Abstract
Recently, DNA pairing analyses showed that Pseudomonas syringae pv. tomato and related pathovars, including P. syringae pv. maculicola, form a genomic species (Pseudomonas tomato) (L. Gardan, H. L. Shafik, and P. A. D. Grimont, p. 445–448, in K. Rudolph, T. J. Burr, J. W. Mansfield, D. Stead, A. Vivian, and J. von Kietzell, ed., Pseudomonas syringae Pathovars and Related Pathogens, 1997). The genetic diversity of 23 strains belonging to this genomic species and 4 outgroup strains was analyzed with randomly amplified polymorphic DNA (RAPD) and amplified fragment length polymorphic (AFLP) techniques. Simple boiling of P. syringae cells was suitable for subsequent DNA amplification to obtain reliable patterns in RAPD and AFLP analyses. In general, the grouping of P. syringae strains by both analysis techniques corresponded well with the classification obtained from an RFLP analysis of ribosomal DNA operons, DNA pairing studies, and an analysis of pathogenicity data. However, two strains of P. syringae pv. maculicola produced distinct DNA patterns compared to the DNA patterns of other P. syringae pv. maculicola strains; these patterns led us to assume that horizontal transfer of DNA could occur between bacterial populations. Both techniques used in this study have high discriminating power because strains of P. syringae pv. tomato and P. syringae pv. maculicola which were indistinguishable by other techniques, including pathogenicity tests on tomato, were separated into two groups by both RAPD and AFLP analyses. In addition, data analysis showed that the AFLP method was more efficient for assessing intrapathovar diversity than RAPD analysis and allowed clear delineation between intraspecific and interspecific genetic distances, suggesting that it could be an alternative to DNA pairing studies. However, it was not possible to distinguish the two races of P. syringae pv. tomato on the basis of an analysis of the data provided by either the AFLP or RAPD technique.
Pseudomonas syringae van Hall (14) is a plant-associated bacterial species which has been divided into more than 50 pathovars. It causes diseases in all major groups of higher plants, producing mainly necrotic lesions on aerial parts of plants. The pathovar concept was introduced by Young et al. (23, 24) in order to provide a nomenclature at the subspecies level based on differences in plant host range and types of symptoms. Such a phenotypically based classification is of practical interest, but it does not reveal the genetic relatednesses between pathovars. Using DNA pairing analysis, Pecknold and Grogan (15) showed that P. syringae is a heterogeneous species. Later, Denny et al. (2) showed that P. syringae pv. tomato strains form a genetically homogeneous group that is clearly distinct from P. syringae pv. syringae strains and that should be considered a genomic species. Furthermore, PCR-restriction fragment length polymorphism (RFLP) analysis of the rrn operon confirmed the heterogeneity and showed that P. syringae pv. tomato and P. syringae pv. maculicola are closely related (12). Recently, nine genomic species were described within P. syringae on the basis of the results of DNA pairing studies. Strains belonging to several pathovars of P. syringae (P. syringae pv. tomato, P. syringae pv. maculicola, P. syringae pv. apii, P. syringae pv. antirrhini, P. syringae pv. delphinii, P. syringae pv. persicae) have been clustered in genomic species III, for which the name Pseudomonas tomato has been proposed. Curiously, one strain of P. syringae pv. maculicola (CFBP 1637) was found not to be a member of genospecies III by DNA pairing studies (7, 18).
From an agronomic point of view, P. syringae pv. tomato is the causal agent of bacterial speck of tomato. This disease is distributed worldwide and is responsible for reductions in the commercial quality of fruits and yield (25). P. syringae pv. tomato is mainly transmitted by infected seeds and plants (9). P. syringae pv. maculicola, including strain CFBP 1637, causes disease on crucifers, but it is also pathogenic on tomato and produces the typical symptoms of tomato speck when it is artificially inoculated (8, 22). Furthermore, strains belonging to the two pathovars have the same biochemical characteristics. Consequently, it has been suggested that these two bacteria should be placed in the same pathovar (20). Against this background, efficient tools for identifying populations within P. syringae pathovars responsible for outbreaks would be very useful for setting up integrated control methods. This group of strains, identified as P. syringae pv. tomato and P. syringae pv. maculicola and belonging to the same genospecies, is therefore a good group for testing the usefulness of techniques at several levels, including the ability of techniques to distinguish bacteria belonging to different genospecies, to group bacteria belonging to the same genospecies, and to identify bacterial strains in epidemiological surveys.
In this paper, two techniques, the randomly amplified polymorphic DNA (RAPD) technique and amplified fragment length polymorphism (AFLP), were used to discriminate pathovars belonging to the genomic species P. tomato and to identify bacterial isolates.
MATERIALS AND METHODS
Bacterial strains.
The bacterial cultures used in this study are listed in Table 1. Most of the strains used belong to the same RFLP group, as determined by an analysis of the internally transcribed sequence between the rrs and rrl genes (ITS1) in the rrn operon (12); the only exceptions were P. syringae pv. delphinii CFBP 2215, P. syringae pv. syringae CFBP 1392, P. syringae pv. phaseolicola CFBP 1390, and Pseudomonas viridiflava CFBP 2107, which were used as outgroups. Most of the test strains which were in RFLP group A belong to genospecies III of P. syringae (P. tomato); the only exception was P. syringae pv. maculicola CFBP 1637, which clustered outside the genospecies (18). Bacterial strains were grown on medium B of King (11) at 27°C and were stored at 4°C. For long-term storage, bacteria were kept at −80°C in YP-glycerol broth (7 g of yeast extract per liter, 7 g of Bacto Peptone [Difco Laboratories, Detroit, Mich.] per liter, 300 g of glycerol per liter; pH 7).
TABLE 1.
Bacterial strains
| Strain | Geographical origin | Host plant | Year of isolation | Pathogenicity for tomato | Genomic speciesa | Groupb |
|---|---|---|---|---|---|---|
| P. syringae pv. tomato strains | ||||||
| CFBP 2212Td | United Kingdom | Lycopersicon esculentum | 1960 | + | III | A |
| CFBP 1321 | Switzerland | Lycopersicon esculentum | ? | + | A | |
| JN16.5 | Portugal | Lycopersicon esculentum | 1991 | + | A | |
| CFBP 1920 | Canada | Lycopersicon esculentum | ? | + | A | |
| TE 951 | Spain | Lycopersicon esculentum | ? | + | A | |
| UCRD | United States | Lycopersicon esculentum | ? | + | A | |
| JN51 | Canada | Lycopersicon esculentum | ? | + | A | |
| JN52 | Canada | Lycopersicon esculentum | ? | + | A | |
| JN53 (race 1)c | Canada | Lycopersicon esculentum | ? | + | A | |
| JN54 (race 1) | Canada | Lycopersicon esculentum | ? | + | A | |
| 1427st | France | Lycopersicon esculentum | ? | + | III | A |
| JN20.1e | France | Lycopersicon esculentum | 1993 | + | A | |
| P. syringae pv. maculicola strains | ||||||
| CFBP 1657T | New Zealand | Brassica oleracea | 1965 | + | III | A |
| CFBP 1637 | United States | Raphanus sativus | 1965 | + | Not III | A |
| CFBP 1738 | United Kingdom | Brassica oleracea | 1965 | + | A | |
| CFBP 1740 | Zimbabwe | Brassica oleracea | 1970 | + | A | |
| ICMP 2744d | United Kingdom | Brassica nigra | 1968 | + | A | |
| JN8.5 | France | Raphanus sativus | 1993 | + | A | |
| JN8.10 | France | Raphanus sativus | 1993 | + | A | |
| P. syringae CFBP 1777 (pathovar undetermined) | New Zealand | Euphorbia pulcherrima | 1972 | + | A | |
| P. syringae pv. antirrhini CFBP 1620T | United Kingdom | Antirrhinum majus | 1965 | − | III | A |
| P. syringae pv. apii CFBP 1726 | United States | Apium gravoleolens | 1975 | + | III | A |
| P. syringae pv. berberidis CFBP 1727T | New Zealand | Berberis sp. | 1972 | − | III | A |
| P. syringae pv. lachrymans CFBP 2440T | United States | Cucumis sativus | 1935 | − | III | A |
| P. syringae pv. passiflorae CFBP 2346T | New Zealand | Passiflora edulis | 1962 | − | III | A |
| P. syringae pv. persicae CFBP 1573T | France | Prunus persicae | 1974 | − | III | A |
| P. syringae pv. delphinii CFBP 2215T | New Zealand | Delphinium sp. | 1957 | − | III | D |
| P. syringae pv. syringae CFBP 1392T | United Kingdom | Syringa vulgaris | 1950 | − | I | M |
| P. syringae pv. phaseolicola CFBP 1390T | Canada | Phaseolus vulgaris | 1949 | − | II | N |
| P. viridiflava CFBP 2107T | Switzerland | Phaseolus sp. | 1927 | − | VI | Q |
Group as determined by PCR-RFLP analysis of the rrn operon (12).
Physiological race determined by inoculation on tomato cultivars bearing the Pto resistance gene (this study).
CFBP, Collection Française de Bactéries Phytopathogènes, Institut National de la Recherche Agronomique, Angers France; ICMP, International Collection of Microorganisms from Plants, Department of Scientific and Industrial Research, Auckland, New Zealand. T = type strain.
JN20-1 was isolated from a tomato field artificially infected 8 months previously with strain 1427st.
Plant inoculation.
The pathogenicity of selected strains was assayed on tomato plants (Lycopersicon esculentum cv. Montfavet 63-4) which were susceptible to both races of P. syringae pv. tomato. Inoculation was accomplished by spraying bacterial suspensions (5 × 108 CFU ml−1) over the foliage of 1-month-old plants. The inoculated plants were then incubated in a growth chamber (16 h of light at 24°C and 8 h of darkness at 20°C per day) with a relative humidity of almost 100%. Symptoms were recorded 7 days after inoculation. Three plants per strain were inoculated.
DNA extraction.
Two methods were used to extract DNA from bacterial cells. In the first method the cetyltrimethylammonium bromide technique of Ausubel et al. (1) was used. The concentration of the DNA was determined spectrophotometrically. Each DNA preparation was diluted in water to obtain a concentration of 2 ng ml−1. The second method involved a partial DNA extraction technique performed as follows. Bacterial suspensions (optical density at 600 nm, 0.3) were boiled for 10 min and immediately placed on ice for 10 min. The insoluble cellular residues were removed by centrifugation at 11,340 × g for 5 min. The supernatants were directly used for PCR or ligation reactions or were stored at 4°C for several weeks.
RAPD analysis.
DNA amplification reactions were carried out in a final volume of 50 μl. A 5-μl portion of a DNA solution, either purified DNA or supernatant from a boiled culture, was added to 45 μl of amplification reaction mixture containing 75 mM Tris-HCl (pH 7.9 at 25°C), 1.5 mM MgCl2, 20 mM (NH4)2SO4, 0.01% (wt/wt) Tween 20, each deoxynucleoside triphosphate at a concentration of 400 mM, 2 U of Red GoldStar DNA polymerase (EUROGENTEC SA, Seraing, Belgium), and 25 ng of primer from either OPERON H and G primer kits (OPERON Technologies, Inc., Alameda, Calif.) or kits 60, 70, and 80 (EUROGENTEC SA). PCR amplifications were performed with a model PTC 100 thermocycler (MJ Research Inc., Watertown, Mass.) by using the following protocol: an initial step consisting of 94°C for 1 min; 45 cycles consisting of denaturation at 94°C for 1 min, annealing at 36°C for 1 s, and extension at 72°C for 1 min; and finally a single extension step consisting of 72°C for 2 min. The PCR amplification products were then maintained at 10°C until electrophoresis analyses were performed.
Simplified AFLP procedure.
The AFLP procedure performed was a simplified version of the AFLP procedure described previously (21, 26). The previously described method was modified in order to provide data for short procaryotic genomes by using standard visualization of PCR products after separation by agarose gel electrophoresis and staining with ethidium bromide. To do this, we used both a single four-base endonuclease (MspI, CCGG) and discriminating primers with three nucleotides downstream from the MspI site which allowed selective amplification of an average of four to five amplified bands; the number of bands depended on the G+C content of the genome. Under these conditions, p(C) = p(G) and p(A) = p(T) were the probabilities of occurrence of the bases at each nucleotide position, p(X1), p(X2), and p(X3) were the probabilities of occurrence of the three nucleotides downstream from the restriction site, and N was the number of base pairs in the genome. The average number of fragments (n) selectively amplified under stringent conditions was n = N × p(C)4 × p(X1)2 × p(X2)2 p(X3)2. As determined with this formula, when N = 5,000,000 bp and p(C) = p(A) = 0.25, n = 5 × 106 × p(0.25)10 = 4.77.
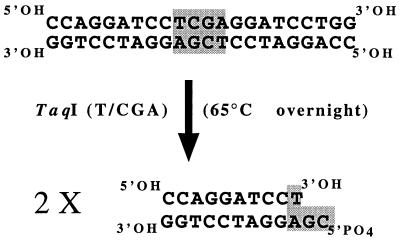
Adaptors were constructed with a 22-bp oligonucleotide (Fig. 1). To do this, 20 μg (2 μg μl−1) of oligonucleotide in TE8 buffer (25 mM Tris-HCl, 5 mM EDTA; pH 8) was heated at 65°C for 10 min and then kept at room temperature for 1 h to allow the oligonucleotide to autohybridize. The resulting double-stranded molecules (20 μg) were then digested with 100 U of TaqI endonuclease (EUROGENTEC SA) at 65°C overnight in order to obtain double-stranded 11-bp oligonucleotides with a GC5′-PO4 cohesive end.
FIG. 1.
Synthesis of 11-bp adaptors by digestion of annealed, symmetrical, 22-bp oligonucleotides with TaqI.
To prepare template DNA, bacterial DNAs were digested with MspI and were ligated to adaptors simultaneously in a single step. Original MspI sites were not restored after the adaptors were ligated, which prevented digestion of ligation products. The following two types of bacterial DNA preparations were used to obtain DNA templates: 500 ng (1 μl) of purified DNA in TE8 buffer and 10 μl of supernatant from bacterial suspensions (108 CFU ml−1) in sterile distilled water which were previously boiled for 10 min and centrifuged at 13,000 × g for 10 min at room temperature. Digestion and ligation were performed for 3 h at 37°C in 60 μl (final volume) of ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 20 mM dithiothreitol, 1 mM ATP; pH 7.9) containing 10 U of MspI (EUROGENTEC SA) and 1 U of T4 DNA ligase (EUROGENTEC SA). The enzymes were inactivated at 65°C for 20 min. The DNA templates were then stored at 4°C until they were used.
MspI restriction fragments tagged with the specific adaptors were used as template DNAs for selective PCR amplification directed by single 16-bp primers with constant peak complementary to the adaptor sequence and the MspI site and a 3-nucleotide variable portion at the 3′-OH end.
DNA amplification reactions were carried out in 50-μl reaction mixtures. A 1-μl portion of ligation products was added to 49 μl of amplification reaction mixture containing 75 mM Tris-HCl (pH 9 at 25°C), 1.5 mM MgCl2, 20 mM (NH4)2SO4, 0.01% (wt/wt) Tween 20, each deoxyribonucleoside triphosphate at a concentration of 200 mM, 2 U of Red GoldStar DNA polymerase (EUROGENTEC SA), and 40 pmol of each of the primers used (EUROGENTEC SA). Amplifications were performed with a model PTC 150 thermocycler (MJ Research) by using the following protocol: an initial step consisting of 94°C for 1 min; and 35 cycles consisting of denaturation at 94°C for 1 min, annealing at 60 or 66°C for 1 min, and extension at 72°C for 1 min. The PCR products were then kept at 10°C until electrophoresis. All PCR amplifications were repeated at least twice with different DNA preparations.
Electrophoresis.
Portions (7 μl) of amplification products were separated by electrophoresis in 2% agarose gels (IDna, FMC Bioproducts, Rockland, Maine) in TAE buffer (40 mM Tris-acetate, 1 mM EDTA; pH 8) for 30 min at 100 V. The gels were then stained with ethidium bromide and photographed under UV light (wavelength, 360 nm).
Data analysis.
Each amplification band was treated as a unit character and was scored as 1 (present) or 0 (absent) for all strains, which allowed us to construct a distance matrix by using the Jaccard coefficient (19). The presence-absence matrices were used for parsimony analyses, and the distance matrices were used to construct dendrograms with the method of Fitch and Margoliash (6), the unweighted pair group method (19), and the neighbor-joining method (17) with the DNAPAR and MIX software of the PHYLIP package (5). The strength of the tree topology was assessed by the bootstrap method with the CONSENSE software of PHYLIP.
RESULTS AND DISCUSSION
In this study, we assessed the suitability of RAPD and AFLP techniques for rapid molecular characterization of plant-pathogenic bacteria, especially at the pathovar and strain levels.
However, the parameters of the reactions must be determined to obtain reliable results. PCR amplification in RAPD analyses and DNA ligation in AFLP analyses were carried out either with a purified DNA solution (2 ng ml−1) or with bacterial suspensions (A260 = 0.3). Simple boiling of bacterial cells produced DNA in suitable quantity and quality to allow PCR amplification in the RAPD procedure. An additional step (centrifugation of bacterial lysates at 13,000 rpm for 5 min) was required to obtain reliable DNA ligations in the AFLP procedure (data not shown). Thus, at least in the case of P. syringae cultivated on King’s medium B, it was not necessary to perform a complex extraction procedure to purify the genomic DNA used as a template for RAPD and AFLP analyses. By reducing the number of manipulations this method limits cross-contamination risks. Moreover, preparations can be obtained quickly and without the use of toxic products, which is useful for fast routine diagnosis performed with numerous strains.
Various primers were tested for efficacy in the RAPD and AFLP methods, and the primers listed in Tables 2 and 3 were selected because they gave readily interpretable and reproducible results. All of these primers had G+C contents greater than 60 mol%, while the genome G+C content of P. syringae is 59 to 60 mol% (3).
TABLE 2.
Selected primers used for the RAPD analysis
| Primer | Sequence |
|---|---|
| Operon G13 | 5′CTCTCCGCCA3′ |
| Operon G19 | 5′GTCAGGGCAA3′ |
| Operon H3 | 5′AGACGTCCAC3′ |
| Operon H13 | 5′GACGCCACAC3′ |
| Genosys 60.20 | 5′GACCGACACG3′ |
| Genosys 70.2 | 5′CAGGGTCGAC3′ |
| Genosys 70.4 | 5′CGCATTCCGC3′ |
| Genosys 70.8 | 5′CTGTACCCCC3′ |
| Genosys 70.9 | 5′TGCAGCACCG3′ |
| Genosys 80.6 | 5′GCACGGAGGG3′ |
| Genosys 80.7 | 5′GCACGCCGGA3′ |
| Genosys 80.8 | 5′CGCCCTCAGC3′ |
TABLE 3.
Selected primers used for the AFLP analysis
| Primer | Sequencea |
|---|---|
| 3 | 5′CCAGGATCCTCGGCCA3′ |
| 4 | 5′CCAGGATCCTCGGCCT3′ |
| 7 | 5′CCAGGATCCTCGGCAG3′ |
| 8 | 5′CCAGGATCCTCGGCAA3′ |
| 10 | 5′CCAGGATCCTCGGCTC3′ |
| 12 | 5′CCAGGATCCTCGGCTA3′ |
| 15 | 5′CCAGGATCCTCGGCGG3′ |
| 18 | 5′CCAGGATCCTCGGAGC3′ |
| 35 | 5′CCAGGATCCTCGGGAG3′ |
| 36 | 5′CCAGGATCCTCGGGAA3′ |
| 38 | 5′CCAGGATCCTCGGGTC3′ |
| 42 | 5′CCAGGATCCTCGGGGC3′ |
Boldface letters indicate discriminating nucleotides located downstream of the MspI recognition site in the template DNA.
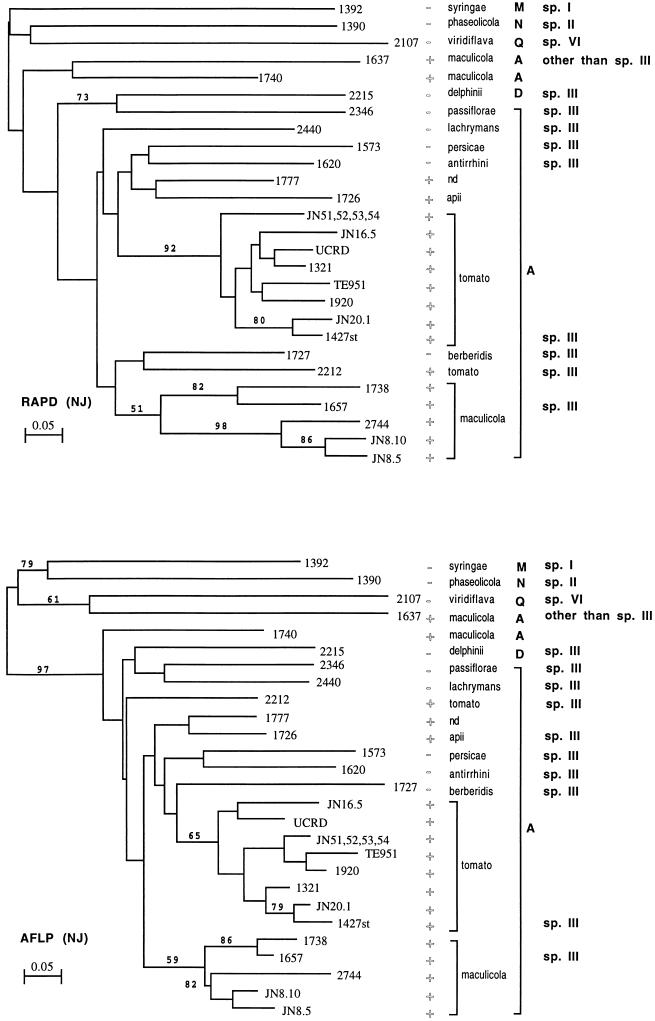
The number of DNA bands observed after electrophoresis of the PCR products obtained with 12 primers varied from 24 to 34 in the RAPD analysis and from 24 to 37 in the AFLP analysis (data not shown). Depending on the strain, 132 and 100 different DNA bands were used for the analyses of RAPD and AFLP results, respectively. The 30 strains produced 27 patterns, and almost every strain produced a unique pattern; the only exceptions were four Canadian strains (JN51, JN52, JN53, and JN54) which produced the same pattern in all of the analyses. The genetic distances between strains were calculated (Table 4). The trees obtained by the distance-based methods (Fitch-Margoliash, neighbor-joining, and unweighted pair group methods), as well as the parsimony method, were very similar in both the RAPD and AFLP analyses. The consensus tree and bootstrap data are shown in Fig. 2.
TABLE 4.
Genetic distances between strains calculated with AFLP and RAPD data
| Strain | Genetic distancesa
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1321 | 1427st | 1920 | 2212 | TE951 | UCRD | JN16-5 | JN20-1 | JN5nb | 1657 | 1637 | 1740 | 1738 | 2744 | JN8-5 | JN8-10 | 1726 | 1392 | 1777 | 1620 | 1727 | 2440 | 2346 | 1573 | 2215 | 1390 | 2107 | |
| 1321 | 0.065 | 0.094 | 0.235 | 0.111 | 0.097 | 0.097 | 0.062 | 0.048 | 0.207 | 0.745 | 0.312 | 0.233 | 0.25 | 0.219 | 0.188 | 0.167 | 0.729 | 0.138 | 0.179 | 0.25 | 0.276 | 0.29 | 0.2 | 0.39 | 0.789 | 0.745 | |
| 1427st | 0.111 | 0.062 | 0.235 | 0.079 | 0.097 | 0.129 | 0.031 | 0.079 | 0.241 | 0.782 | 0.281 | 0.267 | 0.219 | 0.188 | 0.188 | 0.2 | 0.763 | 0.172 | 0.214 | 0.281 | 0.276 | 0.298 | 0.236 | 0.424 | 0.754 | 0.745 | |
| 1920 | 0.062 | 0.079 | 0.229 | 0.046 | 0.062 | 0.062 | 0.061 | 0.046 | 0.233 | 0.789 | 0.303 | 0.258 | 0.242 | 0.212 | 0.182 | 0.194 | 0.738 | 0.167 | 0.241 | 0.242 | 0.267 | 0.288 | 0.263 | 0.41 | 0.729 | 0.754 | |
| 2212 | 0.354 | 0.375 | 0.385 | 0.275 | 0.176 | 0.235 | 0.229 | 0.246 | 0.25 | 0.77 | 0.286 | 0.273 | 0.257 | 0.2 | 0.2 | 0.242 | 0.723 | 0.219 | 0.355 | 0.286 | 0.219 | 0.27 | 0.311 | 0.323 | 0.683 | 0.705 | |
| TE951 | 0.097 | 0.148 | 0.097 | 0.365 | 0.111 | 0.111 | 0.108 | 0.062 | 0.254 | 0.786 | 0.323 | 0.311 | 0.262 | 0.231 | 0.231 | 0.246 | 0.767 | 0.22 | 0.228 | 0.262 | 0.322 | 0.276 | 0.25 | 0.433 | 0.759 | 0.75 | |
| UCRD | 0.048 | 0.129 | 0.111 | 0.344 | 0.148 | 0.065 | 0.094 | 0.111 | 0.207 | 0.782 | 0.25 | 0.233 | 0.188 | 0.156 | 0.156 | 0.167 | 0.729 | 0.138 | 0.214 | 0.219 | 0.241 | 0.263 | 0.236 | 0.356 | 0.719 | 0.745 | |
| JN16.5 | 0.085 | 0.172 | 0.119 | 0.4 | 0.123 | 0.103 | 0.125 | 0.079 | 0.241 | 0.745 | 0.312 | 0.2 | 0.25 | 0.219 | 0.188 | 0.167 | 0.763 | 0.138 | 0.179 | 0.25 | 0.241 | 0.298 | 0.2 | 0.39 | 0.719 | 0.782 | |
| JN20.1 | 0.129 | 0.049 | 0.129 | 0.365 | 0.133 | 0.148 | 0.228 | 0.077 | 0.233 | 0.789 | 0.273 | 0.258 | 0.242 | 0.212 | 0.182 | 0.194 | 0.705 | 0.167 | 0.241 | 0.242 | 0.267 | 0.322 | 0.263 | 0.41 | 0.729 | 0.754 | |
| JN5nb | 0.1 | 0.153 | 0.133 | 0.377 | 0.138 | 0.119 | 0.164 | 0.172 | 0.22 | 0.75 | 0.323 | 0.246 | 0.262 | 0.231 | 0.2 | 0.18 | 0.733 | 0.153 | 0.193 | 0.231 | 0.288 | 0.276 | 0.214 | 0.4 | 0.759 | 0.75 | |
| 1657 | 0.415 | 0.438 | 0.446 | 0.303 | 0.429 | 0.406 | 0.467 | 0.429 | 0.41 | 0.765 | 0.333 | 0.071 | 0.167 | 0.133 | 0.1 | 0.214 | 0.745 | 0.185 | 0.269 | 0.3 | 0.259 | 0.245 | 0.294 | 0.309 | 0.811 | 0.765 | |
| 1637 | 0.742 | 0.77 | 0.774 | 0.683 | 0.8 | 0.77 | 0.754 | 0.767 | 0.828 | 0.778 | 0.614 | 0.736 | 0.754 | 0.754 | 0.754 | 0.698 | 0.846 | 0.725 | 0.714 | 0.825 | 0.765 | 0.76 | 0.708 | 0.692 | 0.84 | 0.708 | |
| 1740 | 0.424 | 0.448 | 0.424 | 0.467 | 0.509 | 0.448 | 0.481 | 0.474 | 0.455 | 0.5 | 0.474 | 0.355 | 0.273 | 0.242 | 0.273 | 0.29 | 0.672 | 0.3 | 0.379 | 0.364 | 0.333 | 0.356 | 0.404 | 0.41 | 0.695 | 0.789 | |
| 1738 | 0.541 | 0.5 | 0.541 | 0.419 | 0.525 | 0.533 | 0.536 | 0.525 | 0.509 | 0.161 | 0.831 | 0.536 | 0.194 | 0.161 | 0.129 | 0.207 | 0.754 | 0.179 | 0.259 | 0.355 | 0.25 | 0.309 | 0.283 | 0.333 | 0.782 | 0.774 | |
| 2744 | 0.517 | 0.474 | 0.517 | 0.525 | 0.536 | 0.509 | 0.509 | 0.5 | 0.481 | 0.39 | 0.786 | 0.509 | 0.273 | 0.061 | 0.091 | 0.258 | 0.705 | 0.233 | 0.276 | 0.303 | 0.233 | 0.254 | 0.333 | 0.344 | 0.763 | 0.754 | |
| JN8-5 | 0.517 | 0.509 | 0.517 | 0.525 | 0.536 | 0.509 | 0.509 | 0.536 | 0.481 | 0.424 | 0.786 | 0.509 | 0.345 | 0.115 | 0.03 | 0.226 | 0.705 | 0.167 | 0.241 | 0.273 | 0.2 | 0.186 | 0.298 | 0.279 | 0.763 | 0.754 | |
| JN8-10 | 0.509 | 0.5 | 0.509 | 0.517 | 0.527 | 0.5 | 0.5 | 0.527 | 0.472 | 0.414 | 0.818 | 0.5 | 0.333 | 0.137 | 0.059 | 0.194 | 0.705 | 0.133 | 0.241 | 0.273 | 0.167 | 0.22 | 0.298 | 0.279 | 0.797 | 0.789 | |
| 1726 | 0.429 | 0.455 | 0.464 | 0.474 | 0.444 | 0.418 | 0.49 | 0.481 | 0.385 | 0.509 | 0.741 | 0.569 | 0.585 | 0.52 | 0.56 | 0.551 | 0.754 | 0.143 | 0.185 | 0.355 | 0.286 | 0.345 | 0.321 | 0.368 | 0.782 | 0.774 | |
| 1392 | 0.821 | 0.782 | 0.786 | 0.719 | 0.778 | 0.782 | 0.804 | 0.815 | 0.769 | 0.789 | 0.778 | 0.765 | 0.811 | 0.88 | 0.88 | 0.878 | 0.75 | 0.745 | 0.736 | 0.705 | 0.745 | 0.815 | 0.885 | 0.714 | 0.593 | 0.731 | |
| 1777 | 0.286 | 0.309 | 0.321 | 0.439 | 0.333 | 0.309 | 0.373 | 0.333 | 0.231 | 0.439 | 0.778 | 0.451 | 0.509 | 0.4 | 0.44 | 0.429 | 0.25 | 0.75 | 0.154 | 0.267 | 0.185 | 0.245 | 0.255 | 0.345 | 0.811 | 0.765 | |
| 1620 | 0.393 | 0.418 | 0.429 | 0.439 | 0.407 | 0.382 | 0.412 | 0.407 | 0.346 | 0.509 | 0.815 | 0.569 | 0.547 | 0.48 | 0.48 | 0.51 | 0.375 | 0.708 | 0.292 | 0.276 | 0.269 | 0.294 | 0.265 | 0.396 | 0.804 | 0.755 | |
| 1727 | 0.355 | 0.344 | 0.355 | 0.27 | 0.3 | 0.344 | 0.368 | 0.333 | 0.31 | 0.23 | 0.833 | 0.509 | 0.322 | 0.464 | 0.464 | 0.455 | 0.444 | 0.704 | 0.37 | 0.37 | 0.3 | 0.288 | 0.333 | 0.41 | 0.695 | 0.789 | |
| 2440 | 0.356 | 0.379 | 0.39 | 0.367 | 0.439 | 0.345 | 0.444 | 0.368 | 0.382 | 0.367 | 0.719 | 0.444 | 0.429 | 0.472 | 0.472 | 0.5 | 0.412 | 0.725 | 0.294 | 0.333 | 0.368 | 0.245 | 0.333 | 0.345 | 0.811 | 0.804 | |
| 2346 | 0.577 | 0.647 | 0.654 | 0.623 | 0.64 | 0.529 | 0.617 | 0.64 | 0.583 | 0.623 | 0.8 | 0.66 | 0.673 | 0.652 | 0.652 | 0.644 | 0.5 | 0.682 | 0.591 | 0.545 | 0.6 | 0.617 | 0.32 | 0.296 | 0.808 | 0.72 | |
| 1573 | 0.424 | 0.448 | 0.458 | 0.5 | 0.439 | 0.414 | 0.444 | 0.474 | 0.382 | 0.5 | 0.86 | 0.593 | 0.571 | 0.509 | 0.509 | 0.462 | 0.373 | 0.765 | 0.333 | 0.333 | 0.404 | 0.481 | 0.617 | 0.385 | 0.8 | 0.833 | |
| 2215 | 0.654 | 0.725 | 0.731 | 0.66 | 0.72 | 0.608 | 0.702 | 0.72 | 0.625 | 0.66 | 0.88 | 0.787 | 0.714 | 0.652 | 0.652 | 0.644 | 0.455 | 0.773 | 0.5 | 0.591 | 0.6 | 0.532 | 0.45 | 0.574 | 0.815 | 0.769 | |
| 1390 | 0.692 | 0.75 | 0.723 | 0.636 | 0.746 | 0.75 | 0.733 | 0.746 | 0.738 | 0.788 | 0.714 | 0.733 | 0.774 | 0.729 | 0.695 | 0.724 | 0.789 | 0.789 | 0.789 | 0.825 | 0.778 | 0.7 | 0.811 | 0.867 | 0.811 | 0.76 | |
| 2107 | 0.909 | 0.938 | 0.939 | 0.91 | 0.938 | 0.908 | 0.902 | 0.938 | 0.935 | 0.851 | 0.781 | 0.902 | 0.81 | 0.8 | 0.833 | 0.831 | 0.897 | 0.931 | 0.897 | 0.862 | 0.906 | 0.869 | 0.889 | 0.869 | 0.815 | 0.821 | |
The values on the upper right are the genetic distances obtained by the AFLP method, and the values on the lower right are the genetic distances obtained by the RAPD method.
Strains JN51, JN52, JN53, and JN54.
FIG. 2.
Dendrograms showing the genetic relatedness of P. syringae and P. viridiflava strains based on RAPD and AFLP analyses. The trees were constructed by the neighbor-joining (NJ) method. A, D, M, N, and Q indicate genomic groups determined by PCR-RFLP analysis of the rrn operon (12), and sp. I, sp. II, sp. III, and sp. VI indicate the genomic species determined by DNA-DNA hybridization (18). The scale indicates the amount of difference. The numbers on the branches indicate the bootstrap percentage at each node. +, strain pathogenic on tomato; −, strain not pathogenic on tomato.
In the present analysis, P. viridiflava CFBP 2107, P. syringae pv. syringae CFBP 1392, and P. syringae pv. phaseolicola CFBP 1390 were used as outgroups in order to check the usefulness of the techniques for separating taxa determined by other analyses since these three strains belong to three different genomic species (18) and three different ribotypes (12). In addition, P. syringae pv. maculicola CFBP 1637, which has been determined to be an atypical strain of P. syringae pv. maculicola by DNA pairing analysis (6a, 18), was also included in the study.
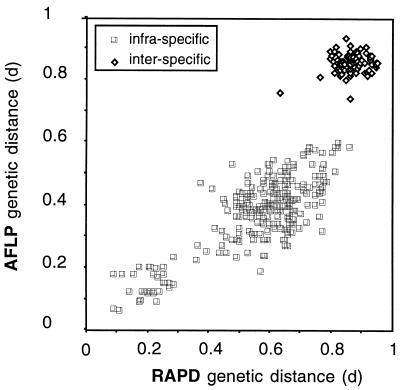
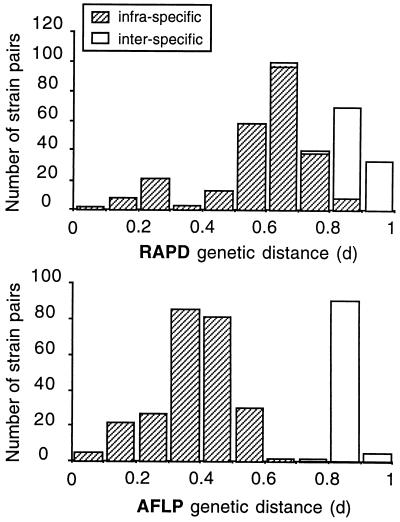
The genetic distances calculated from the AFLP and RAPD data correlated well (Fig. 3), indicating that if a sufficient number of independent markers are considered (100 and 132 bands were used in the AFLP and RAPD analyses, respectively) the two methods provide essentially the same information and are useful for exploring the genetic diversity of P. syringae. Nethertheless, compared to the RAPD method, the AFLP method resulted in clearer delineation between the genetic distances calculated for members of different genospecies and the genetic distances calculated for members of the same genospecies (Fig. 3 and 4). As a consequence, the neighbor-joining method provided a significant delineation (bootstrap value, 97%) of genospecies III (P. tomato) with the AFLP data, while the delineation obtained with the RAPD data was not significant (Fig. 2). Why AFLP data provided better discrimination of intraspecific distances is not known, but we assumed that this finding could be related to the restriction pretreatment which excluded almost all tandem amplification, in contrast to what could happen with the RAPD technique. Strains belonging to the same genospecies were 57 to 100% similar (Table 4) (distance value = 1 − similarity), while the average level of similarity between bacteria belonging to different species was only 25% ± 8.5% (at the P = 0.05 level), with a maximum value of 41% in one instance. These results suggest that the AFLP method, but not the RAPD method, could be used as a rapid and efficient alternative to DNA pairing to verify if a new isolate belongs to the genospecies P. tomato, as has been suggested for other bacterial species (10), even if additional studies are necessary (4).
FIG. 3.
Correlation between the genetic distances calculated with AFLP data and the genetic distances calculated with RAPD data. Data points indicate combinations of genetic distances calculated for pairs of isolates. A total of 27 strains were tested in the study (linear correlation, P < 0.0001, r2 = 0.76).
FIG. 4.
Histograms of intraspecific and interspecific genetic distances calculated with RAPD and AFLP data.
Takikawa et al. (20) could not distinguish P. syringae pv. tomato and P. syringae pv. maculicola with several physiological and biochemical tests. Similarly, we were not able to discriminate between the two pathovars with pathogenicity tests performed with susceptible tomato plants. All strains of P. syringae pv. tomato and P. syringae pv. maculicola, as well as a strain of P. syringae pv. apii and a strain of P. syringae isolated from Euphorbia pulcherrima, gave symptoms typical of bacterial speck on leaves of tomato (L. esculentum cv. Montfavet 63.4) (Table 1). This is consistent with the fact that P. syringae pv. apii was previously found to be very similar to P. syringae pv. tomato (8). RAPD and AFLP techniques have the potential to distinguish these very closely related pathovars. These techniques clustered most strains of P. syringae pv. maculicola in a group designated maculicola and the strains of P. syringae pv. tomato in a group designated tomato (Fig. 2). Our results therefore support the idea that genetic diversity occurs in relation to the host plant, suggesting that groups of strains belonging to genomic species III (P. tomato) evolved independently with the host plants but that pathogenic specialization for the hosts did not occur.
Surprisingly, some strains belonging to P. syringae pv. tomato or P. syringae pv. maculicola were not included in the group, as expected from their pathogenic behavior. Strain CFBP 2212 of P. syringae pv. tomato was not a member of the tomato group, as defined in the present study (Fig. 2). This is rather worrying because strain CFBP 2212 is the type strain of P. syringae pv. tomato. In addition, strains CFBP 1740 and CFBP 1637 of P. syringae pv. maculicola were clearly distinct from all other P. syringae pv. tomato and P. syringae pv. maculicola strains tested (Fig. 2). The differences between strain CFBP 2212 and the other strains of P. syringae pv. tomato were not, however, as pronounced as the differences between CFBP 1740 or CFBP 1637 and the members of the maculicola group; 78% of the CFBP 2212 DNA bands were found in other P. syringae pv. tomato strains in the tomato group, while only 53 and 25% of the CFBP 1740 and CFBP 1637 bands, respectively, were found in other strains in the maculicola group. P. syringae pv. maculicola CFBP 1637 and CFBP 1740 were located outside the lineage containing the tomato and maculicola groups. Interestingly, these two P. syringae pv. maculicola strains grouped with the other P. syringae pv. maculicola strains when an analysis of the diversity within a single locus (the rrn operon) was performed (12). Inconsistencies between the results of the single-locus and multilocus analyses of these bacterial genomes indicate that dramatic rearrangements may have occurred in these strains. We assume that horizontal transfer of genetic elements or large deletions or inversions occurred, and this may explain the unexpected position of strains CFBP 1637 and CFBP 1740. This may indicate that the genospecies P. tomato may not be as clonal as described by Maynard Smith et al. (13) on the basis of the data of Denny et al. (2) when additional strains are considered. Thus, conclusions concerning the clonality of bacterial populations depend on the strains analyzed. In the present case, we had to consider the fact that strains CFBP 1637 and CFBP 1740 were isolated in 1965 and 1970, respectively, and were stored in various collections for about 30 years. They were subcultured numerous times between two periods of conservation by lyophilization and freezing. Thus, selection of bacterial clones with dramatic internal genomic rearrangements might have occurred during their disturbed history, and this may explain their present location outside the maculicola lineage.
RAPD and AFLP techniques allowed very fine discrimination close to the strain level, clearly and reliably differentiating bacterial isolates; the 30 strains tested produced 27 DNA band patterns in both analyses. Most strains were therefore identified by a particular pattern; the notable exceptions were strains JN51, JN52, JN53, and JN54, which grouped together in both analyses (Fig. 2). Strains JN51 and JN52 belong to race 0 while strains JN53 and JN54 belong to race 1, and all of these strains were isolated in Canada. As host specificity is determined by a single locus, avrPto (16), a single genetic modification could be responsible for race differentiation. Thus, we speculate that the race differentiation of these four isolates is a recent event which probably occurred within a poorly differentiated population responsible for outbreaks in Canada.
The two techniques which we used could provide useful epidemiological markers. Strain JN20.1 is a streptomycin-resistant strain isolated from an experimental field plot previously infected with streptomycin-resistant strain 1427st. Strain JN20.1 was assumed to be a reisolate of the inoculated strain since its RAPD and AFLP patterns are very similar to the parental inoculant patterns (98% of the DNA bands in common) and both strains are resistant to streptomycin. Similarly, strains JN8.5 and JN8.10 isolated during the same outbreak from two different radish plants had very similar band patterns (98% of the DNA bands in common).
In conclusion, both techniques which we used generated specific genomic patterns which differentiated closely related strains. Unique fingerprint profiles generated by the RAPD and AFLP techniques can be exploited for strain identification purposes, and they can be useful in epidemiological studies to determine the origin of a bacterial population responsible for a given outbreak. Evolutionary distinctions between P. syringae pv. tomato and P. syringae pv. maculicola can be discerned. Both pathovars might have originated from a single evolutionary line which diverged into two distinct lineages due to the plant environment, including plant dispersal of the pathogen. These results raise questions about the role and the influence of environmental factors on the evolution of the genome. Thus, we suggest that the host plant from which bacteria were isolated could be an important factor in the plasticity of the bacterial genome.
ACKNOWLEDGMENTS
This research was supported by grants from the Institut National de la Recherche Agronomique, France (“AIP Génome des Bactéries Pathogènes”) and from the Bureau des Ressources Génétique, France (BRG) (“Recherches méthodologiques pour l’amélioration des processus de gestion et de conservation des ressources génétiques animales, végétales et microbiennes”).
A.C. received a fellowship from the DEA d’Ecologie Microbienne, Université Claude Bernard, Lyon I. We thank Chrystelle Brin for skillful technical assistance.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates, Wiley Interscience; 1992. [Google Scholar]
- 2.Denny T P, Gilmour M N, Selander R K. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J Gen Microbiol. 1988;134:1949–1960. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- 3.Doudoroff M, Palleroni N J. Genus I. Pseudomonas. In: Buchanan R E, Gibbons N E, editors. Bergey’s manual of determinative bacteriology. 8th ed. Baltimore, Md: The Williams & Wilkins Co.; 1974. pp. 217–243. [Google Scholar]
- 4.Esteve C. Is AFLP fingerprinting a true alternative to the DNA-DNA pairing method to assess genospecies in the genus Aeromonas? Int J Syst Bacteriol. 1997;47:245–246. [Google Scholar]
- 5.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 6.Fitch F W, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 6a.Gardan, L. Personal communication.
- 7.Gardan L, Shafik H L, Grimont P A D. DNA relatedness among pathovars of P. syringae and related bacteria. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 445–448. [Google Scholar]
- 8.Hendson M, Hildebrand D C, Schroth M N. Relatedness of Pseudomonas syringae pv. tomato, Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. antirrhini. J Appl Bacteriol. 1992;73:455–464. [Google Scholar]
- 9.Henis Y, Bashan Y. Epiphytic survival of bacterial leaf pathogens. In: Fokema N N, Van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, United Kingdom: Cambridge University Press; 1985. pp. 252–268. [Google Scholar]
- 10.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 11.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 12.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maynard Smith J M, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palleroni N J. Family I. Pseudomonadaceae. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams and Wilkins Co.; 1984. pp. 141–219. [Google Scholar]
- 15.Pecknold P C, Grogan R G. Deoxyribonucleic acid homology groups among phytopathogenic Pseudomonas species. Int J Syst Bacteriol. 1973;23:111–121. [Google Scholar]
- 16.Ronald P C, Salmeron J M, Carland F M, Staskawicz J B. The cloned avirulence gene avrPto induces disease resistance in tomato cultivar containing the Pto resistance gene. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Shafik H L. Taxonomie des Pseudomonas phytopathogènes du groupe de Pseudomonas syringae: étude phénotypique et génotypique. Ph. D. thesis. Angers, France: University of Angers; 1994. [Google Scholar]
- 19.Sokal R R, Sneath P H A. Principles of numerical taxonomy. San Francisco, Calif: Freeman; 1963. pp. 169–210. [Google Scholar]
- 20.Takikawa Y, Nishiyama N, Okba K, Tsuyumu S, Goto M. Synonymy of Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. tomato. In: Lemattre M, Freigoun S, Rudolph K, Swings J G, editors. Plant pathogenic bacteria. Les Colloques no. 66. Paris, France: INRA éditions; 1994. pp. 199–204. [Google Scholar]
- 21.Vos P, Hogers R, Reijans M, van de Lee T, Hornes M, Freijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new concept for DNA finger printing. Nucleic Acids Res. 1995;21:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiebe W L, Campbell R N. Characterization of Pseudomonas syringae pv. maculicola and comparison with P. s. pv. tomato. Plant Dis. 1993;77:414–419. [Google Scholar]
- 23.Young J M, Dye D W, Bradbury J F, Panagopoulos C G, Robbs C F. Plant pathogenic bacteria. Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, Angers. Beaucouzé, France: Station de Pathologie Végétale et Phytobactériologie, Institut National de la Recherche Agronomique; 1978. The use of the term “pathovar” in the classification of plant pathogenic bacteria; pp. 359–363. [Google Scholar]
- 24.Young J M, Takikawa Y, Gardan L, Stead D E. Changing concepts in the taxonomy of plant pathogenic bacteria. Annu Rev Phytopathol. 1992;30:67–105. [Google Scholar]
- 25.Yunis H, Bashan Y, Okon Y, Henis Y. Weather dependence, yield losses and control of bacterial speck of tomato caused by Pseudomonas tomato. Plant Dis. 1980;64:937–939. [Google Scholar]
- 26.Zabeau M, Vos P. Selective restriction fragment amplification: a general method for DNA fingerprinting. Publication 0 534 858 A1. Munich, Germany: European Patent Office; 1993. [Google Scholar]