Abstract
Artificial intelligence (AI) in radiology is a rapidly developing field with several prospective clinical studies demonstrating its benefits in clinical practice. In 2022, the Korean Society of Radiology held a forum to discuss the challenges and drawbacks in AI development and implementation. Various barriers hinder the successful application and widespread adoption of AI in radiology, such as limited annotated data, data privacy and security, data heterogeneity, imbalanced data, model interpretability, overfitting, and integration with clinical workflows. In this review, some of the various possible solutions to these challenges are presented and discussed; these include training with longitudinal and multimodal datasets, dense training with multitask learning and multimodal learning, self-supervised contrastive learning, various image modifications and syntheses using generative models, explainable AI, causal learning, federated learning with large data models, and digital twins.
Keywords: Artificial intelligence, Challenges, Data privacy, Innovative datasets, Novel techniques
INTRODUCTION
Deep learning (DL), a type of artificial intelligence (AI), is increasingly used in radiology and clinical practice [1]. The effectiveness of DL methods in classifying images and detecting pathological lesions in a supervised manner has been successfully demonstrated in various fields of medicine [2,3,4,5,6,7,8,9,10,11]. Numerous studies have focused on developing and validating DL models in radiology, which includes neurological, thoracic, abdominal, and breast imaging. With an accuracy comparable to that of human experts, convolutional neural networks (CNNs) [12,13] and transformer networks [14] typically classify images based on the presence or absence of disease. In addition, from previously diagnosed lesions, CNNs can extract information regarding morphological characteristics of the lesion, such as size, shape, or outline, as in the case of breast lesions or chest nodules [15,16,17,18]. Therefore, many computer-aided diagnosis (CADx) and detection (CADe) software that utilize DL methods have received approval from the Food and Drug Administration (FDA) and have been successfully commercialized [19]; moreover, the software can offer other advantages, including speed, efficiency, low cost, increased accessibility, and the upkeep of faithful behavior in clinical practice. In prospective studies, early-career radiologists and physicians have noted performance improvements using DL models [20,21,22,23]. Despite the promising results of current AI technology, numerous challenges exist in the development and deployment of supervised learning-based AI systems. First, a major obstacle is the requirement of a huge volume of high-quality medical imaging data to achieve good AI performance [24]. This demanding requirement can be attributed to the high-dimensional nature of the data and the presence of noise and irrelevant information [25], which pose challenges in extracting high-level features. Even expert physicians may encounter difficulties in labeling data with high quality owing to the high cost involved, occasional ambiguity in the ground truth information, tedious nature of the labeling task, and risks to patient privacy. Secondly, although numerous radiologic tasks are performed in actual clinical settings, most radiologic applications are designed for specific clinical situations, and consequently, broad-range radiologic tasks are overlooked. Third, DL models often overfit training data, leading to poor performance when applied to new data [26,27]. Specifically, variations in imaging protocols, manufacturers, and patient populations can lead to significant variability in the appearance and semantics of images [24], making it difficult to develop robust models in response to these variations. In general, most radiology datasets are long-tailed, which can lead to underrepresented classes and biases in the trained model [28]. Fourth, DL models are often considered as “black boxes” [29] regarding model interpretability, which may negatively affect the use of AI in clinical decision-making. Radiology is a rapidly evolving field, and DL models require frequent retraining to keep pace with the latest advancements in imaging techniques and new types of data [24]. This review presents and discusses various potential solutions for addressing the challenges of supervised AI.
A Quick Primer before We Start
Representation Learning
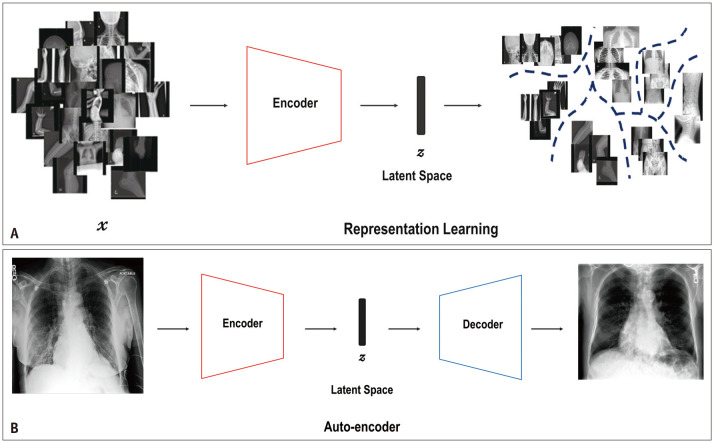
The concepts of representation learning, autoencoder, encoder, decoder, and latent space are presented in Figure 1 using an example. Representation learning allows a model to automatically map the representation space (z) needed for detection or classification of raw data (x). z is a vector in the latent space with smaller dimensions than the input (x). In this example, the model was trained to differentiate between the X-ray images of different body parts, including those of the head, chest, abdomen, spine, pelvis, and upper and lower extremities. Therefore, after training, the model can be used as an encoder to map the input modality (X-ray images, denoted as x) into different latent spaces, where the locations represent the X-ray images of different body parts (Fig. 1A). An early example of representation learning is auto-encoder. An autoencoder is composed of two components—an encoder and a decoder—and can be used to generate synthetic data. For instance, the encoder maps the input modality into the latent vector (z), and then, based on the vector z, the decoder generates a synthetic sample X-ray image of the input body part (Fig. 1B) [30].
Fig. 1. A conceptual diagram of representation learning and auto-encoder. A: Representation learning implies training an encoder to automatically map into the representations space, z, needed for detection or classification from input modality (e.g., X-ray images), x, where z is a vector in the latent space with smaller dimensions than those of input modality, x. This model was trained to differentiate X-ray images of different body parts, including those of the head, chest, abdomen, spine, pelvis, and upper and lower extremities. B: Auto-encoder is mainly composed of two components: an encoder and a decoder. The encoder maps the input modality into a latent vector, z, and then, based on the vector z, the decoder generates a novel sample of the target modality.
Generative Models
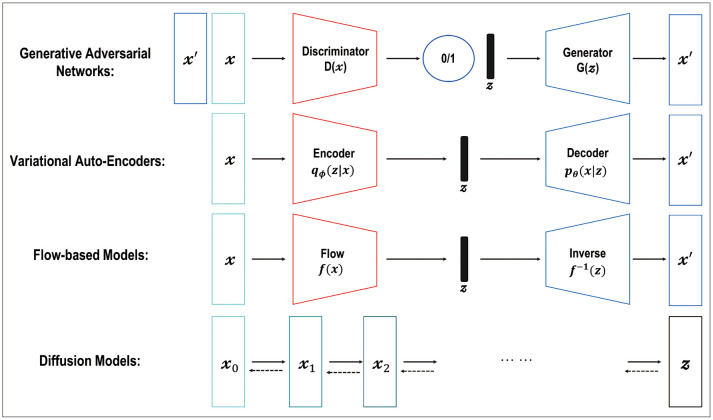
Various types of generative models exist, such as generative adversarial networks (GANs), variational autoencoders (VAEs), flow-based models, and diffusion models, which are a class of machine learning algorithms that aim to learn the underlying data distribution of a given dataset, enabling them to generate new data samples that resemble the original data (Fig. 2) [31,32,33,34,35,36]. Each model is based on different theories—such as adversarial training [31,32,36], maximization of the variational lower bound [35], invertible transformation of distribution [34], and mimicking the Markov chain of diffusion steps to slowly add random noise to the data—and then the models learn to reverse the diffusion process to construct the desired data samples from the noise [33]. Among them, the GAN and diffusion models have been successfully applied in radiology research, such as image denoising [37,38,39], image reconstruction [40,41,42], intermodality image synthesis [43,44,45], improved image segmentation [46,47,48,49], image registration [50,51], classification [52,53], anomaly detection [54,55,56,57,58,59,60,61], and disease progression modeling [62].
Fig. 2. Typical examples of generative models include generative adversarial networks, variational auto-encoder, flow-based models, and diffusion models with different theories—such as adversarial training, maximization of the variational lower bound, invertible transformation of distribution, and mimicking the Markov chain of the diffusion steps to slowly add random noise to data—and then the models learn to reverse the diffusion process to construct desired data samples from the noise.
Overcoming the Challenges
Longitudinal and Multi-Modal Dataset
In real-world radiology settings, images are acquired via multiple modalities and time points that can be combined and analyzed. For example, a multiscale and multimodal deep neural network classifier built with a combination of fluorodeoxyglucose-positron emission tomography (FDG-PET) and structural magnetic resonance imaging (MRI) showed performance improvement [63]. Moreover, a disease progression model using longitudinal data with varying time intervals can improve robustness against missing data and performance in Alzheimer’s disease [64]. Multiple follow-ups with repeated measurements are the norm in clinical practice, thereby affecting an increase in the statistical power [65,66]. In the event of incomplete data and/or limited knowledge of the disease and pathogenesis, multimodal data, such as various types of imaging modalities, demographics, laboratory tests, and electronic medical records (EMRs), may prove crucial to achieve an improved diagnosis [67,68]. Therefore, for better applicability in clinical settings, longitudinal and multimodal datasets should be considered when training AI-based models. However, it is important to acknowledge that although these datasets can enhance model performance, they do not eliminate the need to address the inherent challenges of DL models, such as handling long-tailed distributions and improving model explainability. In addition, the utilization of these datasets can present significant risks to patient privacy. Therefore, it is crucial to utilize these datasets in conjunction with the following strategies aimed at addressing the fundamental challenges of DL models.
Dense Training with Multi-Task Learning and Multi-Modal Learning
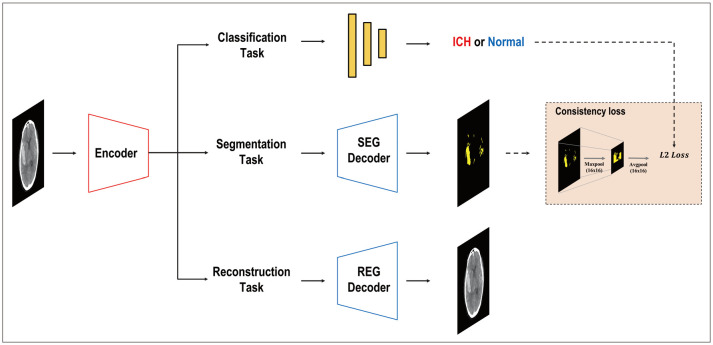
Dense training with multitask learning (MTL) and multimodal learning offers several notable advantages, particularly in the field of radiology. Multimodal learning enables the integration of diverse data sources such as clinical information, imaging modalities, and patient demographics, potentially enhancing the overall predictive power of AI models [69,70,71,72,73,74,75]. By collectively incorporating multiple factors, the accuracy and reliability of the diagnoses can be improved. Furthermore, joint training of models on various tasks using MTL can facilitate the learning of shared representations and leverage correlations among different modalities, which may contribute to improved performance across multiple domains [76]. Additionally, dense training with MTL can offer regularization effects, which helps to prevent overfitting and potentially enhances the generalization capability of the model [77], thereby encouraging the network to learn relevant and robust features simultaneously for multiple tasks, which may translate into better performance on individual tasks. Moreover, this approach may improve the interpretability and explainability. When models are trained to perform multiple tasks, they are forced to capture meaningful and discriminative features, potentially enhancing the transparency of the decision-making processes. Such transparency can be valuable in building trust among radiologists and other healthcare professionals, as it enables them to understand and interpret the underlying factors contributing to the model’s predictions. Recently, different MTL architectures have emerged, and based on where information or features are exchanged within a network, encoder-focused [78,79,80,81] and decoder-focused [82,83,84] designs, among others, are utilized. Many studies have focused on AI in radiology because of the complexities involved in training owing to high-dimensional data and the limitations of data acquisition. These radiology studies demonstrated the superiority of the MTL over other models; for instance, MTL has shown better performance than alternative approaches in various tasks, such as segmenting thoracic organs from computed tomography (CT) slices [85]; object detection, segmentation, and classification in breast cancer diagnosis using full-field digital mammogram datasets [86]; identification and segmentation of coronavirus disease 2019 (COVID-19) lesions from chest CT images [87]; and identification of hemorrhage and segmentation in head and neck CT images [88] (Fig. 3). However, the extent to which multimodality enhances the robustness to distribution shifts, promotes patient privacy, and addresses other limitations remains a topic of ongoing research.
Fig. 3. A typical case of hemorrhage detection with multi-task learning in head and neck computed tomography images, including classification, segmentation, and reconstruction with consistency loss. Modified from Kyung et al. Med Image Anal 2022;81:102489 [88], under the permission from Elsevier. SEG = segmentation, REG = registration, ICH = intracerebral hemorrhage.
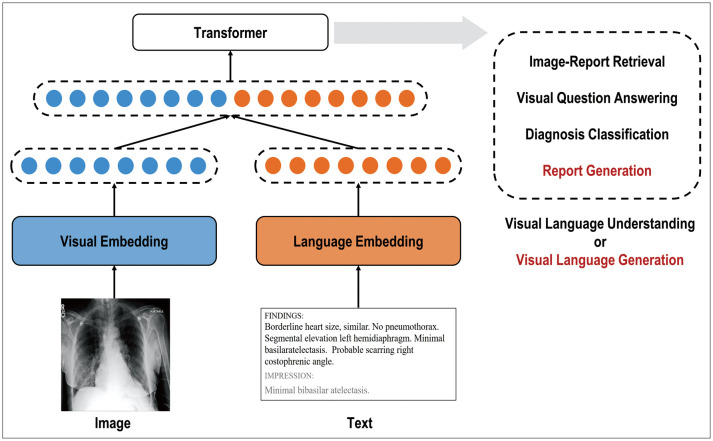
Vision-language (VL) multimodal learning—a prominent area of multimodal research that utilizes radiographic images and their associated free-text descriptions, such as chest radiographs (CXRs) and their reports—is becoming increasingly significant [12,89] (Fig. 4). Images and paired radiology reports provide mutually beneficial semantic information to enhance the training quality of various tasks such as diagnosis classification and report generation in radiology. However, significant technical challenges are encountered in processing both images and clinical reports to learn joint representations. These challenges arise from differences in the high dimensionality, heterogeneity, and systemic biases of multimodal datasets. Recent progress in VL multimodal learning was achieved by extending bidirectional encoder representations from transformer-based architectures (BERT) [90]. Previous studies can be broadly categorized into vision-language understanding (VLU) tasks, which include visual question answering, text-conditioned image retrieval, diagnosis classification, and vision-language generation (VLG) tasks such as image captioning and report generation [91,92]. Numerous studies have explored the applications of VL multimodal learning in radiology. Hsu et al. [93] focused on a VLU task, specifically, image report retrieval, using both supervised and unsupervised methods. Liu et al. [94] focused solely on the VLG task of radiology reports by implementing a CNN-recurrent neural network (RNN) architecture and a hierarchical generation strategy. Liu et al. [95] proposed a transformer encoder-decoder-based approach that uses prior and posterior knowledge of distillation techniques using different modalities. Wang et al. [96] introduced a self-boosting framework with various modules using images and reports based on the collaboration of a primary generation task and an auxiliary image-report matching task. Yang et al. [97] developed MedWriter, which integrates a hierarchical retrieval mechanism to automatically extract reports and sentence-level templates. Recently, large language models (LLMs), such as BERT [90] and generative pre-trained transformer (GPT) [98], have been used to embed high-level semantics or generate expert-level reports in the near future.
Fig. 4. Multi-modal learning using chest X-ray images and clinical reports for visual language understanding or visual language generation tasks. Modified from Moon et al. IEEE J Biomed Health Inform 2022;26:6070-6080 [91], under the permission from IEEE.
Self-supervised Learning with Contrastive Learning as a Foundation Model
A foundation model is any model trained on a broad dataset, typically using large-scale self-supervision. It can consolidate information from various modalities and can subsequently be adapted, for example, through fine-tuning, for a wide range of downstream tasks. Examples of foundation models include the BERT, GPT, and contrastive language–image pre-training (CLIP) [99].
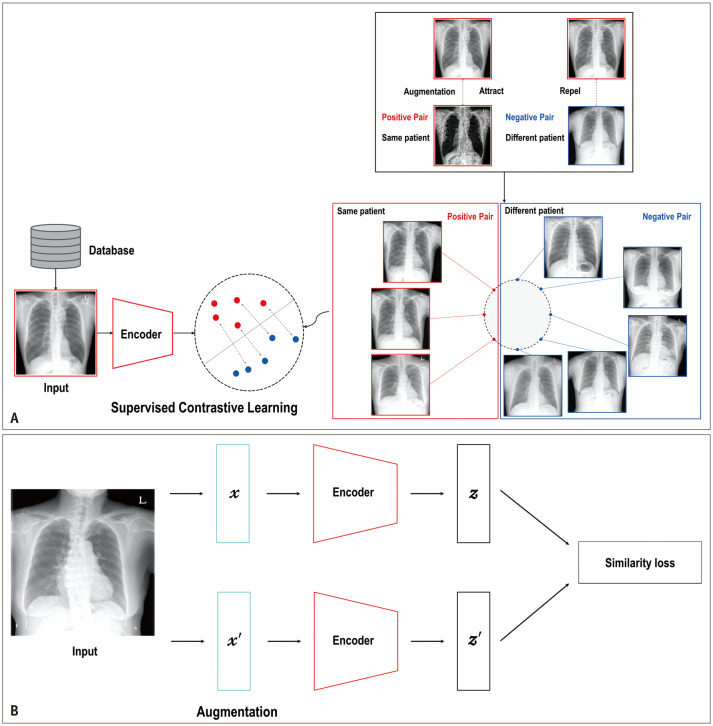
In general, contrastive learning (CL) is an approach used to learn representations in an unsupervised or self-supervised manner with the aim of bringing similar samples closer together while pushing dissimilar ones farther apart in the latent space (Fig. 5A). Automatic labeling without human effort is one of the most powerful approaches to unsupervised or self-supervised CL [100,101]. In medicine, CL can help train an encoder with large-scale medical images or other data, particularly in a self-supervised learning (SSL) manner, because SSL can automatically generate pseudo-labels based on different augmentations of the same or other images without human-annotated labels (Fig. 5B). The encoder can help differentiate between healthy and unhealthy individuals or identify specific diseases or conditions using transfer learning with a relatively small amount of annotated data. This approach is particularly useful for applications, such as CAD and image segmentation, which can assist radiologists in disease diagnosis by analyzing medical images and providing a list of potential diagnoses [102,103].
Fig. 5. A typical example of contrastive learning of supervised and self-supervised learning. A: Supervised contrastive learning with positive pairs of follow-up images of the same patient (red dots) and negative pairs of different patients (blue dots). B: Two different augmentations of medical images are randomly performed. Their embedding vectors are obtained through an image encoder to encourage a model to learn similar representations in the same class and dissimilar representations in different classes with similarity loss in a self-supervised manner.
Using CL, a neural network can identify abnormal brain structures on MRI scans, which can aid in the diagnosis of Alzheimer’s disease, differentiate between different types of images even in the case of image overlap, and improve the accuracy and efficiency of diagnosis by reducing the amount of time and effort required for manual interpretation of images [104]. This technique can aid in the early detection of diseases and provide timely treatment to patients. For medical image segmentation, to separate and identify different structures or tissues within an image, CL can be used to accurately detect segment-specific structures, such as the heart, head, and neck, in CT scans, thereby improving the accuracy of measurements and identifying abnormalities [105,106]. Additionally, contrastive unpaired image translation [107] can be used to synthesize pediatric CXRs, which can help improve the diagnosis of diseases within specific structures based on CT images [108]. Overall, CL has shown great promise as a tool for improving the accuracy and reliability of image analysis and diagnosis in a variety of medical settings.
Denoising, Fast Image Reconstruction, Inter-Modality Synthesis, and Synthetic Data Generation Using Generative Models
In radiology, the use of generative models have shown promising results, improving various aspects such as the image quality, reduction of radiation exposure, and shortened acquisition times. GANs have been particularly effective in CT applications such as denoising [37,38] and metallic artifact removal [39]. In addition, GANs have been explored as a means for artifact correction in accurate radiation therapy planning and as an image guide in cone-beam CT images [109,110]. In MRI, DL techniques have been employed to accelerate image acquisition and improve the image quality. Notably, GANs have demonstrated their superiority over traditional methods, such as compressed sensing MRI, in terms of faster reconstruction and enhanced image quality [40,41,42]. GANs have also been utilized to convert low-magnetic-field MRI into high-magnetic-field MRIs [111], enabling the generation of high-quality images, even with lower magnetic field strengths. This conversion has the potential to reduce the cost and complexity of the MRI systems. Generative models that facilitate the intermodality synthesis offer several advantages. This approach streamlines imaging workups, reduces radiation exposure, and lowers costs by synthesizing images from safer and more cost-effective alternatives such as ultrasound or MRI [44,112,113]. Moreover, intermodality synthesis enhances model performance by enabling training on multiple modalities [114] and fills gaps by imputing missing sequence data [115]. Synthetic CT images generated from MRI data can be used for attenuation correction of PET images [45]. In addition, the use of synthetic data to balance medical datasets is widely recognized as a means of enhancing the performance of models in detecting, segmenting, and predicting medical conditions [116,117,118,119]. However, it is crucial to assess whether synthetic samples accurately capture the complexities and variations in real-world medical data. Further evidence is required to establish the substitutability of synthetic data for real data.
Anomaly Detection Using Generative Models
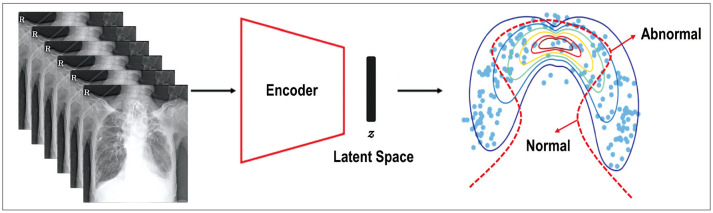
Anomaly detection—a technical term implying the identification of data points in data that do not fit normal patterns—is gaining attention in radiology, because it can identify abnormal lesions on images by learning the data distribution of normal images in semi-supervised learning [120,121] (Fig. 6). It is noteworthy that while anomaly detection can potentially encompass non-pathological findings, such as congenital anomalies, the term is generally used in the medical field to specifically refer to the detection of pathological or clinically significant abnormalities. Therefore, anomaly detection in radiology focuses primarily on identifying abnormal findings that are clinically relevant and may require further investigation or intervention. These algorithms offer several advantages that make them an attractive option, because anomaly detection can be trained without the need for high-quality annotated data, which can be time-consuming and expensive to obtain. This makes anomaly detection a more efficient alternative to supervised learning methods applied to rare or less frequently occurring diseases, which have not been collected enough to train a model in a supervised manner. This approach makes them more clinically applicable, as it helps to detect several conditions rather than just focusing on one or a few specific diseases. Anomaly detection in medical imaging can be approached using various training methods, including supervised, semi-, and un-supervised learning, with several popular network architectures, such as VAEs [122,123,124,125,126,127,128,129,130,131], GANs [55,56,57,58,59,60,61], and diffusion models [54]. Because GANs have the potential to generate high-resolution medical images, GAN-based anomaly detection can reconstruct the closest normal image to a pathologic input image by leveraging normal data from healthy individuals. The reconstruction error between the input (real images) and output (synthetic images) helps to evaluate the anomaly scores for the input data and enables the identification of pathologic anomalies. In the field of radiology, anomaly detection models have been used in various modalities, including CXRs [55,56,122,132,133], mammography [123,124,134], breast ultrasound [57], cardiac CT [135], brain PET-CT [136], brain CT [58,125,126], and brain MRI [59,60,61,127,128,129,130,131,137]. Notably, these studies have proposed the potential utility of anomaly detection for various medical images.
Fig. 6. Anomaly detection with an encoder to map images into a latent space (z), with the data (blue dots) classified as normal or abnormal boundary (red dashed line).
Better Explainability and Validation of Artificial Intelligence
DL networks have remarkably improved their performance with multiple hidden layers of non-linear activation functions; however, this occurs at the cost of interpretability and explainability, thereby aptly earning the nickname of “Black box” [138]. For adequate patient communication and rapport, providing the reasons for decision making in medicine using AI is important. The European Union’s General Data Protection Regulation recommends that automated individual decision-making should be based on the explicit consent of the patient, and meaningful information about the algorithms should be provided to them [139]. However, one atudy [140] pointed out that there can be a “level of opacity” in explainability (i.e., whether the explanations cannot be shared according to the nature of the algorithm or the explanations will not be shared by the developer). Contacting all the authors of individual DL studies and asking them to propose a clear mechanism of action is infeasible and impractical. As the mechanisms underlying some medical discoveries are unclear, further studies are needed to confirm their safety and utility (e.g., the mechanism of action of metformin has not yet been clearly revealed [141], but its safety and utility have been confirmed [142]). Nevertheless, there is still an increasing demand for the explainability of the “black box” in medicine [143].
Medical AI researchers attempt to explain the performance of their models in many ways. Traditional machine learning methods, such as linear regression, support vector machines [144], and tree-based models [145,146], are explainable models. Some algorithms provide textual explanations directly, such as medical visual question answering [147]. However, most researchers in medical DL favor visual explanations such as class activation maps (CAMs) [148] and gradient-weighted CAM (Grad-CAM) [149]. In addition, a local interpretable model-agnostic explanation (LIME) [150] method can be applied to explain the results visually. A review article [151] has shown an increasing trend in the use of explainable AI (XAI) in medical image analyses.
The performance of medical DL should be validated and reported using a more standardized study design, regardless of whether the model is explainable. In particular, handling confounding variables to isolate causality and associations is important in medical research [152,153,154]. Confounding variables should be considered when developing and validating medical DL models. However, exploiting these important confounding variables while developing the DL model can induce shortcut problems [155], because such confounding variables may already be reflected in the results as radiomic features. The identification and exploitation of these shortcuts should be considered when developing robust medical DL models [156]. Nevertheless, exploiting confounding variables to develop DL models and handling them in statistical models may induce overadjustment issues [157]. In addition, factors such as anatomical side markers and image quality can act as confounding variables, which may negatively affect the generalizability and credibility of the research [158]. Furthermore, similar to the comparison of intention-to-treat and per-protocol analyses in clinical research [159], a comparative study on exploitation vs. no exploitation of confounding variables can provide better perspectives for research on DL in medical image analysis. Reporting guidelines for studies on AI in medicine have been developed and registered in the EQUATOR (Enhancing the QUAlity and Transparency Of health Research) Network Library [160,161]. These guidelines include the Consolidated Standards of Reporting Trials–Artificial Intelligence (CONSORT–AI), Standard Protocol Items: Recommendations for Interventional Trials–Artificial Intelligence (SPIRIT–AI), Standards for Reporting of Diagnostic Accuracy Studies–Artificial Intelligence (STARD–AI), Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis–Artificial Intelligence (TRIPOD–AI), Developmental and Exploratory Clinical Investigation of DEcision-support systems driven by Artificial Intelligence (DECIDE-AI), and Clinical Artificial Intelligence Model (CLAIM). These guidelines aimed to improve the transparency, quality, and reproducibility of research in the field of AI in medicine. A recent article pointed out that most research on the use of AI in medical imaging lacks standardized study designs appropriate for real-world clinical settings [162], whereas another study put forth relevant methods for evaluating the clinical performance of medical DL [163].
Causal Learning
Causal learning is an emerging field of machine learning that focuses on the causal relationships between variables in a system [164,165]. The goal of causal learning is to understand the underlying mechanisms governing a system and predict how changes in one variable affect another. Recently, causally-enabled methods for medical analysis that use imaging data have garnered increasing interest [166]. These methods aim to address the challenges in medical imaging using causal inference to better understand and analyze data. Wang et al. [166] proposed a normalizing flow-based causal model, similar to the study by Pawlowski et al. [167] to harmonize heterogeneous medical data. This method was applied to T1 brain MRI to classify Alzheimer's disease to infer counterfactuals, which were then used to harmonize medical data. Pölsterl et al. [168] circumvented the identifiability condition, which states that all confounders should be known; they leveraged the dependencies between causes to determine substitute confounders. This method has been applied in brain neuroimaging to detect Alzheimer’s disease. Zhuang et al. [169] also proposed an alternative to expectation maximization for the dynamic causal modeling of functional magnetic resonance imaging brain scans. They developed an approach based on a multiple-shooting method to estimate the parameters of ordinary differential equations under the noisy observations required for brain causal modeling. They suggested augmentation of the multiple-shooting adjoint method to calculate the loss and gradients of their model. Clivio et al. [170] proposed a neural score-matching method for causal inference in high-dimensional medical images to avoid preprocessing them into a lower-dimensional latent space. da Silva et al. [171] used a generative model to synthesize MR images of brain atrophy to examine and investigate various hypotheses regarding the causes of brain growth and atrophy. Overall, these studies demonstrated the potential of causally enabled methods for medical analysis that utilize imaging data. Causal learning can provide novel insights into the obstacles in machine learning for medical imaging, including the dearth of high-quality annotated data and the mismatch between the training and target datasets. Further research in this field could lead to novel and improved methods for analyzing medical images and provide a better understanding of the underlying causes of diseases.
Federated Learning for Overcoming Privacy Concerns
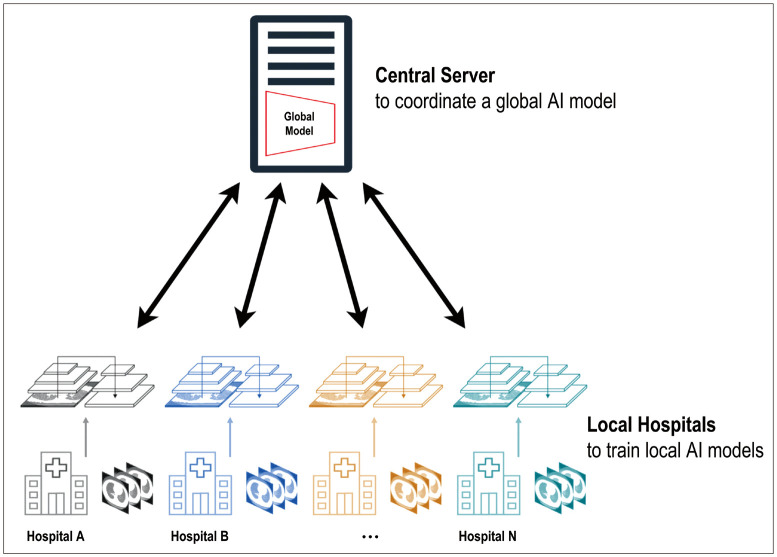
Federated learning presents a promising solution for training models with large and diverse datasets, while preserving patient privacy [172,173]. Enabling multisite collaboration improves generalizability and fosters research on rare diseases that require collective effort for data curation. In federated learning, the weights and parameters of the global model are sent to the different participating institutions. Each institution trains the local AI models using local data and updates the model weights accordingly. After completion of local training, these updated weights are sent back to a central server to coordinate a global AI model. This procedure is repeated until the global model achieves a specific target (Fig. 7). Despite its advantages, federated learning in clinical settings faces several challenges in effective implementation [174]. The primary obstacle is data heterogeneity in each institution. Variations in data types, formats, acquisition protocols, annotation formats, and terminologies across institutions can hinder effective collaboration. Standardization agreements among the institutions involved can help overcome this challenge. Another challenge is technical limitations, such as different computational infrastructures across institutions. Federated learning is a fundamental technique in the future of digital health. Further research and development in federated learning and a concentrated effort in addressing these challenges will contribute to unlocking its full potential in radiology and medical imaging.
Fig. 7. A conceptual diagram of federated learning with training global and local artificial intelligence (AI) models. The weights and parameters of the global model were sent to the different participating institutions. Each institution trains local AI models using local data and updates the model weights accordingly. After completion of local training, these updated weights are sent back to a central server to coordinate a global AI model. This procedure is repeated until the global model achieves a specific target.
Additional Paradigms
Predictive Imaging Biomarkers
Several studies have focused on the extraction of predictive biomarkers from medical images using DL. The development of predictive imaging biomarkers from medical imaging can be approached using various training methods, including supervised, semi-, self-, and un-supervised learning, with several popular network architectures such as CNNs, VAEs, and GANs. Some studies [175,176,177] showed that DL can extract prognostic information from medical images. As these images contain age and sex information, DL-based models can learn demographic information [178,179,180,181]. Studies have shown that neuroimaging-derived age prediction corresponds to the influences of other disorders, such as cognitive impairment and Alzheimer’s disease, and age and sex per se [182,183,184], which have been explored in several brain illnesses. The disparities between predicted and chronological ages may be attributed to the accumulation of age-related alterations in pathological circumstances [185,186,187] or protective factors in brain aging [188,189]. Furthermore, CXR-derived age can be used as an imaging biomarker to indicate the state of the thorax or metabolism [175,176,177] and successfully predict lifespan, mortality, cardiovascular risk, and heart failure prognosis [175,177,190], providing a solid foundation for the imaging biomarker concept. Another approach for extracting relevant prognostic imaging biomarkers involves training a deep survival model using staged binary classifiers of death or incident cancer [175,191,192].
Digital Twin
As simulations have become more prevalent in medicine, the future of precision medicine includes providing tailored diagnoses and treatments for each patient through the development of digital twin (DT) technology. Recently, several studies on DTs in medicine have been published [193,194,195,196,197,198,199,200,201,202], including a wide range of applications such as healthcare management [193], fitness [194], simulating viral infections [195], smart city well-being [196], remote surgery [197], and cardiovascular diseases [200,201,202].
DTs based on image data have significant potential for revolutionizing radiology. These virtual replicas provide radiologists with a comprehensive and dynamic representation of a patient’s anatomy, physiology, and pathology, thereby enabling improved treatment planning and decision-making. By leveraging a patient's radiological imaging data, such as personalized anatomy and imaging biomarkers, DTs offer several benefits in various medical fields. For instance, in orthopedic surgery, DTs created using CT or MRI scans allow surgeons to simulate surgery in advance [203,204]. This enables them to tailor the procedure to the patient's unique anatomy, potentially improving surgical outcomes. By visualizing the joint in a virtual environment, surgeons can plan surgery more accurately, assess different approaches, and anticipate potential complications. In cancer treatment planning, DTs created from a patient's radiological imaging data, such as MRI, PET, or CT scans, can provide a detailed representation of the anatomical and biological characteristics of a tumor [205,206]. This allows medical professionals to test different treatment modalities, such as radiation therapy or chemotherapy, for DTs. By simulating the effects of various treatments, DTs can be used to predict potential effectiveness and side effects. This information aids in the selection of the most suitable and least harmful treatment plan for patients, thereby optimizing the chances of successful outcomes. Similarly, the DTs of a patient's cardiovascular system can be created based on imaging data obtained from angiography, echocardiography, or MRIs, among others [207,208,209]. These DTs can predict the progression of conditions such as atherosclerosis or the risk of events such as myocardial infarctions. They can also be utilized to simulate the effects of interventions, such as stent placement, enabling doctors to plan surgeries more effectively and make informed decisions regarding patient care.
Additionally, DTs offer potential solutions to existing challenges in the AI field. They enable the generation of synthetic yet realistic data through simulations, the augmentation of real-world data, and the enhancement of the robustness of AI training. By incorporating DTs into AI models, the predictive capabilities can be enhanced, because these twins have the potential to dynamically simulate future outcomes based on changing conditions. Furthermore, DTs are based on established physics and human anatomy and provide a more interpretable framework for AI models. This interpretability fosters trust and understanding among healthcare professionals, facilitating AI-driven decision-making.
SUMMARY AND CONCLUSION
Many CADx and CADe software packages with DL methods have received clearance from regulatory agencies and have been successfully commercialized; they offer advantages, including speed, efficiency, low cost, increased accessibility, and the upkeep of faithful behavior in clinical practice. Despite these achievements, several challenges must be addressed in the deployment and development of AI in healthcare; these include ensuring patient privacy, obtaining access to large volumes of high-quality data, achieving generalizability of AI models, and establishing the explainability of the decision-making process. To overcome these issues, some of the various possible solutions are as follows: inclusion of training with longitudinal and multimodal datasets, dense training with multitask and multimodal learning, new generative models including anomaly detection, XAI, self-supervised contrastive and federated learning with large-scale data, causal learning, and DTs. For instance, MTL allows a single AI model to perform multiple tasks simultaneously, thereby increasing its efficiency and adaptability. DL can be used with multi-modality (different types of data) and longitudinal data (data collected over time) to enhance AI's capacity to handle complex medical data. This helps to reduce false positives, leading to more accurate diagnoses and treatments.
Recently spotlighted techniques, such as generative models and federated learning, can provide excellent strategies for improving healthcare outcomes while maintaining patient privacy and data security. Synthetic medical data generated using generative models, such as GANs, can create realistic data samples without compromising patient privacy. Federated learning also allows multiple organizations to collaboratively train a shared AI model without directly sharing sensitive patient data and ensures privacy while still benefiting from a larger, more diverse dataset. In addition, generative models can significantly contribute to radiology by improving various aspects of image processing. These models can aid in fast image acquisition, robust denoising (reducing noise or artifacts), and image reconstruction, resulting in higher-quality images for analysis. Furthermore, generative models can be applied to other research areas in radiology, thereby expanding the possibilities of AI-driven advancements. Semi-supervised and unsupervised learning approaches can save time in AI development and enable broader coverage of various diseases. Specifically, anomaly detection using semisupervised learning can help identify unusual patterns in data and facilitate early diagnosis and intervention for patients. Finally, XAI focuses on developing AI models that provide understandable explanations of predictions and decisions. This approach is crucial for building trust of medical professionals in implementation of AI in clinical practice, to improve their understanding, and to allow the validation of AI-driven recommendations. By enhancing trust and transparency, XAI can accelerate DL research in the medical field, thereby paving the way for more effective and widely adopted solutions. DTs and virtual replicas of real-world objects, systems, or processes can be used to simulate patient conditions, predict treatment outcomes, and optimize clinical decision-making.
More robust technological advancements suggest that significant changes can occur in radiology. Nonetheless, with the integration of better AI into medical imaging, we envision these technological breakthroughs as better cooperative mechanisms designed to alleviate workload and minimize distractions. Currently, DL is used in radiology during the early stages of adolescence. LLMs, such as GPT v4, have recently created a strong impact. However, a crucial factor for AI development and clinical integration into radiology is fostering a comprehensive understanding of the technology, radiology practice, and clinical workflow. Furthermore, the active participation of radiologists, scientists, engineers, and commercials is vital for creating a wide range of radiological applications.
Footnotes
Conflicts of Interest: Joon Beom Seo and Namkug Kim, editorial board members of the Korean Journal of Radiology, were not involved in the editorial evaluation or decision to publish this article. All authors have declared no conflicts of interest.
- Conceptualization: Joon Beom Seo, Namkug Kim, Sang Min Lee.
- Funding acquisition: Gil-Sun Hong.
- Investigation: Gil-Sun Hong, Miso Jang, Sunggu Kyung, Kyungjin Cho, Jiheon Jeong, Grace Yoojin Lee, Keewon Shin, Ki Duk Kim, Seung Min Ryu.
- Project administration: Miso Jang.
- Supervision: Namkug Kim, Sang Min Lee.
- Visualization: Gil-Sun Hong, Namkug Kim.
- Writing—original draft: Gil-Sun Hong, Miso Jang, Sunggu Kyung, Kyungjin Cho, Jiheon Jeong, Grace Yoojin Lee, Keewon Shin, Ki Duk Kim, Seung Min Ryu.
- Writing—review & editing: Joon Beom Seo, Namkug Kim, Sang Min Lee.
Funding Statement: This research was supported by grants from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI21C1148 and HI22C172300).
References
- 1.Kelly BS, Judge C, Bollard SM, Clifford SM, Healy GM, Aziz A, et al. Radiology artificial intelligence: a systematic review and evaluation of methods (RAISE) Eur Radiol. 2022;32:7998–8007. doi: 10.1007/s00330-022-08784-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 3.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, et al. Development and validation of a deep learning system for staging liver fibrosis by using contrast agent–enhanced CT images in the liver. Radiology. 2018;289:688–697. doi: 10.1148/radiol.2018180763. [DOI] [PubMed] [Google Scholar]
- 6.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 8.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 9.Dunnmon JA, Yi D, Langlotz CP, Ré C, Rubin DL, Lungren MP. Assessment of convolutional neural networks for automated classification of chest radiographs. Radiology. 2019;290:537–544. doi: 10.1148/radiol.2018181422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam JG, Park S, Hwang EJ, Lee JH, Jin KN, Lim KY, et al. Development and validation of deep learning–based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology. 2019;290:218–228. doi: 10.1148/radiol.2018180237. [DOI] [PubMed] [Google Scholar]
- 11.Milea D, Najjar RP, Zhubo J, Ting D, Vasseneix C, Xu X, et al. Artificial intelligence to detect papilledema from ocular fundus photographs. N Engl J Med. 2020;382:1687–1695. doi: 10.1056/NEJMoa1917130. [DOI] [PubMed] [Google Scholar]
- 12.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. 2019;1:e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 14.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. NIPS, editors. Advances in neural information processing systems 30. La Jolla: NIPS; 2017. Attention is all you need; pp. 5999–6009. [Google Scholar]
- 15.Li X, Shen L, Xie X, Huang S, Xie Z, Hong X, et al. Multi-resolution convolutional networks for chest X-ray radiograph based lung nodule detection. Artif Intell Med. 2020;103:101744. doi: 10.1016/j.artmed.2019.101744. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Li M, Wang H, Jiang H, Yao Y, Zhang H, et al. Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access. 2019;7:105146–105158. [Google Scholar]
- 17.Yoon JH, Kim EK. Deep learning-based artificial intelligence for mammography. Korean J Radiol. 2021;22:1225–1239. doi: 10.3348/kjr.2020.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu P, Wu F, Peng J, Bao Y, Chen F, Kong D. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J Comput Assist Radiol Surg. 2017;12:399–411. doi: 10.1007/s11548-016-1501-5. [DOI] [PubMed] [Google Scholar]
- 19.U.S. Food and Drug Administration (FDA) Artificial intelligence and machine learning (AI/ML)-enabled medical devices. [accessed on April 16, 2023]. Available at: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices .
- 20.Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, et al. Deep learning-enabled medical computer vision. NPJ Digit Med. 2021;4:5. doi: 10.1038/s41746-020-00376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813–1819. doi: 10.1136/gutjnl-2018-317500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang EJ, Park S, Jin KN, Kim JI, Choi SY, Lee JH, et al. Development and validation of a deep learning–based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open. 2019;2:e191095. doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Ma J, Zhang S, Shao J, Wang Y, Zhou HY, et al. Development and validation of an abnormality-derived deep-learning diagnostic system for major respiratory diseases. NPJ Digit Med. 2022;5:124. doi: 10.1038/s41746-022-00648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35:1153–1159. [Google Scholar]
- 25.Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21:4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Zhang L, Jiang Y. Overfitting and underfitting analysis for deep learning based end-to-end communication systems; Proceedings of the 11th International Conference on Wireless Communications and Signal Processing (WCSP); 2019 Oct 23-25; Xi’an, China. IEEE; 2019. pp. 1–6. [Google Scholar]
- 27.Rice L, Wong E, Kolter Z. Overfitting in adversarially robust deep learning; Proceedings of the 37th International Conference on Machine Learning; 2020 Jul 13-18 (Online); ICML; 2020. pp. 8093–8104. [Google Scholar]
- 28.He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21:1263–1284. [Google Scholar]
- 29.Lipton ZC. The mythos of model interpretability: in machine learning, the concept of interpretability is both important and slippery. Queue. 2018;16:31–57. [Google Scholar]
- 30.Guo W, Wang J, Wang S. Deep multimodal representation learning: a survey. IEEE Access. 2019;7:63373–63394. [Google Scholar]
- 31.Goodfellow I. NIPS 2016 tutorial: generative adversarial networks. [cited January 5, 2023];arXiv: 1701.00160v4 [Preprint] 2016 doi: 10.48550/arXiv.1701.00160. Available at: [DOI] [Google Scholar]
- 32.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. Commun ACM. 2020;63:139–144. [Google Scholar]
- 33.Ho J, Jain A, Abbeel P. Denoising diffusion probabilistic models. Adv Neural Inf Process Syst. 2020;33:6840–6851. [Google Scholar]
- 34.Kingma DP, Dhariwal P. Glow: generative flow with invertible 1x1 convolutions. Adv Neural Inf Process Syst. 2018;31:10215–10224. [Google Scholar]
- 35.Kingma DP, Welling M. Auto-encoding variational bayes. [cited January 5, 2023];arXiv: 1312.6114v11 [Preprint] 2013 doi: 10.48550/arXiv.1312.6114. Available at: [DOI] [Google Scholar]
- 36.Lyu H, Sha N, Qin S, Yan M, Xie Y, Wang R. NIPS, editors. Advances in neural information processing systems 32 (NeurIPS 2019) La Jolla: NIPS; 2019. Manifold denoising by nonlinear robust principal component analysis; pp. 1–11. [PMC free article] [PubMed] [Google Scholar]
- 37.Kang E, Koo HJ, Yang DH, Seo JB, Ye JC. Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med Phys. 2019;46:550–562. doi: 10.1002/mp.13284. [DOI] [PubMed] [Google Scholar]
- 38.Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging. 2017;36:2536–2545. doi: 10.1109/TMI.2017.2708987. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Zhao Y, Noble JH, Dawant BM. Conditional generative adversarial networks for metal artifact reduction in CT images of the ear. Med Image Comput Comput Assist Interv. 2018;11070:3–11. doi: 10.1007/978-3-030-00928-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim KH, Do WJ, Park SH. Improving resolution of MR images with an adversarial network incorporating images with different contrast. Med Phys. 2018;45:3120–3131. doi: 10.1002/mp.12945. [DOI] [PubMed] [Google Scholar]
- 41.Quan TM, Nguyen-Duc T, Jeong WK. Compressed sensing MRI reconstruction using a generative adversarial network with a cyclic loss. IEEE Trans Med Imaging. 2018;37:1488–1497. doi: 10.1109/TMI.2018.2820120. [DOI] [PubMed] [Google Scholar]
- 42.Yang G, Yu S, Dong H, Slabaugh G, Dragotti PL, Ye X, et al. DAGAN: deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE Trans Med Imaging. 2017;37:1310–1321. doi: 10.1109/TMI.2017.2785879. [DOI] [PubMed] [Google Scholar]
- 43.Emami H, Dong M, Nejad-Davarani SP, Glide-Hurst CK. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys. 2018;45:3627–3636. doi: 10.1002/mp.13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei Y, Harms J, Wang T, Liu Y, Shu HK, Jani AB, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys. 2019;46:3565–3581. doi: 10.1002/mp.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. 2019;64:215016. doi: 10.1088/1361-6560/ab4eb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys. 2019;46:2157–2168. doi: 10.1002/mp.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huo Y, Xu Z, Bao S, Bermudez C, Plassard AJ, Liu J, et al. Splenomegaly segmentation using global convolutional kernels and conditional generative adversarial networks. Proc SPIE Int Soc Opt Eng. 2018;10574:1057409. doi: 10.1117/12.2293406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Guo S, Zhang H, He K, Mu S, Guo Y, et al. Accurate colorectal tumor segmentation for CT scans based on the label assignment generative adversarial network. Med Phys. 2019;46:3532–3542. doi: 10.1002/mp.13584. [DOI] [PubMed] [Google Scholar]
- 49.Xue Y, Xu T, Zhang H, Long LR, Huang X. SegAN: adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics. 2018;16:383–392. doi: 10.1007/s12021-018-9377-x. [DOI] [PubMed] [Google Scholar]
- 50.Tanner C, Ozdemir F, Profanter R, Vishnevsky V, Konukoglu E, Goksel O. Generative adversarial networks for MR-CT deformable image registration. [cited January 5, 2023];arXiv: 1807.07349v1 [Preprint] 2018 doi: 10.48550/arXiv.1807.07349. Available at: [DOI] [Google Scholar]
- 51.Yan P, Xu S, Rastinehad AR, Wood BJ. Adversarial image registration with application for MR and TRUS image fusion. [cited January 1, 2023];arXiv: 1804.11024v2 [Preprint] 2018 doi: 10.48550/arXiv.1804.11024. Available at: [DOI] [Google Scholar]
- 52.Madani A, Moradi M, Karargyris A, Syeda-Mahmood T. Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation; Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018 Apr 4-7; Washington, DC, USA. IEEE; 2018. pp. 1038–1042. [Google Scholar]
- 53.Xie Y, Zhang J, Xia Y. Semi-supervised adversarial model for benign-malignant lung nodule classification on chest CT. Med Image Anal. 2019;57:237–248. doi: 10.1016/j.media.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Wolleb J, Bieder F, Sandkühler R, Cattin PC. In: Medical image computing and computer assisted intervention – MICCAI 2022. Lecture notes in computer science, vol 13438. Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Cham: Springer; 2022. Diffusion models for medical anomaly detection; pp. 35–45. [Google Scholar]
- 55.Wolleb J, Sandkühler R, Cattin PC. In: Medical image computing and computer assisted intervention – MICCAI 2020. Lecture notes in computer science, vol 12264. Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga MA, Zhou SK, et al., editors. Cham: Springer; 2020. Descargan: disease-specific anomaly detection with weak supervision; pp. 14–24. [Google Scholar]
- 56.Nakao T, Hanaoka S, Nomura Y, Murata M, Takenaga T, Miki S, et al. Unsupervised deep anomaly detection in chest radiographs. J Digit Imaging. 2021;34:418–427. doi: 10.1007/s10278-020-00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujioka T, Kubota K, Mori M, Kikuchi Y, Katsuta L, Kimura M, et al. Efficient anomaly detection with generative adversarial network for breast ultrasound imaging. Diagnostics (Basel) 2020;10:456. doi: 10.3390/diagnostics10070456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee S, Jeong B, Kim M, Jang R, Paik W, Kang J, et al. Emergency triage of brain computed tomography via anomaly detection with a deep generative model. Nat Commun. 2022;13:4251. doi: 10.1038/s41467-022-31808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Hespen KM, Zwanenburg JJM, Dankbaar JW, Geerlings MI, Hendrikse J, Kuijf HJ. An anomaly detection approach to identify chronic brain infarcts on MRI. Sci Rep. 2021;11:7714. doi: 10.1038/s41598-021-87013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khosla M, Jamison K, Kuceyeski A, Sabuncu MR. In: Machine learning in medical imaging (MLMI 2019). Lecture notes in computer science, vol 11861. Suk HI, Liu M, Yan P, Lian C, editors. Cham: Springer; 2019. Detecting abnormalities in resting-state dynamics: an unsupervised learning approach; pp. 301–309. [Google Scholar]
- 61.Han C, Rundo L, Murao K, Noguchi T, Shimahara Y, Milacski ZÁ, et al. MADGAN: unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinformatics. 2021;22(Suppl 2):31. doi: 10.1186/s12859-020-03936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bowles C, Gunn R, Hammers A, Rueckert D. In: Medical imaging 2018: image processing (vol 10574) Angelini ED, Landman BA, editors. Bellingham, WA: SPIE; 2018. Modelling the progression of Alzheimer’s disease in MRI using generative adversarial networks; pp. 397–407. [Google Scholar]
- 63.Lu D, Popuri K, Ding GW, Balachandar R, Beg MF Alzheimer’s Disease Neuroimaging Initiative. Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci Rep. 2018;8:5697. doi: 10.1038/s41598-018-22871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehdipour Ghazi M, Nielsen M, Pai A, Cardoso MJ, Modat M, Ourselin S, et al. Training recurrent neural networks robust to incomplete data: application to Alzheimer’s disease progression modeling. Med Image Anal. 2019;53:39–46. doi: 10.1016/j.media.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Goulet MA, Cousineau D. The power of replicated measures to increase statistical power. Adv Methods Pract Psychol Sci. 2019;2:199–213. [Google Scholar]
- 66.Ma Y, Mazumdar M, Memtsoudis SG. Beyond repeated-measures analysis of variance: advanced statistical methods for the analysis of longitudinal data in anesthesia research. Reg Anesth Pain Med. 2012;37:99–105. doi: 10.1097/AAP.0b013e31823ebc74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat Med. 2022;28:1773–1784. doi: 10.1038/s41591-022-01981-2. [DOI] [PubMed] [Google Scholar]
- 68.Vickers AJ. How many repeated measures in repeated measures designs? Statistical issues for comparative trials. BMC Med Res Methodol. 2003;3:22. doi: 10.1186/1471-2288-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barros V, Tlusty T, Barkan E, Hexter E, Gruen D, Guindy M, et al. Virtual biopsy by using artificial intelligence-based multimodal modeling of binational mammography data. Radiology. 2023;306:e220027. doi: 10.1148/radiol.220027. [DOI] [PubMed] [Google Scholar]
- 70.Goto S, Mahara K, Beussink-Nelson L, Ikura H, Katsumata Y, Endo J, et al. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat Commun. 2021;12:2726. doi: 10.1038/s41467-021-22877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang SC, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med. 2020;3:136. doi: 10.1038/s41746-020-00341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kline A, Wang H, Li Y, Dennis S, Hutch M, Xu Z, et al. Multimodal machine learning in precision health: a scoping review. NPJ Digit Med. 2022;5:171. doi: 10.1038/s41746-022-00712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin W, Gao Q, Yuan J, Chen Z, Feng C, Chen W, et al. Predicting Alzheimer’s disease conversion from mild cognitive impairment using an extreme learning machine-based grading method with multimodal data. Front Aging Neurosci. 2020;12:77. doi: 10.3389/fnagi.2020.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tiulpin A, Klein S, Bierma-Zeinstra SMA, Thevenot J, Rahtu E, Meurs JV, et al. Multimodal machine learning-based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci Rep. 2019;9:20038. doi: 10.1038/s41598-019-56527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venugopalan J, Tong L, Hassanzadeh HR, Wang MD. Multimodal deep learning models for early detection of Alzheimer’s disease stage. Sci Rep. 2021;11:3254. doi: 10.1038/s41598-020-74399-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vandenhende S, Georgoulis S, Van Gansbeke W, Proesmans M, Dai D, Van Gool L. Multi-task learning for dense prediction tasks: a survey. IEEE Trans Pattern Anal Mach Intell. 2021;44:3614–3633. doi: 10.1109/TPAMI.2021.3054719. [DOI] [PubMed] [Google Scholar]
- 77.Caruana R. Multitask learning. Mach Learn. 1997;28:41–75. [Google Scholar]
- 78.Misra I, Shrivastava A, Gupta A, Hebert M. Cross-stitch networks for multi-task learning; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, USA. IEEE; 2016. pp. 3994–4003. [Google Scholar]
- 79.Gao Y, Ma J, Zhao M, Liu W, Yuille AL. NDDR-CNN: layerwise feature fusing in multi-task cnns by neural discriminative dimensionality reduction; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 3205–3214. [Google Scholar]
- 80.Liu S, Johns E, Davison AJ. End-to-end multi-task learning with attention; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 1871–1880. [Google Scholar]
- 81.Kisling LA, Das JM. Prevention strategies. Treasure Island, FL: StatPearls Publishing; 2023. [PubMed] [Google Scholar]
- 82.Xu D, Ouyang W, Wang X, Sebe N. PAD-net: multi-tasks guided prediction-and-distillation network for simultaneous depth estimation and scene parsing; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2018 Jun 18-23; Salt Lake City, UT, USA. IEEE; 2018. pp. 675–684. [Google Scholar]
- 83.Zhang Z, Cui Z, Xu C, Yan Y, Sebe N, Yang J. Pattern-affinitive propagation across depth, surface normal and semantic segmentation; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 4106–4115. [Google Scholar]
- 84.Zhang Z, Cui Z, Xu C, Jie Z, Li X, Yang J. Joint task-recursive learning for semantic segmentation and depth estimation; Proceedings of the European Conference on Computer Vision (ECCV); 2018 Sep 8-14; Munich, Germany. ECCV; 2018. pp. 235–251. [Google Scholar]
- 85.He T, Guo J, Wang J, Xu X, Yi Z. Multi-task learning for the segmentation of thoracic organs at risk in CT images; Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI); 2019 Apr 8-11; Venice, Italy. IEEE; pp. 10–13. [Google Scholar]
- 86.Gao F, Yoon H, Wu T, Chu X. A feature transfer enabled multi-task deep learning model on medical imaging. Expert Syst Appl. 2020;143:112957 [Google Scholar]
- 87.Amyar A, Modzelewski R, Li H, Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput Biol Med. 2020;126:104037. doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kyung S, Shin K, Jeong H, Kim KD, Park J, Cho K, et al. Improved performance and robustness of multi-task representation learning with consistency loss between pretexts for intracranial hemorrhage identification in head CT. Med Image Anal. 2022;81:102489. doi: 10.1016/j.media.2022.102489. [DOI] [PubMed] [Google Scholar]
- 89.Wang X, Peng Y, Lu L, Lu Z, Summers RM. TieNet: text-image embedding network for common thorax disease classification and reporting in chest X-rays; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2018 Jun 18-23; Salt Lake City, UT, USA. IEEE; 2018. pp. 9049–9058. [Google Scholar]
- 90.Devlin J, Chang MW, Lee K, Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding. [cited January 5, 2023];arXiv: 1810.04805v2 [Preprint] 2018 doi: 10.48550/arXiv.1810.04805. Available at: [DOI] [Google Scholar]
- 91.Moon JH, Lee H, Shin W, Kim YH, Choi E. Multi-modal understanding and generation for medical images and text via vision-language pre-training. IEEE J Biomed Health Inform. 2022;26:6070–6080. doi: 10.1109/JBHI.2022.3207502. [DOI] [PubMed] [Google Scholar]
- 92.Park S, Lee ES, Lee JE, Ye JC. Self-supervised multi-modal training from uncurated image and reports enables zero-shot oversight artificial intelligence in radiology. [cited January 5, 2023];arXiv: 2208.05140v4 [Preprint] 2022 doi: 10.48550/arXiv.2208.05140. Available at: [DOI] [Google Scholar]
- 93.Hsu TMH, Weng WH, Boag W, McDermott M, Szolovits P. Unsupervised multimodal representation learning across medical images and reports. [cited January 5, 2023];arXiv: 1811.08615v1 [Preprint] 2018 doi: 10.48550/arXiv.1811.08615. Available at: [DOI] [Google Scholar]
- 94.Liu G, Hsu TMH, McDermott M, Boag W, Weng WH, Szolovits P, et al. Clinically accurate chest X-ray report generation; Proceedings of the 4th Machine Learning for Healthcare Conference; 2019 Aug 9-10; Ann Arbor, Michigan, MI, USA. PMLR; 2019. pp. 249–269. [Google Scholar]
- 95.Liu F, Wu X, Ge S, Fan W, Zou Y. Exploring and distilling posterior and prior knowledge for radiology report generation; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 13753–13762. [Google Scholar]
- 96.Wang Z, Zhou L, Wang L, Li X. A self-boosting framework for automated radiographic report generation; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 2433–2442. [Google Scholar]
- 97.Yang X, Ye M, You Q, Ma F. Association for Computational Linguistics, editors. Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (volume 1: long papers) Stroudsburg: Association for Computational Linguistics; 2021. Writing by memorizing: hierarchical retrieval-based medical report generation; pp. 5000–5009. [Google Scholar]
- 98.OpenAI. GPT-3.5. [accessed on February 16, 2023]. Available at: https://platform.openai.com/docs/models/gpt-3-5 .
- 99.Bommasani R, Hudson DA, Adeli E, Altman R, Arora S, von Arx S, et al. On the opportunities and risks of foundation models. [cited January 5, 2023];arXiv: 2108.07258v3 [Preprint] 2021 doi: 10.48550/arXiv.2108.07258. Available at: [DOI] [Google Scholar]
- 100.Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 101.Oord AVD, Li Y, Vinyals O. Representation learning with contrastive predictive coding. [cited January 5, 2023];arXiv: 1807.03748v2 [Preprint] 2018 doi: 10.48550/arXiv.1807.03748. Available at: [DOI] [Google Scholar]
- 102.Ouyang J, Zhao Q, Adeli E, Zaharchuk G, Pohl KM. Self-supervised learning of neighborhood embedding for longitudinal MRI. Med Image Anal. 2022;82:102571. doi: 10.1016/j.media.2022.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu Y, Zeng D, Wang Z, Shi Y, Hu J. Distributed contrastive learning for medical image segmentation. Med Image Anal. 2022;81:102564. doi: 10.1016/j.media.2022.102564. [DOI] [PubMed] [Google Scholar]
- 104.Seyfioğlu MS, Liu Z, Kamath P, Gangolli S, Wang S, Grabowski T, et al. In: Medical image computing and computer assisted intervention – MICCAI 2022. Lecture notes in computer science, vol 13431. Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Cham: Springer; 2022. Brain-aware replacements for supervised contrastive learning in detection of Alzheimer’s disease; pp. 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chaitanya K, Erdil E, Karani N, Konukoglu E. In: Advances in neural information processing systems 33 (NeurIPS 2020) Larochelle H, Ranzato M, Hadsell R, Balcan M, Lin H, editors. La Jolla, CA: NIPS; 2020. Contrastive learning of global and local features for medical image segmentation with limited annotations; pp. 12546–12558. [Google Scholar]
- 106.Wang J, Li X, Han Y, Qin J, Wang L, Qichao Z. Separated contrastive learning for organ-at-risk and gross-tumor-volume segmentation with limited annotation. Proc AAAI Conf Artif Intell. 2022;36:2459–2467. [Google Scholar]
- 107.Park T, Efros AA, Zhang R, Zhu JY. In: Computer vision – ECCV 2020 (vol 12354) Vedaldi A, Bischof H, Brox T, Frahm JM, editors. Cham: Springer; 2020. Contrastive learning for unpaired image-to-image translation; pp. 319–345. [Google Scholar]
- 108.Cho K, Seo J, Kyung S, Kim M, Hong GS, Kim N. Bone suppression on pediatric chest radiographs via a deep learning-based cascade model. Comput Methods Programs Biomed. 2022;215:106627. doi: 10.1016/j.cmpb.2022.106627. [DOI] [PubMed] [Google Scholar]
- 109.Liang X, Chen L, Nguyen D, Zhou Z, Gu X, Yang M, et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. 2019;64:125002. doi: 10.1088/1361-6560/ab22f9. [DOI] [PubMed] [Google Scholar]
- 110.Harms J, Lei Y, Wang T, Zhang R, Zhou J, Tang X, et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. 2019;46:3998–4009. doi: 10.1002/mp.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, et al. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 2018;65:2720–2730. doi: 10.1109/TBME.2018.2814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yao Z, Luo T, Dong Y, Jia X, Deng Y, Wu G, et al. Virtual elastography ultrasound via generative adversarial network for breast cancer diagnosis. Nat Commun. 2023;14:788. doi: 10.1038/s41467-023-36102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk-Schulz IM, et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol. 2018;63:185001. doi: 10.1088/1361-6560/aada6d. [DOI] [PubMed] [Google Scholar]
- 114.Lei Y, Dong X, Tian Z, Liu Y, Tian S, Wang T, et al. CT prostate segmentation based on synthetic MRI-aided deep attention fully convolution network. Med Phys. 2020;47:530–540. doi: 10.1002/mp.13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conte GM, Weston AD, Vogelsang DC, Philbrick KA, Cai JC, Barbera M, et al. Generative adversarial networks to synthesize missing T1 and FLAIR MRI sequences for use in a multisequence brain tumor segmentation model. Radiology. 2021;299:313–323. doi: 10.1148/radiol.2021203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Al Khalil Y, Amirrajab S, Lorenz C, Weese J, Pluim J, Breeuwer M. On the usability of synthetic data for improving the robustness of deep learning-based segmentation of cardiac magnetic resonance images. Med Image Anal. 2023;84:102688. doi: 10.1016/j.media.2022.102688. [DOI] [PubMed] [Google Scholar]
- 117.Chung M, Kong ST, Park B, Chung Y, Jung KH, Seo JB. Utilizing synthetic nodules for improving nodule detection in chest radiographs. J Digit Imaging. 2022;35:1061–1068. doi: 10.1007/s10278-022-00608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jayachandran Preetha C, Meredig H, Brugnara G, Mahmutoglu MA, Foltyn M, Isensee F, et al. Deep-learning-based synthesis of post-contrast T1-weighted MRI for tumour response assessment in neuro-oncology: a multicentre, retrospective cohort study. Lancet Digit Health. 2021;3:e784–e794. doi: 10.1016/S2589-7500(21)00205-3. [DOI] [PubMed] [Google Scholar]
- 119.Sandfort V, Yan K, Pickhardt PJ, Summers RM. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci Rep. 2019;9:16884. doi: 10.1038/s41598-019-52737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goldstein M, Uchida S. A comparative evaluation of unsupervised anomaly detection algorithms for multivariate data. PLoS One. 2016;11:e0152173. doi: 10.1371/journal.pone.0152173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tschuchnig ME, Gadermayr M. In: Data science – Analytics and applications. Haber P, Lampoltshammer TJ, Leopold H, Mayr M, editors. Wiesbaden: Springer Vieweg; 2022. Anomaly detection in medical imaging - A mini review; pp. 33–38. [Google Scholar]
- 122.Zhang J, Xie Y, Pang G, Liao Z, Verjans J, Li W, et al. Viral pneumonia screening on chest X-rays using confidence-aware anomaly detection. IEEE Trans Med Imaging. 2020;40:879–890. doi: 10.1109/TMI.2020.3040950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei Q, Ren Y, Hou R, Shi B, Lo JY, Carin L. In: Medical imaging 2018: computer-aided diagnosis (vol 10575) Petrick N, Mori K, editors. Bellingham: SPIE; 2018. Anomaly detection for medical images based on a one-class classification; pp. 375–380. [Google Scholar]
- 124.Tlusty T, Amit G, Ben-Ari R. Unsupervised clustering of mammograms for outlier detection and breast density estimation; Proceedings of the 24th International Conference on Pattern Recognition (ICPR); 2018 Aug 20-24; Beijing, China. IEEE; 2018. pp. 3808–3813. [Google Scholar]
- 125.Sato D, Hanaoka S, Nomura Y, Takenaga T, Miki S, Yoshikawa T, et al. In: Medical imaging 2018: computer-aided diagnosis (vol 10575) Petrick N, Mori K, editors. Bellingham: SPIE; 2018. A primitive study on unsupervised anomaly detection with an autoencoder in emergency head CT volumes; pp. 388–393. [Google Scholar]
- 126.Pawlowski N, Lee MC, Rajchl M, McDonagh S, Ferrante E, Kamnitsas K, et al. Unsupervised lesion detection in brain CT using bayesian convolutional autoencoders. [cited January 5, 2023];OpenReview [Preprint] 2018 Available at: https://openreview.net/forum?id=S1hpzoisz . [Google Scholar]
- 127.Zimmerer D, Isensee F, Petersen J, Kohl S, Maier-Hein K. In: Medical image computing and computer assisted intervention – MICCAI 2019. Lecture notes in computer science, vol 11767. Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, et al., editors. Cham: Springer; 2019. Unsupervised anomaly localization using variational auto-encoders; pp. 289–297. [Google Scholar]
- 128.Heer M, Postels J, Chen X, Konukoglu E, Albarqouni S. The OOD blind spot of unsupervised anomaly detection; Proceedings of the 4th Medical Imaging with Deep Learning; 2021 Jul 7-9; Lübeck, Germany. PMLR; 2021. pp. 286–300. [Google Scholar]
- 129.Chen X, You S, Tezcan KC, Konukoglu E. Unsupervised lesion detection via image restoration with a normative prior. Med Image Anal. 2020;64:101713. doi: 10.1016/j.media.2020.101713. [DOI] [PubMed] [Google Scholar]
- 130.Baur C, Wiestler B, Muehlau M, Zimmer C, Navab N, Albarqouni S. Modeling healthy anatomy with artificial intelligence for unsupervised anomaly detection in brain MRI. Radiol Artif Intell. 2021;3:e190169. doi: 10.1148/ryai.2021190169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alaverdyan Z, Jung J, Bouet R, Lartizien C. Regularized siamese neural network for unsupervised outlier detection on brain multiparametric magnetic resonance imaging: application to epilepsy lesion screening. Med Image Anal. 2020;60:101618. doi: 10.1016/j.media.2019.101618. [DOI] [PubMed] [Google Scholar]
- 132.Zhao H, Li Y, He N, Ma K, Fang L, Li H, et al. Anomaly detection for medical images using self-supervised and translation-consistent features. IEEE Trans Med Imaging. 2021;40:3641–3651. doi: 10.1109/TMI.2021.3093883. [DOI] [PubMed] [Google Scholar]
- 133.Kim CM, Hong EJ, Park RC. Chest X-ray outlier detection model using dimension reduction and edge detection. IEEE Access. 2021;9:86096–86106. [Google Scholar]
- 134.Quellec G, Lamard M, Cozic M, Coatrieux G, Cazuguel G. Multiple-instance learning for anomaly detection in digital mammography. IEEE Trans Med Imaging. 2016;35:1604–1614. doi: 10.1109/TMI.2016.2521442. [DOI] [PubMed] [Google Scholar]
- 135.Wong KC, Karargyris A, Syeda-Mahmood T, Moradi M. In: Medical image computing and computer assisted intervention – MICCAI 2017. Lecture notes in computer science, vol 10435. Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins D, Duchesne S, editors. Cham: Springer; 2017. Building disease detection algorithms with very small numbers of positive samples; pp. 471–479. [Google Scholar]
- 136.Choi H, Ha S, Kang H, Lee H, Lee DS Alzheimer’s Disease Neuroimaging Initiative. Deep learning only by normal brain PET identify unheralded brain anomalies. EBioMedicine. 2019;43:447–453. doi: 10.1016/j.ebiom.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baur C, Graf R, Wiestler B, Albarqouni S, Navab N. In: Medical image computing and computer assisted intervention – MICCAI 2020. Lecture notes in computer science, vol 12262. Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga MA, Zhou SK, et al., editors. Cham: Springer; 2020. SteGANomaly: inhibiting CycleGAN steganography for unsupervised anomaly detection in brain MRI; pp. 718–727. [Google Scholar]
- 138.Watson DS, Krutzinna J, Bruce IN, Griffiths CE, McInnes IB, Barnes MR, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ. 2019;364:l886. doi: 10.1136/bmj.l886. [DOI] [PubMed] [Google Scholar]
- 139.European Council. The general data protection regulation. [accessed on April 21, 2023]. Available at: https://www.consilium.europa.eu/en/policies/data-protection/data-protection-regulation .
- 140.Cohen IG. Informed consent and medical artificial intelligence: what to tell the patient? Georgetown Law J. 2020;108:1425–1469. [Google Scholar]
- 141.Rena G, Hardie DG, Pearson ER. The mechanisms of action of metformin. Diabetologia. 2017;60:1577–1585. doi: 10.1007/s00125-017-4342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johansen K. Efficacy of metformin in the treatment of NIDDM. Meta-analysis. Diabetes Care. 1999;22:33–37. doi: 10.2337/diacare.22.1.33. [DOI] [PubMed] [Google Scholar]
- 143.Jia X, Ren L, Cai J. Clinical implementation of AI technologies will require interpretable AI models. Med Phys. 2020;47:1–4. doi: 10.1002/mp.13891. [DOI] [PubMed] [Google Scholar]
- 144.Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 145.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 146.Safavian SR, Landgrebe D. A survey of decision tree classifier methodology. IEEE Trans Syst Man Cybern. 1991;21:660–674. [Google Scholar]
- 147.Lin Z, Zhang D, Tao Q, Shi D, Haffari G, Wu Q, et al. Medical visual question answering: a survey. Artif Intell Med. 2023;143:102611. doi: 10.1016/j.artmed.2023.102611. [DOI] [PubMed] [Google Scholar]
- 148.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, USA. IEEE; 2016. pp. 2921–2929. [Google Scholar]
- 149.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization; Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. IEEE; 2017. pp. 618–626. [Google Scholar]
- 150.Ribeiro MT, Singh S, Guestrin C. “Why should I trust you?”: explaining the predictions of any classifier; Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016 Aug 13-17; San Francisco, CA, USA. ACM; 2016. pp. 1135–1144. [Google Scholar]
- 151.van der Velden BHM, Kuijf HJ, Gilhuijs KGA, Viergever MA. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Image Anal. 2022;79:102470. doi: 10.1016/j.media.2022.102470. [DOI] [PubMed] [Google Scholar]
- 152.Robinson LD, Robert Dorroh J, Lien D, Tiku ML. The effects of covariate adjustment in generalized linear models. Commun Stat Theory Methods. 1998;27:1653–1675. [Google Scholar]
- 153.Shpitser I, VanderWeele T, Robins JM. On the validity of covariate adjustment for estimating causal effects. [cited January 5, 2023];arXiv: 1203.3515v1 [Preprint] 2012 doi: 10.48550/arXiv.1203.3515. Available at: [DOI] [Google Scholar]
- 154.Kahlert J, Gribsholt SB, Gammelager H, Dekkers OM, Luta G. Control of confounding in the analysis phase - an overview for clinicians. Clin Epidemiol. 2017;9:195–204. doi: 10.2147/CLEP.S129886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Geirhos R, Jacobsen JH, Michaelis C, Zemel R, Brendel W, Bethge M, et al. Shortcut learning in deep neural networks. Nat Mach Intell. 2020;2:665–673. [Google Scholar]
- 156.Brown A, Tomasev N, Freyberg J, Liu Y, Karthikesalingam A, Schrouff J. Detecting shortcut learning for fair medical AI using shortcut testing. Nat Commun. 2023;14:4314. doi: 10.1038/s41467-023-39902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schisterman EF, Cole SR, Platt RW. Overadjustment bias and unnecessary adjustment in epidemiologic studies. Epidemiology. 2009;20:488–495. doi: 10.1097/EDE.0b013e3181a819a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kim KD, Cho K, Kim M, Lee KH, Lee S, Lee SM, et al. Enhancing deep learning based classifiers with inpainting anatomical side markers (L/R markers) for multi-center trials. Comput Methods Programs Biomed. 2022;220:106705. doi: 10.1016/j.cmpb.2022.106705. [DOI] [PubMed] [Google Scholar]
- 159.Smith VA, Coffman CJ, Hudgens MG. Interpreting the results of intention-to-treat, per-protocol, and as-treated analyses of clinical trials. JAMA. 2021;326:433–434. doi: 10.1001/jama.2021.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Klontzas ME, Gatti AA, Tejani AS, Kahn CE., Jr AI reporting guidelines: how to select the best one for your research. Radiol Artif Intell. 2023;5:e230055. doi: 10.1148/ryai.230055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Park SH, Han K, Jang HY, Park JE, Lee JG, Kim DW, et al. Methods for clinical evaluation of artificial intelligence algorithms for medical diagnosis. Radiology. 2023;306:20–31. doi: 10.1148/radiol.220182. [DOI] [PubMed] [Google Scholar]
- 162.Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20:405–410. doi: 10.3348/kjr.2019.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 164.Luo Y, Peng J, Ma J. When causal inference meets deep learning. Nat Mach Intell. 2020;2:426–427. [Google Scholar]
- 165.Vlontzos A, Rueckert D, Kainz B. A review of causality for learning algorithms in medical image analysis. [cited January 5, 2023];arXiv: 2206.05498v2 [Preprint] 2022 doi: 10.48550/arXiv.2206.05498. Available at: [DOI] [Google Scholar]
- 166.Wang R, Chaudhari P, Davatzikos C. Harmonization with flow-based causal inference. Med Image Comput Comput Assist Interv. 2021;12903:181–190. doi: 10.1007/978-3-030-87199-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pawlowski N, Coelho de Castro D, Glocker B. In: Advances in neural information processing systems 33 (NeurIPS 2020) Larochelle H, Ranzato M, Hadsell R, Balcan M, Lin H, editors. La Jolla: NIPS; 2020. Deep structural causal models for tractable counterfactual inference; pp. 857–869. [Google Scholar]
- 168.Pölsterl S, Wachinger C. In: Information processing in medical imaging. IPMI 2021. Lecture notes in computer science, vol 12729. Feragen A, Sommer S, Schnabel J, Nielsen M, editors. Cham: Springer; 2021. Estimation of causal effects in the presence of unobserved confounding in the Alzheimer’s continuum; pp. 45–57. [Google Scholar]
- 169.Zhuang J, Dvornek N, Tatikonda S, Papademetris X, Ventola P, Duncan JS. In: Information processing in medical imaging. IPMI 2021. Lecture notes in computer science, vol 12729. Feragen A, Sommer S, Schnabel J, Nielsen M, editors. Cham: Springer; 2021. Multiple-shooting adjoint method for whole-brain dynamic causal modeling; pp. 58–70. [Google Scholar]
- 170.Clivio O, Falck F, Lehmann B, Deligiannidis G, Holmes C. Neural score matching for high-dimensional causal inference; Proceedings of The 25th International Conference on Artificial Intelligence and Statistics; 2022 Mar 28-30; Valencia, Spain. PMLR; 2022. pp. 7076–7110. [Google Scholar]
- 171.da Silva M, Garcia K, Sudre CH, Bass C, Cardoso MJ, Robinson E. Biomechanical modelling of brain atrophy through deep learning. [cited January 5, 2023];arXiv: 2012.07596v1 [Preprint] 2020 doi: 10.48550/arXiv.2012.07596. Available at: [DOI] [Google Scholar]
- 172.McMahan B, Moore E, Ramage D, Hampson S, y Arcas BA. Communication-efficient learning of deep networks from decentralized data; Proceedings of the 20th International Conference on Artificial Intelligence and Statistics (AISTATS 2017); 2017 Apr 20-22; Ft. Lauderdale, FL, USA. PMLR; 2017. pp. 1273–1282. [Google Scholar]
- 173.Mothukuri V, Parizi RM, Pouriyeh S, Huang Y, Dehghantanha A, Srivastava G. A survey on security and privacy of federated learning. Future Gener Comput Syst. 2021;115:619–640. [Google Scholar]
- 174.Joshi M, Pal A, Sankarasubbu M. Federated learning for healthcare domain-pipeline, applications and challenges. ACM Trans Comput Healthc. 2022;3:1–36. [Google Scholar]
- 175.Lu MT, Ivanov A, Mayrhofer T, Hosny A, Aerts HJWL, Hoffmann U. Deep learning to assess long-term mortality from chest radiographs. JAMA Netw Open. 2019;2:e197416. doi: 10.1001/jamanetworkopen.2019.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lu MT, Raghu VK, Mayrhofer T, Aerts HJWL, Hoffmann U. Deep learning using chest radiographs to identify high-risk smokers for lung cancer screening computed tomography: development and validation of a prediction model. Ann Intern Med. 2020;173:704–713. doi: 10.7326/M20-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Raghu VK, Weiss J, Hoffmann U, Aerts HJWL, Lu MT. Deep learning to estimate biological age from chest radiographs. JACC Cardiovasc Imaging. 2021;14:2226–2236. doi: 10.1016/j.jcmg.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sabottke CF, Breaux MA, Spieler BM. Estimation of age in unidentified patients via chest radiography using convolutional neural network regression. Emerg Radiol. 2020;27:463–468. doi: 10.1007/s10140-020-01782-5. [DOI] [PubMed] [Google Scholar]
- 179.Li D, Lin CT, Sulam J, Yi PH. Deep learning prediction of sex on chest radiographs: a potential contributor to biased algorithms. Emerg Radiol. 2022;29:365–370. doi: 10.1007/s10140-022-02019-3. [DOI] [PubMed] [Google Scholar]
- 180.Yi PH, Wei J, Kim TK, Shin J, Sair HI, Hui FK, et al. Radiology “forensics”: determination of age and sex from chest radiographs using deep learning. Emerg Radiol. 2021;28:949–954. doi: 10.1007/s10140-021-01953-y. [DOI] [PubMed] [Google Scholar]
- 181.Yang CY, Pan YJ, Chou Y, Yang CJ, Kao CC, Huang KC, et al. Using deep neural networks for predicting age and sex in healthy adult chest radiographs. J Clin Med. 2021;10:4431. doi: 10.3390/jcm10194431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gaser C, Franke K, Klöppel S, Koutsouleris N, Sauer H Alzheimer’s Disease Neuroimaging Initiative. BrainAGE in mild cognitive impaired patients: predicting the conversion to Alzheimer’s disease. PLoS One. 2013;8:e67346. doi: 10.1371/journal.pone.0067346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Franke K, Gaser C, Manor B, Novak V. Advanced BrainAGE in older adults with type 2 diabetes mellitus. Front Aging Neurosci. 2013;5:90. doi: 10.3389/fnagi.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Löwe LC, Gaser C, Franke K Alzheimer’s Disease Neuroimaging Initiative. The effect of the APOE genotype on individual BrainAGE in normal aging, mild cognitive impairment, and Alzheimer’s disease. PLoS One. 2016;11:e0157514. doi: 10.1371/journal.pone.0157514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cole JH, Annus T, Wilson LR, Remtulla R, Hong YT, Fryer TD, et al. Brain-predicted age in Down syndrome is associated with beta amyloid deposition and cognitive decline. Neurobiol Aging. 2017;56:41–49. doi: 10.1016/j.neurobiolaging.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cole JH, Underwood J, Caan MW, De Francesco D, van Zoest RA, Leech R, et al. Increased brain-predicted aging in treated HIV disease. Neurology. 2017;88:1349–1357. doi: 10.1212/WNL.0000000000003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cole JH, Poudel RPK, Tsagkrasoulis D, Caan MWA, Steves C, Spector TD, et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage. 2017;163:115–124. doi: 10.1016/j.neuroimage.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 188.Steffener J, Habeck C, O'Shea D, Razlighi Q, Bherer L, Stern Y. Differences between chronological and brain age are related to education and self-reported physical activity. Neurobiol Aging. 2016;40:138–144. doi: 10.1016/j.neurobiolaging.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luders E, Cherbuin N, Kurth F. Forever Young(er): potential age-defying effects of long-term meditation on gray matter atrophy. Front Psychol. 2015;5:1551. doi: 10.3389/fpsyg.2014.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ieki H, Ito K, Saji M, Kawakami R, Nagatomo Y, Koyama S, et al. Deep learning-based chest X-ray age serves as a novel biomarker for cardiovascular aging. [cited January 5, 2023];bioRxiv [Preprint] 2021 doi: 10.1101/2021.03.24.436773. Available at: [DOI] [Google Scholar]
- 191.Li Z, Li W, Yan W, Zhang R, Xie S. Data-driven learning to identify biomarkers in bipolar disorder. Comput Methods Programs Biomed. 2022;226:107112. doi: 10.1016/j.cmpb.2022.107112. [DOI] [PubMed] [Google Scholar]
- 192.Nam JG, Kang HR, Lee SM, Kim H, Rhee C, Goo JM, et al. Deep learning prediction of survival in patients with chronic obstructive pulmonary disease using chest radiographs. Radiology. 2022;305:199–208. doi: 10.1148/radiol.212071. [DOI] [PubMed] [Google Scholar]
- 193.Shengli W. Is human digital twin possible? Comput Methods Programs Biomed Update. 2021;1:100014 [Google Scholar]
- 194.Barricelli BR, Casiraghi E, Gliozzo J, Petrini A, Valtolina S. Human digital twin for fitness management. IEEE Access. 2020;8:26637–26664. [Google Scholar]
- 195.Laubenbacher R, Sluka JP, Glazier JA. Using digital twins in viral infection. Science. 2021;371:1105–1106. doi: 10.1126/science.abf3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Laamarti F, Badawi HF, Ding Y, Arafsha F, Hafidh B, El Saddik A. An ISO/IEEE 11073 standardized digital twin framework for health and well-being in smart cities. IEEE Access. 2020;8:105950–105961. [Google Scholar]
- 197.Laaki H, Miche Y, Tammi K. Prototyping a digital twin for real time remote control over mobile networks: application of remote surgery. IEEE Access. 2019;7:20325–20336. [Google Scholar]
- 198.Pang J, Huang Y, Xie Z, Li J, Cai Z. Collaborative city digital twin for the COVID-19 pandemic: a federated learning solution. Tsinghua Sci Technol. 2021;26:759–771. [Google Scholar]
- 199.Liu Y, Zhang L, Yang Y, Zhou L, Ren L, Wang F, et al. A novel cloud-based framework for the elderly healthcare services using digital twin. IEEE Access. 2019;7:49088–49101. [Google Scholar]
- 200.Martinez-Velazquez R, Gamez R, El Saddik A. Cardio twin: a digital twin of the human heart running on the edge; Proceedings of the 2019 IEEE International Symposium on Medical Measurements and Applications (MeMeA); 2019 Jun 26-28; Istanbul, Turkey. IEEE; 2019. pp. 1–6. [Google Scholar]
- 201.Jones G, Parr J, Nithiarasu P, Pant S. Machine learning for detection of stenoses and aneurysms: application in a physiologically realistic virtual patient database. Biomech Model Mechanobiol. 2021;20:2097–2146. doi: 10.1007/s10237-021-01497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hirschvogel M, Jagschies L, Maier A, Wildhirt SM, Gee MW. An in silico twin for epicardial augmentation of the failing heart. Int J Numer Method Biomed Eng. 2019;35:e3233. doi: 10.1002/cnm.3233. [DOI] [PubMed] [Google Scholar]
- 203.Ahmadian H, Mageswaran P, Walter BA, Blakaj DM, Bourekas EC, Mendel E, et al. A digital twin for simulating the vertebroplasty procedure and its impact on mechanical stability of vertebra in cancer patients. Int J Numer Method Biomed Eng. 2022;38:e3600. doi: 10.1002/cnm.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Aubert K, Germaneau A, Rochette M, Ye W, Severyns M, Billot M, et al. Development of digital twins to optimize trauma surgery and postoperative management. A case study focusing on tibial plateau fracture. Front Bioeng Biotechnol. 2021;9:722275. doi: 10.3389/fbioe.2021.722275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Batch KE, Yue J, Darcovich A, Lupton K, Liu CC, Woodlock DP, et al. Developing a cancer digital twin: supervised metastases detection from consecutive structured radiology reports. Front Artif Intell. 2022;5:826402. doi: 10.3389/frai.2022.826402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wu C, Jarrett AM, Zhou Z, Elshafeey N, Adrada BE, Candelaria RP, et al. MRI-based digital models forecast patient-specific treatment responses to neoadjuvant chemotherapy in triple-negative breast cancer. Cancer Res. 2022;82:3394–3404. doi: 10.1158/0008-5472.CAN-22-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Coorey G, Figtree GA, Fletcher DF, Redfern J. The health digital twin: advancing precision cardiovascular medicine. Nat Rev Cardiol. 2021;18:803–804. doi: 10.1038/s41569-021-00630-4. [DOI] [PubMed] [Google Scholar]
- 208.Coorey G, Figtree GA, Fletcher DF, Snelson VJ, Vernon ST, Winlaw D, et al. The health digital twin to tackle cardiovascular disease-a review of an emerging interdisciplinary field. NPJ Digit Med. 2022;5:126. doi: 10.1038/s41746-022-00640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Corral-Acero J, Margara F, Marciniak M, Rodero C, Loncaric F, Feng Y, et al. The ‘digital twin’ to enable the vision of precision cardiology. Eur Heart J. 2020;41:4556–4564. doi: 10.1093/eurheartj/ehaa159. [DOI] [PMC free article] [PubMed] [Google Scholar]