Abstract
Understanding the molecular and phenotypic heterogeneity of cancer is a prerequisite for effective treatment. For chronic lymphocytic leukemia (CLL), recurrent genetic driver events have been extensively cataloged, but this does not suffice to explain the disease’s diverse course. Here, we performed RNA sequencing on 184 CLL patient samples. Unsupervised analysis revealed two major, orthogonal axes of gene expression variation: the first one represented the mutational status of the immunoglobulin heavy variable (IGHV) genes, and concomitantly, the three-group stratification of CLL by global DNA methylation. The second axis aligned with trisomy 12 status and affected chemokine, MAPK and mTOR signaling. We discovered non-additive effects (epistasis) of IGHV mutation status and trisomy 12 on multiple phenotypes, including the expression of 893 genes. Multiple types of epistasis were observed, including synergy, buffering, suppression and inversion, suggesting that molecular understanding of disease heterogeneity requires studying such genetic events not only individually but in combination. We detected strong differentially expressed gene signatures associated with major gene mutations and copy number aberrations including SF3B1, BRAF and TP53, as well as del(17)(p13), del(13)(q14) and del(11)(q22.3) beyond dosage effect.
Our study reveals previously underappreciated gene expression signatures for the major molecular subtypes in CLL and the presence of epistasis between them.
Introduction
Chronic lymphocytic leukemia (CLL) etiology has been linked to abnormal B-cell receptor (BCR) activation and gene mutations targeting multiple pathways, including DNA damage pathways (TP53, ATM), NOTCH signaling (NOTCH1, FBXW7, MED12)1-6 and the spliceosome (SF3B1).7,8 In addition, the immunoglobulin heavy variable (IGHV) gene mutation status, the result of a physiological mutation and maturation process, reflects the tumor’s cell of origin and is one of the strongest predictors of clinical behavior.9 Several genetic subgroups of CLL are known to show profound differences in clinical course, presentation and outcome,10,11 although considerable variability remains within subgroups. Gene expression profiling can provide a better understanding of the functional role of mutations and may help dissect disease heterogeneity. Indeed, previous studies of CLL transcriptomes found substantial variability,12-17 however, it has been a surprise how little of that variability could be associated with the genetic subgroups or other properties of the disease. For instance, Ferreira et al.12,13 found only a few robust gene expression changes associated with the major cancer driver mutations of CLL. IGHV mutation status only accounted for 1.5% of the overall variance in their study. Their study reported two gene expression-based subgroups, termed C1/C2, as a predictor of clinical outcome independent of the known genetic disease groups. However, a later re-analysis of the data suggested a relation of C1/C2 to sample processing.18 Overall, the relations between prominent genetic events that have significant impact on disease course and the gene expression programs of CLL have remained unclear. Among possible explanations for this scarcity of associations are small sample sizes, confounding effects of multiple cytogenetic abnormalities or technical limitations. More recent studies have thus collected larger cohorts with focus on a particular genetic aberration. Abruzzo et al.15 identified a set of dysregulated and potentially targetable pathways in CLL with trisomy 12. Herling et al.16 developed a 17-gene signature that can identify a subset of treatment-naive patients with IGHV-unmutated CLL (U-CLL) who might substantially benefit from treatment with FCR (fludarabine, cyclophosphamide and rituximab) chemoimmunotherapy. These findings underline the importance of transcriptional changes in CLL. Since they were based on focused studies limited to specific, selected subtypes of CLL, one may expect a more systematic picture and additional insights from a comprehensive RNA sequencing-based survey.
In order to understand the impact of genetic and epigenetic subgroups of CLL on gene expression, we profiled 184 CLL samples using RNA sequencing. After careful control of technical variations, and of possible confounding between genetic variants, we searched for transcriptomic signatures and pathway activity changes associated with the major recurrent genetic alterations in CLL. Furthermore, as a step towards gaining a better understanding of functional interdependencies between mutations in a tumor, we used a quantitative model of genetic interactions to identify nonadditive effects of mutations on gene expression profiles.
Methods
Data acquisition
RNA sequencing
We selected 184 CLL patient samples for RNA sequencing. One hundred and twenty-three of these patients were used in a prior study.19 The current study is an extension, designed specifically to increase sample sizes of major molecular subgroups and focus on gene expression. The majority of patients (177 of 184) showed the typical CLL phenotype, and five patients were diagnosed with atypical CLL. Ninety-two patients had undergone prior treatment. Patient characteristics are shown in the Online Supplementary Table S1. Total RNA was isolated from blood samples (CD19+ purified n=161) and sequenced using Illumina HiSeq. Sequenced reads were mapped to the Ensembl human reference genome (Homo sapiens GRCh37.75) using STAR20 version 2.5.2a. Mapped reads were summarized in per gene counts using htseq-count.21 version 0.9.0. Further details are provided in the Online Supplementary Appendix.
Somatic variants
Mutation calls for 66 distinct gene mutations and 22 structural variants were generated through targeted sequencing, whole-exome sequencing and whole-genome sequencing as described previously.19 Statistical analyses were restricted to variants found in ≥5 patients, i.e., to 14 gene mutations (BRAF, NOTCH1, SF3B1, TP53, KRAS, ATM, MED12, EGR2, KLHL6, ACTN2, MGA, NFKBIE, PCLO, XPO1), and nine copy number aberrations (CNA): trisomy 12, del(11)(q22.3), del(13)(q14), del(17)(p13), del(8)(p12), gain(8)(q24), gain(2)(p25.3), del(15)(q15.1), gain(14)(q32) (Online Supplementary Figure S2B). In addition, the somatic hypermutation status of the IGHV and a CLL subtype classification defined by global patterns of CpG methylation level22,23 were recorded. Here, we discuss results for variants with >200 differentially expressed genes detected: four CNA, three gene mutations and IGHV mutation status. The somatic mutation information is available in our online repository: https://github.com/almutlue/transcriptome_cll and summarized in the Online Supplementary Table S2.
Drug response profiling
For 113 of 184 samples, drug response profiles were reported in a previous study.24
Statistical analysis
Statistical analyses were performed using R25 version 3.6. We performed quality controls including batch effect estimation26 (Online Supplementary Figure S1), exploratory data analysis and differential gene expression analysis using the Gamma-Poisson generalized linear modeling (GLM) approach of DESeq2, version 1.16.1.27,28 Genetic interactions were identified by testing for an interaction term in the regression of gene expression data on the two variables IGHV mutation status and trisomy 12 using DESeq2. For the validation study, we analyzed microarray data from Abruzzo et al.15 using the R package limma version 3.50.1.29 Gene set enrichment analysis30 was performed using the R package clusterProfiler31 version 3.12.0. Hallmark and KEGG gene set collections version 4.0 were downloaded from MsigDB.32 Transcription factor target genes sets were downloaded from Harmonizome.33 All P values were adjusted for multiple testing using the method of Benjamini and Hochberg.34 Further details are provided in the Online Supplementary Appendix.
Study approval
The study was approved by the Ethics Committee Heidelberg (University of Heidelberg, Germany; S-206/2011; S-356/2013) and Zurich, Switzerland (2019-01744).
Results
Unsupervised analysis reveals major drivers of gene expression variability
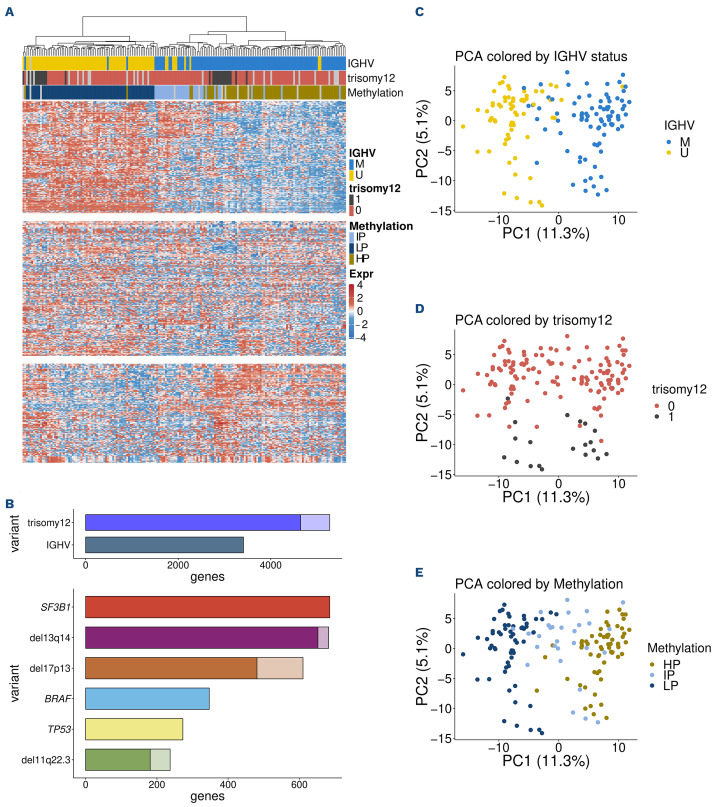
We generated RNA-sequencing data from 184 CLL patient samples. In order to obtain a first overview of patterns of gene expression variability in CLL, we performed an unsupervised clustering analysis based on the 500 most variable genes (Figure 1A). This analysis showed a separation of distinct subgroups that coincided very well with IGHV mutation status/methylation epitype and the presence of trisomy 12. The role of IGHV mutation status and trisomy 12 was also reflected in the number of differentially expressed (DE) genes (>3,000 DE genes) (Figure 1B). A similar separation was seen in a principal component analysis (PCA) (Figure 1C, D). The first principal component, which represented 11% of the variance, was associated with IGHV mutation status, while the second component separated samples based on trisomy 12. These results indicate that these two genetic variables shape gene expression in CLL to a previously underappreciated extent. We also considered a classification of CLL based on global DNA methylation levels into three groups according to previous studies,23,24 a refinement of the binary grouping by IGHV mutation status (Figure 1E). The first principal component arranged the DNA methylation subgroups in the consistent order: low, intermediate and high programmed (LP, IP, HP). These results indicate that even though the three-group classification was discovered using DNA methylation data, it is now also apparent at the level of gene expression. Indeed, the global gene expression patterns shown in Figure 1 imply a further refinement into major groups, namely LP, IP and HP each with and without trisomy 12.
The results of our unsupervised clustering analysis differ from those of a previous gene expression study, which also used unsupervised clustering of RNA-sequencing data to find novel subgroups of CLL termed C1/C2, marked by 600 differentially expressed genes and associated with BCR activation and outcome.12 In our data, hierarchical clustering of the samples based on the measurements of these 600 genes only, indeed resulted in two main clusters. However, most of these genes showed low variability across samples and only 26 of them were among the 500 most variable genes (Online Supplementary Figure S2).
Mutations modulate gene expression in chronic lymphocytic leukemia
We performed differential expression analysis to explore the effect of 23 recurrent genetic aberrations and the IGHV mutation status (Online Supplementary Figure S3). In total, we found six additional variants (besides trisomy 12 and the IGHV status) associated with more than 200 differentially expressed genes. These were del(13)(q14), del(17)(p13), del(11)(q22), and SF3B1, TP53 and BRAF mutations (Figure 1B). Complete tables are provided in the computational analysis transcript. We compared previous findings from the literature for single genetic aberrations with differentially expressed genes in this study.
Mutations in the splicing factor SF3B1 gene showed more than 600 associations. Gene sets enriched in CLL with SF3B1 mutations included “Cytokine-cytokine receptor interaction” and “Phosphatidylinositol signaling system” (Online Supplementary Figure S4A). Among differentially expressed genes, we found the chaperone gene UQCC1 (Online Supplementary Figure S4B), which has already been linked to SF3B1 mutations by differential isoform usage.35 Indeed, a differential exon usage analysis using DEXSeq27 showed that UQCC1 had both differential expression and differential exon usage (Online Supplementary Figure S5A, B; Online Supplementary Table S3). There were also instances of genes that had differential exon usage, but for which no gene level differential expression was detected (Online Supplementary Figure S5C, E). These included BRD9, a tumor suppressor whose splicing has also been reported to be regulated by SF3B1.36,37 A recent proteomic study in CLL confirmed the downregulation of BRD9 in samples with SF3B1 mutations.37 A potential explanation is that the SF3B1 mutation leads to a mis-spliced version of the BRD9 transcript whose level is not detectably altered in gene level RNA-sequencing analysis but whose translation is impaired. Conversely, there were genes detected by gene level differential expression analysis but not by differential exon usage analysis, including PSD2, SRRM5 and TNXB (Online Supplementary Figure S4C-E).
TP53 mutations are recurrent in CLL and are associated with poor prognosis.1 Differentially expressed genes in samples with TP53 mutation were enriched in “Oxidative phosphorylation” and “p53 signaling pathway” (Online Supplementary Figure S6A). The transcriptional regulator CDK12 is upregulated in TP53-mutated samples (Online Supplementary Figure S6C). In order to understand the overlap of genes deregulated by 17p deletion and p53 mutation we analyzed the overlap between those two variants (Online Supplementary Figure S6B). In total, 76 of 272 differentially expressed genes associated with TP53 mutation were also differentially expressed in samples with del(17)(p13). As del(17)(p13) includes the region of the TP53 gene, the overlap is to be expected. Further associations with TP53 include PGBD2 and HYPK (Online Supplementary Figure S6D, E)).
Immunoglobulin heavy variable gene mutation status is linked to distinct gene expression changes
The second highest number of differentially expressed genes was found in the comparison between IGHV-mutated CLL (M-CLL) and IGHV-unmutated CLL (U-CLL): 3,410 genes. This result is in agreement with the PCA of Figure 1C and shows that IGHV mutation status is the main determinant of gene expression variability in CLL. It implies a much larger impact of IGHV status on the transcriptome than previously detected (11.3% variance instead of 1.5%12 explained by the associated principal component in PCA) is in line with the key impact of IGHV mutation status on clinical course and biology of disease.9-11 Our observation is also consistent with recent reports that IGHV status is one of the major determinants of the protein expression landscape in CLL.37, 3 8 Genes previously found to be markers related to IGHV mutation status, including CD38, LPL, ZAP70, SEPT10 and ADAM29,39-41 were also detected in our analysis (Online Supplementary Table S4).
Figure 1.
Gene expression variability in chronic lymphocytic leukemia. (A) Heatmap of the gene expression counts. Samples (columns) are ordered in agreement with the hierarchical clustering based on the 500 most variable genes. Gene (rows) counts are row-centered log-transformed (base 2) and split into the 3 main hierarchical clusters. Immunoglobulin heavy variable (IGHV) gene mutation status, methylation subgroups and trisomy 12 align with the clustering result. (B) Number of differentially expressed genes (adjusted [adj] P values <0.01) for genomic markers in chronic lymphocytic leukemia (CLL). Lighter colors indicate genes located on the same chromosome as the respective genetic lesion (potential dosage effects). (C) IGHV status is associated with the first principal component, which explains 11.3% of the variance. (D) Trisomy 12 is associated with thesecond principal component (PC2), which explains 5.1% of the variance. (E) Methylation subgroups split up along principal component 1 (PC1).
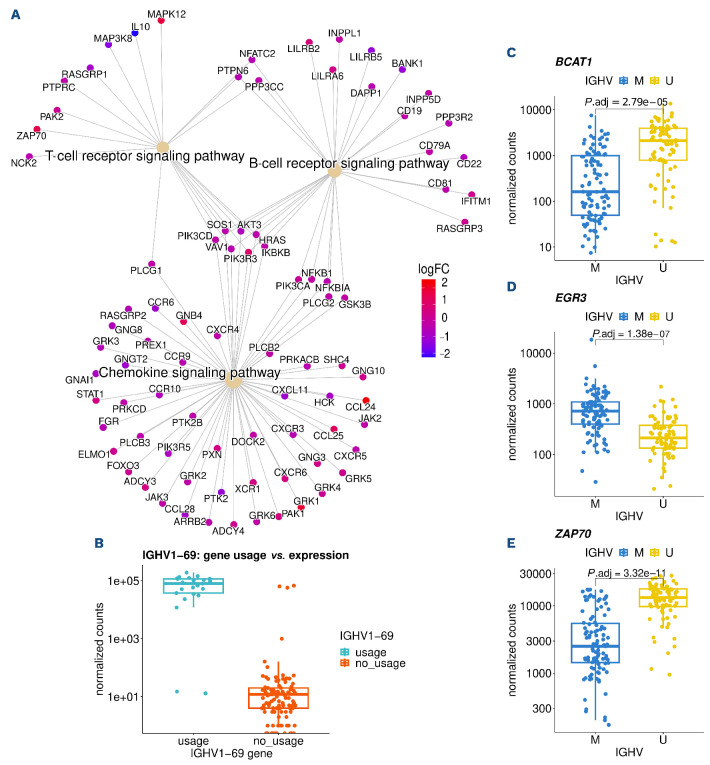
In order to understand which pathways were differentially engaged between U-CLL and M-CLL, we performed gene set enrichment analysis. Differentially expressed genes between IGHV groups were enriched in BCR signaling, T-cell receptor signaling and chemokine signaling pathways (Figure 2A). Within the BCR signaling gene set, we identified cell surface molecules (CD19, CD22, CD81) and NFAT and NF-κB to be downregulated in U-CLL. From the “T-cell receptor signaling” gene set, ZAP70, PAK2 and MAPK12 were upregulated in U-CLL, while IL10 and MAP3K8 were down-regulated. Within chemokine signaling pathways, we found downregulation of CXCR3 and CXCR5 in U-CLL, while a set of cytokines (CCL24, CCL25) were upregulated.
IGHV genes were also found among the most differentially expressed genes, but showed heterogeneous expression within the U-CLL and M-CLL groups. As expected, commonly used IGHV genes (IGHV1-69 or IGHV4-34) were associated with U-CLL and M-CLL, respectively. Gene expression showed a strong relation to IG gene usage and its variant’s expression (Figure 2B). These data show that RNA sequencing can be used to assess IG gene usage. Further genes associated with IGHV groups were BCAT1, EGR3 and ZAP70 (Figure 2C-E)
In summary, our data are in line with the major biological role of IGHV mutation status in CLL and provide a resource to identify deregulated pathways in the disease.
Intermediate programmed methylation group forms an independent gene expression cluster
Based on the global DNA methylation pattern, the stratification of CLL by IGHV mutation status has been refined by introducing a categorization into LP, IP, and HP programmed samples, which are thought to represent the cell of ori-gin.22,23 Based on gene expression data, we found these three groups along the first principle component. The IP group was placed between the LP and HP groups (Figure 1E). Thus, even though the groups were discovered on the basis of DNA methylation, a strong separation was found on the basis of unsupervised PCA of the gene expression data. Previous analysis of methylation groups in CLL suggested a disease-specific role of the transcription factors EGR, NFAT, AP1 and EGF by establishing aberrant methylation patterns.22,23 In line with this, we found NFATC1 and EGR1 among genes whose expression patterns were associated with methylation groups (Online Supplementary Figure S7A, B). A detailed analysis of the intermediate methylated subgroup revealed multiple genes including SOX11, that were specific for this subgroup (Online Supplementary Figure S7C). SOX11 is a transcription factor whose expression has been associated with adverse prognostic markers in CLL.43
Expression signature in chronic lymphocytic leukemia with trisomy 12
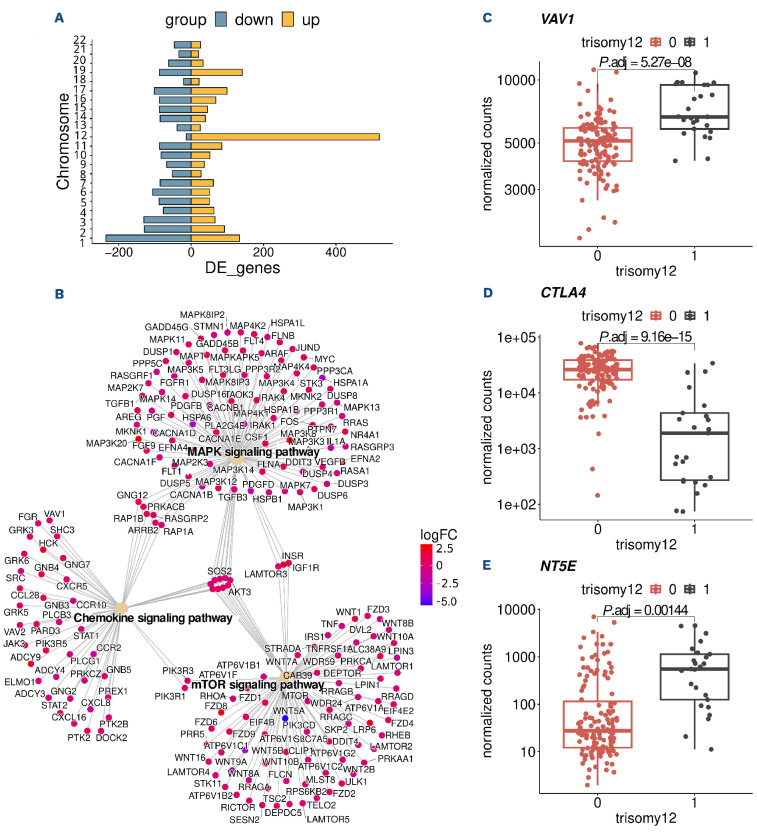
We identified over 5,000 differentially expressed genes (with adjusted P value <0.05) in CLL with trisomy 12 (Figure 1B). Even though chromosome 12 harbors many upregulated genes, the majority of differentially expressed genes were on other chromosomes and, therefore, cannot be ascribed to a simple gene dosage effect (Figure 3A).
Among the differentially expressed genes, we found numerous genes involved in chemokine signaling such as VAV1 (Figure 3B, C). Chemokine signaling pathways are induced by chemokine binding and activate MAPK signaling.44,45 In line with this, we identified differentially expressed genes enriched in MAPK signaling. We also detected an enrichment for the mTOR-signaling pathway, a known modulator of chemokine signaling46 (Figure 3B). Consistent with previous reports, integrins like ITGAM, ITGB2 and ITGA4 were also upregulated in trisomy 12 samples15 (Online Supplementary Table S4). We also found the immune checkpoint gene CTLA4 (Figure 3D) downregulated in trisomy 12 samples. CTLA4 has previously been linked to CLL and is associated with increased proliferation and tumor progres-sion.47, 4 8 By comparison with published protein expression data, we found the protein expressions of VAV1 and ITGB2 were also strongly upregulated in trisomy 12 CLL (Online Supplementary Figure S8). Chemokine signaling and mTOR-signaling pathways in trisomy 12 CLL have also been shown to be upregulated on protein level.38
A known mechanism of tumor cells to escape the immune system is to inhibit tumor-specific T cells, and support the conversion of anti-tumor type 1 macrophages to pro-tumor type 2 macrophages by upregulation of ecto-5′-nucleoti-dase (NT5E), which is necessary to convert extracellular ATP into adenosine. A previous study on gene expression in trisomy 12 patients inferred NT5E to be an important element in a trisomy 12 expression network model.15 This inference was indirect, as the microarray-based study only quantified selected transcripts, excluding NT5E. Here, we directly observed higher expression of NT5E in trisomy 12 and thus can confirm the hypothesis of Abruzzo et al.15 (Figure 3E). Another study of Tsagiopolou et al.49 studied epigenetic regulatory elements in CLL with trisomy 12 and found several transcription factors in particular RUNX3, which is a master regulator of gene expression during development and oncogene in cancer, to be up regulated. We tested differentially expressed genes in trisomy 12 for enrichment of transcription factor target genes sets and found target genes of RUNX3 to be among the top enriched genes sets (Online Supplementary Figure S9).
Figure 2.
Gene expression changes between immunoglobulin heavy variable gene subgroups. (A) Differentially expressed genes in enriched KEGG pathways for immunoglobulin heavy variable (IGHV) genes. (B) IGHV1-69 expression by corresponding IGHV1-69 gene usage determined by IG gene analysis. (C-E) Normalized gene counts for BCAT1, EGR3 and ZAP70 separated by IGHV mutation status.
Altogether, these results confirm and expand on the results of existing studies. They suggest that modulation of MAPK signaling through chemokine signaling and mTOR signaling are important mechanisms in trisomy 12 tumorigenesis.
Epistatic interaction of immunoglobulin heavy variable gene mutation status and trisomy 12
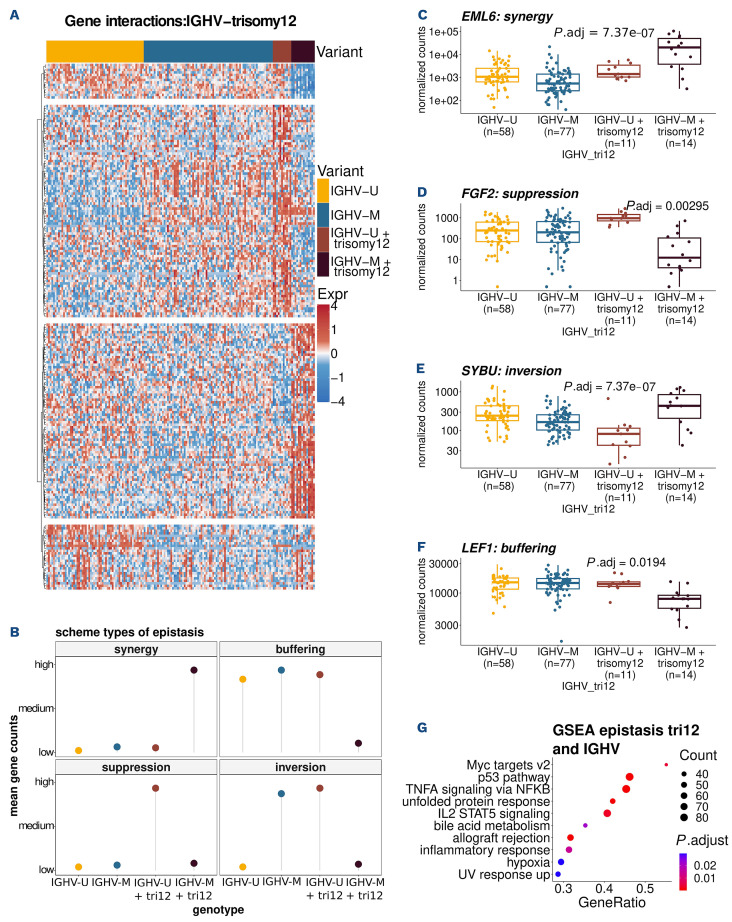
Epistasis describes a phenomenon where the effect of a genetic variant depends on the presence or absence of another genetic variant.50 There is almost no data on epistasis between cancer mutations. Because of the large effects of each of these variants individually, we asked whether and to what extent epistatic interactions existed between IGHV mutation status and trisomy 12. We fit a generalized linear model (DESeq2, see Methods) with main and interaction effects for these co-variates. Significant interactions were detected for 893 genes at 10% false discovery rate (FDR) (Figure 4A; Online Supplementary Table S5). For these genes, the effect of trisomy 12 was different in U-CLL and M-CLL. We observed four distinct types of epistatic interactions51,52 and classified the 893 genes into according categories: synergy, where the samples with both variants, i.e., M-CLL samples with trisomy 12, showed a stronger up-regulation than expected from the single variants; bufering, when the presence of both variants led to a strong reduction of gene expression; inversion, when the effects in the single variants were reversed in the double variant; suppression, when a strong expression change (up- or downregulation) of a gene in a single variant was absent in samples with both variants (Figure 4B). Figure 4C-F shows the count data for exemplary genes. EMAP-like 6 (EML6) is upregulated in all trisomy 12 cases, but this effect is on average about 1,000 times stronger in M-CLL patients compared to U-CLL patients (synergy) (Figure 4C). While fibroblast growth factor 2 (FGF2) is consistently up-regulated in trisomy 12 cases with U-CLL, this effect is reversed in M-CLL (suppression) (Figure 4D). SYBU shows an inverse expression pattern in M-CLL cases with trisomy 12 compared to U-CLL cases with trisomy 12 (Figure 4E). Lymphoid enhancer binding factor 1 (LEF-1) shows a stable gene expression across samples and has been suggested and tested as a clinical marker for CLL.53 While the presence of either one of the genetic variants (IGHV-M or trisomy 12) does not seem to have an effect, samples with both of them express consistently lower levels of LEF1 (buffering) (Figure 4F). These effects cannot be explained by looking at the genotypes independently or by modeling an additive effect of the variants.
Figure 3.
Gene expression in chronic lymphocytic leukemia with trisomy 12. (A) Role of dosage effect: chromosomal distribution of differentially expressed (DE) genes in chronic lymphocytic leukemia (CLL) with trisomy 12. Chromosome 12 has the highest number of DE genes, but the majority of DE genes is distributed across all chromosomes and cannot be ascribed to a dosage effect. (B) DE genes in enriched KEGG pathways of trisomy 12. (C-E) Normalized gene counts of VAV1, CTLA4 and NT5E.
We next asked about the biological functions underlying the epistatic interaction between IGHV status and trisomy 12. We used gene set enrichment analysis on the combined set of all 893 genes. The overall set of genes with an epistasis expression pattern was enriched in TNF α signaling via NF-κB, MYC targets, IL2/STAT5 signaling and G2M checkpoint pathway (Figure 4G). A recent study linked NFκB expression with reduced levels of B-cell signaling.54 Both IGHV status and trisomy 12 are known to affect BCR signaling, but the above data may suggest that signaling in IGHV-mutated CLL is mediated by BCR plus additional survival signals. In order to further investigate these findings, we used an independent cohort of samples from 47 patients with known trisomy 12 and IGHV status assayed on an Illumina microarray with 47,231 probes15 and again screened for epistatic interaction by testing for interaction effects in the linear model. In line with our results, we found multiple probes (100 probes with adjusted P value ≤0.17) that show the same epistatic interaction patterns in their expression for each of the four types of epistasis (Online Supplementary Figure S10A). Furthermore, we investigated the expression of the above-mentioned genes with epistatic interaction (EML6, FGF2, SYBU, LEF-1) in particular (Online Supplementary Figure S10B-E). We found significant epistatic expression patterns for FGF2, SYBU and could assign both genes to the same epistasis groups as in our cohort. While there was no significant epistatic interaction in the expression pattern of EML6 and LEF-1 their expression trends were in line with the expression of the epistatic groups (synergy, buffering) these genes were assigned to in our cohort. The protein expressions of two of those genes, FGF2 and LEF-1, could also be detected in our proteomics dataset38 and were found to have epistatic interactions (Online Supplementary Figure S11).
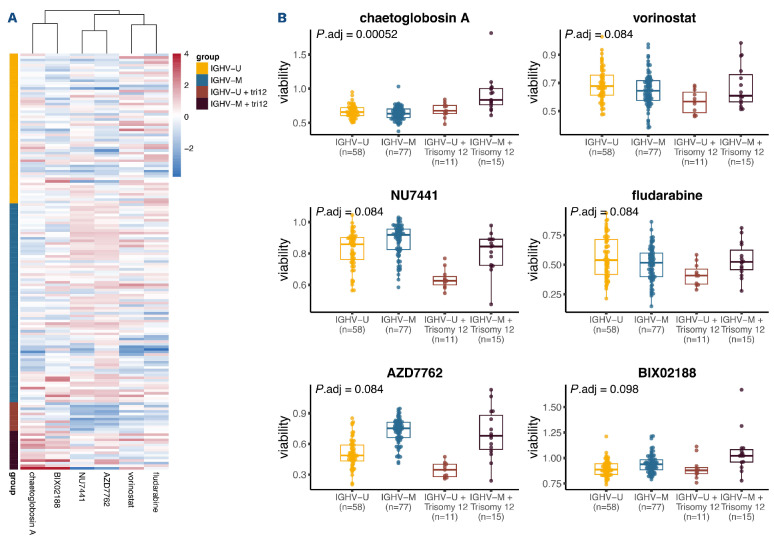
The epistatic interaction between immunoglobulin heavy variable gene mutation status and trisomy 12 affects ex vivo drug response in chronic lymphocytic leukemia
Ex vivo sensitivity to drugs reflects pathway dependencies of CLL cells. We asked whether the epistatic interaction between IGHV mutation status and trisomy 12 on expression level also affected the drug response phenotype. Previously, we measured the ex vivo sensitivity, as measured by cell viability, of our 184 CLL samples towards 63 compounds.24 Again using two-way ANOVA with an interaction term, we identified six drugs for which there was a significant (10% FDR) interaction between IGHV mutation status and trisomy 12 (Figure 5). For four drugs, namely, vorinostat, NU7441, fludarabine and AZD7762, we observed a suppression effect, where trisomy 12 led to increased drug sensitivity in the U-CLL, but not the M-CLL samples. For the other two drugs, chaetoglobosin A and BIX02188, the samples with trisomy 12 showed decreased sensitivity particularly in M-CLL, but not in U-CLL. The four drugs with the suppression phenotype directly or indirectly target DNA: NU7441 inhibits DNA-dependent protein kinase (DNA-PK) and, therefore, potentiate DNA double-strand breaks;55 AZD7762 is a checkpoint kinase (CHEK) inhibitor, which can impair DNA repair and increases cell death;56 fludarabine directly inhibits DNA synthesis and disrupt cell cycle;57 vorinostat, a histone deacetylase (HDAC) inhibitor, was also reported to induce reactive oxygen species and DNA damage in leukemia cells.58 As 42 of 161 patients in the drug screening dataset were treated prior to the acquisition of the samples, we also repeated these analyses only in untreated patient samples. We found consistent results (Online Supplementary Figure S12), suggesting no substantial effect of prior treatment on our findings.
Discussion
We analyzed 184 CLL transcriptomes and identified gene expression signatures for the most prevalent genetic aberrations. We show that these can be used to capture underlying pathways. The IGHV mutation status, the three DNA methylation subgroups and trisomy 12 were found as the main drivers of gene expression variability in CLL. This is evident both in unsupervised analyses (clustering, PCA) and in supervised differential-expression analysis. We revealed a much higher impact of IGHV mutation status on the CLL transcriptome than previous reports.12 A further refinement of the disease stratification by IGHV status is provided by the three DNA methylation subgroups. We identified genes whose expression follows an apparent continuum from LP, IP to HP in CLL. This finding supports the biological relevance of these three groups and suggests that although they were discovered based on DNA methylation, they are similarly evident at the level of gene expression. These results highlight the potential of gene expression profiling to increase our understanding of CLL.
Figure 4.
Mixed epistasis of trisomy 12 and immunoglobulin heavy variable gene mutation status. (A) Heatmap showing the expression of genes affected by the epistatic interactions between trisomy 12 and immunoglobulin heavy variable (IGHV) genemutated chronic lymphocytic leukemia (M-CLL) (adjusted [adj] P value <0.1). (B) Schematic classification of epistasis. (C-F) Types of gene expression epistasis: EML6 (synergy), FGF2 (suppression), SYBU (inversion), LEF1 (buffering). (G) Enriched pathways in genes with epistasis expression pattern.
Figure 5.
Ex vivo drug response phenotype related to the epistatic interaction between immunoglobulin heavy variable gene status and trisomy 12. (A) Heatmap plot showing the responses of chronic lymphocytic leukemia (CLL) samples (rows) towards the 6 drugs (columns) for which there was a significant interaction between immunoglobulin heavy variable (IGHV) gene mutation status and trisomy 12. The coloring encodes the column-wise z-scores of sample viabilities after drug treatment. (B) Boxplots of the viabilities (normalized to dimethyl sulfoxide [DMSO] controls) of CLL samples, stratified by their IGHV and trisomy 12 status, towards the 6 drugs shown in the heatmap. The IGHV-unmutated (IGHV-U) and IGHV-mutated (IGHV-M) groups contain unmutated-CLL (U-CLL) and mutated-CLL (M-CLL) samples, respectively, without trisomy 12. The IGHV-U+tri12 and IGHV-M+tri12 groups contain U-CLL and M-CLL samples, respectively, with trisomy 12.
In order to avoid potential confounding effects of multiple aberrations, other studies have focused on samples with single abnormalities. While this approach was successfully used to understand trisomy 12 specific gene expression, it is limited to only a subset of the disease and to selected variants.15 Here, we demonstrate an improved approach that employs differential expression analysis with multi-variate generalized linear models and blocking factors, and that is able to use the full range of CLL and to investigate a larger number of genetic aberrations.
Genetic interactions, or epistasis, where the effect of one mutation depends on the presence or absence of another mutation, is a well known concept in genetics. However, there is surprisingly little data on such phenomena in cancer. Hence, we used the opportunity to study the combinatorial effects of trisomy 12 and the IGHV mutation status on gene expression variability. We identified numerous genes (~900) whose expression depended on the presence or absence of these two aberrations in a non-additive manner. We categorized these genes into four categories: buffering, synergy, suppression and inversion, each of which contained dozens to hundreds of genes. This means that there is not a single, simple epistasis phenotype between these two aberrations, but a complex, “mixed” pattern. Mixed epistasis of the gene expression phenotypes of pairs of gene alterations has been described in a yeast model system.52 We employed the genetic interactions between IGHV mutation status and trisomy 12 on gene expression phenotypes to identify pathways, including TNF α signaling via NF-κB and the G2M checkpoint pathway, that mediate the effects of these variants. We reproduced this finding in an independent cohort of 47 patients.15
Our results raise the question whether the pathobiology of trisomy 12 may be different between U-CLL and M-CLL. Based on previous studies, the additional copy of chromosome 12 in CLL cells seems to directly enhance BCR signaling, especially in U-CLL samples: for example, Abruzzo et al. found that NFAT is overexpressed in trisomy 12 CLL, indicating the hyperactivation of calcineurin/NFAT signaling, which is central to BCR signaling.15 On proteomic levels, our previous study suggested that proteins upregulated in trisomy 12 CLL are enriched in BCR signaling pathway.38 In addition, genes located on chromosome 12 and upregulated in trisomy 12 CLL can also give rise to proteins that form protein complexes with BCR components.38 This evidence indicates that trisomy 12 status directly or indirectly modulates BCR signaling. As IGHV status is also a determinant of BCR signaling in CLL, it is reasonable that trisomy 12 regulate the downstream of BCR signaling differentially in IGHV mutated and unmutated CLL cells, i.e., epistasis. We also observed that the epistatic interaction between trisomy 12 and IGHV had an impact on the ex vivo responses to drugs, including those targeting DNA damage response. The effect on drug response levels is currently unclear, but may be in line with the observation on transcriptomic levels that the G2M checkpoint pathway is affected by the epistatic interaction.
Further mechanistic studies are needed to clarify the combinatorial effects of IGHV status and trisomy 12 on the transcriptome, on drug response phenotypes, and the links between these. For studies on CLL biology, the interaction of IGHV and trisomy 12 status need to be considered when gene expression profiles are investigated.
This study provides evidence of epistatic interactions in human cancer. Both the phenomenon of epistasis between cancer drivers, and the observation that it can be ‘mixed’ (follow different patterns) for different phenotypes form a new layer of complexity in CLL and in tumor biology more generally. Hence, our study highlights the inherent limitations of studying individual cancer genetic lesions, and points to the need to also map out and understand how they interact and modify each other’s effects.
Supplementary Material
Acknowledgments
We thank members of the Huber and Zenz research teams for valuable discussions. We thank Hanno Glimm, Stefan Fröhling, Daniela Richter, Roland Eils, Peter Lichter, Stephan Wolf, Katja Beck, and Janna Kirchhof for infrastructure and program development within DKFZ- HIPO and NCT POP. For technical support and expertise, we thank the DKFZ Genomics and Proteomics Core Facility.
Funding Statement
Funding: The work was supported by the European Union (Horizon 2020 project SOUND under grant agreement number 633974) and the German Federal Ministry of Education and Research (TRANSCAN project GCH-CLL 143 under grant agreement number 01KT1610 and CompLS project MOFA under grant agreement number 031L0171A). TZ was supported by the UZH Clinical Research Priority Program “Next Generation Drug Response Profiling for Personalized Cancer Care”, the Swiss Cancer League (KFS-4439-02-2018), The LOOP Zurich (INTERCEPT) and the Monique-Dornonville-dela-Cour Stiftung. RR received funding from the Swedish Cancer Society, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, and Radiumhemmets Forskningsfonder, Stockholm.
References
- 1.Campo E, Cymbalista F, Ghia P, et al. TP53 aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018;103(12):1956-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi D, Gaidano G. ATM and chronic lymphocytic leukemia: mutations, and not only deletions, matter. Haematologica. 2012;97(1):5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi D, Rasi S, Fabbri G, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119(2):521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118(16):4313-4320. [DOI] [PubMed] [Google Scholar]
- 5.Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28(1):108-117. [DOI] [PubMed] [Google Scholar]
- 6.Wu B, Słabicki M, Sellner L, et al. MED12 mutations and NOTCH signalling in chronic lymphocytic leukaemia. Br J Haematol. 2017;179(3):421-429. [DOI] [PubMed] [Google Scholar]
- 7.Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37-50. [DOI] [PubMed] [Google Scholar]
- 8.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3):145-162. [DOI] [PubMed] [Google Scholar]
- 9.Rosenquist R, Ghia P, Hadzidimitriou A, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: updated ERIC recommendations. Leukemia. 2017;31(7):1477-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840-1847. [PubMed] [Google Scholar]
- 11.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848-1854. [PubMed] [Google Scholar]
- 12.Ferreira PG, Jares P, Rico D, et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014;24(2):212-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194(11):16391647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haslinger C, Schweifer N, Stilgenbauer S, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22(19):3937-3949. [DOI] [PubMed] [Google Scholar]
- 15.Abruzzo LV, Herling CD, Calin GA, et al. Trisomy 12 chronic lymphocytic leukemia expresses a unique set of activated and targetable pathways. Haematologica. 2018;103(12):2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herling CD, Coombes KR, Benner A, et al. Time-to-progression after front-line fludarabine, cyclophosphamide, and rituximab chemoimmunotherapy for chronic lymphocytic leukaemia: a retrospective, multicohort study. Lancet Oncol. 2019;20(11):1576-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloehdorn J, Braun A, Taylor-Weiner A, et al. Multi-platform profiling characterizes molecular subgroups and resistance networks in chronic lymphocytic leukemia. Nat Commun. 2021;12(1):5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvinge H, Ries RE, Ilagan JO, et al. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci USA. 2014;111(47):16802-16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietrich S, Oleś M, Lu J, et al. Drug-perturbation-based stratification of blood cancer. J Clin Invest. 2018;128(1):427-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin A, Gingeras TR. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics. 2015;51:11.14.1-11.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W. HTSeq - a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakes CC, Seifert M, Assenov Y, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48(3):253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236-1242. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich S, Oleś M, Sellner L, et al. Drug perturbation based stratification of lymphoproliferative disorders. Hematol Oncol. 2017;35(S2):56-56. [Google Scholar]
- 25.R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/ Accessed 12 June 2023. [Google Scholar]
- 26.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11(10):733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouillard AD, Gundersen GW, Fernandez NF, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford). 2016;2016:baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289-300. [Google Scholar]
- 35.Reyes A, Blume C, Pelechano V, et al. Mutated SF3B1 is associated with transcript isoform changes of the genes UQCC and RPL31 both in CLLs and uveal melanomas. BioRxiv. 2013. July 13. doi: 10.1101/000992 [preprint, not peer-reviewed]. [DOI] [Google Scholar]
- 36.Inoue D, Chew G-L, Liu B, et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019;574(7778):432-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbst SA, Vesterlund M, Helmboldt AJ, et al. Proteogenomics refines the molecular classification of chronic lymphocytic leukemia. Nat Commun. 2022;13(1):6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier-Abt F, Lu J, Cannizzaro E, et al. The protein landscape of chronic lymphocytic leukemia. Blood. 2021;138(24):2514-2525. [DOI] [PubMed] [Google Scholar]
- 39.Kienle D, Benner A, Läufle C, et al. Gene expression factors as predictors of genetic risk and survival in chronic lymphocytic leukemia. Haematologica. 2010;95(1):102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rassenti LZ, Jain S, Keating MJ, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112(5):1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benedetti D, Bomben R, Dal-Bo M, et al. Are surrogates of IGHV gene mutational status useful in B-cell chronic lymphocytic leukemia? The example of Septin-10. Leukemia. 2008;22(1):224-226. [DOI] [PubMed] [Google Scholar]
- 42.Beekman R, Chapaprieta V, Russiñol N, et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018;24(6):868-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roisman A, Stanganelli C, Nagore VP, et al. SOX11 expression in chronic lymphocytic leukemia correlates with adverse prognostic markers. Tumour Biol. 2015;36(6):4433-4440. [DOI] [PubMed] [Google Scholar]
- 44.Soriano SF, Serrano A, Hernanz-Falcón P, et al. Chemokines integrate JAK/STAT and G-protein pathways during chemotaxis and calcium flux responses. Eur J Immunol. 2003;33(5):1328-1333. [DOI] [PubMed] [Google Scholar]
- 45.Cuesta-Mateos C, López-Giral S, Alfonso-Pérez M, et al. Analysis of migratory and prosurvival pathways induced by the homeostatic chemokines CCL19 and CCL21 in B-cell chronic lymphocytic leukemia. Exp Hematol. 2010;38(9):756–64,764. [DOI] [PubMed] [Google Scholar]
- 46.Munk R, Ghosh P, Ghosh MC, et al. Involvement of mTOR in CXCL12 mediated T cell signaling and migration. PLoS One. 2011;6(9):e24667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittal AK, Chaturvedi NK, Rohlfsen RA, et al. Role of CTLA4 in the proliferation and survival of chronic lymphocytic leukemia. PLoS One. 2013;8(8):e70352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh YM, Kwon YE, Kim JM, et al. Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat Cell Biol. 2009;11(3):295302. [DOI] [PubMed] [Google Scholar]
- 49.Tsagiopoulou M, Chapaprieta V, Duran-Ferrer M, et al. Chronic lymphocytic leukemias with trisomy 12 show a distinct DNA methylation profile linked to altered chromatin activation. Haematologica. 2020;105(12):2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher RA. The Correlation between relatives on the supposition of mendelian inheritance. Trans R Soc Edinb. 1919;52(02):399-433. [Google Scholar]
- 51.van Wageningen S, Kemmeren P, Lijnzaad P, et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010;143(6):991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sameith K, Amini S, Groot Koerkamp MJA, et al. A high-resolution gene expression atlas of epistasis between gene-specific transcription factors exposes potential mechanisms for genetic interactions. BMC Biol. 2015;13:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menter T, Trivedi P, Ahmad R, et al. Diagnostic utility of lymphoid enhancer binding factor 1 immunohistochemistry in small B-cell lymphomas. Am J Clin Pathol. 2017;147(3):292-300. [DOI] [PubMed] [Google Scholar]
- 54.Meijers RWJ, Muggen AF, Leon LG, et al. Responsiveness of chronic lymphocytic leukemia cells to B-cell receptor stimulation is associated with low expression of regulatory molecules of the nuclear factor-κB pathway. Haematologica. 2020;105(1):182-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong J, Ren Y, Zhang T, et al. Inactivation of DNA-PK by knockdown DNA-PKcs or NU7441 impairs non-homologous end-joining of radiation-induced double strand break repair. Oncol Rep. 2018;39(3):912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7(9):2955-2966. [DOI] [PubMed] [Google Scholar]
- 57.Ricci F, Tedeschi A, Morra E, Montillo M. Fludarabine in the treatment of chronic lymphocytic leukemia: a review. Ther Clin Risk Manag. 2009;5(1):187-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petruccelli LA, Dupéré-Richer D, Pettersson F, et al. Vorinostat induces reactive oxygen species and DNA damage in acute myeloid leukemia cells. PLoS One. 2011;6(6):e20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.