Abstract
BIOLOG GN plates are increasingly used to characterize microbial communities by determining the ability of the communities to oxidize various carbon sources. Studies were done to determine whether the BIOLOG GN plate assay accurately reflects the catabolic potential of the inoculum used. To gain insight into which populations of microbial communities contribute to the BIOLOG patterns, denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis (TGGE) were used to assess the diversity of ribotypes in the inocula and individual wells of BIOLOG plates following incubation. These studies were done with microbial communities from the rhizosphere of potatoes and an activated sludge reactor fed with glucose and peptone. TGGE analyses of BIOLOG wells inoculated with cell suspensions from the potato rhizosphere revealed that, compared with the inoculum, there was a decrease in the number of 16S rRNA gene fragments obtained from various wells, as well as a concomitant loss of populations that had been numerically dominant in the inoculum. The dominant fragments in TGGE gels could be assigned to the γ subclass of the class Proteobacteria, suggesting that fast-growing bacteria adapted to high substrate concentrations were numerically dominant in the wells and may have been primarily responsible for the patterns of substrate use that were observed. Similarly, the community structure changed in wells inoculated with cells from activated sludge; one or more populations were enriched, but all dominant populations of the inoculum could be detected in at least one well. This study showed that carbon source utilization profiles obtained with BIOLOG GN plates do not necessarily reflect the functional potential of the numerically dominant members of the microbial community used as the inoculum.
BIOLOG GN microtiter plates were originally developed for classification of bacterial isolates based on the ability of the isolates to oxidize 95 different carbon sources (1). However, this method has also been adapted and used to characterize the functional potential of microbial communities (8). When BIOLOG GN plates are used for this purpose, the inoculum is a mixture of organisms obtained from a microbial community rather than a cell suspension from an axenic culture, but otherwise the procedures are similar. A bacterial cell suspension is used to inoculate wells of a microtiter plate in which each well contains a different carbon source, nutrients, and a tetrazolium dye. The plate is then incubated for a suitable period of time, and oxidation of the substrate is periodically monitored by measuring the concomitant reduction of the tetrazolium dye. Previous studies have shown that microbial communities produce habitat-specific and reproducible patterns of carbon source oxidation, and so the method can be used to discern temporal and spatial differences among microbial communities from bulk soils (2, 13, 28, 30, 31), rhizospheres (10–12), and subsurface cores (20). In other studies workers have used the BIOLOG GN method to determine the effect of nonindigenous bacterial species or transgenic plants on microbial communities in soil (4, 6, 26), in plant litter (19, 24), or in the phyllosphere (5, 17).
Studies done to elucidate the basis for the patterns of substrate oxidation obtained with BIOLOG GN plates have shown that oxidation of the substrates depends on both the composition and the density of the inoculum used (9). Bacterial growth occurs in the wells during the course of the assay (8, 9, 13, 29, 30), and hence the pattern of substrate use observed may only reflect the functional characteristics of organisms that are able to grow in the BIOLOG GN plate wells under the assay conditions used. For each study published so far it remains unclear what fraction of the microbial population in a given community actually contributed to the observed pattern of carbon source oxidation. However, several studies of model communities have shown that it is likely that not all constituents contributed to the patterns (13, 17, 29).
Carbon source utilization patterns obtained from BIOLOG GN plates are being used as part of a polyphasic approach to investigate the potential effects of T4 lysozyme expressed by transgenic potatoes on the microbial communities of the rhizosphere and phyllosphere that are associated with the plants (15, 17). When the effects of environmental perturbations or transgenic organisms on microbial communities are estimated, it is important to determine whether the patterns of sole carbon source utilization were caused by a limited number of populations and whether these populations were numerically dominant in the community at the time of sampling or became dominant during the course of the assay. To gain insight into this, we used temperature gradient gel electrophoresis (TGGE) and denaturing gradient gel electrophoresis (DGGE) profiles of the 16S rRNA gene (16S rDNA) fragments amplified from the microbial communities of different BIOLOG wells (16, 22). With both methods separation of the PCR-amplified 16S rDNA fragments is based on the differential melting behavior of double-stranded DNA fragments as they migrate through a linearly increasing gradient of denaturants or temperature (21). These methods are essentially equivalent (16, 18) and offer the distinct advantage that they do not require cultivation of the community members. The findings obtained for the microbial community of potato rhizospheres were compared to the findings obtained for samples taken from an activated sludge reactor that had been continuously fed with glucose and peptone. The latter community was chosen because of its better adaptation to high substrate input and higher growth rates. It was hypothesized that the BIOLOG patterns obtained for such a community should more closely reflect the functional potential of the inoculum.
MATERIALS AND METHODS
Sampling and recovery of bacterial communities.
Rhizosphere samples (roots with adhering soil particles; approximately 3 g) from greenhouse-grown potatoes were placed in sterile Stomacher bags (Seward Medical Limited, London, United Kingdom) and treated twice at high speed (260 rpm), once with 20 ml of a solution containing phosphate-buffered saline and 0.3 g of Chelex-100 (Bio-Rad, Hercules, Calif.) and once with 10 ml of NDP (0.02% sodium deoxycholate and 0.5% polyethylene glycol in 100 ml of sterile water), by using a protocol slightly modified from the protocol described by Herron and Wellington (14). The bacterial fraction of the treated sample was recovered by differential centrifugation as follows: (i) soil and root particles were removed by centrifugation at 500 × g for 2 min; (ii) the bacterial fraction was recovered from the supernatant by centrifugation at 10,000 × g for 20 min; (iii) the resulting bacterial pellet was resuspended in 10 ml of sterile saline and centrifuged at 10,000 × g for 20 min; and (iv) the resulting cell pellet was resuspended in 40 ml of sterile saline.
The activated sludge samples were obtained from a sequential batch reactor operated in a fill-and-draw mode by using a mixture of glucose (0.1%) and peptone (1.0%) as a sole carbon source and a mean cell residence time of about 7 days. The optical density at 600 nm (OD600) of the sludge was adjusted to 0.5 with sterile saline.
BIOLOG GN assays.
Each suspension of bacterial cells in saline was used to inoculate BIOLOG GN plates (150 μl per well), which were then incubated at room temperature for 48 h. Samples (10 μl) were removed from selected wells of each plate at specific times during the incubation period. The OD595 was measured with a microtiter plate reader (model Vmax; Molecular Devices Corp., Menlo Park, Calif.) after 24 and 48 h of incubation.
Characterization of BIOLOG GN communities by TGGE or DGGE.
The BIOLOG wells analyzed by TGGE or DGGE were wells whose substrates had been used by the microbial community. The wells used for the time course study were selected on the basis of differences in the patterns of fragments produced from samples after 48 h of incubation. Well B6 (α-d-glucose) was included in the time course study of activated sludge because the sludge reactor had been fed glucose. Cells in samples from BIOLOG GN wells were lysed by three cycles of freezing and boiling and were used as templates for PCR amplification of 16S rDNA fragments spanning regions V6 to V8. Each PCR was performed with a model 480 DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). Regions within 16S rDNA genes were amplified by using 25-μl reaction mixtures containing 2.5 U of the Taq polymerase Stoffel fragment (Perkin-Elmer), each deoxynucleoside triphosphate at a concentration of 0.2 mM, 3.75 mM MgCl2, 0.2 μM primer F968 (5′GC-clamp-AAC GCG AAG AAC CTT AC-3′), 0.2 μM primer R1401 (5′CGG TGT GTA CAA GGC CCGGGA ACG 3′), and 1 μl of DNA template. A GC-clamp sequence (5′CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG G3′) had been added to the 5′ end of F968 (18). PCR amplification was done by using an initial denaturation step (94°C for 5 min), followed by 35 cycles consisting of 1 min of denaturation at 94°C, 1 min of primer annealing at 54°C, and 1 min of primer extension at 72°C. To reduce the proportion of single-stranded DNA, 10 cycles consisting of 0.5 min at 54°C and 1 min at 72°C were included. The resulting PCR products were analyzed on a 1% agarose gel to confirm their sizes and the yield by the standard protocols described by Sambrook et al. (25). The standard used for DGGE consisted of PCR products that were generated with genomic DNAs of the following strains as templates and were combined in equal proportions: Clostridium pasteurianum, Erwinia carotovora, Agrobacterium tumefaciens, Pseudomonas fluorescens, Rhizobium leguminosarum, Pantoea agglomerans, Burkholderia gladioli, Streptomyces aureofaciens, and a Nocardia sp. The standard used for TGGE did not contain the PCR products of the two last strains.
TGGE and DGGE analyses.
Analysis of the amplification products by TGGE and DGGE was done by using previously described procedures (18). A temperature gradient from 38 to 52°C was used for TGGE, and a 40 to 60% denaturing gradient (7 M urea plus 40% [wt/vol] deionized formamide) was used for DGGE.
Plating and characterization of bacterial isolates.
The viable counts of the cell suspensions used as inocula for BIOLOG GN plates and samples taken after 24 and 48 h of incubation from selected wells of BIOLOG GN plates were determined by plating serial dilutions on plate count agar (Merck, Darmstadt, Germany) for the potato rhizosphere samples and on R2A (Difco Laboratories, Detroit, Mich.) for the activated sludge samples. The number of CFU was determined after incubation of the plates at room temperature for 48 h. The dominant culturable bacteria isolated from the potato rhizosphere inoculum or after BIOLOG incubation were chosen based on differences in colony morphology. Representative colonies were purified and characterized by repetitive extragenic palindromic PCR with primers based on repetitive extragenic palindromic elements (27) and by TGGE analysis. The isolates were taxonomically classified based on cellular fatty acid methyl ester profiles (MIDI, Newark, N.J.) and with API strips (bioMérieux, Charbonnières les Bains, France) and BIOLOG plates.
Probes based on the V6 region of rDNA genes of dominant bacterial isolates.
Probes for strains were generated by PCR by using the reaction conditions described above, except that the primers used (primer F971 [5′GCG AAG AAC CCT ACC 3′] and primer R1057 [5′CAT GCA GCA CCT GT 3′]) flanked the V6 region of the 16S rRNA gene and the deoxynucleoside triphosphate mixture contained 0.13 mM dTTP and 0.07 mM digoxigenin-labeled dUTP instead of 0.2 mM dTTP. Amplification was performed after an initial denaturation step consisting of 5 min at 94°C; each of the subsequent 35 cycles consisted of 1 min of denaturation at 94°C, 1 min of primer annealing at 46°C, and 1 min of primer extension at 72°C. The specificity of the probes was tested by dot blot hybridization of PCR fragments amplified from axenic cultures of bacteria isolated from the potato rhizosphere with primers F968 and R1401.
Southern hybridization.
TGGE gels were blotted onto Hybond N+ membranes (Amersham Buchler GmbH, Braunschweig, Germany) for 1 h at 0.4 A with a Trans-Blot SD apparatus (Bio-Rad, Hercules, Calif.) according to the instructions of the manufacturer. Hybridization of electroblotted or dot-blotted DNA with digoxigenin-labeled V6 fragments as probes was done under high-stringency conditions (7). The blots were washed, and the DNA hybrids were detected by the protocol provided by Boehringer (Mannheim, Germany).
RESULTS
Growth in BIOLOG GN wells after inoculation with microbial communities.
A suspension of bacterial cells from the rhizosphere of greenhouse-grown potatoes was used to inoculate BIOLOG GN plates. The final cell density in the wells at the beginning of the assay was 7 × 105 CFU per g. After 24 and 48 h of incubation the total numbers of CFU in wells A1 (no substrate), A12 (cellobiose), D6 (d-galacturonic acid), E1 (p-hydroxyphenylacetic acid), and H2 (inosine) were determined by plating serial dilutions on plate count agar. Growth occurred in all wells, including the no-substrate control well (well A1), and after 48 h of incubation, the total numbers of CFU ranged from 9 × 107 CFU/ml (well A1) to 8 × 108 CFU/ml (well A12). The OD595 increased for all wells analyzed after 48 h of incubation, indicating that the substrates were oxidized by the potato rhizosphere populations; the OD595 values after 24 and 48 h of incubation were 1.3 and 1.63, respectively, for well A12, 1.15 and 1.43, respectively, for well D6, 0.46 and 1.41, respectively, for well E1, and 0.6 and 1.34, respectively, for well H2. Similar results were obtained from BIOLOG GN plates inoculated with activated sludge, in which on average the cell densities also increased approximately 2 orders of magnitude.
Analyses of BIOLOG GN plate wells by TGGE and DGGE.
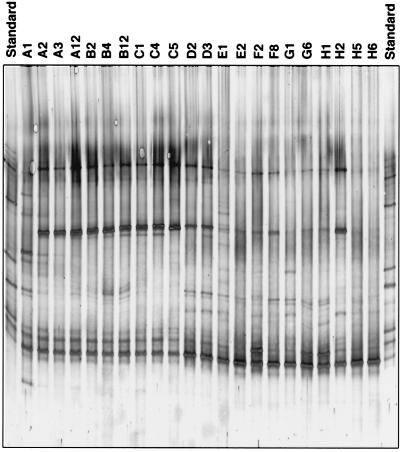
TGGE and DGGE analyses of the mixed bacterial populations in BIOLOG GN wells were done to determine how many different populations contributed to the patterns of substrate use in any given well, whether these populations corresponded to the dominant populations of the inoculum, and whether certain populations were common to most or all wells of a plate. Cell lysates from wells of BIOLOG GN plates inoculated with the potato rhizosphere bacterial community were analyzed by TGGE, whereas cell lysates from activated sludge were analyzed by DGGE. The TGGE profiles of 22 different wells of a BIOLOG GN plate inoculated with the potato rhizosphere bacterial community were determined after 48 h of incubation (Fig. 1). The TGGE pattern of the no-substrate control well (well A1) had at least 11 bands (Fig. 1). In contrast, the profiles of many wells (e.g., wells A2, A3, C5, and D2) had only four prominent bands that differed in electrophoretic mobility. Interestingly, at least one of the dominant bands in the profiles of wells with carbon sources was not present in the profile of the no-substrate well (well A1). Similarly, the TGGE profiles of other wells (wells E1, H5, and H6) had only one dominant band. Although the substrate-containing wells were similar to one another, there were differences in the number and mobility of the bands in the profiles. These data indicate that during the 48-h incubation period specific bacterial populations were enriched in wells that contained a substrate, and this resulted in decreased diversity among the numerically dominant populations. Perhaps even more important was the fact that substrate oxidation was most likely effected by comparatively few populations, not all of which were found in high numbers in the inoculum.
FIG. 1.
16S rDNA TGGE profiles of the bacterial potato rhizosphere community present in different wells of a BIOLOG plate after 48 h of incubation at room temperature. Lane A1, no C source; lane A2, α-cyclodextrin; lane A3, dextrin; lane A12, cellobiose; lane B2, d-fructose; lane B4, d-galactose; lane B12, d-mannose; lane C1, d-melibiose; lane C4, d-raffinose; lane C5, l-rhamnose; lane D2, cis-aconitic acid; lane D3, citric acid; lane E1, p-hydroxyphenylacetic acid; lane E2, itaconic acid; lane F2, succinamic acid; lane F8, l-asparagine; lane G1, l-histidine; lane G6, l-proline; lane H1, urocanic acid; lane H2, inosine; lane H5, phenyl ethylamine; lane H6, putrescine.
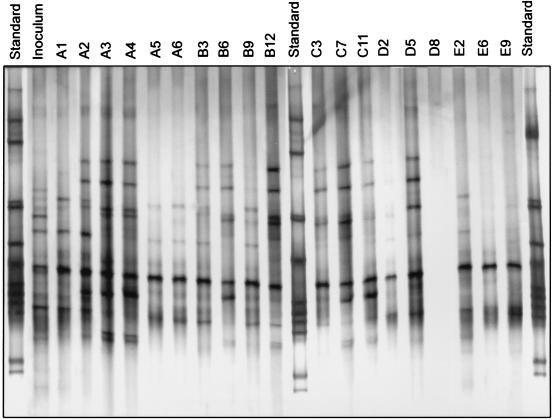
Figure 2 shows the DGGE profiles of BIOLOG GN wells 24 h after inoculation with the activated sludge bacterial community. The DGGE profiles of the activated sludge inoculum (Fig. 2) had 4 dominant fragments and approximately 10 faint fragments. The DGGE profile of the no-substrate control well (well A1) after 24 h of incubation was similar to the profiles of the inoculum. For several substrates (e.g., wells A2, A3, and A4) the number of dominant fragments was even higher than the number of dominant fragments observed for the inoculum and well A1. All dominant community members from the activated sludge reactor were enriched in at least one of the BIOLOG wells following incubation, indicating that all of the dominant community members of the inoculum probably contributed to the utilization of the substrates analyzed. The DGGE profiles of wells containing multiple fragments differed from well to well, but all contained a prominent fragment from a population that was also dominant in the inoculum. In both habitats a single population in the inoculum dominated in nearly all of the wells after BIOLOG incubation, and this population probably was to a large extent responsible for the BIOLOG patterns observed.
FIG. 2.
16S rDNA DGGE profiles of the bacterial sludge communities present in different BIOLOG wells after 48 h of incubation at room temperature. Lane A1, no C source; lane A2, α-cyclodextrin; lane A3, dextrin; lane A4, glycogen; lane A5, Tween 40; lane A6, Tween 80; lane B3, l-fucose; lane B6, α-d-glucose; lane B9, lactulose; lane B12, d-mannose; lane C3, d-psicose; lane C7, sucrose; lane C11, methyl pyruvate; lane D2, cis-aconitic acid; lane D5, d-galactonic acid lactone; lane E2, itaconic acid; lane E6, dl-lactic acid; lane E9, quinic acid.
Population dynamics in wells of BIOLOG GN plates during incubation.
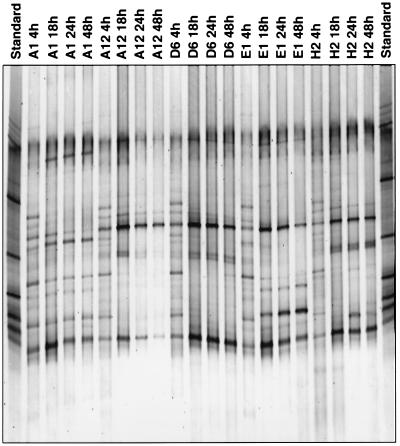
Previous studies have shown that the numbers of bacterial cells in BIOLOG GN wells often increase following inoculation (13), particularly if the density of the inoculum is less than 108 cells/ml. Obviously, this provides an opportunity for shifts in the structure of the bacterial community to occur as a result of differential growth of populations that are able to utilize the sole carbon sources. To determine if this was the case, wells A1 (no substrate), A12 (cellobiose), D6 (galacturonic acid), E1 (p-hydroxyphenylacetic acid), and H2 (inosine) of BIOLOG GN plates inoculated with the bacterial community from potato rhizospheres were sampled after various periods of incubation and analyzed by TGGE (Fig. 3). The profiles obtained for these wells were identical after 4 h of incubation and contained approximately 15 fragments. In the no-substrate control well, five bands were lost from the profile and one band emerged and became dominant during the ensuing hours of incubation. The latter band was not observed in the profiles of wells that contained substrates. Overall, the number of bands in the no-substrate control (well A1) decreased from 15 to 11. Wells that contained substrates (wells A12, D6, E1, and H2) showed an even more dramatic reduction in the number of bands, indicating that shifts in the structure of the bacterial communities in the BIOLOG GN wells occurred. In these wells many of the dominant bands that were apparent at the beginning of the incubation period were lost from the profile, while other bands became more pronounced after long periods of incubation. Moreover, in well E1 (Fig. 3) a dominant band present in the profile after 48 h of incubation was not present in the profile at the start of incubation. Similarly, there were some bands that were present in profiles of wells obtained after short periods of incubation that increased in intensity in samples taken later.
FIG. 3.
16S rDNA TGGE profiles of the bacterial potato rhizosphere communities present after different incubation times in BIOLOG wells. Lanes A1, no C source; lanes A12, cellobiose; lanes D6, d-galacturonic acid; lanes E1, p-hydroxyphenylacetic acid; lanes H2, inosine.
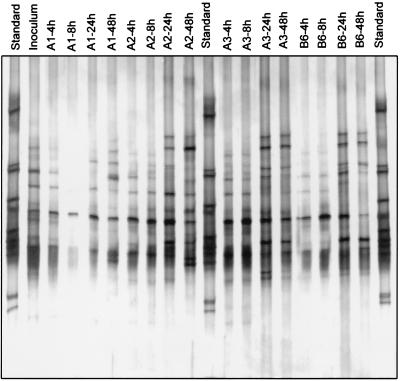
Shifts in the DGGE pattern also occurred in BIOLOG GN wells A1 (no substrate), A2 (cyclodextrin), A3 (dextrin), and B6 (α-d-glucose) after they had been inoculated with an activated sludge bacterial community and incubated for 48 h (Fig. 4). In this case the number of bands in the DGGE patterns of different wells did not decrease during the course of incubation. Instead, there was a general increase in the number and intensity of bands in the profiles, indicating that enrichment of the strains occurred. All of the dominant bands of the inoculum occurred in at least one of the wells after enrichment.
FIG. 4.
16S rDNA DGGE profiles of the bacterial sludge communities present after different incubation times in BIOLOG wells. Lanes A1, no C source; lanes A2, α-cyclodextrin; lanes A3, dextrin; lanes B6, α-d-glucose.
Classification of dominant bacterial populations in wells of BIOLOG GN plates.
We attempted to determine if the organisms that gave rise to the dominant bands in the TGGE profiles of BIOLOG wells were among the organisms which could be cultured from the wells. This was done by comparing the electrophoretic mobilities of fragments amplified from isolates to the electrophoretic mobilities of bands in the TGGE profiles of the BIOLOG wells. In addition, we used probes based on the V6 region of the 16S rDNA gene (16) derived from dominant rhizosphere bacteria to probe electroblotted TGGE gels on which PCR products from BIOLOG wells had been analyzed alongside similar PCR products from bacterial isolates.
Most of the bands present in the profiles from wells at the start of incubation could not be correlated to bands of cultivated bacteria from the potato rhizosphere inoculum. However, one dominant band assigned to Brevundimonas vesicularis (a member of the α subclass of the class Proteobacteria) was observed in the profiles of all wells at the start of incubation yet was absent in the profiles of substrate-containing wells after incubation. In some cases, the probes derived from dominant isolates hybridized to more than one fragment (data not shown). However, when the data were used in combination with differences in electrophoretic mobility, it was possible (with one exception) to correlate bands in TGGE gels with strains which had been cultivated from the same sample. The major fragments in the TGGE profiles of BIOLOG GN wells after incubation were identified as fragments from Enterobacter cloacae, Pantoea agglomerans, Salmonella spp., and Pseudomonas putida, all of which are members of the γ subclass of the Proteobacteria. Only one dominant band present in the fingerprint of well E1 could not be assigned to a strain that had been cultivated. Thus, nearly all of the numerically dominant bacterial strains in substrate-containing wells could be cultured; this was in contrast to what was found with the numerically dominant strains in the inoculum.
DISCUSSION
Because substantial bacterial growth occurs during the course of the assays performed with BIOLOG GN plates, it is unlikely that the carbon source utilization profiles obtained reflect the in situ function of the microbial communities used as inocula. Although this is generally recognized, it is still assumed that the observed profile of carbon sources metabolized reflects the catabolic potential of a community. This is reasonable only if the community structure remains unchanged from the community structure of the inoculum. However, if there is a significant change in the species composition or an alteration in the relative proportions of populations in the wells of the microtiter plate, then the ability to extrapolate the findings of the assay to the microbial community being studied is compromised since the profile obtained does not necessarily reflect the functional potential of the numerically dominant members of the microbial community used as the inoculum.
In this study, we used TGGE and DGGE to analyze 16S rDNA gene sequences that had been amplified from total microbial community DNA isolated from inocula, as well as various wells of BIOLOG GN plates after various periods of incubation. This enabled us to monitor any shifts in the microbial community structure that occurred during incubation of the plates.
The patterns obtained for individual BIOLOG GN wells were different than those obtained for the inocula used, indicating that changes in the structure of the microbial community had occurred during the assay. With each inoculum used the profiles of the communities in the wells of BIOLOG GN plates had several prominent bands, suggesting that more than one population contributed to carbon source oxidation. However, it was striking that some numerically dominant bacterial populations in the potato rhizosphere community were not represented in the TGGE profiles of any of the BIOLOG wells following incubation. For example, Brevundimonas vesicularis (a member of the α subclass of the Proteobacteria) was found to be a dominant member of the potato rhizosphere community but was absent from the profiles of all substrate-containing wells that were examined. Interestingly, this organism was not evident in the profile of well A12, which contained cellobiose, even though an axenic culture of Brevundimonas vesicularis was shown to metabolize cellobiose. Similarly, the relative proportions of the numerically dominant populations in activated sludge were altered during incubation in the wells of BIOLOG plates, and there were differences among the profiles of the various wells analyzed.
Since the structures of the microbial communities in various wells that received the same inoculum were not identical, it is apparent that various subsets of populations from the microbial communities had been selected. Whether a bacterial population becomes numerically dominant and “wins” in BIOLOG GN wells is undoubtedly dependent on numerous factors, including the ability of the organisms to oxidize a carbon source at the concentration provided and their competitiveness under the cultivation conditions used. The competitiveness of populations is determined by a variety of factors, including their nutritional requirements and their generation times under the prevailing conditions, as well as antagonistic and synergistic interactions among the populations. Thus, it seems likely that the patterns of substrate oxidation observed were not caused by the numerically dominant populations in the community at the time of inoculation but instead reflected the altered community structure that developed during incubation of the BIOLOG GN plates. Those bacterial populations that commonly become dominant in various wells of BIOLOG GN plates are putatively versatile with respect to the ability to use different carbon sources and are competitive under the conditions of the assay.
Whether the bacterial populations that are numerically dominant in the inoculum remain dominant in the wells of BIOLOG GN plates may in part depend on the history of the microbial community. For example, there was no significant decrease in the number of populations in wells inoculated with bacteria recovered from an activated sludge reactor. This might be explained by the fact that the activated sludge reactor had been continuously fed with glucose and peptone and thus the dominant bacteria in this community had been selected for rapid growth on readily utilizable carbon sources. All of the bands present in the inoculum also occurred as dominant bands in at least one substrate-containing well. Therefore, the BIOLOG GN patterns generated by activated sludge communities may more closely resemble those of the inoculum and the catabolic potential of the numerically dominant bacteria in activated sludge. In contrast, the dominant populations of the potato rhizosphere community had been selected in an environment that contained diverse substrates that were present at lower concentrations than the substrate concentrations in the activated sludge reactor. In this case, the dominant populations of the inoculum were displaced in the wells of the BIOLOG plates by other, apparently more competitive populations.
When the numbers of bands in TGGE or DGGE profiles are used to estimate the structural diversity of the most abundant populations in the microbial community analyzed, the following issues should be taken into consideration. Due to sequence heterogeneities of 16S rDNA operons (23), more than one fragment can be PCR amplified from any one bacterial strain. This can lead to an overestimate of the number of populations present in the community. Conversely, the PCR products of phylogenetically related strains may have very similar or identical electrophoretic mobilities because the sequences of the amplified region of the 16S rDNA genes are identical or nearly so (3). Finally, the primers used for PCR amplification are not truly “universal,” and other biases in primer annealing and amplification are possible. In some cases it may be possible to obtain a more accurate estimate of species richness by correlating the fragments derived from axenic cultures (cultivated from the wells) to the fragments found in the profiles of the communities by hybridization or by DNA sequence comparisons. This approach was successfully used in this study to characterize the dominant populations of BIOLOG wells after incubation but was not effective for populations in the potato rhizosphere inoculum because they could not be cultivated.
Based on the findings of this study, it appears that while the carbon source utilization patterns of BIOLOG GN plates may be habitat specific, the pattern of a community does not necessarily reflect the functional potential of the community at the time of inoculation. Although the use of carbon utilization profiles to characterize microbial communities has clear limitations, this rapid technique remains a valuable tool for comparison of microbial communities, provided the data are cautiously interpreted.
ACKNOWLEDGMENTS
This work was supported by BMBF grants 0310582 and 0311295 to K.S., by a grant from the Procter & Gamble Corporation to L.F., and by grant BIR 9120006 from the National Science Foundation (United States) to the Center for Microbial Ecology, which provided a Distinguished Visiting Scientist Fellowship to K.S.
We are grateful to the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany, for identifying several strains by the API method.
REFERENCES
- 1.Bochner B. Breathprints at the microbial level. ASM News. 1989;55:536–539. [Google Scholar]
- 2.Bossio D H, Scow K M. Impact of carbon and flooding on the metabolic diversity of microbial communities in soils. Appl Environ Microbiol. 1995;61:4043–4050. doi: 10.1128/aem.61.11.4043-4050.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brim, H. Personal communication.
- 4.Donegan K K, Palm C J, Fieland V J, Porteous L A, Ganio L M, Schaller D L, Bucao L Q, Seidler R J. Changes in levels, species and DNA profiles of soil microorganisms associated with cotton expressing the Bacillus thuringiensis var. kurstaki endotoxin. Appl Soil Ecol. 1995;2:111–124. [Google Scholar]
- 5.Ellis R J, Thompson I P, Bailey M J. Metabolic profiling as a means of characterizing plant-associated microbial communities. FEMS Microbiol Ecol. 1995;16:9–18. [Google Scholar]
- 6.England L S, Lee H, Trevors J T. Recombinant and wild-type Pseudomonas aureofaciens strains introduced into soil microcosms: effect on decomposition of cellulose and straw. Mol Ecol. 1995;4:221–230. doi: 10.1111/j.1365-294x.1995.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 7.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garland J L, Mills A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garland J L, Mills A L. A community-level physiological approach for studying microbial communities. In: Ritz K, Dighton J, Giller K E, editors. Beyond the biomass: compositional and functional analysis of soil microbial communities. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1994. pp. 77–83. [Google Scholar]
- 10.Garland J L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem. 1996;28:213–221. [Google Scholar]
- 11.Garland J L. Patterns of potential C source utilization by rhizosphere communities. Soil Biol Biochem. 1996;28:223–230. [Google Scholar]
- 12.Grayston S J, Campbell C D, Vaughan D. Microbial diversity in the rhizospheres of different tree species. In: Pankhurst C E, Doube B M, Gupta W S R, Grace P R, editors. Soil biota—management in sustainable farming systems. Adelaide, Australia: CSIRO Press; 1994. pp. 155–157. [Google Scholar]
- 13.Haack S K, Garchow H, Klug M J, Forney L J. Analysis of factors affecting the accuracy, reproducibility, and interpretation of microbial community carbon source utilization patterns. Appl Environ Microbiol. 1995;61:1458–1468. doi: 10.1128/aem.61.4.1458-1468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herron P H, Wellington E M H. New method for extraction of streptomycete spores from soil and application to the study of lysogeny in sterile amended and nonsterile soil. Appl Environ Microbiol. 1990;56:1406–1412. doi: 10.1128/aem.56.5.1406-1412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuer H, Hartung K, Engelen B, Smalla K. Studies on microbial communities associated with potato plants by BIOLOG GN and TGGE patterns. Ninth Forum for Applied Biotechnology Proc Med Fac Landbouww Univ Gent. 1995;60/4b:2639–2645. [Google Scholar]
- 16.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In: van Elsas J D, Wellington E M H, Trevors J T, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker Inc.; 1997. pp. 353–373. [Google Scholar]
- 17.Heuer H, Smalla K. Evaluation of community level catabolic profiling using BIOLOG GN microplates to study microbial community changes in potato phyllosphere. J Microbiol Methods. 1997;30:49–61. [Google Scholar]
- 18.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Insam H, Amor K, Renner M, Crepaz C. Changes in functional abilities of the microbial community during composting of manure. Microb Ecol. 1996;31:77–87. doi: 10.1007/BF00175077. [DOI] [PubMed] [Google Scholar]
- 20.Lehman R M, Colwell F S, Ringelberg D B, White D C. Combined microbial community-level analyses for quality assurance of terrestrial subsurface cores. J Microbiol Methods. 1995;22:263–281. [Google Scholar]
- 21.Lerman L S, Fischer S G, Hurley I, Silverstein K, Lumelsky N. Sequence-determined DNA separations. Annu Rev Biophys Bioeng. 1984;13:399–423. doi: 10.1146/annurev.bb.13.060184.002151. [DOI] [PubMed] [Google Scholar]
- 22.Muyzer G, de Waal E C, Uitterlinden A. Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfender W F, Fieland V P, Ganio L M, Seidler R J. Microbial community structure and activity in wheat straw after inoculation with biological control organisms. Appl Soil Ecol. 1996;3:69–78. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Vahjen W, Munch J-C, Tebbe C C. Carbon source utilization of soil extracted microorganisms as a tool to detect the effects of soil supplemented with genetically engineered and non-engineered Corynebacterium glutamicum and a recombinant peptide at the community level. FEMS Microbiol Ecol. 1995;18:317–328. [Google Scholar]
- 27.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 28.Winding A. Fingerprinting bacterial soil communities using BIOLOG microtitre plates. In: Ritz K, Dighton J, Giller K E, editors. Beyond the biomass: compositional and functional analysis of soil microbial communities. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1994. pp. 85–94. [Google Scholar]
- 29.Winding A, Hendriksen N B. Biolog substrate utilization assay for metabolic fingerprints of soil bacteria: incubation effects. In: Insam H, Rangger A, editors. Microbial communities: function versus structural approaches. Berlin, Germany: Springer-Verlag; 1997. pp. 195–205. [Google Scholar]
- 30.Wünsche L, Brüggemann L, Babel W. Determination of substrate utilization patterns of soil microbial communities: an approach to assess population changes after hydrocarbon pollution. FEMS Microbiol Ecol. 1995;17:295–306. [Google Scholar]
- 31.Zak J C, Willig M R, Moorhead D L, Wildman H G. Functional diversity of microbial communities: a quantitative approach. Soil Biol Biochem. 1994;26:1101–1108. [Google Scholar]