Abstract
A method was developed to study the biodegradation and oxidative biodehalogenation of fluorinated phenols by 19F nuclear magnetic resonance (NMR). Characterization of the 19F NMR spectra of metabolite profiles of a series of fluorophenols, converted by purified phenol hydroxylase, catechol 1,2-dioxygenase, and/or by the yeast-like fungus Exophiala jeanselmei, provided possibilities for identification of the 19F NMR chemical shift values of fluorinated catechol and muconate metabolites. As an example, the 19F NMR method thus defined was used to characterize the time-dependent metabolite profiles of various halophenols in either cell extracts or in incubations with whole cells of E. jeanselmei. The results obtained for these two systems are similar, except for the level of muconates observed. Altogether, the results of the present study describe a 19F NMR method which provides an efficient tool for elucidating the metabolic pathways for conversion of fluorine-containing phenols by microorganisms, with special emphasis on possibilities for biodehalogenation and detection of the type of fluorocatechols and fluoromuconates involved. In addition, the method provides possibilities for studying metabolic pathways in vivo in whole cells.
The aerobic microbial degradation of halogenated aromatics can proceed through formation of intermediate halophenols that can be converted to catechols suitable for ring cleavage reactions and further conversion of the compounds to substrates for normal carbon metabolism. This conversion of halophenols to tricarboxylic acid cycle intermediates often requires that at some point in the catabolic pathway, the halogen atoms are removed from the molecule, which results in formation of nonhalogenated metabolites. In many cases, these dehalogenation steps are difficult and/or result in metabolic intermediates that need specific enzymes for their further degradation through, in most cases, the 3-oxoadipate pathway (6–8, 20, 21, 24, 30).
Up to now, many studies of possibilities for biodehalogenation have focused on the reactions catalyzed by purified enzymes (7, 8, 10, 20, 24, 28). However, the relative importance of the various pathways for the in vivo conversion of a specific halophenol remains to be investigated for each organism and each halophenol separately. Therefore, the development of a technique that can efficiently characterize the relative importance of the various biodehalogenation pathways for the conversion of a specific halophenol by a specific microorganism would be of great value.
Of all nuclear magnetic resonance (NMR)-observable isotopes, 19F is the one perhaps most convenient for studies of biodegradation of environmental pollutants (3, 15, 22). This originates from several advantages of the 19F isotope compared to those of other nuclei. First, the intrinsic sensitivity of the 19F nucleus is high and is almost comparable to that of 1H. Obviously, sensitivity is an important factor, because xenobiotics and their metabolites are usually present in relatively low concentrations. Second, for fluorine, the sensitivity is further increased because of the absence of background signals, because biological systems do not contain NMR-visible fluorinated endogenous compounds. This implies that all resonances observed can be unambiguously ascribed to the xenobiotic compound and its biodegradation products. Third, the 19F nucleus is known to have a broad chemical shift range of about 500 ppm. This is large compared to the chemical shift range of, for example, 1H resonances, 15 ppm, and that of 13C, 250 ppm. Thus, the chemical shift of a 19F nucleus is highly sensitive to its molecular surroundings, resulting in relatively large changes in chemical shift as a result of biochemical modification of a xenobiotic. This reduces the chances of peak overlap.
The results of the present study demonstrate that 19F NMR provides a tool with which to efficiently characterize the first steps in the catabolic pathway of fluorophenols by whole cells and/or cell extracts. Special emphasis is on the characterization of the various fluorocatechols and fluoromuconates, since identification of their 19F NMR resonances is required before the 19F NMR method can be used as an analytical tool. Thus, by using purified phenol hydroxylase and catechol 1,2-dioxygenase, as well as whole cells and cell extracts from Exophiala jeanselmei, the 19F NMR chemical shift values of fluorocatechols and, especially, fluoromuconates were identified. E. jeanselmei CBS 658.76 was chosen for the biodegradation and metabolic identification studies because this yeast-like fungus has previously been reported to be able to degrade and grow on an extremely large series of benzene compounds, but not on halogenated derivatives (17). This implies that this species provides good possibilities for identification and demonstration of the various fluorocatechol and fluoromuconate intermediates expected to accumulate in the metabolic pathways of the various fluorophenols.
MATERIALS AND METHODS
Chemicals.
Phenol was purchased from Merck (Darmstadt, Germany). 2-Fluoro-, 3-fluoro-, and 4-fluorophenol were obtained from Janssen Chimica (Beerse, Belgium). 2,4-Difluoro-, 2,5-difluoro-, 3,4-difluoro-, and 2,4,5-trifluorophenol and 2,3,4,5-tetrafluorophenol were purchased from Aldrich (Steinheim, Germany). 2,3-Difluoro-, 2,6-difluoro-, 3,5-difluoro-, 2,3,4-trifluoro-, 2,3,5-trifluoro-, 2,3,6-trifluoro-, and 3,4,5-trifluorophenol were obtained from Fluorochem (Derbyshire, United Kingdom). Pentafluorophenol and catechol were obtained from Sigma (St. Louis, Mo.). Fluorocatechols were prepared as previously described (19) from the corresponding fluorophenols with purified phenol hydroxylase from Trichosporon cutaneum. Fluoromuconates were prepared by incubating the fluorocatechols thus formed with catechol 1,2-dioxygenase from Pseudomonas arvilla C-1.
Incubations with purified enzymes.
Phenol hydroxylase was isolated from the yeast T. cutaneum as described previously (19). Catechol 1,2-dioxygenase from P. arvilla C-1 was purified essentially according to the method of Nakai et al. (18).
Incubations with purified phenol hydroxylase, in the absence or presence of catechol 1,2-dioxygenase, for 19F NMR measurements were carried out at 30°C in closed reaction vessels to prevent evaporation of the phenolic substrate. Incubation media contained (final concentrations) 0.1 M potassium phosphate (pH 7.6); 0.7 mM halophenol, added as 1% (vol/vol) of a 70 mM stock in dimethyl sulfoxide; 10 μM FAD; 1 mM ascorbic acid; and 1 mM NADPH. The incubations were started by the addition of the purified phenol hydroxylase and, in case of biosynthesis of the fluoromuconates, the purified catechol 1,2-dioxygenase. Different amounts of the enzyme preparations and incubation times were used for the different substrates, depending on the reactivity of the substrate.
Organism and growth conditions for biodegradation studies.
Strain CBS 658.76 of the yeast-like fungus E. jeanselmei was used and cultured as previously described (17). In short, the cells were grown at 30°C on an orbital shaker in a synthetic medium containing (per liter): KH2PO4, 9 g; K2HPO4, 1 g; MgSO4 · 7H2O, 0.5 g; NH4Cl, 2 g; CaCl2, 0.1 g; FeCl3 · 6H2O, 2 mg; H3BO3, 0.5 mg; CuSO4 · 5H2O, 0.1 mg; MnCl2, 0.4 mg; KI, 0.1 mg; NaMoO4, 0.2 mg; ZnSO4, 0.4 mg; myo-inositol, 0.01 g; 4-aminobenzoic acid, 0.3 mg; d-biotin, 0.02 mg; Ca-pantothenate, 2 mg; nicotinic acid, 5 mg; pyridoxine, 1 mg; riboflavin, 0.1 mg; thiamine, 0.2 mg; the pH was adjusted to 5.5. For growth on phenol, the substrate was added four times at 1 to 2 mM (final concentration), over a period of 2 days.
The yeast strain was maintained at 4°C on agar slants of the same medium, containing glucose (2 g/liter) as the carbon source.
Degradation of fluorophenols by whole cells.
The fluorophenol test compounds were added to a final concentration of 0.7 mM (each) to 80 ml of a phenol-grown culture after phenol had been exhausted (as checked by high-performance liquid chromatography (HPLC). To monitor the conversion of these substrates, samples were taken at different time intervals. Before freezing the samples into liquid nitrogen, 10 mM ascorbic acid was added to prevent possible autoxidation of catechol metabolites. Samples were stored at −20°C until analyzed. Before 19F NMR analysis, samples were unfrozen and centrifuged (5 min, 13,000 × g, 0°C) to remove cell debris.
Conversion of fluorophenols by cell extracts.
Cells of the phenol-adapted culture (1 liter) were harvested by centrifugation (10,000 × g, 15 min); washed twice in 50 mM potassium phosphate (pH 7.6), containing 2.0 μM FAD, 1 mM dithiothreitol, and 0.1 mM EDTA; and resuspended in a final total volume of 50 ml of the same buffer, to which 1.0 mM phenylmethylsulfonyl fluoride was added. The cells were disrupted through a precooled French press, and the extract was clarified by centrifugation (10,000 × g, 15 min). Microscopic analysis showed that over 90% of the cells were destroyed. During this cell extract preparation step, the relative concentration factor compared to that of the cell cultures was 20 (i.e., the amount of enzyme expected per ml was 20 times higher in the cell extract than in the cell cultures). Incubations with cell extracts were carried out as described above for the incubations with purified enzymes. Instead of the enzymes, cell extract (50 μl of cell extract in 1 ml of incubation mixture) was added. Samples were taken at 15-min intervals, frozen in liquid nitrogen, and stored at −20°C until analyzed. During the experiment, 1 mM ascorbic acid was added to the incubation mixture every few hours to prevent autoxidation of the catechols formed.
19F NMR measurements.
19F NMR measurements were performed with a Bruker DPX 400 NMR, and some measurements were performed with a Bruker AMX 300 spectrometer as previously described (29). The temperature of the measurements was 7°C. A dedicated 10-mm-diameter 19F NMR probe head was used with both NMR instruments. The spectral width used for the 19F NMR measurements was between 20,000 and 50,000 Hz, depending on the sample measured. The number of data points used for data acquisition was 65,536. Pulse angles of 30° were used. Between 2,000 and 60,000 scans were recorded, depending on the concentrations of the fluorine-containing compounds and the signal-to-noise ratio required. The sample volume was 1.72 ml, containing 1.4 ml of sample; 200 μl of 0.8 M potassium phosphate (pH 7.6); 100 μl of 2H2O, used as a deuterium lock; and 20 μl of 8.4 mM 4-fluorobenzoate, added as an internal standard. Concentrations of the various metabolites were calculated by comparison of the integrals of the 19F NMR resonances of the metabolites to the integral of the 4-fluorobenzoate resonance. Chemical shifts are reported relative to CFCl3. The resonance of the internal standard, 4-fluorobenzoate, was set at −114.2 ppm with respect to CFCl3. The Lorentzian line shape of the resonances was converted to a Gaussian-type line shape in order to improve resolution for careful determination of the coupling constants. In the cases in which the coupling constants could be determined directly from the data sets, the resolution was at least 0.2 Hz. In cases in which AA′XX′ spectra were obtained, the splitting patterns were carefully analyzed by using a formal calculation as described by Günther (9).
1H decoupling was achieved with a Waltz16 decoupling sequence. 19F NMR chemical shift values of the various fluorine-containing compounds were identified on the basis of added authentic reference compounds for fluoride anions and all fluorophenols. The resonances of the different fluorocatechol metabolites were identified previously (19), and those of the fluoromuconates were identified as described in Results.
HPLC analysis.
HPLC analysis was conducted with a LiChrosorb RP18 column (100 by 3 mm). Elution was carried out at a flow rate of 1.0 ml/min, starting with 100% water for 1 min, and then a linear gradient of 0 to 80% of methanol in 14 min was applied, followed by 1 min of 80% methanol. Detection was performed at 280 or 295 nm with a Waters 996 photo diode array detector. Metabolite peaks were identified on the basis of added authentic reference compounds.
RESULTS
Conversion of 2-fluorophenol by whole cells and cell extracts as determined by 19F NMR.
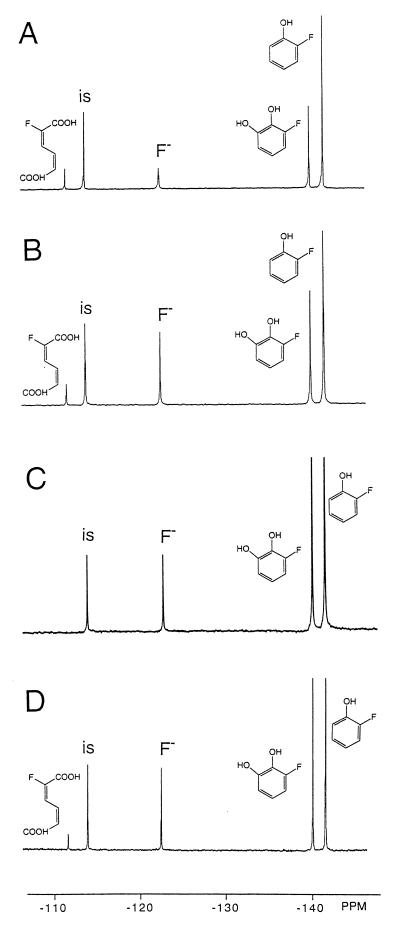
Figure 1 presents, as an example, the 19F NMR spectra of the 1-h incubations of whole cells (Fig. 1A) and of cell extracts (Fig. 1B) of E. jeanselmei incubated with 2-fluorophenol. Formation of three metabolites with 19F NMR chemical shift values of −140.4, −111.8, and −123.0 ppm were observed to an extent that accounts for the decrease in the amount of 2-fluorophenol. The resonance at −123.0 ppm originates from fluoride anions, indicating that biodehalogenation has occurred. The HPLC chromatogram of the 1-h incubation of cell extracts with 2-fluorophenol (not shown) indicates formation of nonfluorinated catechol, pointing at oxidative dehalogenation of 2-fluorophenol by a phenol hydroxylase. The amount of nonfluorinated catechol formed equals the amount of fluoride anion formed (corrected for the amount of fluoride anion already present in blank incubations). Thus, all fluoride anions formed can be accounted for by the amount of nonhalogenated catechol formed from 2-fluorophenol by phenol hydroxylase, showing that no significant other pathways for biodehalogenation occur. This implies that oxidative dehalogenation by phenol hydroxylase appears to be the main pathway for biodehalogenation of 2-fluorophenol in E. jeanselmei. The resonances of the other two main metabolites in the 19F NMR spectra in Fig. 1A and B represent 3-fluorocatechol and 2-fluoromuconate, the latter resulting from ring cleavage of the 3-fluorocatechol by a catechol 1,2-dioxygenase. These chemical shift values were identified on the basis of experiments in which fluorophenols were incubated with purified phenol hydroxylase and/or purified catechol 1,2-dioxygenase (Fig. 1C and D). Figure 1C and D show that incubation of 2-fluorophenol with purified phenol hydroxylase (Fig. 1C) or purified phenol hydroxylase and catechol 1,2-dioxygenase (Fig. 1D) gives rise to formation of the metabolites with chemical shift values of −140.4 and −111.8 ppm, respectively. These results indicate that the metabolite at −140.4 ppm is 3-fluorocatechol and that the metabolite at −111.8 ppm is 2-fluoromuconate.
FIG. 1.
19F NMR spectra of culture medium after 1 h of incubation of whole cells of E. jeanselmei with 2-fluorophenol at 30°C (A), cell extracts from E. jeanselmei incubated for 1 h with 2-fluorophenol at 30°C (B), incubation of purified phenol hydroxylase with 2-fluorophenol (C), and incubation of purified phenol hydroxylase plus purified catechol 1,2-dioxygenase with 2-fluorophenol (D). 19F NMR chemical shift values were identified as described previously (19) and on the basis of results from the present study (see text and Table 1). The resonance marked “is” is from the internal standard, 4-fluorobenzoate.
Altogether, the results in Fig. 1 indicate that biodegradation of 2-fluorophenol by E. jeanselmei is blocked at the level of catechol 1,2-dioxygenase and muconate cycloisomerase, leading to accumulation of 3-fluorocatechol and 2-fluoromuconate as the ultimate reaction products.
Assignment of the chemical shift values of fluoromuconates by 1H-(de)coupled 19F NMR.
Additional experiments of the present paper describe the identification of the 19F NMR signals of an extended series of fluorine-containing catechol and muconate derivatives.
Experiments in which purified phenol hydroxylase and/or purified catechol 1,2-dioxygenase was incubated with 2-fluoro-, 3-fluoro-, 4-fluoro-, 2,3-difluoro-, 2,4-difluoro-, 2,5-difluoro-, 2,6-difluoro-, 3,4-difluoro, 3,5-difluoro-, 2,3,4-trifluoro-, 2,3,5-trifluoro-, 2,3,6-trifluoro-, 2,4,5-trifluoro-, 3,4,5-trifluoro-, 2,3,5,6-tetrafluoro-, and pentafluorophenol gave rise to 19F NMR spectra containing the resonances of the various fluorocatechol and fluoromuconate metabolites derived from the different fluorophenol derivatives. The 19F NMR chemical shift values of the fluorophenols, except for 2,4,5-trifluorophenol, were previously reported (19). In the present study, the 19F NMR chemical shift values of 2,4,5-trifluorophenol were identified at −147.1 (F-2), −153.5 (F-4), and −143.7 ppm (F-5), since the compound was now commercially available. The parts-per-million values observed for the fluorocatechols were identical to those previously described and identified (19). The parts-per-million values of the fluoromuconates prepared in the present study with purified phenol hydroxylase and catechol 1,2-dioxygenase are listed in Table 1. Table 1 also presents the results of 1H-coupled 19F NMR measurements, which were performed to unequivocally identify the various 19F NMR chemical shift values of the fluoromuconates. The F-H and F-F coupling patterns derived from the 1H-coupled 19F NMR measurements identify the position of the fluorine substituents in the different muconates. Although at first glance the coupling constants for the various muconates might seem inconsistent at some points, comparison to J(F-H) and J(F-F) values reported in the literature for similar compounds (11, 35) support the present assignments. This comparison to the literature data (11, 27, 35) also illustrates that the configuration and conformation of the muconate strongly affect the size of the coupling constants observed. Thus, especially the relatively large 3J(F-F) coupling constant observed in 2,3,5-trifluoromuconate seems to deviate from the much smaller 3J(F-F) coupling constant in 2,3-difluoromuconate and 2,3,4-trifluoromuconate. Based on the very large difference in such 3J(F-F) coupling constants for other cis and trans RFC=CFR′ isomers (35), one might suggest that, especially in the 2,3,5-trifluoromuconate, cis-trans isomerization had occurred. Alternatively the relatively large 3J(F-F) in 2,3,5-trifluoromuconate may point to a very unusual geometry of the cis isomer (11). However, this phenomenon was not further investigated in the present study.
TABLE 1.
Chemical shift values of fluorinated muconate metabolites formed from fluorophenols by purified phenol hydroxylase and purified catechol 1,2-dioxygenasea
| Muconate | Chemical structure | Fluorophenol(s) muconate formed from | Chemical shift (ppm)b | Coupling assignment | Coupling constant (Hz) |
|---|---|---|---|---|---|
| 2-Fluoromuconate | 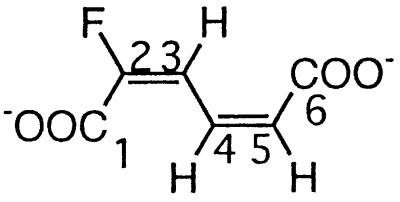 |
2- and 3-Fluorophenol and 2,3- and 2,6difluorophenol | −111.8 (F-2) | 3J (2-F, 3-H) | 20.7 |
| 3-Fluoromuconate | 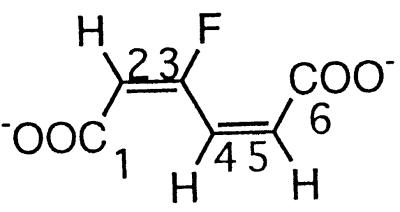 |
3- and 4-Fluorophenol and 2,4- and 2,5difluorophenol | −108.2 (F-3) | 3J (3-F, 2-H) | 21.7 |
| 3J (3-F, 4-H) | 32.9 | ||||
| 2,3-Difluoromuconate | 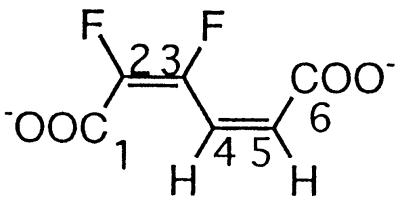 |
2,3- and 3,4-Difluorophenol and 2,3,6-trifluorophenol | −134.5 (F-3) | 3J (3-F, 2-F) | 2.7 |
| 3J (3-F, 4-H) | 31.6 | ||||
| −142.3 (F-2) | 3J (2-F, 3-F) | 2.7 | |||
| 2,4-Difluoromuconate | 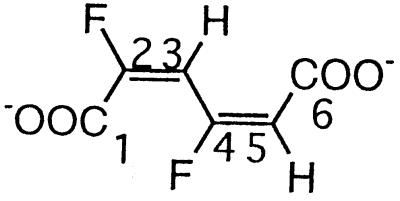 |
2,4- and 3,5-Difluorophenol and 2,3,5-trifluorophenol | −102.5 (F-4) | 3J (4-F, 3-H) | 25.2 |
| 3J (4-F, 5-H) | 21.4 | ||||
| −107.4 (F-2) | 3J (2-F, 3-H) | 21.0 | |||
| 2,5-Difluoromuconate | 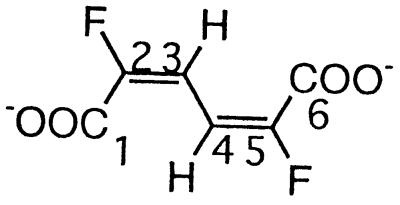 |
2,5-Difluorophenol and 2,3,6-trifluorophenol | −111.1 (F-2/5) | 3J (2/5-F, 3/4-H) | 11.5 |
| 4J (2/5-F, 4/3-H) | 6.8 | ||||
| 3,4-Difluoromuconatec | 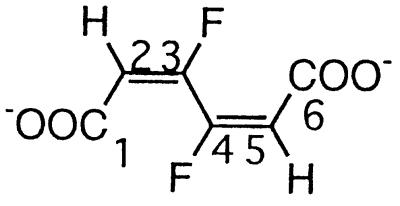 |
3,4-Difluorophenol and 2,4,5-trifluorophenol | −121.1 (F-3/4) | 3J (3/4-F, 2/5-H) | 15 |
| 3J (3-F, 4-F) | 8 | ||||
| 4J (3/4-F, 5/2-H) | 3 | ||||
| 2,3,4-Trifluoromuconate | 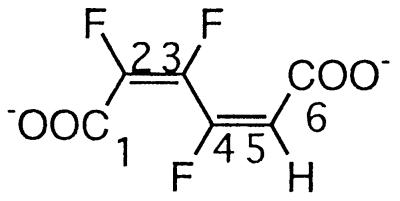 |
2,3,4- and 3,4,5-Trifluorophenol | −106.2 (F-4) | 3J (4-F, 3-F) | 39.0 |
| 3J (4-F, 5-H) | 16.0 | ||||
| 4J (4-F, 2-F) | 7.6 | ||||
| −131.4 (F-3) | 3J (3-F, 2-F) | 7.3 | |||
| 3J (3-F, 4-F) | 39.0 | ||||
| −137.0 (F-2) | 3J (2-F, 3-F) | 7.3 | |||
| 4J (2-F, 4-F) | 7.6 | ||||
| 2,3,5-Trifluoromuconate | 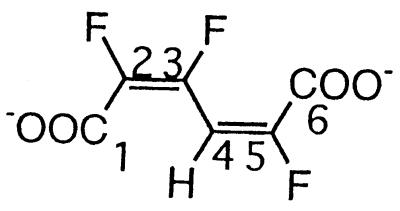 |
2,3,5- and 2,4,5-Trifluorophenol and 2,3,5,6-tetrafluorophenol | −106.9 (F-3) | 3J (3-F, 2-F) | 30.6 |
| 3J (3-F, 4-H) | 20.7 | ||||
| −127.5 (F-5) | 3J (5-F, 4-H) | 23.7 | |||
| −141.9 (F-2) | 3J (2-F, 3-F) | 30.6 | |||
| Tetrafluoromuconated | 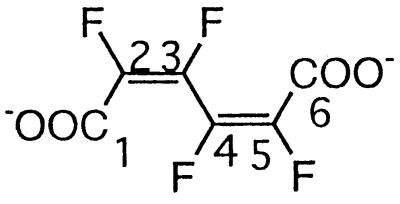 |
Pentafluorophenol | −130.2 (F-3/4) | 3J (3-F, 2-F) | Small |
| 3J (3-F, 4-F) | Small | ||||
| 4J (3-F, 5-F) | Small | ||||
| 3J (4-F, 5-F) | Small | ||||
| 4J (4-F, 2-F) | Small | ||||
| −134.5 (F-2/5) | 5J (2-F, 5-F) | Small |
Values were determined in 0.1 M potassium phosphate (pH 7.6) at 7°C.
Chemical shift relative to CFCl3.
The 19F NMR (1H decoupled) NMR spectrum is of the AA′XX′ type. The splitting pattern has been analyzed by using a formal description as described by Günther (9). The coupling constants have, as a result, been determined to be less accurate.
The 19F NMR spectra of this compound with and without 1H decoupling are identical. A formal calculation of the observed splitting pattern reveals that the F-F coupling constants all must be very small.
Conversion of fluorophenol derivatives in cell extracts and incubations with whole cells as determined by 19F NMR.
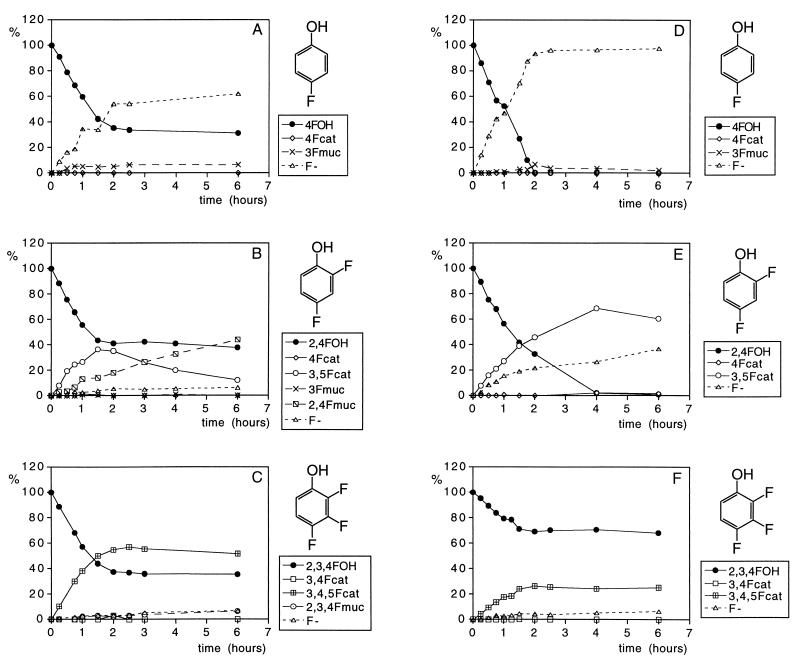
Together, the data presented in the literature and Table 1 provide a basis with which to study the conversion of fluorinated phenolic derivatives by 19F NMR in cell extracts as well as their conversion in vivo in whole-cell cultures. Results obtained to illustrate this are presented in Fig. 2. Although Fig. 2 only presents the metabolic profiles of three of the seven fluorophenols studied, the metabolic profiles presented are representative examples also for the other fluorophenols. Thus, the time-dependent changes in the metabolic profiles of 2-fluorophenol are similar to those presented for 2,4-difluorophenol (Fig. 2B and E), those of 3-fluorophenol and 3,4-difluorophenol are similar to those presented for 4-fluorophenol (Fig. 2A and D), and those of 3,4,5-trifluorophenol resemble the patterns presented for 2,3,4-trifluorophenol (Fig. 2C and F). From the degradation patterns obtained with cell extracts (see Fig. 2A to C for representative examples), it can be derived that the rate of conversion of fluorophenols is linear in time for at least 1 h and ceases after 2 h of incubation, which appeared to be due to NADPH limitation. As a result, catechol formation by the NADPH-dependent phenol hydroxylase stops, whereas conversion of the catechols to the muconates, catalyzed by the NADPH-independent catechol 1,2-dioxygenase, is still observed for at least 6 h. In addition to biodegradation by cell extracts, degradation patterns of the fluorophenols by whole cells of E. jeanselmei were determined. Some representative metabolite profiles obtained are shown in Fig. 2 D to F. In these studies, no lack of cofactor is observed, and the phenol degradation continues beyond the hour. Comparison of the metabolic profiles obtained with cell extracts and whole-cell cultures shows that the metabolic profiles in both systems are similar, except for the level of muconates that can be detected. Clearly cells do not efficiently excrete the muconate intermediates into the culture medium, resulting in relatively lower levels of muconates in culture media than those observed in incubations with cell extracts.
FIG. 2.
Time-dependent changes in the metabolic profile of incubations with 4-fluorophenol (A and D), 2,4-difluorophenol (B and E), and 2,3,4-trifluorophenol (C and F), where panels A to C show the profiles of incubations with cell extracts and panels D to F show the profiles of incubations with whole cells of E. jeanselmei. Metabolites were identified and quantified by 19F NMR. 4FOH, 4-fluorophenol; 4Fcat, 4-fluorocatechol; 3Fmuc, 3-fluoromuconate; F−, fluorine.
From the data in Fig. 2, it can also be derived that for E. jeanselmei, degradation of the fluorophenols becomes increasingly difficult with an increasing number of fluorine substituents.
Moreover, the metabolic profiles also indicate that for phenols with a fluorine substituent ortho with respect to the hydroxyl moiety, and/or phenols providing possibilities for formation of an ortho fluorocatechol, the biodegradation results in accumulation of the respective catechol and muconate. Only for 3-fluoro-, 4-fluoro-, and 3,4-difluorophenol, in which the main catechol metabolite does not contain a fluorine ortho with respect to the hydroxyl groups of the catechol, is swift biodehalogenation observed. This results in fluoride anion formation and the absence of significant accumulation of the catechol and muconate intermediates. The fact that, in spite of this efficient biodehalogenation, no significant growth of E. jeanselmei is detected on 3-fluoro-, 4-fluoro-, and 3,4-difluorophenol points at cometabolism.
Thus, the present 19F NMR method provides information on, not only catechol and muconate metabolites formed and the metabolic pathway involved, but also the extent of dehalogenation (i.e., the amount of fluoride anion formation). Clearly this is an advantage of the 19F NMR method over HPLC analysis, in which formation of the halide anion is not registered simultaneously. Furthermore, this method provides a possibility for studying metabolic pathways with intact cells in vivo.
DISCUSSION
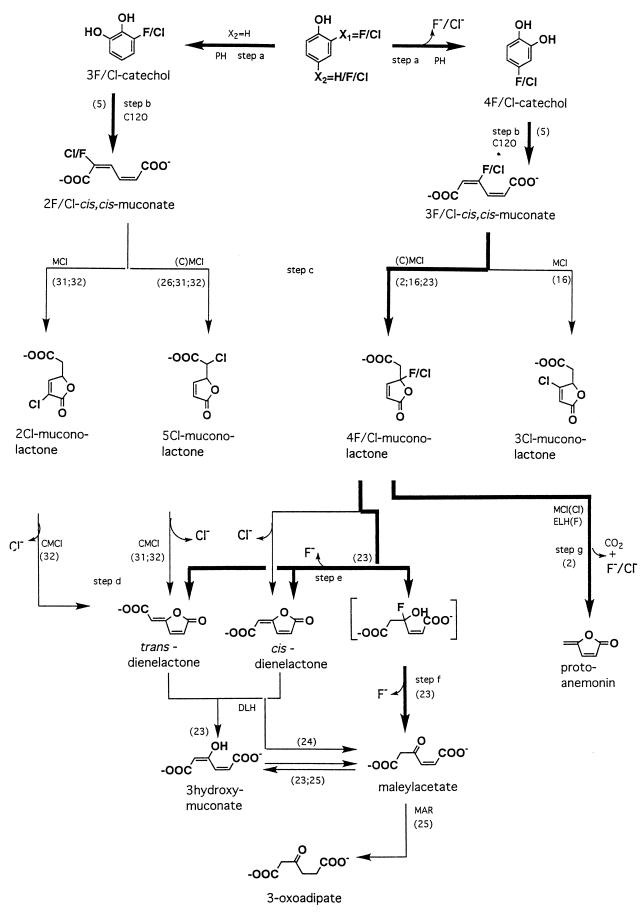
Most studies of the microbial degradation of halogenated phenolic compounds have been performed with bacteria. Up to now, only two yeast strains, T. cutaneum and Candida tropicalis, have been shown to metabolize monosubstituted phenols by the initial steps of the 3-oxoadipate pathway, analogous to bacterial degradation of monochlorophenols (8, 12, 14). The present study, however, focuses on the biodegradation of fluorophenols as measured by 19F NMR, and it is important to stress that the routes for fluorophenol degradation may vary considerably from those responsible for chlorophenol degradation by the same organism (2, 6, 8, 16, 23–25). Figure 3 summarizes the various metabolic routes described in the literature for biodehalogenation of halophenols, with special emphasis on the routes identified so far for fluorophenols, as well as the routes proven to be relevant for conversion of fluorophenols by E. jeanselmei in the present study.
FIG. 3.
Pathways for the biodegradation and oxidative biodehalogenation of phenol and its halogenated analogs. Numbers in parentheses refer to literature references. The enzymes involved are indicated as follows: PH, phenol hydroxylase; C12O, catechol 1,2-dioxygenase; (C)MCl, (chloro)muconate cycloisomerase; DLH, dienelactone hydrolase; ELH, enol-lactone hydrolase; MAR, maleylacetate reductase. Pathways identified so far for fluorophenols, either in the literature or in the present study for E. jeanselmei, are indicated in boldface.
E. jeanselmei revealed a broad substrate specificity and was capable of transforming phenols, containing up to four fluorine substituents, to their corresponding catechols. However, when one or both of the carbon atoms positioned ortho with respect to the hydroxyl moieties of the catechol formed were fluorinated, the further biodegradation of the catechol and muconates was inhibited. Thus, catechol 1,2-dioxygenase of E. jeanselmei exhibited rather high activities only towards 4-fluoro- and 4,5-difluorocatechols, but low-level activities towards 3-fluoro-, 3,4-difluoro-, 3,5-difluoro-, and 3,4,5-trifluorocatechols. This impairment of ring cleavage by the presence of a halogen substituent ortho with respect to the hydroxyl moieties of a ring cleavage metabolite is similar to the inhibition of protocatechuate dioxygenase by ortho-halogenated protocatechuates (33) and of catechol 2,3-dioxygenase by ortho-halogenated catechols (1). The phenomenon was reported to originate from formation of an abortive complex between the 3-halocatechol and the dioxygenase (34).
The 19F NMR data of the present study also provide information on the various possibilities and pathways chosen by the yeast-like fungus to take care of the biodehalogenation of fluorophenols. When the fluorine was in the ortho position with respect to the hydroxyl moiety of the phenolic substrate (i.e., 2-fluoro-, 2,4-difluoro-, and 2,3,4-trifluorophenol), the phenol hydroxylase of E. jeanselmei catalyzed C-2 hydroxylation accompanied by dehalogenation and fluoride anion formation in addition to hydroxylation at the nonfluorinated C-6 position. A similar reactivity was reported previously for conversion of C-2-fluorinated phenols by phenol hydroxylase of T. cutaneum (19). In case of hydroxylation at C-6, biodegradation ceases at the level of the 3-fluorocatechol and/or 2-fluoromuconate. In contrast, when oxidative dehalogenation occurred, further metabolism of the catechols could result in release of additional fluoride anions as long as the fluorine substituents were not at a position ortho with respect to the hydroxyl moieties of the catechol formed.
The conversion of fluorophenols that gave rise to preferential formation of catechols without ortho fluorine substituents (i.e., 3-fluoro-, 4-fluoro-, and 3,4-difluorophenol) showed only transient formation of fluorinated catechols and muconates. Instead of bioaccumulation of the fluorocatechols and fluoromuconates, swift accumulation of stoichiometric amounts of fluoride anions was detected, apparently due to fluoride anion elimination in reactions following ring cleavage by the catechol 1,2-dioxygenase. Such dehalogenation reactions could be either one of the steps c, d, e, f, or g, (Fig. 3) proven previously to occur in most cases, especially for the chloromuconate analogs. For 3,4-difluorophenol, such a dehalogenation of the corresponding difluoromuconate implies the formation of a fluorinated muconolactone and/or fluoromaleylacetate. However, neither fluoromuconolactone nor fluoromaleylacetate was detected in the incubation media, apparently due to elimination of the fluorine substituent from 3-fluoromaleylacetate. Although maleylacetate reductase is known to catalyze the dehalogenation of 2-fluoromaleylacetate (13), the exact pathway for dehalogenation of 3-fluoromaleylacetate remains to be elucidated.
Clearly the only fluorophenol for which efficient dehalogenation was observed in the present study, was 4-fluorophenol. However, in spite of the fact that 80 to 100% of the 4-fluorophenol lost could be accounted for by fluoride anions, indicating efficient biodehalogenation, no growth of E. jeanselmei on 4-fluorophenol was observed. This indicates that the biodegradation pathway of 4-fluorophenol is also blocked at some level. When 4-fluorophenol is degraded, the fluorine most probably is eliminated in the reaction steps leading to formation of a cis and/or trans dienelactone (2, 16, 23, 24) (step c or e, Fig. 3). Subsequent conversion to 3-oxoadipate requires enzymes like dienelactone hydrolase and maleylacetate reductase that are apparently absent in E. jeanselmei.
Altogether, the results presented in this paper clearly illustrate that 19F NMR analysis can provide valuable information about the microbial degradation and biodehalogenation of fluorinated aromatic compounds. This corroborates the findings in a previous, rather preliminary, 19F NMR study of the degradation of 2,5-difluoro- and 3,5-difluorobenzoate by Pseudomonas putida JT103 (4). The present work provides a large series of 19F NMR chemical shift data on fluorophenols, fluorocatechols, and fluoromuconates and new data on the degradation and (co)metabolism of fluorophenols by the yeast-like fungus E. jeanselmei. Furthermore, the results clearly show that 19F NMR analysis is effective (i) in identification of fluorinated intermediates and/or dead-end products of biodegradation pathways; (ii) in providing not only qualitative but also quantitative information, with a sensitivity up to a 1 μM concentration of the fluorinated compounds; (iii) in following the pathway and extent of biodehalogenation, giving, in contrast to HPLC, information about the amount of the eliminated halogen anion; (iv) in determination of the rate-limiting steps in microbial pathways of fluoroaromatic conversion; and (v) in providing possibilities for in vivo measurements in whole cells, which may be especially of interest for organisms with labile biodegradation enzymes.
ACKNOWLEDGMENTS
This work was supported by the EU large-scale WNMRC facility (grant ERBCHGECT 940061), the Research School of Environmental Chemistry and Toxicology (M&T), the EU-COPERNICUS program (grant IC15-CT96-0103), and NATO grant ENVIR.LG960907.
REFERENCES
- 1.Bartels I, Knackmuss H-J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco R, Wittich R-M, Mallavarapu M, Timmis K N, Pieper D H. From xenobiotic to antibiotic, formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J Biol Chem. 1995;270:29229–29235. doi: 10.1074/jbc.270.49.29229. [DOI] [PubMed] [Google Scholar]
- 3.Cabanac S, Marlet-Martino M C, Bon M, Martino R, Nedelec J F, Dimicoli J L. Direct 19F NMR spectroscopic observation of 5-fluorouracil metabolism in the isolated perfused mouse liver model. NMR Biomed. 1988;1:113–120. doi: 10.1002/nbm.1940010303. [DOI] [PubMed] [Google Scholar]
- 4.Cass A E G, Ribbons D W, Rossiter J T, Williams S R. Biotransformation of aromatic compounds. Monitoring fluorinated analogues by NMR. FEBS Lett. 1987;220:353–357. doi: 10.1016/0014-5793(87)80845-1. [DOI] [PubMed] [Google Scholar]
- 5.Dorn E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol-1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem J. 1978;174:73–84. doi: 10.1042/bj1740073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engesser K H, Fischer P. Degradation of haloaromatic compounds. In: Betts W B, editor. Biodegradation: natural and synthetic materials. London, United Kingdom: Springer-Verlag; 1991. pp. 15–54. [Google Scholar]
- 7.Evans W C, Smith B S W, Moss P, Fernley H N. Bacterial metabolism of 4-chlorophenoxyacetate. Biochem J. 1971;122:509–517. doi: 10.1042/bj1220509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaal A, Neujahr H Y. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J Bacteriol. 1979;137:13–21. doi: 10.1128/jb.137.1.13-21.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Günther H. NMR spectroscopy. Basic principles, concepts, and applications in chemistry. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1995. pp. 136–198. [Google Scholar]
- 10.Haigler B E, Nishino S F, Spain J C. Degradation of 1,2-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1988;54:294–301. doi: 10.1128/aem.54.2.294-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes D W, Bain A D. NMR analysis of fluorinated corticosteriods related to fluocinonide: detection of long-range 1H-19F couplings using the heteronuclear equivalent of the COSY experiment. Magn Reson Chem. 1991;29:387–397. [Google Scholar]
- 12.Ivoilov V S, Karasevich Y N. Monochlorophenols as substrates of the enzymes of preparatory phenol metabolism in the yeast Candida tropicalis. Mikrobiologiya. 1983;52:956–961. [PubMed] [Google Scholar]
- 13.Kaschabek S R, Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: specificity of substrate conversion and halide elimination. J Bacteriol. 1995;177:320–325. doi: 10.1128/jb.177.2.320-325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krug M, Straube G. Degradation of phenolic compounds by the yeast Candida tropicalis HP 15. Some properties of the first two enzymes of the degradation pathway. J Basic Microbiol. 1986;26:271–281. doi: 10.1002/jobm.3620260505. [DOI] [PubMed] [Google Scholar]
- 15.Malet-Martino M C, Martino R. The application of nuclear magnetic resonance spectroscopy to drug metabolism studies. Xenobiotica. 1989;19:583–607. doi: 10.3109/00498258909042297. [DOI] [PubMed] [Google Scholar]
- 16.Mazur P, Pieken W A, Budihas S R, Williams S E, Wong S, Kozarich J W. Cis,cis-muconate lactonizing enzyme from Trichosporon cutaneum: evidence for a novel class of cycloisomerases in eucaryotes. Biochemistry. 1994;33:1961–1970. doi: 10.1021/bi00173a045. [DOI] [PubMed] [Google Scholar]
- 17.Middelhoven W J. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi. A literature review and an experimental approach. Antonie Leeuwenhoek. 1993;63:125–144. doi: 10.1007/BF00872388. [DOI] [PubMed] [Google Scholar]
- 18.Nakai C, Nakazawa T, Nozaki M. Purification and properties of catechol 1,2-dioxygenase (pyrocatechase) from Pseudomonas putida mt-2 in comparison with that from Pseudomonas arvilla C-1. Arch Biochem Biophys. 1988;267:701–713. doi: 10.1016/0003-9861(88)90079-3. [DOI] [PubMed] [Google Scholar]
- 19.Peelen S, Rietjens I M C M, Boersma M G, Vervoort J. Conversion of phenol derivatives to hydroxylated products by phenol hydroxylase from Trichosporon cutaneum. A comparison of regioselectivity and rate of conversion with calculated molecular orbital substrate characteristics. Eur J Biochem. 1995;227:284–291. doi: 10.1111/j.1432-1033.1995.tb20386.x. [DOI] [PubMed] [Google Scholar]
- 20.Reineke W, Knackmuss H-J. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol. 1984;47:395–402. doi: 10.1128/aem.47.2.395-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reineke W, Knackmuss H-J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- 22.Rietjens I M C M, Cnubben N H P, de Jager P A, Boersma M G, Vervoort J. Application of NMR in biotransformation studies. In: Weitzer M I, editor. Toxicology: molecules to morals. Melksham, United Kingdom: Redwood Press; 1993. pp. 94–109. [Google Scholar]
- 23.Schlömann M, Fischer P, Schmidt E, Knackmuss H-J. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J Bacteriol. 1990;172:5119–5129. doi: 10.1128/jb.172.9.5119-5129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Biochem J. 1980;192:339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seibert V, Stadler-Fritzsche K, Schlömann M. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4) J Bacteriol. 1993;175:6745–6754. doi: 10.1128/jb.175.21.6745-6754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solyanikova I P, Maltseva O V, Vollmer M D, Golovleva L A, Schlömann M. Characterization of muconate and chloromuconate cycloisomerase from Rhodococcus erythropolis 1CP: indications for functionally convergent evolution among bacterial cycloisomerases. J Bacteriol. 1995;177:2821–2826. doi: 10.1128/jb.177.10.2821-2826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spahic B, Schlosser M. Die Stereochemie der fragmentierenden Enthalogenierung von 1-Chlor-1-fluor-2-(1-halogenalkyl)cyclopropanan. Helv Chim Acta. 1980;63:1242–1256. [Google Scholar]
- 28.Spain J C, Nishino S F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987;53:1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vervoort J, de Jager P A, Steenbergen J, Rietjens I M C M. Development of a 19F-NMR method for studies on the in vivo and in vitro metabolism of 2-fluoroaniline. Xenobiotica. 1990;20:657–670. doi: 10.3109/00498259009046882. [DOI] [PubMed] [Google Scholar]
- 30.Vollmer M D, Stadler-Fritzsche K, Schlömann M. Conversion of 2-chloromaleylacetate in Alcaligenes eutrophus JMP134. Arch Microbiol. 1993;159:182–188. doi: 10.1007/BF00250280. [DOI] [PubMed] [Google Scholar]
- 31.Vollmer M D, Fischer P, Knackmuss H-J, Schlömann M. Inability of muconate cycloisomerases to cause dehalogenation during conversion of 2-chloro-cis,cis-muconate. J Bacteriol. 1994;176:4366–4375. doi: 10.1128/jb.176.14.4366-4375.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vollmer M D, Schlömann M. Conversion of 2-chloro-cis,cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerases of pJP4 and pAC27. J Bacteriol. 1995;177:2938–2941. doi: 10.1128/jb.177.10.2938-2941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh T A, Ballou D P. Halogenated protocatechuates as substrates for protocatechuate dioxygenase from Pseudomonas cepacia. J Biol Chem. 1983;258:14413–14421. [PubMed] [Google Scholar]
- 34.Walsh T A, Ballou D P, Mayer R, Que L., Jr Rapid reaction studies on the oxygenation reactions of catechol dioxygenase. J Biol Chem. 1983;258:14422–14427. [PubMed] [Google Scholar]
- 35.Wray V. Fluorine-19 nuclear magnetic resonance spectroscopy. Annu Rep NMR Spectrosc. 1983;14:179–191. [Google Scholar]