Abstract
Background
Tiger nut (Cyperus esculentus) is widely known as an additional source of food, oil and feed worldwide. The agricultural production of tiger nut has been greatly hindered by drought stress, reducing both yield and quality. Protein phosphatase 2 C (PP2Cs) plays an important role in plant responses to drought stress however, the molecular mechanism of PP2Cs in tiger nuts still unclear.
Results
In this study, we identified a putative group A PP2C-encoding gene (CePP2C19) from tiger nut using transcriptome analysis, which is highly induced by drought stress. The transient expression assay suggested that CePP2C19 was localized to nucleus. Furthermore, the interaction between CePP2C19 and CePYR1, a coreceptor for ABA signaling, was first detected using a yeast two-hybrid assay and then verified using a bimolecular fluorescence complementation (BiFC) analysis. In addition, the transgenic Arabidopsis lines overexpressing CePP2C19 exhibited extreme tolerance to ABA and mannitol stresses during seed germination and root growth. At the mature stage, overexpression of CePP2C19 resulted in a higher tolerance to drought stress in transgenic Arabidopsis, as confirmed by a visible phenotype and several physiological parameters. Noticeably, the silencing of CePP2C19 by virus-induced gene silencing (VIGS) showed obvious reduction in drought tolerance in tiger nut plants.
Conclusions
The CePP2C19 emerges as a pivotal gene involved in the ABA signaling pathway, which likely reduce ABA sensitivity and thus enhances drought tolerance in Cyperus esculentus.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-023-04522-2.
Keywords: Protein phosphatase 2 C (PP2Cs), ABA signaling, Drought stress, Arabidopsis, Tiger nut
Introduction
Plants are often subjected to various biotic and abiotic stresses during their growth and development processes [1]. Drought conditions pose a severe threat to plant life as they impede growth and interfere with photosynthesis. This disruption subsequently retards the rate of carbon dioxide absorption, nutrient utilization, seed germination, and ultimately leads to yield loss [2, 3]. During extended periods of drought stress, plants exhibit a primary adaptive mechanism by accumulating osmotic compounds such as soluble sugars, proline, and glycine betaine. This accumulation serves the vital purpose of upholding cellular osmotic potential [4, 5]. A number of antioxidant enzymes, including malondialdehyde (MDA), superoxide dismutase (SOD), catalase (CAT), etc. have been demonstrated to undergo variations in their activity in response to drought stress. However, changes in plant physiological signals lead to variation in the expression of numerous drought-resistant genes [6, 7].
Protein kinases and phosphatases in plants play indispensable roles in the transmission of stress signals within plant cells. Based on the amino acid residues they dephosphorylate; this significant class of phosphatases is generally divided into serine/threonine (Ser/Thr) and tyrosine (Tyr) phosphatases. There are several subclasses of serine/threonine phosphatases, including PP1, PP2A, PP2B, and PP2C [8–10]. Prior studies have demonstrated that protein phosphatase 2Cs (PP2Cs) family is the major class of phosphatases with a plethora of biological processes related to ABA signaling cascade, environmental stresses, and plant defense [11, 12]. It is well-established that the phytohormone ABA plays a crucial role in the coordination of responses to abiotic stressors, particularly drought stress. During stress conditions, the process of ABA biosynthesis induces, and act as a dual negative regulatory mechanism. Group A PP2Cs, which consists of ABI1, ABI2, Hab1, Hab2, AHG1, and PP2CA, is assumed to be a suppressor of ABA signaling in Arabidopsis thaliana [13, 14]. Previous researches have shown that osmolarity-induced ABA alterations in plants may led to either conformational changes in ABA receptors such as pyrabactin resistance 1 (PYR1)/PYR1-like (PYL) or regulatory elements of ABA receptors such as (RCARs). As a result, PP2Cs dissociate from sucrose non-fermenting 1 (SNF1)-related protein kinase 2 (SnRK2s), allowing SnRK2s to autophosphorylate and activate downstream substrates such as bZIP transcription factor under stress condition [15–17]. Phosphorylation and dephosphorylation of target proteins are two essential mechanisms for efficiently activating and inhibiting key biochemical pathways. Positive modulators of ABA and osmotic response comprise subclass III SnRK2 kinases, which include SnRK2.2, SnRK2.3, and SnRK2.6 [18–20]. Nevertheless, in the absence of ABA, PP2Cs inhibit the activity of SnRK2 proteins through physical interaction and dephosphorylation.
Prior to the discovery of a new ABA receptor (ABAR) in plants, PP2Cs were considered to be crucial players in ABA signal transduction [8]. Arabidopsis contains 14 members of ABARs, and 13 of which are involved in ABA sensing [21–23]. Several molecular interactions between Arabidopsis ABARs and PP2Cs have been discovered. For instance, members of group A PP2Cs such as (ABI1, HAB1, HAB2, and PP2CA) were de-activated in response to ABA, as shown by the functions of ABA receptors including PYR1, PYL1, PYL2, and PYR3 [24–26]. Interactions involving ABAR and PP2Cs have been found in several other higher plants than Arabidopsis, including rice, soybean, and maize. Rice ABA receptor OsPYL/RCAR5 was found to interact with group A PP2C, OsPP2C30, and a core ABA signaling which also comprised of a Ser/Thr kinase SAPK2 [27–29]. In maize, the overexpression of ZmPP2C55 resulted in improved drought stress as well as tolerance to salt, high temperature, and exogenous ABA responses [30]. In contrast, of the overexpression of OsPP108 demonstrated substantial improvements in physiological indices such as water loss, fresh weight, chlorophyll content, and photosynthetic potential (Fv/Fm) when subjected to salt or drought stress. These findings suggested that the overexpression of OsPP108 both positively influences abiotic stress responses and inversely modulates abscisic acid signaling [31].
Tiger nut is an herbaceous oil crop with rich oil content in its tubers and high nutritional value [32]. However, drought conditions greatly affect the yield and production of tiger nut, and therefore, it is of great practical importance to study the drought tolerance mechanisms in this plant. In this study, we identified a total 146 PP2C genes by transcriptome sequencing of tiger nut samples under different drought stress conditions. Based on the significant differential expression analysis, we identified a putative group A PP2C encoding gene (CePP2C19) from tiger nut. Further functional validation was conducted by cloning the full length of CePP2C19 gene and then subcellular localization analysis, yeast two-hybrid and BiFC assay, overexpression in Arabidopsis thaliana, and gene silencing experiments, respectively. Altogether, our results demonstrated that the overexpression of CePP2C19-encoding gene leads to a high degree of drought tolerance in tiger nut by negatively regulating ABA signaling. The establishment of this platform will pave the wave in understanding the regulatory mechanism of a putative drought tolerant CePP2C19 gene in tiger nut.
Materials and methods
Plant material and growth conditions
The plant material of tiger nut (Cyperus esculentus Hebei 01 cultivar, deposited in Engineering Research Center of Bioreactor and Pharmaceutical Development, Ministry of Education, JLAU) used in this study was identified by Jia Li. Seeds were soaked in water and placed in a constant temperature incubator at 37 °C to germinate. After 7 days, the uniformly growing seedlings were moved to 1/4 Hoagland nutrient solution for 10 days, and the hydroponic solution was changed every two days. Tobacco (Nicotiana benthamiana) seeds were planted and cultured in a soil mixture (peat moss, vermiculite, 1:1, v/v). Arabidopsis seeds (ecotype Col-0) were planted at 4 °C for 2 d after vernalization and cultured in a soil mixture (peat moss, vermiculite, 3:7, v/v). Overexpression of CePP2C19 Arabidopsis seeds was grown in whole vermiculite after 2 d of vernalization at 4 °C. The pp2c19 mutant is achieved by conducting the AT2G29380 gene mutation in wild Arabidopsis thaliana (SALK-033011). The pp2c19 mutant Arabidopsis seeds were grown in whole vermiculite after 2 d of vernalization at 4 °C under control conditions.
Detection of physiological indicators of tiger nut and Arabidopsis
The leaves and roots were taken at 0 h, 12 h, 24 h, 48 h, and 72 h of stress treatment with 30% PEG6000 (Beijing Solarbio Science &Technology Co.,Ltd) and immediately placed in liquid nitrogen and stored in -80℃ refrigerator. The SOD, MDA, CAT, and soluble sugar contents were determined using physiological index test kits (Beijing Boxbio Science Technology Co., Ltd). The determination of ABA in tiger nut was performed using Shanghai Youxuan ABA ELISA kit. Fresh weight (Fw) of wild-type, mutant, and overexpression lines of CePP2C19 in the control group and drought treatment group was measured and recorded. The Arabidopsis plants were de-greenized at 105℃ for 15 min and dried at 80℃ to constant weight (about 3 days) in the oven. Then, the dry weight (Dw) was weighed and recorded. Similarly, the water content of wild type and overexpression of CePP2C19 Arabidopsis lines were calculated using the formula of water content (Fw - Dw)/ Dw *100%. Each group was tested for three biological replicates. In order to study the stomatal changes of Arabidopsis leaves after drought stress, Arabidopsis leaves were decolorized and fixed in ethanol: acetic acid (6:1) solution for 24 h, 30 min was treated with 70% ethanol dehydration, and anhydrous ethanol was treated 3 times, each time 30 min. Then the leaves were removed and transferred to the mixed solution of water and chloral [water and trichloro acetal: water: glycerol(w/v1/v2) = 8:1:2] for 8 h, and the stomata were observed by laser confocal microscope (Leica TCS SP8, Ger-many). Measurement of stomatal area was carried by ImageJ software.
Differential expression analysis
Total RNA was obtained using the Fast-Pure Plant Total RNA Isolation Kit (Vazyme). Then, the paired-end sequencing using an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA) was conducted using three independent biological replicates for two samples. Following this, an assessment of the quality of the raw reads was performed using FastQC with the default settings. Additionally, the removal of adapter sequences was carried out using CutAdapt in order to obtain reads of high quality. The Trinity program was employed for the assembly of the transcriptome of tiger nuts. The contigs that were assembled were subjected to clustering based on the common reads of transcripts using Corset. From each cluster, representative sequences were identified and designated as unigenes. Furthermore, the functional classification within the Gene Ontology (GO) was conducted using WeGo. Additionally, for comprehensive functional annotation of unigenes, distinct sequences were mapped against multiple protein and nucleotide databases, including Nr, Nt, Pfam, COG, and Swiss-Prot. The calculation of transcript expression levels utilized the FPKM (Fragments Per Kilobase of transcript sequence per Millions of base pairs sequenced) method [33]. To control for false positives in multiple testing, the False Discovery Rate (FDR) correction was applied using DEseq2 [34]. Differentially expressed genes (DEGs) were identified based on criteria of |log2 (Fold Change) | > 1 and FDR < 0.05.
Phylogenetic analysis of the PP2C gene family in tiger nut
Using tiger nut transcriptome data as well as rice and Arabidopsis public data, a phylogenetic tree was constructed by N-J method using MEGAX software with Bootstrap parameter set to 1000. The PP2C gene family of tiger nut was classified according to the Arabidopsis PP2C family gene classification pattern as well as rice. Tree files were visualized and modified using the online tools available at (https://itol.embl.de/).
RNA isolation and qRT-PCR
Total RNA extraction was performed according to our previous method [35]. Then, the first-strand cDNA synthesis was carried out using the MonScript™ RTIII All-in-One Mix included in the dsDNase (Monad) kit. Gene primers for the tiger nut and Arabidopsis genes were generated with Premier 6 software (Premier Biosoft, Palo Alto, CA, USA) using the genomic sequence of each gene. To ensure the specificity of the primers, each primer pair was designed to target regions outside of the conserved domains of the genes. Subsequently, qRT-PCR was conducted using the MonAmp™ ChemoHS qPCR Mix from Monad Biotechnology Co. Ltd. (China) on a Stratagene MX3000P real-time PCR machine. In accordance with the manufacturer’s guidelines, a 20 µL reaction mixture was prepared, comprising 2.0 µL of cDNA template, 0.2 µM of primers (F/R), 0.2 µL of ROX dye (100x), 10 µL of master mix, and 7 µL of RNase-free water. Relative transcript abundance values were computed using the 2−ΔΔCt method, with the tiger nut ADF7 (CeADF7) and Arabidopsis Actin3 (AtACT3) genes serving as standard controls for normalization.
Drought and ABA treatments
Tiger nut were treated with 30% PEG6000 stress treatment. To examine CePP2C19 expression in tiger nut. In the Arabidopsis plate experiment, 1/2 MS medium was added with 300 mM mannitol and 5 µM ABA.
Yeast two-hybrid assay
The coding regions of CePP2C19 and CePYR1 were separately inserted into the pGBKT7 (BD) or pGADT7 (AD) vector. These construct pairs were then co-transfected into Y2HGOLD cells of the yeast strain. To evaluate the reliability of the protein interactions, we cultured yeast cells transformed with various concentrations (100, 10−1, 10−2, and 10−3 dilutions) on SD medium deficient in Leu, Trp, His, and Ade (SD/−LTHA) supplemented with 5-bromo-4-chloro-3-indolyl-alpha-D-galactopyranoside (X-α-gal). After an incubation period of 3–4 days, we captured photographs of the resulting yeast growth. STRING website https://cn.string-db.org/.
Bimolecular fluorescence complementation (BiFC) assay
The pxy104-CePYR1 recombinant vector and pxy106-CePP2C19 recombinant vector were constructed, and the recombinant plasmids were transferred into GV3101 (pSoup-p19) Agrobacterium receptor state, infiltrated with three-week-old leaves of Nicotiana benthamiana, cultured in dark for 24 h and then placed in an artificial climate chamber for normal growth, and GFP fluorescence was observed using confocal microscopy after 48 h.
Subcellular localization
We conducted subcellular localization analysis by transiently expressing CePP2C19-GFP proteins in the leaves of Nicotiana benthamiana. The recombinant plasmids constructed using FlyCut (TransGen Biotech) including pGDG-CePP2C19-GFP, pGDG-CePYR1-GFP, pGDG-GFP, and mCherry-NLS were introduced into Agrobacterium tumefaciens GV3101(pSoup-19) competent cells using the freeze-thaw method, individually. The leaves of Nicotiana benthamiana were subjected to transient expression through the injection of Trans-formed GV3101(pSoup-19). Following a 48 h incubation period under long-day conditions consisting of 16 h of light exposure, the cells were subjected to observation using a laser confocal microscope (Leica TCS SP8, Germany).
Stress tolerance experiment of transgenic yeast
The INVSc1 yeast strain (Invitrogen, USA) was transformed with the expression vector and an empty pYES2 control plasmid using the lithium acetate technique [36]. The transformed cells were subsequently chosen for growth on uracil-deficient synthetic complete (SC) media supplemented with 2% (w/v) galactose at a temperature of 30 °C for a duration of 36 h. The assessment of osmotic tolerance was conducted in accordance with the previously published methodologies [37].
Acquisition of overexpressing Arabidopsis thaliana
The pCAMBIA3301-CePP2C19 recombinant plasmid was constructed, and the recombinant plasmid was transferred into the EHA105 receptor state and infested with wild-type Arabidopsis thaliana in Colombia using the inflorescence infestation method (Floral-Dip) [38]. Screening pure strains for further research.
Germination rate and soil phenotype analysis of overexpressed Arabidopsis thaliana
Arabidopsis seeds of the overexpression strain, mutant, and wild-type seeds were vernalized in a refrigerator at 4 °C, planted on 1/2 MS medium with ABA and mannitol stress, and placed in an incubator (16 h light, 8 h dark incubation) to observe changes in germination rate and root length within 6 days. The vernalized seeds, planted in whole vermiculite, were watered with 1/2 Hoagland solution when Arabidopsis grew to four rosette leaves, and drought stress was applied after three weeks.
Virus-induced gene silencing assay
We employed the tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system to generate plants with silenced CePP2C19. Specifically, we transformed Agrobacterium rhizogenic strain GV3101 with pTRV1, pTRV2-CePP2C19, and pTRV2:00 (used as a negative control), and subsequently, these Agrobacterium cultures were introduced into tiger nut leaves.
Statistical analysis
All experiments mentioned were conducted in triplicate, and the resulting data were subjected to statistical analysis using the t-test. A significant difference was defined as *P < 0.05 and **P < 0.01 and *** P < 0.001.
Results
Changes in the phenotypic and physiological indicators under drought stress in tiger nut
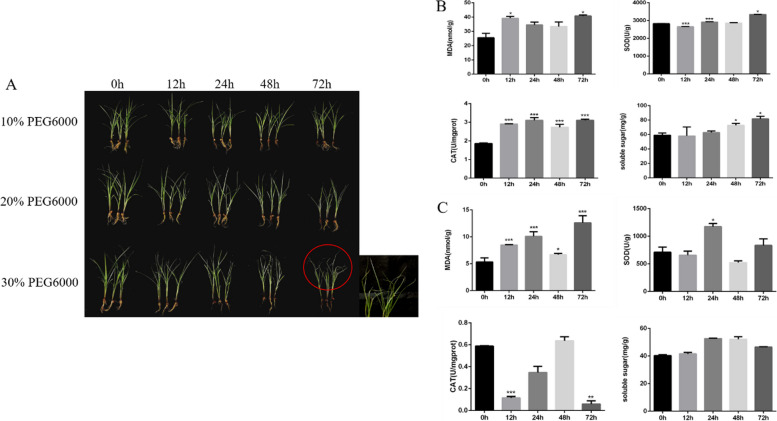
To investigate the effect of drought stress on tiger nut, simulated drought conditions were introduced using different concentrations of PEG6000 (10%, 20%, and 30%) at different time intervals (0 h, 12 h, 24 h, 48 h, and 72 h). Under 30% PEG6000 stress for 12 h, the leaves of tiger nut exhibited noticeable phenotypic changes including folding, wilting and water deficiency. Consequently, we assessed the physiological indicators of both the leaves and roots of tiger nuts under 30% PEG6000 stress (Fig. 1A). MDA content is an important index reflecting the degree of damage to the membrane system and the stress resistance of plants. The MDA content in leaves increased significantly (P < 0.05) after 30% PEG6000 treatment for 12 h. Moreover, the SOD activity also of different treatment times showed a significant difference. The activity of SOD was significantly reduced after 12 h of treatment but increased after 24 h of treatment. Similarly, the activity of CAT in the leaves was significantly enhanced at all time periods compared to the control treatment (P < 0.001). Furthermore, the content of soluble sugars in the leaves was significantly increased than the control group (P < 0.05) after 48 and 72 h of treatment (Fig. 1B). In the same way, these physiological indicators were investigated in the roots of tiger nut under 30% PEG6000 stress. The MDA content in roots was increased significantly (P < 0.05) compared to the control plants at all time points (Fig. 1C). SOD activity in the root tissues was significantly increased at 24 h treatment times. Moreover, the CAT activity in the roots was significantly decreased at 12 h (P < 0.001) and 72 h (P < 0.01) treatment times. The soluble sugar content in roots was slightly increased at all treatment times than the control group (Fig. 1C). Overall, the tiger nut leaves subject to 30% PEG6000 stress for 12 h treatment time was used for further analysis.
Fig. 1.
Determination of phenotypic and physiological indicators of tiger nut under drought stress. A The phenotype of tiger nut under different drought stress conditions at different time intervals (B) The contents of MDA, SOD, CAT, and soluble sugar in the leaves of tiger nut under 30% PEG6000 stress. Error bars represent the standard deviation. * Significant difference at P < 0.05 *** significant difference at P < 0.001. C The contents of MDA, SOD, CAT and soluble sugar in the roots of tiger nut under 30% PEG6000 stress. Error bars represent the standard deviation. * Significant difference at P < 0.05 **significant difference at P < 0.01 *** significant difference at P < 0.001
Transcriptome data analysis
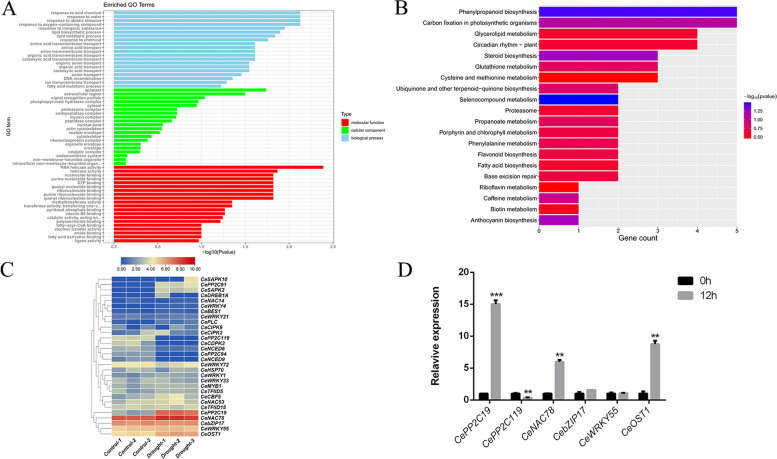
To investigate the molecular mechanisms of plant interactions and coordination under drought stress, transcriptome sequencing of tiger nut leaves after 30% PEG6000 stress was performed. A total of 44,965 genes were annotated and 288 differentially expressed genes (DEGs) were identified with differential expression level in tiger nut. Of these DEGs, 196 were up-regulated and 92 were down-regulated in the leaves of tiger nut. We performed GO enrichment analysis of these DEGs, which classified them into three functional categories including biological process (BP), molecular function (MF), and cellular component (CC) (Fig. 2A). In the biological process category, the most significant enriched terms include response to water, response to abiotic stimulus, and response to oxygen-containing compounds. Similarly, the molecular function category showed the enrichment of the terms such as RNA-helicase activity, methyltransferase activity, ribonucleoside binding and catalytic activity. In case of cellular component, the go terms of apoplast, extracellular region, signal recognition particles and cytosol were mostly enriched. In addition, the top 20 functional KEGG enriched pathways of the drought-induced leaves of tiger nut were screened. Most number of DEGs were significantly enriched into the functional pathways including phenylpropanoid biosynthesis, selenocompound metabolism, carbon fixation in photosynthetic organisms, steroid biosynthesis, circadian rhythm plant, and anthocyanin biosynthesis (Fig. 2B).
Fig. 2.
Transcriptome data analysis and functional annotation of DEGs identified in drought-induced stress of tiger nut. A GO enrichment analysis of the total DEGs were classified into three main categories: MF, CC, and BP. The names of the GO categories are listed along the y-axis. The degree of GO enrichment is represented by the -log10(P-value) and the number of transcripts enriched in each category. B Scatterplot of enriched KEGG pathways (www.kegg.jp/kegg/kegg1.html) of DEGs identified from drought stressed tiger nut leaves. Only the top 20 most strongly represented pathways are displayed. The color of the bar represents the range of the -log10(P-value). C Expression pattern of drought related genes. The colors indicate the abundance of transcripts calculated as Log2 (FPKM) in the control and drought-stressed plants (see color key). D Expression patterns of six genes in tiger nut leaves after treatment with 30% PEG6000 for 12 h. Error bars represent the standard deviation. ** significant difference at P < 0.01 *** significant difference at P < 0.001
Furthermore, thirty genes most likely related to drought stress tolerance were screened from the transcriptome data, and their differential expression pattern was investigated (Fig. 2C). Most of these genes such as CePP2C94, CePP2C119, CeNAC14, CeWRKY21, CeWRKY33, CeMYB1, CeHSP70, and CeCIPK9 were down-regulated under drought stress conditions. However, genes encoding CePP2C19, CeNAC78, CebZIP17, CeWRKY55 and CeOST1 showed up-regulation under drought conditions when compared with the normal treatment group. To validate the integrity and stability of transcriptome data, we further conducted qRT-PCR assay of six genes under control and drought stress treatment. The results showed that the expression level of CebZIP17, and CeWRKY55 was not significantly induced under both control and stressed condition. Similarly, gene-encoding CePP2C119 also showed down-regulation in both control and stressed plants. Noticeably, the expression level of CePP2C19 gene was high and significantly up-regulated under drought stress (Fig. 2D). Altogether, the analysis of transcriptome data and functional annotations of DEGs in drought-induced leaves of tiger nut suggested that CePP2C19 could be the candidate gene in alleviating drought stress.
Phylogenetic analysis of the CePP2C family
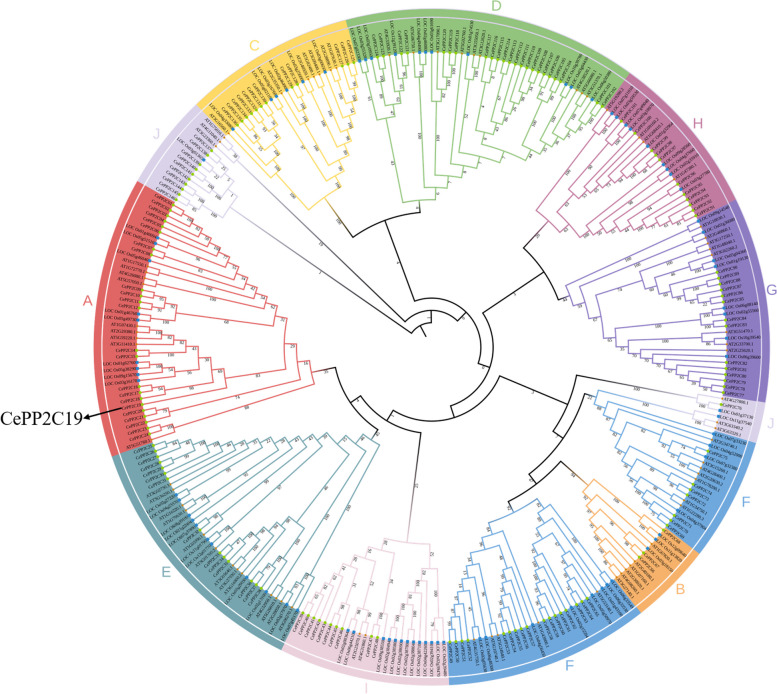
To explore evolutionary divergence and conserved relationships with other plant species, a phylogenetic tree of PP2C proteins was constructed among 146 Cyperus esculentus PP2C proteins, 76 Arabidopsis thaliana PP2C proteins, and 85 Oryza sativa PP2C proteins (Fig. 3). According to the classification pattern of the Arabidopsis family into A-J. Most of them belong to group A, group D and group F. Studies have shown that group A PP2C plays a decisive role in ABA signaling pathway. The phylogenetic analysis indicate that the tiger nut CePP2C19 belongs to the group A subfamily.
Fig. 3.
Phylogenetic analysis of PP2C members from Arabidopsis thaliana, Oryza sativa, and Cyperus esculentus. The phylogenetic tree was constructed by MEGA 6.0 with the neighbor-joining (NJ) method and 1000 bootstrap replications. Values less than 50 were removed. Members in the same clade with a unique color belong to the same subgroup
CePP2C19 interacts with the key interactor partner CePYR1 in ABA signaling pathway
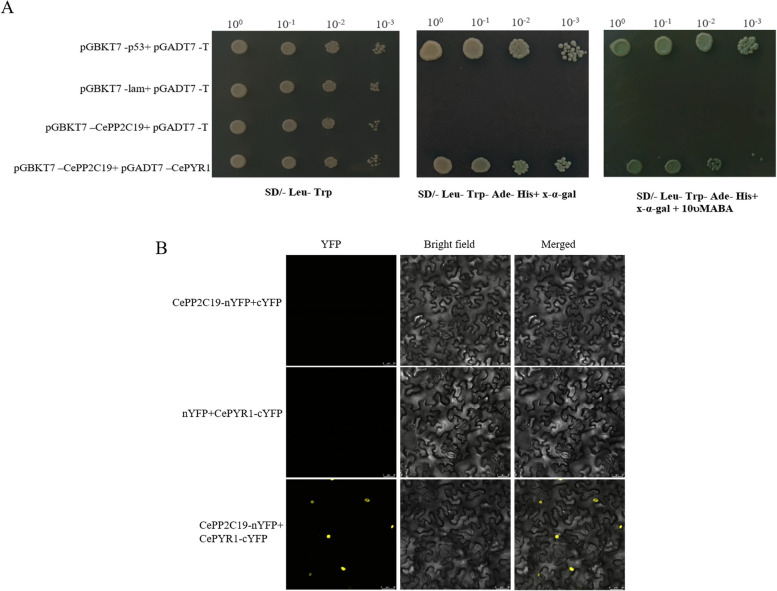
To determine whether CePP2C19 is a component of the ABA pathway, The primary predictions of protein interaction network was conducted using the online webserver of STRING. The results showed that CePP2C19 is directly interacting with PYR1, a member of the widely known family of PYR/PYL/RCAR-ABA receptors. The expression of CePYR1 gene was up-regulated under drought stress (Figure S1). To determine whether the interaction of CePP2C19 with CePYR1 is dependent on exogenous ABA, the ABA solution was added to SD/-Leu-Trp-Ade-His medium. The initial evidence from yeast two-hybrid assay demonstrated the possible interaction between CePP2C19 and PYR1 that is unaffected by exogenous ABA (Fig. 4A). To further validate this interaction relationship, we performed a BiFC assay in tobacco leaves using YFP fusion (CePP2C19-YFP) construct. We produced DNA constructs in which CePP2C19 and CePYR1 were fused separately to the N-terminal (nYFP) and C-terminal (cYFP) parts of YFP containing vector. The results showed the appearance of yellow fluorescence in the nucleus, which further indicated that CePP2C19 and CePYR1 interact in the nucleus (Fig. 4B).
Fig. 4.
The interaction of CePP2C19 with CePYR1 receptor during ABA signaling. A Yeast-two-hybrid assay of CePP2C19 and CePYR1. B BiFC analysis. The fluorescence resulted from the complementation of the N-terminal portion of YFP fused to CePP2C19 (CePP2C19-nYFP) with the C-terminal portion of YFP fused to CePYR1. Fluorescence was observed in tobacco leaf epidermal cells. Scale bars = 50μm
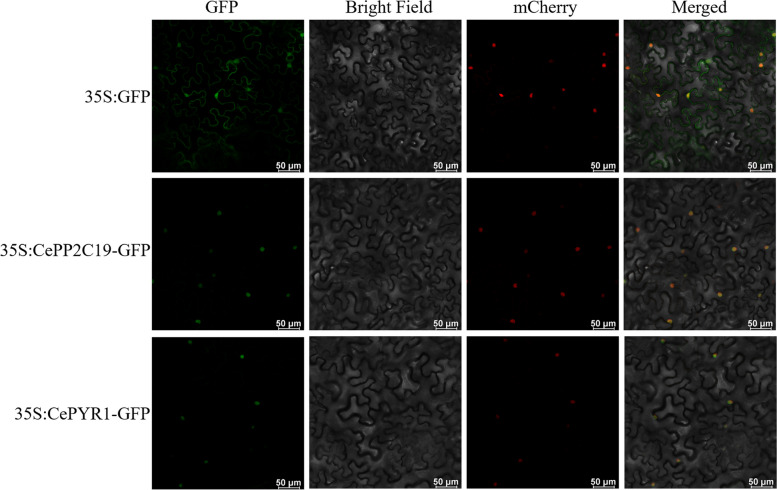
Subcellular localization of CePP2C19 and CePYR1
The subcellular localization of CePP2C19 and CePYR1 were determined by utilizing the transient transformation system in tobacco leaves using the CePP2C19-GFP and CePYR1-GFP fusion construct. Confocal laser microscopy was used to examine the pattern of GFP fluorescence in the tobacco leaves. The subcellular localization of CePP2C19 and CePYR1 were examined through transient expression of CePP2C19-GFP and CePYR1-GFP construct in leaves of Nicotiana benthamiana plants with expressing a red nuclear location signal protein mCherry-NLS. The CePP2C19-GFP and CePYR1-GFP fusion were microscopically observed to be solely localized in nucleus, which was co-localized with a red fluorescent protein mCherry-NLS. In contrast, GFP alone was seen to be ubiquitously distributed throughout the cell without specific localization. These results indicate that CePP2C19 and CePYR1 are a nucleus-localized protein (Fig. 5).
Fig. 5.
Subcellular localization of CePP2C19-GFP fusion proteins. Fusion proteins were transiently expressed under the control of the CaMV35S promoter in tobacco leaves and the results were observed under a laser scanning confocal microscope. The green color is the green fluorescent protein (GFP) signal. The red coloris a fluorescent protein mCherry-NLS. Scale Bars = 50 μm
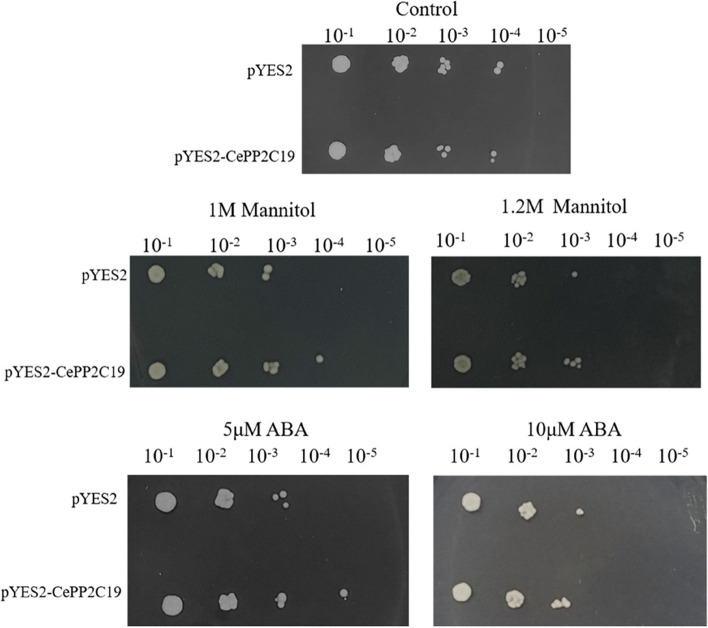
Functional verification of CePP2C19 in yeast
The heterologously overexpressed CePP2C19 gene in the yeast strain INVSc1 using the pYES2 vector was further investigated for its possible role under the exposure of mannitol and ABA stress. The results showed no significant difference in the survival rates between the pYES2 and CePP2C19 transgenic yeast cells under non-stress conditions. However, after 36 h of incubation in 1 M mannitol and 1.2 M mannitol, the transformed lines survived better than the control lines. Similarly, the transformed lines also survived better than the control lines after 36 h of incubation in the medium supplemented with ABA stress (Fig. 6). The results suggest that CePP2C19 protein may confer drought tolerance and ABA tolerance to yeast cells.
Fig. 6.
Phenotypic growth assays of Saccharomyces cerevisiae INVSc1 cells transformed with the pYES2 empty vector, pYES2-CePP2C19 under drought stress and ABA stress. Yeast cells transformed with the pYES2 empty vector, pYES2-CePP2C19, were spotted on SC-Ura medium 10-fold serially diluted (10−1, 10−2, 10−3,10−4, and 10−5) cultures and were then incubated at 30 °C for 36 h
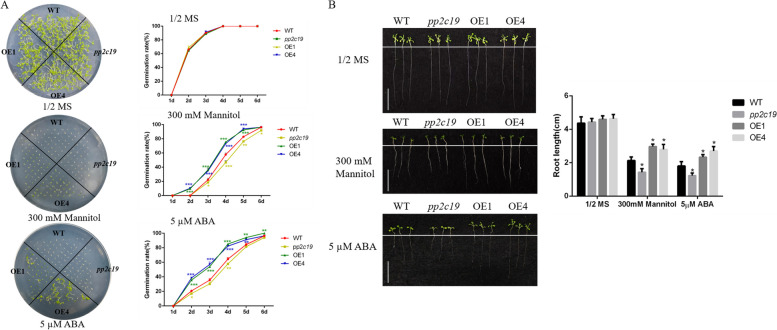
Overexpression of CePP2C19 confers abiotic stress tolerance in Arabidopsis seedlings
We performed seed germination and root growth assays of wild-type (WT), pp2c19 mutant and two overexpression lines (OE1 and OE4) of CePP2C19 on 1/2 MS medium supplemented with mannitol and ABA. Our statistical results showed that mannitol and ABA supply leads to an inhibitory effect on seed germination rate of transgenic lines. As described in Fig. 7A, no obvious difference was detected in the germination rates of WT, pp2c19 mutant, and overexpression strains on 1/2 MS medium alone. However, the overexpression lines started to germinate on the second day, while the WT and pp2c19 mutant lines did not germinate under mannitol stress. Noticeably, the germination rate of the overexpression lines was significantly faster than that of the WT. Importantly, the germination rate of the overexpression lines was faster than that of the WT and the mutant at 2–5 days under ABA stress, and the germination rate of the mutant was lower than that of the WT. Furthermore, the overexpression lines exhibited a significant increase in root length compared to the WT and mutant lines under mannitol stress conditions (Fig. 7B). In the same way, the root length of the overexpression lines was longer than that of the WT and mutant lines under ABA exposure. Collectively, these results suggested that the overexpression of CePP2C19 enhance drought tolerance and decrease ABA sensitivity in Arabidopsis, which might play a kay role during the regulation of ABA signaling at the germination stages.
Fig. 7.
Overexpression of CePP2C19 improved the tolerance of Arabidopsis seedlings to drought and ABA stress. A Seed germination under the treatment of mannitol and ABA. The surface-sterilized Arabidopsis seeds of wild-type and overexpression lines were sown on solid media of 1/2 MS containing mannitol (0, 300 mM) and ABA (5 µM). The status of seed germination was taken by photos after the treatment of stress for 6 days, respectively, while the germination rate of seeds was counted every day. WT (wild type); pp2c19 (mutant); OE1, OE4 (overexpression lines). Error bars represent the standard deviation. * significant difference at P <0.05 ** significant difference at P < 0.01 *** significant difference at P < 0.001. B Seedlings under the treatment of mannitol and ABA. WT (wild type); pp2C19 (mutant); OE1, OE4 (overexpression lines). Error bars represent the standard deviation. * significant difference at P <0.05
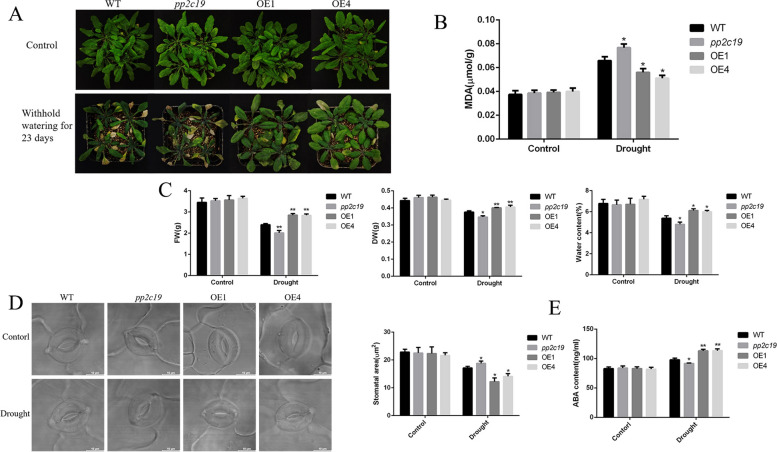
Overexpression of CePP2C19 confers drought tolerance in mature Arabidopsis plants
To investigate the effect of drought stress on mature transgenic Arabidopsis plants, we conducted phenotypic and physiological analysis of CePP2C19 overexpression lines, mutant and WT lines. After a period of 23 days of drought treatment, it was observed that the leaves of both WT and mutant plants had become dry, yellowed, and wilted. However, the overexpression lines exhibited a healthier appearance with less yellowing of the leaves (Fig. 8A). In addition, the amount of MDA accumulation in transgenic Arabidopsis was found to be lower than that of WT Arabidopsis after drought stress (Fig. 8B). However, the fresh weight, dry weight, and water content of CePP2C19 transgenic lines were more than that of WT plants (Fig. 8C). Moreover, under drought stress, the stomatal area of transgenic Arabidopsis was significantly smaller compared with WT (Fig. 8D). The ABA content of transgenic Arabidopsis lines was significantly higher than WT Arabidopsis under drought stress (Fig. 8E). These results showed that the transgenic Arabidopsis demonstrated less damage and showed considerable ability to resist drought stress.
Fig. 8.
Overexpression of CePP2C19 enhances drought tolerance in mature Arabidopsis plants. A The phenotypes of CePP2C19 transgenic lines tolerant to drought stress. B The determination of MDA content in the WT (wild type); pp2C19 (mutant); OE1, OE4 (overexpression lines), Error bars represent the standard deviation. * Significant difference at P < 0.05. C The determination of fresh weight, dry weight, and moisture content. Arabidopsis seeds of WT,mutant and CePP2C19 transgenic lines of the T3 generation were sown in vermiculite and cultured in the chamber room. WT (wild type); pp2C19 (mutant); OE1, OE4 (overexpression lines). Error bars represent the standard deviation. The asterisk indicates * significant difference at P < 0.05 **significant difference at P < 0.01. D Stomatal image and stomatal area of Arabidopsis thaliana leaves under drought stress. WT (wild type); pp2C19 (mutant); OE1, OE4 (overexpression lines). Scale Bars = 10 µm. Error bars represent the standard deviation. * significant difference at P < 0.05 E Determination of ABA content in Arabidopsis. WT (wild type); pp2C19 (mutant); OE1, OE4 (overexpression lines) Error bars represent the standard deviation. *Significant difference at P < 0.05. **significant difference at P < 0.01
Reduced drought tolerance in CePP2C19-silenced tiger nut
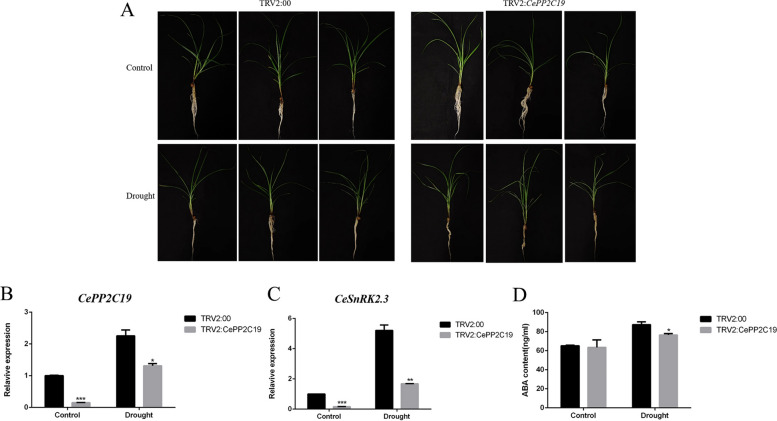
As the stable transformation system of tiger nut has not yet been fully developed, we performed the phenotypic analysis of CePP2C19-silenced plants using virus-induced gene silencing (VIGS). We subjected TRV2:CePP2C19 (silenced) containing plants and TRV2:00 (control) to 30% PEG6000 stress. Notably, the TRV2:CePP2C19 exhibited a more pronounced withering phenotype compared to the TRV2:00 plants (Fig. 9A). Likewise, the expression level of CePP2C19 was down-regulated under drought stress in the CePP2C19-silenced tiger nut (TRV2:CePP2C19) as opposed to the control tiger nut (TRV2:00) (Fig. 9B). Furthermore, the expression level of SnRK2 gene was observed in TRV2:CePP2C19 plants and TRV2:00 plants. The SnRK2 gene, known as a central gene in drought, salt, and ABA signaling, served as a positive control. Prior to drought exposure, TRV2:CePP2C19 plants displayed a noteworthy reduction in CeSnRK2.3 expression in comparison to TRV2:00 plants. However, both plant lines exhibited an upregulation of CeSnRK2.3 expression post drought treatment. Notably, the expression levels of CeSnRK2.3 were considerably lower in TRV2:CePP2C19 plants compared to TRV2:00 plants (Fig. 9C). Additionally, we quantified the ABA content in TRV2:CePP2C19 and TRV2:00 plants after drought stress, the results indicated a lower ABA content in TRV2:CePP2C19 plants compared to TRV2:00 plants following drought stress (Fig. 9D). Collectively, these results indicated that CePP2C19 could be involved in the molecular regulatory mechanism of drought stress tolerance by exploiting ABA signaling pathway in tiger nut.
Fig. 9.
Reduced drought tolerance of TRV2:CePP2C19 in tiger nut. A Drought stress with 30% PEG6000 on TRV2:CePP2C19 and TRV2:00 plants. B qRT-PCR analysis of CePP2C19 expression in leaves of TRV2:00 and TRV2:CePP2C19 plants exposed to drought stress. Relative expression level of each gene was normalized to that of the tiger nut ADF7 (CeADF7) as a standard control. Error bars represent the standard deviation. * Significant difference at P < 0.05 *** significant difference at P < 0.001. C qRT-PCR analysis of CeSnRK2.3 expression in leaves of TRV2:00 and TRV2:CePP2C19 plants exposed to drought stress. Relative expression level was normalized to that of the tiger nut ADF7 (CeADF7) as a standard control. Error bars represent the standard deviation. **significant difference at P < 0.01 *** significant difference at P < 0.001 (D) Determination of ABA content in TRV2:00 and TRV2:CePP2C19. Error bars represent the standard deviation. *Significant difference at P < 0.05
Discussion
Drought is an important factor limiting the yield of Cyperus esculentus (tiger nut). Therefore, understanding the molecular and genetic basis of drought resistance in tiger nut is an urgent need for breeding drought tolerant varieties. By triggering the activation of a multitude of stress-related genes, ABA orchestrates the plant’s adaptive response to abiotic stress, particularly in conditions of water scarcity. When it comes to cell differentiation and stress signaling, PP2Cs are the crucial regulators among others [39]. Many PP2C genes have now been discovered throughout plant taxa. Several members of group A PP2Cs play important roles in regulating ABA signaling in Arabidopsis, especially when the plant is subjected to abiotic stressors. A parallel signaling cascade was emerged in rice and maize [40–42]. Admittedly, there is a lack of publicly available research on tiger nut and it is unclear what role it plays under different stress conditions. In this study, we provide the first report of in planta functional characterization of a group A PP2Cs from tiger nuts, which likely play important role in a signaling module involved in drought stress and ABA signaling.
We identified a total of 146 PP2C members from the transcriptome data of tiger nuts. Phylogenetic analysis showed that CePP2C19 belongs to group A of tiger nut PP2C, and its sequence is highly homologous to members of the ABA-related Arabidopsis group A PP2C. Moreover, subcellular localization in Nicotiana benthamiana suggested that CePP2C19 was a nuclear protein. Consistent with the previous study, the rice PP2C subfamily group A member OsPP108 was likewise shown to be nuclear-localized [31]. Furthermore, the qRT-PCR analysis distinctly illustrates the induced expression of CePP2C19 genes to drought stress. This additional substantiation significantly underscores the assertion that CePP2C19 genes actively contribute to enhancing stress tolerance mechanisms. Other components such as the complex of ABA signaling components (PYR/PYL receptors, PP2Cs, and Ser/Thr Kinase) which is formed in the nucleus, also regulate ABA-mediated gene expression [43]. The interaction between PP2C and the ABA receptor PYL is the key step that triggers the downstream signaling genes to evoke ABA signaling [44, 45]. This signaling network has also been reported in plant species such as Arabidopsis, maize, tomato, and cotton. Many studies have shown that group A PP2C can interact with the ABA receptor family. For example, ZmPLY3 and ZmPP2C16 proteins in maize interact in response to ABA induction, demonstrating that they may be the most likely members of the second component and receptor of the ABA signaling pathway [46]. The HAI PP2Cs interacted with PYL5 and PYL7-10 in Arabidopsis [47]. AHG3 interacts with PYL12 in response to ABA and functions specifically in seed germination and early seedling growth [48]. The Cotton GhPYLs are effective against the phosphatase activity of GhHAI2, GhAHG3, and GhABI2 [49]. We also found that CePP2C19 interacts with CePYR1, a member of the ABA receptor family, and provided further evidence that CePP2C19 is pivotal to ABA signaling cascade.
We performed a preliminary validation of the CePP2C19 gene for drought tolerance and ABA tolerance in a yeast system. Previously, the overexpression of four transcription factors from vetch plant in transgenic yeast enhanced yeast tolerance to osmotic stress [37]. In our further findings, the overexpression of CePP2C19 gene in Arabidopsis showed drought tolerance and ABA sensitivity on MS medium. Under stress, the transgenic lines showed significant differences compared with WT, with the transgenic lines showing a faster germination rate and longer root length than WT. In addition, phenotypic experiments in soil experiments similarly demonstrated this difference. Under water deficit stress, complicated regulation processes are activated in plants, including at the biochemical, physiological, and molecular levels [50]. MDA content is an indicator of lipid peroxidation. MDA was measured and the MDA values of the transgenic strains were all less than WT, indicating that the membranes were damaged to a lesser extent and were drought tolerant. Fresh weight, dry seeds, and water content are also important indicators of drought tolerance of plants. The dry and fresh weights and water contents of the transgenic strains were higher than those of WT. The VIGS method has been widely used in many plants to interfere with genes to obtain mutant plants [51]. We used VIGS genetic analysis in tiger nut to further understand the function of CePP2C19 gene. In our phenotypic experiments, plants with CePP2C19- silenced showed a sensitivity to drought, which was followed by a reduction in ABA levels. Taken together, these data led to the conclusion that low CePP2C19 expression reduces the plant’s ability to withstand drought. Reducing the expression levels of GhHAI2, GhAHG3, and GhABI2 in cotton has been shown to increase the plant tolerance to osmotic stress [49]. Group A PP2Cs inhibit the ABA signaling pathway by dephosphorylating SnRK2 type kinases, which leads to the degradation of these proteins [52]. The conducted qRT–PCR analysis was undertaken with the specific aim of elucidating the causal relationship between variations in the CePP2C19 gene and the ensuing changes in the downstream gene SnRK2.3. The results yielded from this investigation distinctly demonstrate that the presence of the CePP2C19 gene does exert a discernible influence on the alterations observed in the downstream gene cascade. This observation substantiates the intricate interplay between these genes within the broader regulatory network. This pivotal role is testament to the intricate web of molecular interactions that orchestrate a plant’s response to drought stress, with CePP2C19 occupying a central position in this intricate regulatory framework.
Conclusion
The comprehensive understanding of a nuclear-localized CePP2C19 unfolds crucial insights into the intricate defense mechanisms that tiger nuts deploy when confronted with drought stress. Notably, the augmentation of CePP2C19 overexpression distinctly influences seed germination patterns and augments drought tolerance, especially during the early seedling stage, accompanied by a notable reduction in ABA sensitivity. This multifaceted response underscores the critical role of CePP2C19 as a key mediator of stress adaptation. Envisaging the broader agricultural context, the strategic overexpression of CePP2C19 emerges as a promising avenue to mitigate the vulnerability of plants to adverse conditions in their natural habitat. This approach holds the potential to foster elevated crop yields and enhanced productivity, thereby contributing significantly to the sustainability and resilience of agricultural systems in challenging environments.
Supplementary Information
Additional file 1: Figure S1. qRT-PCR analysis of CePYR1 expression in leaves of tiger nut exposed to drought stress. Error bars represent the standard deviation. **significant difference at P < 0.01
Acknowledgements
Thanks to professor Wenxian Sun, School of Plant Protection, Jilin Agricultural University for giving pGDG vector and mCherry-NLS.
Abbreviations
- PP2C
Protein phosphatase 2C
- MDA
Malondialdehyde
- SOD
Superoxide Dismutase
- CAT
CATalase
- DEGs
Differentially expressed genes
- BiFC
Bimolecular fluorescence complementation
- VIGS
Virus-induced gene silencing
Authors' contributions
Jia Li, Xinyi Liu and Naveed Ahmad wrote the manuscript and interpreted the data. Hengshuo Ge, Yifei Wang, Yijin Wang, Weican Liu, Nan Wang and Xiaowei Li contributed to the analysis and interpretation of data. Jia Li, Fawei Wang and Yuanyuan Dong designed the work and revised the manuscript. All authors read and approved the manuscript.
Funding
This research was funded by The Science and Technology Development Project of Jilin province (20220101354JC) and the Science and Technology Research Project of the Education Department of Jilin Province (JJKH20220325KJ).
Availability of data and materials
The RNA sequencing data for both drought stress and non-drought stress tiger nut samples were deposited in the SRA database at the NCBI under accession number PRJNA975975. All other data and material analyzed in the current study are included in the manuscript and the supplementary information.
Declarations
Ethics approval and consent to participate
JL undertook the formal identification of the plant material used in our study. The authors confirm that all the experimental methods complied with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fang Y, Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci. 2015;72(4):673–689. doi: 10.1007/s00018-014-1767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubey A, Kumar A, Malla MA, Chowdhary K, Singh G, Ravikanth G, Harish, Sharma S, Saati-Santamaria Z, Menéndez E, et al. Approaches for the amelioration of adverse effects of drought stress on crop plants. Front Biosci (Landmark Ed) 2021;26(10):928–947. doi: 10.52586/4998. [DOI] [PubMed] [Google Scholar]
- 3.Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL. Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci. 2016;7:1029. doi: 10.3389/fpls.2016.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad N, Jianyu L, Xu T, Noman M, Jameel A, Na Y, Yuanyuan D, Nan W, Xiaowei L, Fawei W. Overexpression of a novel cytochrome P450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes. 2019;10(10):756. doi: 10.3390/genes10100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo R, Yang Z, Li F, Yan C, Zhong X, Liu Q, Xia X, Li H, Zhao L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015;15:170. doi: 10.1186/s12870-015-0546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goswami A, Banerjee R, Raha S. Drought resistance in rice seedlings conferred by seed priming: role of the anti-oxidant defense mechanisms. Protoplasma. 2013;250(5):1115–1129. doi: 10.1007/s00709-013-0487-x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Ahmad N, Li Z, He J, Wang N, Naeem M, Jin L, Yao N, Liu X. CtCYP71A1 promotes drought stress tolerance and lignin accumulation in safflower and arabidopsis. Environ Experimental Bot. 2023;213:105430. doi: 10.1016/j.envexpbot.2023.105430. [DOI] [Google Scholar]
- 8.Singh A, Pandey A, Srivastava AK, Tran LS, Pandey GK. Plant protein phosphatases 2 C: from genomic diversity to functional multiplicity and importance in stress management. Crit Rev Biotechnol. 2016;36(6):1023–1035. doi: 10.3109/07388551.2015.1083941. [DOI] [PubMed] [Google Scholar]
- 9.Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 10.Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2 C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140(1):115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas PE, Ronald PC. Rice XB15, a protein phosphatase 2 C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol. 2008;6(9):e231. doi: 10.1371/journal.pbio.0060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tähtiharju S, Palva T. Antisense inhibition of protein phosphatase 2 C accelerates cold acclimation in Arabidopsis thaliana. Plant J. 2001;26(4):461–470. doi: 10.1046/j.1365-313X.2001.01048.x. [DOI] [PubMed] [Google Scholar]
- 14.Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot. 2011;62(8):2615–2632. doi: 10.1093/jxb/erq446. [DOI] [PubMed] [Google Scholar]
- 15.Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9(5):236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462(7273):660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Li L, ShangGuan G, Jia C, Deng S, Noman M, Liu Y, Guo Y, Han L, Zhang X. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-72390-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong S, Lim CW, Lee SC. CaADIP1-dependent CaADIK1-kinase activation is required for abscisic acid signalling and drought stress response in Capsicum annuum. New Phytol. 2021;231(6):2247–2261. doi: 10.1111/nph.17544. [DOI] [PubMed] [Google Scholar]
- 19.Lee SC, Lim CW, Lan W, He K, Luan S. ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol Plant. 2013;6(2):528–538. doi: 10.1093/mp/sss078. [DOI] [PubMed] [Google Scholar]
- 20.Hirayama T, Umezawa T. The PP2C-SnRK2 complex: the central regulator of an abscisic acid signaling pathway. Plant Signal Behav. 2010;5(2):160–163. doi: 10.4161/psb.5.2.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Guzmán M, Rodríguez L, Lorenzo-Orts L, Pons C, Sarrión-Perdigones A, Fernández MA, Peirats-Llobet M, Forment J, Moreno-Alvero M, Cutler SR, et al. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J Exp Bot. 2014;65(15):4451–4464. doi: 10.1093/jxb/eru219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24(6):2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Andrade J, González B, Gonzalez-Guzman M, Rodriguez PL, Vera P. The role of ABA in Plant Immunity is mediated through the PYR1 receptor. Int J Mol Sci. 2020;21(16):5852. doi: 10.3390/ijms21165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161(2):931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013;18(5):259–266. doi: 10.1016/j.tplants.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2 C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot. 2012;63(2):1013–1024. doi: 10.1093/jxb/err338. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Ali S, Zhang T, Wang W, Xie L. Identification, evolutionary and expression analysis of PYL-PP2C-SnRK2s gene families in soybean. Plants (Basel) 2020;9(10):1356. doi: 10.3390/plants9101356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Zhao Y, Li Z, Hsu CC, Liu X, Fu L, Hou YJ, Du Y, Xie S, Zhang C, et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell. 2018;69(1):100–112e106. doi: 10.1016/j.molcel.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang Y, Sun X, Gao S, Qin F, Dai M. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates Drought Tolerance of Maize Seedlings. Mol Plant. 2017;10(3):456–469. doi: 10.1016/j.molp.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Singh A, Jha SK, Bagri J, Pandey GK. ABA inducible rice protein phosphatase 2 C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE. 2015;10(4):e0125168. doi: 10.1371/journal.pone.0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Jing M, Ahmad N, Wang Y, Wang Y, Li J, Li X, Liu W, Wang N, Wang F, et al. Tracing key molecular regulators of lipid biosynthesis in Tuber Development of Cyperus esculentus using transcriptomics and Lipidomics Profiling. Genes (Basel) 2021;12(10):1492. doi: 10.3390/genes12101492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Ge H, Ahmad N, Li J, Wang Y, Liu X, Liu W, Li X, Wang N, Wang F. Genome-wide identification of MADS-box family genes in Safflower (Carthamus tinctorius L.) and functional analysis of CtMADS24 during flowering. Int J Mol Sci. 2023;24(2):1026. doi: 10.3390/ijms24021026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai S, Hashimoto W, Murata K. Transformation of Saccharomyces cerevisiae and other fungi: methods and possible underlying mechanism. Bioeng Bugs. 2010;1(6):395–403. doi: 10.4161/bbug.1.6.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min X, Lin X, Ndayambaza B, Wang Y, Liu W. Coordinated mechanisms of leaves and roots in response to drought stress underlying full-length transcriptome profiling in Vicia sativa L. BMC Plant Biol. 2020;20(1):165. doi: 10.1186/s12870-020-02358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 39.Robert N, Merlot S, N’Guyen V, Boisson-Dernier A, Schroeder JI. A hypermorphic mutation in the protein phosphatase 2 C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 2006;580(19):4691–4696. doi: 10.1016/j.febslet.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140(1):127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatnagar N, Min MK, Choi EH, Kim N, Moon SJ, Yoon I, Kwon T, Jung KH, Kim BG. The protein phosphatase 2 C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol Biol. 2017;93(4–5):389–401. doi: 10.1007/s11103-016-0568-2. [DOI] [PubMed] [Google Scholar]
- 42.He Z, Wu J, Sun X, Dai M. The Maize Clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int J Mol Sci. 2019;20(14):3573. doi: 10.3390/ijms20143573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 44.Dupeux F, Antoni R, Betz K, Santiago J, Gonzalez-Guzman M, Rodriguez L, Rubio S, Park SY, Cutler SR, Rodriguez PL, et al. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2 C allele. Plant Physiol. 2011;156(1):106–116. doi: 10.1104/pp.110.170894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Shi C, Sun D, He Y, Lai C, Lv P, Xiong Y, Zhang L, Wu F, Tian C. The HAB1 PP2C is inhibited by ABA-dependent PYL10 interaction. Sci Rep. 2015;5:10890. doi: 10.1038/srep10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YG, Yu HQ, Zhang YY, Lai CX, She YH, Li WC, Fu FL. Interaction between abscisic acid receptor PYL3 and protein phosphatase type 2 C in response to ABA signaling in maize. Gene. 2014;549(1):179–185. doi: 10.1016/j.gene.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012;160(1):379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, Elias A, Rabissi A, Lumbreras V, González-Guzmán M, et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell. 2013;25(10):3871–3884. doi: 10.1105/tpc.113.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shazadee H, Khan N, Wang L, Wang X. GhHAI2, GhAHG3, and GhABI2 negatively regulate osmotic stress tolerance via ABA-Dependent pathway in cotton (Gossypium hirsutum L) Front Plant Sci. 2022;13:905181. doi: 10.3389/fpls.2022.905181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rampino P, De Pascali M, De Caroli M, Luvisi A, De Bellis L, Piro G, Perrotta C. Td4IN2: a drought-responsive durum wheat (Triticum durum Desf.) Gene coding for a resistance like protein with serine/threonine protein kinase, nucleotide binding site and leucine rich domains. Plant Physiol Biochem. 2017;120:223–231. doi: 10.1016/j.plaphy.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Rössner C, Lotz D, Becker A. VIGS goes viral: how VIGS transforms our understanding of Plant Science. Annu Rev Plant Biol. 2022;73:703–728. doi: 10.1146/annurev-arplant-102820-020542. [DOI] [PubMed] [Google Scholar]
- 52.Ali A, Pardo JM, Yun DJ. Desensitization of ABA-Signaling: the Swing from activation to degradation. Front Plant Sci. 2020;11:379. doi: 10.3389/fpls.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. qRT-PCR analysis of CePYR1 expression in leaves of tiger nut exposed to drought stress. Error bars represent the standard deviation. **significant difference at P < 0.01
Data Availability Statement
The RNA sequencing data for both drought stress and non-drought stress tiger nut samples were deposited in the SRA database at the NCBI under accession number PRJNA975975. All other data and material analyzed in the current study are included in the manuscript and the supplementary information.