010 [Google Scholar]
- 31.Jackman DM, Jegede O, Zauderer MG, Mitchell EP, Zwiebel JA, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams P, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. A phase 2 study of defactinib (VS-6063) in patients with NF2 altered tumors: Results from NCI-MATCH (EAY131) Subprotocol U. J Clin Oncol. 2021; 39(15). ABSTRACT 3087 [Google Scholar]
- 32.Gien LT, Song Z, Poklepovic A, Collisson EA, Mitchell EP, Zwiebel JA, Harris P, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of Sunitinib in tumors with c-KIT mutations: Results from the NCI-MATCH ECOG-ACRIN trial (EAY131) Subprotocol V. Presented at: 34th EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; October 26–28, 2022; Barcelona, Spain. Abstract 238 [Google Scholar]
- 33.Chae Y, Hong F, Vaklavas C, Cheng HH, Hammerman PS, Mitchell EP, Zwiebel JA, Ivy SP, Gray RJ, Li S, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Mansfield AS, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of AZD4547 in patients with tumors harboring aberrations in the FGFR pathway: Results from the NCI-MATCH Trial (EAY131) Subprotocol W. J Clin Oncol. 2020; 38(21): 2407–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinsky KM, Hong F, McCourt CK, Sachdev JC, Mitchell EP, Zwiebel JA, Doyle LA, McShane LM, Li S, Gray RJ, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, O’Dwyer PJ, Harris LN, Arteaga CL, Chen AP, Flaherty KT. Effect of Capivasertib in patients with an AKT1 E17K-mutated tumor: NCI-MATCH subprotocol EAY131-Y nonrandomized trial. JAMA Oncology. 2020: e206741. [DOI] [PMC free article] [PubMed]
- 35.Cleary JM, Wang X, Heist RS, Kopetz ES, Mitchell EP, Zwiebel JA, Kapner KS, Chen HX, Li S, Gray RJ, McShane LM, Rubinstein LV, Patton DR, Meric-Bernstam F, Dillmon MS, Williams M, Hamilton SR, Conley BA, Aguirre A, O’Dwyer PJ, Harris LN, Arteaga CL, Chen AP, Flaherty KT. Differential outcomes in codon 12/13 and codon 61 NRAS-mutated cancers in the phase 2 NCI-MATCH trial of binimetinib in patients with NRAS-mutated tumors. Clin Cancer Res. 2021; 27(11): 2996–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark AS, Hong F, Finn RS, DeMichele AM, Mitchell EP, Zwiebel JA, Arnaldez FI, McShane L, Li S, Gray RJ, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Copur MS, Kasbari SS, Thind R, Conley BA, O’Dwyer PJ, Harris LN, Arteaga C, Chen AP, Flaherty KT. Molecular analysis for therapy choice (NCI-MATCH, EAY131) Arm Z1B: Phase II trial of palbociclib for CCND1, 2 or 3 amplified tumors. Cancer Res. 2020; 79(13S). ABSTRACT LB-010 [Google Scholar]
- 37.Azad NS, Gray RJ, Overman MJ, Schoenfeld JD, Mitchell EP, Zwiebel JA, Sharon E, Streicher HZ, Li S, McShane L, Rubinstein LV, Patton DR, Williams P, Coffey B, Hamilton SR, Bahary N, Suga JM, Hatoum H, Abrams JS, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Nivolumab is effective in mismatch repair-deficient noncolorectal cancers: Results from arm Z1D-A subprotocol of the NCI-MATCH (EAY131) Study. J Clin Oncol. 2020; 38(3): 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damodaran S, Zhao F, Deming DA, Mitchell EP, Wright JJ, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Suga JM, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of Copanlisib in patients with tumors with PIK3CA mutations: Results from the NCI-MATCH ECOG-ACRIN trial (EAY131) Sub-protocol Z1F. J Clin Oncol. 2022; 40(14): 1552–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janku F, Wei Z, Davies MA, Mitchell EP, Wright JJ, Doyle LA, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams M, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Phase II study of PI3K inhibitor copanlisib in patients with cancers with deleterious PTEN sequencing results and retained PTEN protein expression: Results from the NCI-MATCH Trial (EAY131) Sub-protocol Z1H. Ann Oncol. 2021; 32(55): S595–S596. ABSTRACT [Google Scholar]
- 40.Kalinsky KM, Wei Z, McCourt CK, Mitchell EP, Wright JJ, Doyle LA, Gray RJ, Wang V, McShane LM, Rubinstein LV, Patton DR, Williams PM, Hamilton SR, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. Ipatasertib in Patients with Tumors with AKT Mutations: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) Sub-protocol Z1K. Presented at: 34th EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; October 26–28, 2022; Barcelona, Spain. Abstract 11 [Google Scholar]
- 41.Subbiah V, Zhao F, Kudchadkar RR, Sullivan RJ, Mitchell EP, Wright JJ, Chen HX, Gray RJ, Wang X, McShane LM, Rubinstein LV, Patton DR, Williams M, Hamilton SR, Sundaresan TK, Conley BA, Arteaga CL, Harris LN, O’Dwyer PJ, Chen AP, Flaherty KT. BVD-523FB (Ulixertinib) in Patients with Tumors with BRAF Fusions, or with Non-V600E, Non-V600K BRAF Mutations: Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) Sub-protocol EAY131-Z1L. Proceedings from the American Association for Cancer Research. 2022; 82(12). ABSTRACT CT160 [Google Scholar]
- 42.Tuveson DA, Willis NA, Jacks T, Griffin JD, Singer S, Fletcher CD, Fletcher JA, Demetri GD. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene. 2001. Aug 16;20(36):5054–8. doi: 10.1038/sj.onc.1204704. [DOI] [PubMed] [Google Scholar]
- 43. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/rare-cancer .
- 44.Parsons DW, Janeway KA, Patton DR, Winter CL, Coffey B, Williams PM, Roy-Chowdhuri S, Tsongalis GJ, Routbort M, Ramirez NC, Saguilig L, Piao J, Alonzo TA, Berg SL, Fox E, Hawkins DS, Abrams JS, Mooney M, Takebe N, Tricoli JV, Seibel NL; NCI-COG Pediatric MATCH Team. Actionable Tumor Alterations and Treatment Protocol Enrollment of Pediatric and Young Adult Patients With Refractory Cancers in the National Cancer Institute-Children’s Oncology Group Pediatric MATCH Trial. J Clin Oncol. 2022. Jul 10;40(20):2224–2234. doi: 10.1200/JCO.21.02838. Epub 2022 Mar 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Chi P. Basket trial of TRK inhibitors demonstrates efficacy in TRK fusion-positive cancers. J Hematol Oncol. 2018. Jun 7;11(1):78. doi: 10.1186/s13045-018-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gatta G, Capocaccia R, Botta L, Mallone S, De Angelis R, Ardanaz E, Comber H, Dimitrova N, Leinonen MK, Siesling S, van der Zwan JM, Van Eycken L, Visser O, Žakelj MP, Anderson LA, Bella F, Kaire I, Otter R, Stiller CA, Trama A; RARECAREnet working group. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet-a population-based study. Lancet Oncol. 2017. Aug;18(8):1022–1039. doi: 10.1016/S1470-2045(17)30445-X. [DOI] [PubMed] [Google Scholar]
- 47.Hoes LR, van Berge Henegouwen JM, van der Wijngaart H, Zeverijn LJ, van der Velden DL, van de Haar J, Roepman P, de Leng WJ, Jansen AML, van Werkhoven E, van der Noort V, Huitema ADR, Gort EH, de Groot JWB, Kerver ED, de Groot DJ, Erdkamp F, Beerepoot LV, Hendriks MP, Smit EF, van der Graaf WTA, van Herpen CML, Labots M, Hoeben A, Morreau H, Lolkema MP, Cuppen E, Gelderblom H, Verheul HMW, Voest EE. Patients with Rare Cancers in the Drug Rediscovery Protocol (DRUP) Benefit from Genomics-Guided Treatment. Clin Cancer Res. 2022. Apr 1;28(7):1402–1411. doi: 10.1158/1078-0432.CCR-21-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A, Onstenk W, Roepman P, Voda M, Bloemendal HJ, Tjan-Heijnen VCG, van Herpen CML, Labots M, Witteveen PO, Smit EF, Sleijfer S, Voest EE, Cuppen E. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019; 575:210–216. doi: 10.1038/s41586-019-1689-y. Epub 2019 Oct 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheeler DA, Takebe N, Hinoue T, Hoadley KA, Cardenas MF, Hamilton AM, Laird PW, Wang L, Johnson A, Dewal N, Miller V, Piñeyro D, Castro de Moura M, Esteller M, Shen H, Zenklusen JC, Tarnuzzer R, McShane LM, Tricoli JV, Williams PM, Lubensky I, O’Sullivan-Coyne G, Kohn EC, Little RF, White J, Malik S, Harris L, Weil C, Chen AP, Karlovich C, Rodgers B, Shankar L, Jacobs P, Nolan T, Hu J, Muzny DM, Doddapaneni H, Korchina V, Gastier-Foster J, Bowen J, Leraas K, Edmondson EF, Doroshow JH, Conley BA, Ivy SP, Staudt LM. Molecular Features of Cancers Exhibiting Exceptional Responses to Treatment. Cancer Cell. 2021. Jan 11;39(1):38–53.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adashek JJ, Menta AK, Reddy NK, Desai AP, Roszik J, Subbiah V. Tissue Agnostic Activity of BRAF plus MEK inhibitor in BRAF V600 mutant tumors. Mol Cancer Ther. 2022. Apr 12:molcanther.0950.2021. doi: 10.1158/1535-7163. [DOI] [PMC free article] [PubMed]
- 51.Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, Mariotto AB, Lowy DR, Feuer EJ. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N Engl J Med. 2020. Aug 13;383(7):640–649. doi: 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kahlon N, Doddi S, Yousif R, Najib S, Sheikh T, Abuhelwa Z, Burmeister C, Hamouda DM. Melanoma Treatments and Mortality Rate Trends in the US, 1975 to 2019. JAMA Netw Open. 2022. Dec 1;5(12):e2245269. doi: 10.1001/jamanetworkopen.2022.45269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long GV, Hauschild A, Santinami M, Atkinson V, Mandalà M, Chiarion-Sileni V, Larkin J, Nyakas M, Dutriaux C, Haydon A, Robert C, Mortier L, Schachter J, Schadendorf D, Lesimple T, Plummer R, Ji R, Zhang P, Mookerjee B, Legos J, Kefford R, Dummer R, Kirkwood JM. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med. 2017. Nov 9;377(19):1813–1823. doi: 10.1056/NEJMoa1708539. Epub 2017 Sep 10. [DOI] [PubMed] [Google Scholar]
- 54.Meric-Bernstam F, Ford JM, O’Dwyer PJ, Shapiro GI, McShane LM, Freidlin B, O’Cearbhaill RE, George S, Glade Bender J, Lyman GH, Tricoli JV, Patton D, Hamilton SR, Gray RJ, Hawkins DS, Ramineni B, Flaherty KT, Grivas P, Yap TA, Berlin J, Doroshow JH, Harris LN, Moscow JA. National Cancer Institute Combination Therapy Platform Trial with Molecular Analysis for Therapy Choice (ComboMATCH). Clin Cancer Res. 2023. Jan 20:CCR-22–3334. doi: 10.1158/1078-0432.CCR-22-3334. [DOI] [PMC free article] [PubMed]
- 55.Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, Yu K, Rubanova Y, Macintyre G, Demeulemeester J, Vázquez-García I, Kleinheinz K, Livitz DG, Malikic S, Donmez N, Sengupta S, Anur P, Jolly C, Cmero M, Rosebrock D, Schumacher SE, Fan Y, Fittall M, Drews RM, Yao X, Watkins TBK, Lee J, Schlesner M, Zhu H, Adams DJ, McGranahan N, Swanton C, Getz G, Boutros PC, Imielinski M, Beroukhim R, Sahinalp SC, Ji Y, Peifer M, Martincorena I, Markowetz F, Mustonen V, Yuan K, Gerstung M, Spellman PT, Wang W, Morris QD, Wedge DC, Van Loo P; PCAWG Evolution and Heterogeneity Working Group and the PCAWG Consortium. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021. Apr 15;184(8):2239–2254.e39. doi: 10.1016/j.cell.2021.03.009. Epub 2021 Apr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahn WC, Bader JS, Braun TP, Califano A, Clemons PA, Druker BJ, Ewald AJ, Fu H, Jagu S, Kemp CJ, Kim W, Kuo CJ, McManus M, B Mills G, Mo X, Sahni N, Schreiber SL, Talamas JA, Tamayo P, Tyner JW, Wagner BK, Weiss WA, Gerhard DS; Cancer Target Discovery and Development Network. An expanded universe of cancer targets. Cell. 2021. Mar 4;184(5):1142–1155. doi: 10.1016/j.cell.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choueiri TK, Powles T, Burotto M, Escudier B, Bourlon MT, Zurawski B, Oyervides Juárez VM, Hsieh JJ, Basso U, Shah AY, Suárez C, Hamzaj A, Goh JC, Barrios C, Richardet M, Porta C, Kowalyszyn R, Feregrino JP, Żołnierek J, Pook D, Kessler ER, Tomita Y, Mizuno R, Bedke J, Zhang J, Maurer MA, Simsek B, Ejzykowicz F, Schwab GM, Apolo AB, Motzer RJ; CheckMate 9ER Investigators. Nivolumab plus Cabozantinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2021. Mar 4;384(9):829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiacci E, De Carolis L, Simonetti E, Capponi M, Ambrosetti A, Lucia E, Antolino A, Pulsoni A, Ferrari S, Zinzani PL, Ascani S, Perriello VM, Rigacci L, Gaidano G, Della Seta R, Frattarelli N, Falcucci P, Foà R, Visani G, Zaja F, Falini B. Vemurafenib plus Rituximab in Refractory or Relapsed Hairy-Cell Leukemia. N Engl J Med. 2021. May 13;384(19):1810–1823. [DOI] [PubMed] [Google Scholar]
- 59.Mandelker D, Zhang L. The emerging significance of secondary germline testing in cancer genomics. J Pathol. 2018. Jan; 244 (5): 610–615. doi. 10.1002/path.5031. [DOI] [PubMed] [Google Scholar]