Abstract
Degradation of long-chain alkanes by Acinetobacter sp. strain ADP1 involves rubredoxin and rubredoxin reductase. We complemented a mutant deficient in alkane utilization and sequenced four open reading frames (ORFs) on the complementing DNA. Each of these ORFs was disrupted by insertional mutagenesis on the chromosome. As determined from sequence comparisons, ORF1 and ORF4 seem to encode a rotamase of the PpiC type and an acyl coenzyme A dehydrogenase, respectively. Disruption of these ORFs does not affect alkane utilization. In contrast, the two other ORFs, alkR and alkM, are essential for growth on alkanes as sole carbon sources. alkR encodes a polypeptide with extensive homology to AraC-XylS-like transcriptional regulators. It is located next to alkM, which encodes the terminal alkane hydroxylase, but is in the opposite orientation. Sequence homologies with other bacterial integral-membrane hydrocarbon hydroxylases suggest that AlkM may be the first member of a new protein family. The genes identified here are not linked to the rubredoxin- and rubredoxin reductase-encoding genes on the Acinetobacter sp. strain ADP1 chromosome.
Degradation of n-alkanes is a widespread trait among bacteria. Despite the apparent environmental impact of this process, very little is known about the genetics of alkane utilization. Extensive genetic and biochemical studies have been conducted on alkane utilization in Pseudomonas oleovorans, which can grow on medium-chain-length alkanes ranging from hexane to dodecane (27). The alk genes, encoding conversion of alkanes to acyl coenzyme A (acyl-CoA), are located in two different regions of the OCT plasmid. The alkBFGHJKL genes are cotranscribed from the alk promoter and encode the alkane hydroxylase, two rubredoxins, an aldehyde dehydrogenase, an alcohol dehydrogenase, an acyl-CoA synthetase, and an outer membrane protein whose function is not known (27). The other region contains alkS and alkT, which encode a LuxR-UhpA-like regulator of alk operon expression and rubredoxin reductase (27). Of special interest is the initial oxidation of the inert alkanes catalyzed in P. oleovorans by a three-component alkane monooxygenase complex that is composed of alkane hydroxylase, rubredoxin, and rubredoxin reductase and leads to the primary alcohol (27).
Several alkane oxidation pathways have been found in Acinetobacter spp. (1). Biochemical data have suggested that cytochrome P-450 is the terminal hydroxylase in some Acinetobacter strains which grow on medium-chain-length and long-chain alkanes (2). Recently, an alkane-oxidizing enzyme with putative dioxygenase activity involved in degradation of long-chain alkanes has been described (17). It has been postulated that there is a rubredoxin- and rubredoxin reductase-dependent terminal alkane hydroxylase involved in long-chain alkane oxidation in Acinetobacter calcoaceticus 69-V (1) and Acinetobacter sp. strain ADP1 (7, 24).
We describe here an alkane hydroxylase-encoding gene, alkM, and its regulatory gene, alkR, which are essential for growth of Acinetobacter sp. strain ADP1 on alkanes. The genetic organization of these alk genes is completely different from the arrangement found in P. oleovorans. Moreover, the similarity of AlkR to AraC-XylS-like transcriptional regulators implies that a different mechanism for regulating alkane utilization is present in Acinetobacter sp. strain ADP1. The sequence homologies of AlkM and other bacterial integral-membrane hydrocarbon hydroxylases suggest that these molecules constitute a novel protein family.
MATERIALS AND METHODS
General methods, bacterial strains, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and Acinetobacter sp. strain ADP1 were transformed as described previously (18, 21). Acinetobacter cells were electroporated with a gene pulser (Bio-Rad Laboratories, Munich, Germany). Total DNA was prepared and small-scale preparations of plasmids were obtained as described previously (7), and large-scale preparations of plasmids were obtained with a Nucleobond kit (Macherey-Nagel, Düren, Germany). Nucleotide sequence analysis and Southern hybridizations were performed as described previously (7). All other general methods and DNA manipulations were performed as described previously (21).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genetic marker(s) | Reference or source |
|---|---|---|
| Strains | ||
| Acinetobacter sp. strain ADP1 (=BD413) | Wild type | 13 |
| Acinetobacter sp. strain WH364 | alk | 7 |
| Acinetobacter sp. strain WH405 | alkM::lacZ, Kmr | This study |
| Acinetobacter sp. strain WH406 | alkR(ΔMluI-StyI), Cmr | This study |
| Acinetobacter sp. strain WH408 | ORF1::lacZ, Kmr | This study |
| Acinetobacter sp. strain WH409 | ORF4::lacZ, Kmr | This study |
| E. coli DH5α | recA1 endA1 supE44 gyrA96 thi hsdR17(rK− mK+) relA1 φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | 21 |
| Plasmids | ||
| pACYC184 | Cmr Tcr | 20 |
| pBluescript II SK+ | Apr | Stratagene |
| pKOK6.1 | Apr Kmr, promoterless lacZ | 15 |
| pWH767 | Apr | This study |
| pWH773 | Apr Kmr | This study |
| pWH777 | Apr Cmr | This study |
| pWH779 | Apr Kmr | This study |
| pWH782 | Apr | This study |
| pWH785 | Apr | This study |
| pWH786 | Apr | This study |
| pWH787 | Apr | This study |
| pWH788 | Apr | This study |
| pWH789 | Apr | This study |
| pWH790 | Apr | This study |
| pWH791 | Apr Kmr | This study |
| pWH1274 | Apr Tcr, shuttle vector | 12 |
Media and growth conditions.
Acinetobacter sp. strains were grown at 28°C on Luria broth (LB) (21) plates. Selectivity was achieved by adding ampicillin (300 mg/liter), kanamycin (10 mg/liter), or chloramphenicol (5 mg/liter). Selection for growth on specific carbon sources was performed on minimal medium supplemented with metal solution 44 and solidified with 1.5% agar (Noble agar; Difco Laboratories, Detroit, Mich.) as described previously (19). Alkanes and dodecanol were supplied through the gas phase by placing 200 μl of the compound (>99%) onto the center of a sterile filter paper disk placed in the lid of an inverted petri dish. If required, ampicillin was added at a concentration of 200 mg/liter. E. coli was grown at 37°C in LB and was grown under selective conditions with ampicillin (100 mg/liter), kanamycin (30 mg/liter), or chloramphenicol (20 mg/liter). Cultures used for preparation of total or plasmid DNA were grown in LB supplemented with an antibiotic if appropriate.
Nucleotide sequence accession number.
The 5,958-bp nucleotide sequence of Acinetobacter sp. strain ADP1 DNA cloned into pWH767 has been deposited in the EMBL data bank under accession no. AJ002316.
RESULTS
Characterization and complementation of a mutant deficient in alkane utilization.
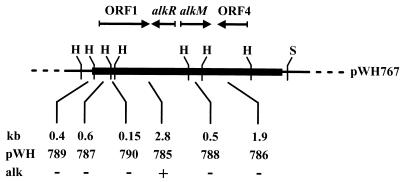
Ethyl methanesulfonate mutagenesis of Acinetobacter sp. strain ADP1 (7) yielded strain WH364, which does not grow on dodecane or hexadecane or longer-chain alkanes, but does grow on lauric acid. A gene library was constructed from strain ADP1 containing partially Sau3AI-digested DNA cloned into the BamHI site of pWH1274. The ligation mixture was electroporated into WH364. Plasmids from candidates growing on minimal medium supplemented with hexadecane as the sole carbon source were prepared, passed through E. coli DH5α, and retransformed into WH364. Five independently isolated plasmids conferred the ability to grow with hexadecane as the sole carbon source. Restriction analyses revealed insert lengths between 4.0 and 7.3 kb and the presence of common DNA fragments. The inserts represented continuous chromosomal fragments from Acinetobacter sp. strain ADP1, as confirmed by Southern hybridization (data not shown). The genetic organization of the 7.3-kb insert in pWH767 is shown in Fig. 1.
FIG. 1.
Genetic organization of pWH767. The gene bank insert from Acinetobacter is indicated by the solid box. The dashed lines indicate the vector part of the plasmid. Relevant restriction sites are shown. The distances between two HindIII sites and the resulting plasmids after subcloning of the fragments are indicated. The abilities of the plasmids to cause WH364 to revert to the wild-type phenotype are indicated. The arrows indicate the orientations of ORF1, alkR, alkM, and ORF4. Abbreviations: H, HindIII; S, SphI; alk, phenotype of transformed WH364 on hexadecane.
Subcloning and nucleotide sequence analysis of the complementing DNA.
pWH767 was cleaved with HindIII, which yielded six fragments from the insert. Each fragment was subcloned into the HindIII site of pBluescript II SK+. pWH785, carrying a 2.8-kb HindIII fragment, allowed WH364 to grow with hexadecane as the sole carbon source (Fig. 1). The complete nucleotide sequences of all subfragments were determined on both strands, and these sequences revealed four open reading frames (ORFs) (Fig. 1). These ORFs are typical of Acinetobacter genes with respect to their G+C contents (39 to 44.2% mol%) and codon usage (28).
A comparison of the deduced amino acid sequences with data bank entries revealed homology between the polypeptide encoded by ORF1 and peptidyl-prolyl cis-trans isomerases of the PpiC type. No functionally characterized protein in the data banks exhibited homology to the full-length ORF1 gene product. A rotamase of the cyclophilin type has been found in Acinetobacter sp. strain BD413 (synonymous with ADP1). Since a deletion of this gene is not lethal, it has been proposed that another gene encoding an enzyme with a similar function should be present in this strain (14).
alkR encodes a putative new member of the XylS-AraC family of transcriptional regulators, as indicated by the presence of the characteristic sequence motif (Prosite accession no. PS00041) in the conserved C-terminal region (6) and homology to typical members of this protein family. The highest level of similarity (percentage of identical residues) to AlkR (306 amino acids [aa]) was 22%; this value was obtained with RhaS (277 aa) from Salmonella typhimurium (SwissProt accession no. P27029) and XylS (321 aa) from the TOL plasmid of Pseudomonas putida (SwissProt accession no. P07859). AraC (292 aa) from E. coli (SwissProt accession no. P03021) had 19% identical residues.
AlkM (408 aa) exhibited homology to AlkB (401 aa) (41% of the amino acids are identical; 377 aa overlap) from P. oleovorans (SwissProt accession no. P12691). Only the following four other homologous proteins were found: the xylene monooxygenase XylM (369 aa) (25% of the amino acids are identical; 289 aa overlap) from P. putida (SwissProt accession no. P21395), the p-cymene monooxygenase CymAa (376 aa) (25% of the amino acids are identical; 253 aa overlap) from P. putida (EMBL accession no. U24215), the alkane hydroxylase AlkB (372 aa) (22% of the amino acids are identical; 355 aa overlap) from Pseudomonas maltophilia (GenBank accession no. U40233), and the deduced amino acid sequence of an ORF whose function is not known (416 aa) (45% of the amino acids are identical; 381 aa overlap) from Mycobacterium tuberculosis (EMBL accession no. Z95121). A hydrophobicity analysis revealed a pattern for AlkM that was very similar to that found for AlkB (data not shown). AlkB was previously shown to be an integral cytoplasmic membrane protein with six membrane-spanning regions (26).
The polypeptide encoded by ORF4 (401 aa) exhibited homology to acyl-CoA dehydrogenases. The highest level of residue identity (36%) was found with the medium-chain-length-specific acyl-CoA dehydrogenase from Homo sapiens (SwissProt accession no. P11310).
Inactivation of alkR, alkM, ORF1, and ORF4.
Growth of mutant WH364 on alkanes was restored by the nonreplicating plasmid pWH785, indicating that the original mutation was located on the 2.8-kb HindIII chromosomal fragment, leaving ORF1, alkR, and alkM as potentially essential genes for alkane utilization (Fig. 1). Therefore, we disrupted these genes on the Acinetobacter chromosome to analyze their contribution to alkane degradation.
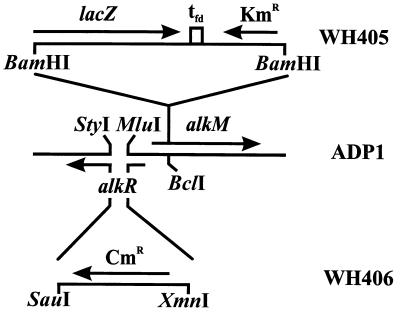
pWH785 was cleaved with MluI and StyI (Fig. 2), and the protruding ends were filled in. A Cmr cassette, excised with SauI and XmnI from pACYC184, was inserted, yielding pWH777. pWH777 was digested with SacII and ApaI, and the linear fragments were transformed into Acinetobacter sp. strain ADP1. Selection for chloramphenicol resistance resulted in alkR mutant strain WH406.
FIG. 2.
Genetic organization of constructed alk mutant strains WH405 and WH406. The lacZ-Kmr cassette from pKOK6.1 was inserted into alkM, yielding strain WH405, and the Cmr cassette from pACYC184 was inserted into alkR, yielding strain WH406. Genes and markers are indicated by arrows. The strain designations are indicated on the right. For details concerning construction, see the text.
Inactivation of alkM was achieved by cleavage of pWH785 with BclI. A Kmr cassette, excised with BamHI from pKOK6.1, was inserted after the protruding ends were filled in (Fig. 2). The resulting plasmid was called pWH773. Cleavage of pWH773 with SacII and ApaI followed by transformation into Acinetobacter sp. strain ADP1 and selection for kanamycin resistance yielded strain WH405.
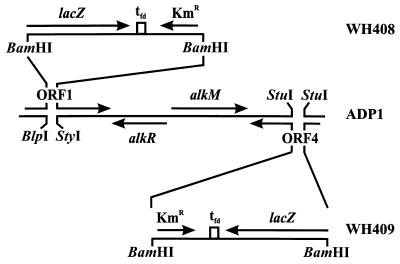
ORF1 was cleaved from pWH767 with EarI and BseRI. The protruding ends were filled in, and the fragment was inserted into the SmaI site of pBluescript II SK+, yielding pWH782. The Kmr cassette from pKOK6.1 was excised with BamHI, and this was followed by insertion into BlpI- and StyI-digested pWH782, after the respective protruding ends were filled in (Fig. 3). The resulting plasmid, pWH791, was cleaved with SacII and ApaI, and the linear fragments were transformed into Acinetobacter sp. strain ADP1. Selection for kanamycin resistance yielded strain WH408.
FIG. 3.
Genetic organization of constructed alk+ mutant strains WH408 and WH409. The lacZ-Kmr cassette from pKOK6.1 was inserted into ORF1 and ORF4, yielding strains WH408 and WH409, respectively. Genes and markers are indicated by arrows. The strain designations are indicated on the right. For details concerning construction, see the text.
Restriction of pWH786 with StuI resulted in a deletion within ORF4 (Fig. 3). The same Kmr cassette from pKOK6.1 was inserted, which yielded pWH779. Cleavage of pWH779 with SacII and ApaI, followed by transformation into Acinetobacter sp. strain ADP1 and selection for kanamycin resistance, resulted in strain WH409 (Fig. 3).
The inactivation of alkR, alkM, ORF1, and ORF4 as shown in Fig. 2 and 3 was confirmed by Southern blotting (data not shown). Each mutant was studied to determine its ability to grow on alkanes as sole carbon and energy sources. WH408 and WH409 did not have the alk phenotype, whereas WH405 and WH406 did not grow on alkanes but grew well on dodecanol.
DISCUSSION
Alkane utilization is a common feature of Acinetobacter species. The biochemistry of alkane oxidation by members of this genus has been intensely studied, but previously nothing was known about the nature of the terminal alkane hydroxylase and its regulation (1). AlkR is similar to XylS-AraC, which is characterized by a conserved C-terminal domain that mediates DNA binding via an α-helix–turn–α-helix motif (6). The nonconserved N-terminal domain is responsible for oligomerization and effector binding (3, 6). We concluded from the indispensability of alkR for alkane degradation that this gene encodes an activator of alk expression. Genes encoding regulators of the XylS-AraC family are typically in orientations opposite to those of their target genes (6), which is consistent with the opposite orientations of alkR and alkM (Fig. 1). Since AlkR shows no similarity to the LuxR-UhpA-like regulator AlkS from P. oleovorans, alk regulation appears to be different in the two organisms.
AlkM is essential for growth on alkanes, but not for growth on the corresponding alkanols, suggesting that it is the terminal alkane hydroxylase. Interestingly, the relationships among the different homologous hydroxylases are not uniform, as deduced from a pairwise comparison of the proteins. Most of the hydrocarbon hydroxylases compared in this study are homologous to each other; the only exception is AlkB from P. maltophilia. This protein is not homologous to XylM and CymAa, as indicated by low RDF2 comparison scores. Moreover, the alcohol dehydrogenase from Schizosaccharomyces pombe (SwissProt accession no. P00332) exhibits the best match with AlkB from P. maltophilia, ahead of AlkB from P. oleovorans and followed by more than 15 other alcohol dehydrogenases from prokaryotic and eukaryotic organisms. In contrast, we found no homology between the other hydrocarbon hydroxylases and alcohol dehydrogenases. Another feature is the presence of the eight-histidine motif, which is characteristic of nonheme integral-membrane desaturases and hydroxylases (23). The histidyl residues are thought to bind 1 to 3 mol of iron per mol of enzyme as a cofactor and to be part of the active sites of these enzymes (22, 23). They are completely conserved in AlkM and in all of the other homologous hydroxylases except AlkB from P. maltophilia, in which a His-to-Ile change occurs at the last position in the first (HXXXH) block (Fig. 4). These enzymes seem to be integral-membrane proteins, as shown previously for AlkB from P. oleovorans (26). We suggest that the different integral-membrane hydrocarbon hydroxylases originated from a common ancestor and that they evolved into proteins with different functions. On the one hand, this resulted in the hydroxylase unit of three-component alkane monooxygenase complexes, like AlkM, AlkB from P. oleovorans (27), AlkB from P. maltophilia (16), and maybe ORFX from M. tuberculosis. On the other hand, it resulted in the hydroxylase unit of two-component monooxygenases, like XylM (25) and CymAa (5). We propose a new family of bacterial integral-membrane hydrocarbon hydroxylases. This family should not depend only on the presence of the eight-histidine motif, because this pattern seems to be functionally rather than evolutionarily determined. This motif has also been found in integral-membrane proteins like desaturases, hydroxylases, oxidases, and decarbonylases from prokaryotic and eukaryotic organisms, which are not necessarily related to each other, and also occurs in some soluble proteins (22). The amino acid signature deduced from the most highly conserved region without gaps in the multiple sequence alignment shown in Fig. 4 provides a more specific classification, which could help identify additional members of this family. The unique position of AlkB from P. maltophilia within this family might be explained by a greater evolutionary distance from the other proteins of this family.
FIG. 4.
Part of a multiple sequence alignment of homologous hydrocarbon hydroxylases. The presence of four or more identical amino acids at a given position is indicated by a white letter on a black background. Two of the three conserved histidine boxes of the eight-histidine motif (23) are indicated above the sequences; H indicates a conserved histidyl residue, and X indicates a spacing residue. The numbers before each sequence indicate absolute positions within the primary structure. The deduced protein signature is given below the sequence alignment. The letters represent residues that are conserved in all sequences (uppercase letters) or in the majority of sequences (lowercase letters); completely conserved S or T at one position is designated O or (if the residues occur in the majority of sequences) o; a percent sign indicates an aliphatic residue; an ampersand indicates a bulky aliphatic or aromatic residue; a section sign indicates a charged or polar residue; and a dot indicates any residue. Abbreviations: AlkM_Ac, AlkM from Acinetobacter sp. strain ADP1; AlkB_Po, AlkB from P. oleovorans; AlkB_Pm, AlkB from P. maltophilia; XylM_Pp, XylM from P. putida; CymA_Pp, CymAa from P. putida; OrfX_Mt, deduced amino acid sequence of a gene with unknown function from M. tuberculosis.
The alk genes are tightly clustered on the OCT plasmid in P. oleovorans (27). The genetic organization in Acinetobacter sp. strain ADP1 is completely different. The alk genes characterized so far are neither grouped in large operons nor clustered or localized on a plasmid. alkM is a distinct gene, which is in the orientation opposite to that of alkR (Fig. 1). Both genes are located on the chromosome about 369 kb from the rubA and rubB genes, which encode rubredoxin and rubredoxin reductase, respectively (10). In contrast to the P. oleovorans genes, the rubAB genes are arranged in a putative operon (7). Moreover, genes for alcohol and aldehyde dehydrogenases have not been found in the vicinity of these alk genes (Fig. 1) (8, 9). The apparent lack of linkage of genes involved in the same biochemical pathway could indicate the originality of this system. It has been demonstrated that xcpR, a gene encoding a subunit of the general secretory pathway, is required for alkane utilization in Acinetobacter sp. strain ADP1 (19). The general secretory pathway is also present in pseudomonads (4), and it might be involved in alkane utilization, because a secreted protein participates in this degradative pathway in Pseudomonas aeruginosa (11).
Our results suggest that alkane degradation is not uniform in different Acinetobacter strains. Cytochrome P-450 in some Acinetobacter strains, which grow on medium-chain-length and long-chain alkanes (2), and the dioxygenase activity involved in long-chain alkane degradation in Acinetobacter sp. strain M-1 (17) might be alternative systems for alkane oxidation. It is, however, clear that none of the enzymes mentioned above is present in ADP1, as indicated by the indispensability of alkM for alkane utilization.
ACKNOWLEDGMENTS
We thank E. Pook for critically reading the manuscript. pKOK6.1 was kindly provided by W. Lotz.
This work was supported by the Fonds der chemischen Industrie.
REFERENCES
- 1.Asperger O, Kleber H-P. Metabolism of alkanes by Acinetobacter. In: Towner K J, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 323–351. [Google Scholar]
- 2.Asperger O, Naumann A, Kleber H-P. Occurrence of cytochrome P-450 in Acinetobacter strains after growth on n-hexadecane. FEMS Microbiol Lett. 1981;11:309–312. [Google Scholar]
- 3.Bustos S, Schleif R. Functional domains of the AraC protein. Proc Natl Acad Sci USA. 1993;90:5638–5642. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot A, Krijger J-J, Filloux A, Tommassen J. Characterization of type II protein secretion (xcp) genes in the plant growth-stimulating Pseudomonas putida strain WCS358. Mol Gen Genet. 1996;250:491–501. doi: 10.1007/BF02174038. [DOI] [PubMed] [Google Scholar]
- 5.Eaton R W. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol. 1997;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallegos M-T, Michan C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geissdörfer W, Frosch S C, Haspel G, Ehrt S, Hillen W. Two genes encoding proteins with similarities to rubredoxin and rubredoxin reductase are required for conversion of dodecane to lauric acid in Acinetobacter calcoaceticus ADP1. Microbiology. 1995;141:1425–1432. doi: 10.1099/13500872-141-6-1425. [DOI] [PubMed] [Google Scholar]
- 8.Geissdörfer W, Ratajczak A, Hillen W. Nucleotide sequence of a putative periplasmic Mn superoxide dismutase from Acinetobacter calcoaceticus ADP1. Gene. 1997;186:305–308. doi: 10.1016/s0378-1119(96)00728-7. [DOI] [PubMed] [Google Scholar]
- 9.Geissdörfer W, Ratajczak A, Hillen W. Transcription of ppK from Acinetobacter sp. strain ADP1 encoding a putative polyphosphate kinase is induced by phosphate starvation. Appl Environ Microbiol. 1998;64:896–901. doi: 10.1128/aem.64.3.896-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 11.Hardegger M, Koch A K, Ochsner U A, Fiechter A, Reiser J. Cloning and heterologous expression of a gene encoding an alkane-induced extracellular protein involved in alkane assimilation from Pseudomonas aeruginosa. Appl Environ Microbiol. 1994;60:3679–3687. doi: 10.1128/aem.60.10.3679-3687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunger M, Schmucker R, Veerebrahma K, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87:45–51. doi: 10.1016/0378-1119(90)90494-c. [DOI] [PubMed] [Google Scholar]
- 13.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kok R G, Christoffels V M, Vosman B, Hellingwerf K J. A gene of Acinetobacter calcoaceticus BD413 encodes a periplasmic peptidyl-prolyl cis-trans isomerase of the cyclophilin sub-class that is not essential for growth. Biochim Biophys Acta. 1994;1219:601–606. doi: 10.1016/0167-4781(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 15.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promotor probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 16.Lee N R, Hwang M O, Jung G H, Kim Y S, Min K H. Physical structure and expression of alkBA encoding alkane hydroxylase and rubredoxin reductase from Pseudomonas maltophilia. Biochem Biophys Res Commun. 1996;218:17–21. doi: 10.1006/bbrc.1996.0004. [DOI] [PubMed] [Google Scholar]
- 17.Maeng J H, Sakai Y, Tani Y, Kato N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J Bacteriol. 1996;178:3695–3700. doi: 10.1128/jb.178.13.3695-3700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmen R, Vosman B, Buijsman P, Breek C K, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 19.Parche S, Geissdörfer W, Hillen W. Identification and characterization of xcpR encoding a subunit of the general secretory pathway necessary for dodecane degradation in Acinetobacter calcoaceticus ADP1. J Bacteriol. 1997;179:4631–4634. doi: 10.1128/jb.179.14.4631-4634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose R E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Shanklin J, Achim C, Schmidt H, Fox B G, Munck E. Mossbauer studies of alkane omega-hydroxylase: evidence for a diiron cluster in an integral-membrane enzyme. Proc Natl Acad Sci USA. 1997;94:2981–2986. doi: 10.1073/pnas.94.7.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanklin J, Whittle E, Fox B G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 24.Strätz M, Mau M, Timmis K N. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol Microbiol. 1996;22:207–215. doi: 10.1046/j.1365-2958.1996.00099.x. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki M, Hayakawa T, Shaw J P, Rekik M, Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991;173:1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Beilen J B, Penninga D, Witholt B. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J Biol Chem. 1992;267:9194–9201. [PubMed] [Google Scholar]
- 27.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 28.White P J, Hunter I S, Fewson C A. Codon usage in Acinetobacter structural genes. In: Towner K J, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 251–257. [Google Scholar]