Abstract
The use of PCR to amplify a specific virA gene fragment serves as a highly specific and sensitive method to detect virulent bacteria of the genus Shigella and enteroinvasive Escherichia coli. Amplification of a 215-bp DNA band was obtained by using isolated genomic DNA of Shigella, individual cells of Shigella dysenteriae, and mayonnaise contaminated with S. dysenteriae. Moreover, a multiplex PCR with specific (virA) and bacterium-restricted (16S ribosomal DNA) primers generated an amplification product of approximately 755 bp for all bacteria tested and an additional 215-bp product for Shigella and enteroinvasive E. coli.
Shigella constitutes a genus of gram-negative, nonmotile, non-spore-forming rod-shaped bacteria with four species or serotypes, namely, S. boydii, S. dysenteriae, S. flexneri, and S. sonnei (12). Virulent Shigella organisms cause the human illness known as bacillary dysentery, as do enteroinvasive Escherichia coli (EIEC) strains. Bacillary dysentery (shigellosis) causes mild diarrhea, fever, abdominal cramps, and severe fluid loss (25). All of the virulent strains mentioned above harbor a 120- to 230-kb plasmid named the virulence plasmid (7), which was first described for S. flexneri 2a (16). It was established that the loss of the virulence plasmid results in avirulent strains (21) and that the genes implicated in virulent functions are localized not only in the virulence plasmid but also in the chromosome (a complete review of chromosome and plasmid virulence genes is presented in reference 6). The virA gene has been identified in the virulence plasmid of S. flexneri 2a, and it has been implicated in invasion and intercellular spreading (23).
By means of human transmission, Shigella can contaminate several kinds of foods, including raw vegetables, milk, poultry, and some dairy products (24). Therefore, as with other pathogenic microorganisms, it is important that the presence of Shigella be detected in foods. Traditionally, the detection test of food-borne microorganisms (hazard test) is made by plating a food homogenate on highly selective media, although in the case of some bacteria a preenrichment step is required. After several days of incubation, the presence or absence of the microorganism or the number of colonies is determined (9). This plating technique, based on the phenotype of the bacteria, is labor-intensive and can take several weeks to obtain results (11). On the other hand, rapid, highly sensitive, and specific techniques based on genetic characteristics have been developed recently. DNA probe hybridization and PCR are the best known of these techniques and are used as hazard tests for the detection and identification of food-borne microorganisms (2, 10, 18, 19).
In this paper, we describe the highly sensitive and specific detection of virulent Shigella organisms and EIEC by PCR combined with DNA hybridization. The virA gene is the target chosen for the PCR. The applicability of this PCR method for detection of these organisms in mayonnaise is demonstrated.
MATERIALS AND METHODS
Genomic DNA isolation from bacterial strains.
The bacterial strains used in this work are listed in Table 1. Bacteria were grown overnight in a liquid medium (5 g of tryptone per liter, 2.5 g of yeast extract per liter, 1 g of glucose per liter), sedimented, and lysed with detergent to release DNA, which was extracted with phenol-chloroform (25:24, vol/vol) and precipitated with ethanol (1). The contaminating RNA was degraded by suspending the DNA sample in TER (10 mM Tris-HCl [pH 8], 0.1 mM EDTA [pH 8], 1 μg of RNase A per ml).
TABLE 1.
PCR results and origin of the DNA samples
| Species | Sourcee | PCR results
|
||
|---|---|---|---|---|
| virA (215 bp) |
virA plus 16S rDNA
|
|||
| 215 bp | 755 bp | |||
| Shigella boydii serovar 10 | CECT 583 | + | + | + |
| Shigella dysenteriae serovar 1 | CECT 584 | + | + | + |
| Shigella flexneri serovar 2a | CECT 585 | + | + | + |
| Shigella sonnei serovar a | CECT 542 | + | + | + |
| Shigella sonnei | CECT 457 | + | + | + |
| Shigella boydii 238a | Clinical sample | + | + | + |
| Shigella dysenteriae 193a | Clinical sample | + | + | + |
| Shigella dysenteriae 300a | Clinical sample | + | + | + |
| Shigella flexneri 295a | Clinical sample | + | + | + |
| Shigella flexneri 299a | Clinical sample | + | + | + |
| Shigella sonnei 296a | Clinical sample | + | + | + |
| Shigella sonnei 298a | Clinical sample | + | + | + |
| Escherichia coli serovar O1 | CECT 515 | − | − | + |
| EIEC 41a | Clinical sample | + | + | + |
| EIEC 42a | Clinical sample | + | + | + |
| EIEC 120a | Clinical sample | + | + | + |
| EIEC 121a | Clinical sample | + | + | + |
| EPEC E2348/69b | Clinical sample | − | − | + |
| EPEC B171b | Clinical sample | − | − | + |
| Bacillus sp. | CECT 450 | − | − | + |
| Enterobacter aerogenes | CECT 684 | − | − | + |
| Enterococcus faecalis | CECT 481 | − | − | + |
| Lactobacillus cellobiosus | CECT 562 | − | − | + |
| Lactobacillus sake | CECT 906 | − | − | + |
| Micrococcus luteus | CECT 241 | − | − | + |
| Mycobacterium phlei | CECT 3009 | − | − | + |
| Proteus vulgaris | CECT 484 | − | − | + |
| Pseudomonas fluorescens | CECT 378 | − | − | + |
| Salmonella dublinc | Clinical sample | − | − | + |
| Salmonella enteritidisc | Clinical sample | − | − | + |
| Salmonella montevideoc | Clinical sample | − | − | + |
| Salmonella panamac | Clinical sample | − | − | + |
| Salmonella typhimuriumc | Clinical sample | − | − | + |
| Serratia marcescens | CECT 159 | − | − | + |
| Staphylococcus aureus | CECT 240 | − | − | + |
| Yersinia enterocolitica serovar O:3 biovar 4d | Clinical sample | − | − | + |
| Yersinia pestis EV76f | Clinical sample | − | − | + |
| Cryptococcus sp. | Lab stock | − | − | − |
| Rhodotorula sp. | Lab stock | − | − | − |
| Saccharomyces cerevisiae | Lab stock | − | − | − |
| Human | Boehringer | − | − | − |
Strain kindly donated by G. Prats, Hospital Universitario Sant Pau, Barcelona, Spain.
Strain kindly donated by M. Donnenberg, University of Maryland School of Medicine, Baltimore. EPEC, enteropathogenic E. coli.
Strain kindly donated by J. C. Palomares, Hospital Universitario Virgen Macarena, Seville, Spain.
Strain kindly donated by G. Kapperud, National Institute of Public Health, Oslo, Norway.
CECT, Spanish type culture collection.
Strain kindly donated by E. Carniel, Institut Pasteur, Paris, France.
Genomic DNA isolation from yeast strains.
The yeast strains used in this work are listed in Table 1. Yeasts were grown overnight in a liquid medium (20 g of peptone per liter, 10 g of yeast extract per liter, 20 g of glucose per liter), precipitated, and lysed with 50 μg Zymolase 20T per ml plus detergent to release DNA, which was extracted with phenol-chloroform (25:24, vol/vol) and precipitated with ethanol (15). Finally, the DNA was suspended in TER.
Food sample preparation.
A 5-g sample of commercial mayonnaise (Ybarra, Seville, Spain) was diluted to 50 ml with buffered peptone water (Merck) and mixed to complete homogenization. Diluted mayonnaise was also prepared and externally contaminated with S. dysenteriae serovar 1 at (88 ± 11) × 104 cells per ml of mayonnaise. Samples (10 μl) of mayonnaise prepared in these two ways were used in the PCR.
Determination of CFU.
Bacteria were grown on plate count agar (Oxoid) to achieve isolated colonies. One colony was suspended in 1 ml of buffered peptone water, and 10-fold serial dilutions were made. Aliquots (10 μl) of selected dilutions were made up to 1 ml with buffered peptone water and spread on plate count agar. After incubation, the colonies were counted. Aliquots (10 μl) of selected dilutions were used in the PCR.
DNA amplifications (PCR).
Amplifications were made in a 50-μl reaction mixture which contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate, 25 pmol of each primer, and 2.5 U of Taq polymerase (Boehringer), with either isolated DNA, a bacterial dilution, or a diluted mayonnaise sample. Temperature conditions were as follows: denaturation for 45 s at 94°C, hybridization for 30 s at 65°C, and polymerization for 30 s at 72°C. Thirty-five cycles were carried out as mentioned above. The sequences of primers used to amplify virA and 16S ribosomal DNA (rDNA) gene sequences are shown in Table 2.
TABLE 2.
Sequences of primers used in PCR
| Primer | Sequence |
|---|---|
| virA | |
| Forward | 5′-CTG CAT TCT GGC AAT CTC TTC ACA TC-3′ |
| Reverse | 5′-TGA TGA GCT AAC TTC GTA AGC CCT CC-3′ |
| 16S rDNA | |
| Forward | 5′-AGA CTG CTA CGG GAG GCA GCA GT-3′ |
| Reverse | 5′-GTT GCG CTC GTT GCG GGA CTT AA-3′ |
Agarose gel electrophoresis.
Aliquots (10 or 25 μl) of the amplification reaction solutions were run on a 1% (wt/vol) agarose gel (SeaKem; FMC) stained with 0.8 mg of ethidium bromide (Amresco) per ml. The DNA was observed by irradiating the gel with UV light at 264 nm. When a negative amplification was obtained, a new PCR was done and 25 μl of the resultant solution was run on an agarose gel to confirm the first result.
Cloning and sequencing of the virA fragment.
A 50-ng sample of the amplified virA fragment of S. dysenteriae serovar 1 was ligated at 16°C in a 10-μl reaction mixture containing 30 mM Tris-HCl of (pH 7.5), 10 mM MgCl2, 10 mM dithiotreithol, 1 mM ATP, 50 ng of pGEM-T, and 1 U of T4 ligase (Promega). The transformation of the ligated DNA was carried out in XL1-Blue MRF′ bacterial strain (Stratagene), as previously described (14). DNA was sequenced by the chain termination method, modified to use universal primers labeled with digoxigenin (Dig) and Taq polymerase (20). A good separation of DNA bands was achieved with MWG-Biotech’s direct-blotting electrophoresis system (3).
DNA-DNA hybridization.
Amplification reaction solution volumes of 10 or 25 μl were run on an agarose gel and transferred to a nylon membrane (Hybond; Amersham) with a vacuum blotter (model 785; Bio-Rad) at a pressure of 1.72 × 104 Pa applied over 90 min in 0.5 M NaOH–0.6 M NaCl. The membrane was hybridized (22) against a Dig probe at 65°C in 5× SSC (1× SSC is 15 mM sodium citrate and 150 mM NaCl)–0.1% (wt/vol) sodium dodecyl sulfate (SDS)–1% (wt/vol) blocking reagent (Boehringer). The cloned and sequenced virA fragment of S. dysenteriae serovar 1 was used as a probe in the hybridization. This probe was labeled by PCR as described above, except that deoxynucleoside triphosphate was replaced by a Dig labeling mix (Boehringer) containing Dig-dUTP (17). After hybridization, the membrane was washed (high-stringency conditions) twice at 65°C over a period of 15 min in 2× SSC–0.1% (wt/vol) SDS and then twice at room temperature over a period of 15 min in 0.1× SSC–0.1% (wt/vol) SDS. The Dig probe was detected by color by using an anti-Dig antibody coupled to alkaline phosphatase (Boehringer), as described elsewhere (13).
Nucleotide sequence accession number.
The virA sequence of S. dysenteriae serovar 1 will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AF010147.
RESULTS
Specificity of PCR with virA primers.
We have designed two specific primers flanking a 215-bp region of the S. flexneri virA gene (accession no. D26468). These primers amplified not only isolated DNA from S. flexneri but also those isolated from all the other Shigella and EIEC strains tested (Table 1). All amplification-generated products were of the expected size (approximately 215 bp) on agarose gel electrophoresis. Isolated DNA from microorganisms other than Shigella and EIEC produced no amplification product (Table 1), even though there was sufficient DNA (100 ng) to detect a single-copy sequence of virA.
Sensitivity of PCR with virA primers.
When S. dysenteriae serovar 1 was used, as little as 25 fg of isolated DNA (Fig. 1A), 10 μl of a bacterial dilution containing between 1 and 10 CFU, i.e. 100 to 1,000 cells/ml (data not shown), and deliberately contaminated mayonnaise (data not shown) gave positive amplifications, as revealed on an agarose gel.
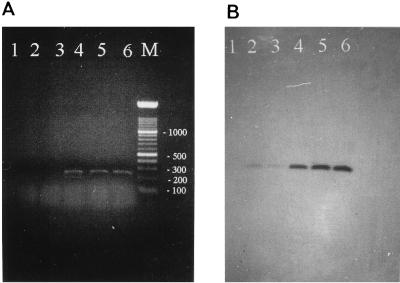
FIG. 1.
PCR with virA primers and different amounts of DNA of S. dysenteriae serovar 1. (A) Twenty-five microliters of the amplification reaction solutions containing different amounts of DNA subjected to agarose gel electrophoresis; (B) same solutions as those in panel A after filter DNA hybridization against the virA Dig-labeled probe. Lane M contains a 100-bp marker, with numbers indicating the size in base pairs. Lanes 1 to 6 contain 0, 1, 5, 25, 50, and 100 fg of template DNA, respectively.
The amplification product of S. dysenteriae serovar 1 was cloned in a plasmid vector. A representative clone was sequenced in both directions. This sequence (accession no. AF010147) was 215 nucleotides long and identical to that of virA from S. flexneri. This representative clone was labeled with Dig and used as a highly specific probe to hybridize against the amplification products obtained with different amounts of DNA from S. dysenteriae serovar 1. Positive hybridization was observed for all lanes in which template DNA (1 to 100 fg) had been added to the PCR (Fig. 1B); i.e., the sensitivity was 1 fg of template DNA.
Multiplex PCR.
The 16S rDNA primers were designed by using conserved regions of 16S rDNA of bacteria and the corresponding sequence of E. coli (accession no. J01695). They should produce an amplification product of around 755 bp whenever bacteria are present. PCR experiments using both virA and 16S rDNA primers (multiplex PCR) produced in all bacteria tested one DNA band of approximately 755 bp (Table 1) and an additional DNA band of approximately 215 bp in Shigella and EIEC (Table 1 and Fig. 2A). On the other hand, the few samples of eukaryotic DNA that were tested did not produce either the 755-bp DNA band or the 215-bp DNA band (Table 1). Moreover, the 215-bp product could be differentiated from that of 755 bp not only by gel electrophoresis but also by hybridization against the virA probe (Fig. 2B).
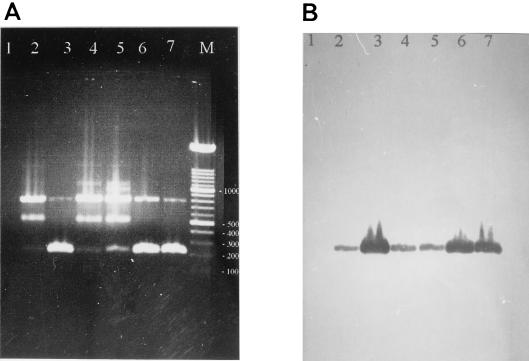
FIG. 2.
Multiplex PCR with DNA from Shigella spp. or EIEC. (A) Ten microliters of amplification reaction solutions with 100 ng of DNA from various bacteria after agarose gel electrophoresis; (B) the same solutions as those in panel A after filter DNA hybridization against the virA Dig-labeled probe. Lane M, 100-bp marker, with numbers indicating the size in base pairs. Lanes 1 to 7, control without DNA, S. boydii serovar 10, S. dysenteriae serovar 1, S. flexneri serovar 2a, S. sonnei serovar a, EIEC 41, and EIEC 121, respectively.
DISCUSSION
Epidemiological studies on Shigella have established that 10 cells are sufficient to be an infective dose (24). This amount of bacteria could easily be present in contaminated food. The results presented in this work showed that PCR with virA primers could be a useful hazard test because its sensitivity, similar to that reported elsewhere (4, 5), would allow 1 fg of DNA or 1 to 10 cells in 10 μl of sample to be detected. Moreover, the high annealing temperature between primers and target and the nature of the target itself gave the desirable specificity for strains containing the virulence plasmid, Shigella and EIEC.
Legislation in many countries requires the absence of Shigella in 25-g amounts of foods (9), such as mayonnaise. Since it is not possible to directly carry out PCR on 25 g of mayonnaise, an enrichment step is necessary. Despite this additional step, the PCR is faster and also more sensitive than conventional methods. A positive result with the virA primers does not conclusively demonstrate that a virulent organism is present in the sample. Given that the genes implicated in virulence are located on both the chromosome and the plasmid and that virA is on a plasmid, which could be transferred to other bacteria, it is theoretically possible that other bacteria can carry virA and be nonpathogenic.
The applicability of PCR with the virA primers for detection of Shigella and EIEC in mayonnaise was demonstrated since a positive amplification was obtained with mayonnaise diluted with peptone water that had been deliberately contaminated with Shigella. The absence of a PCR product when an enrichment medium is used as the source of template DNA could be due to either (i) no contamination (absence of target DNA) or (ii) a failure in the reaction due to the presence of inhibitors and/or the unavailability of DNA, e.g., no bacterial lysis. Failure to detect contamination by PCR could be confirmed by using multiplex PCR to verify the absence of the 755-bp product both in a food sample and, as a positive control, in a food sample contaminated with exogenous bacteria.
Cost factors are likely to be considered when selecting the method for detection of Shigella in foods, and PCR seems to be more expensive than the conventional method (if confirmation of positive results is excluded). However, it should be kept in mind that the latter method does not include an enrichment medium intended for Shigella (8). Also, the conventional method is time-consuming, requiring food to be stored for a long period. Therefore, a comparison of the costs associated with storage and those of the PCR itself should be made.
ACKNOWLEDGMENTS
We are very grateful to M. Donnenberg, G. Kepperud, J. C. Palomares, and G. Prats for providing bacterial strains and to J. L. Algeciras and E. Trujillo for providing Ybarra mayonnaise. Our thanks to A. Villalobo, C. Díaz-Ramos, and P. Pérez-Romero for their critical reading of the manuscript.
Eduardo Villalobo is the recipient of a predoctoral fellowship from the Ministerio de Educación y Ciencia (España).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 2.4.1–2.4.5. [Google Scholar]
- 2.Batt C A. Molecular diagnostic for dairy-borne pathogens. J Dairy Sci. 1997;80:220–229. doi: 10.3168/jds.S0022-0302(97)75931-9. [DOI] [PubMed] [Google Scholar]
- 3.Beck S, Pohl F M. DNA sequencing with direct blotting electrophoresis. EMBO J. 1984;3:2905–2909. doi: 10.1002/j.1460-2075.1984.tb02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bej A K, Steffan R J, DiCesare J, Haff L, Atlas R M. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990;56:307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bej A K, DiCesare J, Haff L, Atlas R M. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol. 1991;57:1013–1017. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale T L. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale T L, Sansonetti P J, Schad P A, Austin S, Formal S B. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect Immun. 1983;40:340–350. doi: 10.1128/iai.40.1.340-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrigan W F, McCance M E. Detection of pathogenic and enterotoxigenic microorganisms: Salmonella and Shigella. In: Gibbs B M, Skinner F A, editors. Methods in food and dairy microbiology. New York, N.Y: Academic Press, Inc.; 1966. pp. 142–146. [Google Scholar]
- 9.Hayes P R. Food microbiology and hygiene. London, United Kingdom: Elsevier Applied Science Publishers; 1985. [Google Scholar]
- 10.Hill W E. The polymerase chain reaction: applications for the detection of foodborne pathogens. Crit Rev Food Sci Nutr. 1996;36:123–173. doi: 10.1080/10408399609527721. [DOI] [PubMed] [Google Scholar]
- 11.Hill W E, Ferreira J L, Payne W L, Jones V M. Probability of recovering pathogenic Escherichia coli from foods. Appl Environ Microbiol. 1985;49:1374–1378. doi: 10.1128/aem.49.6.1374-1378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt J G, Kreig N R, Sneath H A, Staley J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. pp. 187–188. [Google Scholar]
- 13.Holtke H J, Seibl R, Burg J, Muhlegger K, Kessler C. Nonradioactive labeling and the detection of nucleic acids. II. Optimization of the digoxigenin system. Biol Chem Hoppe-Seyler. 1990;371:929–938. doi: 10.1515/bchm3.1990.371.2.929. [DOI] [PubMed] [Google Scholar]
- 14.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser C S, Michaelis S, Mitchell A, editors. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1994. pp. 139–140. [Google Scholar]
- 16.Kopecko D J, Washington O, Formal S B. Genetic and physical evidence of plasmids from virulent and spontaneously occurring avirulent colonial variants of Shigella flexneri. Infect Immun. 1980;29:207–214. doi: 10.1128/iai.24.2.580-582.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanzillo J J. Preparation of digoxigenin-labeled probes by the polymerase chain reaction. Biotechniques. 1990;8:620. [PubMed] [Google Scholar]
- 18.Notermans S, Wernars K. Evaluation and interpretation of data obtained with immunoassays and DNA-DNA hybridization techniques. Int J Food Microbiol. 1990;11:35–49. doi: 10.1016/0168-1605(90)90038-7. [DOI] [PubMed] [Google Scholar]
- 19.Olsen J E, Aabo S, Hill W, Notermans S, Wernars K, Granum P E, Popovic T, Rasmussen H N, Olsvik O. Probes and polymerase chain reaction for detection of food-borne bacterial pathogens. Int J Food Microbiol. 1995;28:1–78. doi: 10.1016/0168-1605(94)00159-4. [DOI] [PubMed] [Google Scholar]
- 20.Peterson M G. DNA sequencing using Taq polymerase. Nucleic Acids Res. 1988;16:10915. doi: 10.1093/nar/16.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sansonetti P J, Kopecko D J, Formal S B. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 23.Uchiya K, Tobe T, Komatsu K, Suzuki T, Watarai M, Fukuda I, Yoshikawa M, Sasakawa C. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol Microbiol. 1995;17:241–250. doi: 10.1111/j.1365-2958.1995.mmi_17020241.x. [DOI] [PubMed] [Google Scholar]
- 24.Wachsmuth K, Morris G K. Shigella. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 448–660. [Google Scholar]
- 25.Ward D, Hart K. HACCP: hazard analysis and critical control point training curriculum. Raleigh, N.C: North Carolina State University; 1996. Hazards found in seafood; pp. 163–180. [Google Scholar]