Abstract
Objectives:
Behçet’s disease tends to be more severe in men than women. This study was undertaken to investigate sex-specific genetic effects in Behçet’s disease.
Methods:
A total of 1762 male and 1216 female patients with Behçet’s disease from six diverse populations were studied, with the majority of patients of Turkish origin. Genotyping was performed using an Infinium ImmunoArray-24 BeadChip, or extracted from available genotyping data. Following imputation and extensive quality control measures, genome-wide association analysis was performed comparing male to female patients in the Turkish cohort, followed by a meta-analysis of significant results in all six populations. In addition, a weighted genetic risk score for Behçet’s disease was calculated and compared between male and female patients.
Results:
Genetic association analysis comparing male to female patients with Behçet’s disease from Turkey revealed an association with male sex in HLA-B/MICA within the HLA region with a GWAS level of significance (rs2848712, OR = 1.46, P = 1.22 × 10−8). Meta-analysis of the effect in rs2848712 across six populations confirmed these results. Genetic risk score for Behçet’s disease was significantly higher in male compared to female patients from Turkey. Higher genetic risk for Behçet’s disease was observed in male patients in HLA-B/MICA (rs116799036, OR = 1.45, P = 1.95 × 10−8), HLA-C (rs12525170, OR = 1.46, P = 5.66 × 10−7), and KLRC4 (rs2617170, OR = 1.20, P = 0.019). In contrast, IFNGR1 (rs4896243, OR = 0.86, P = 0.011) was shown to confer higher genetic risk in female patients.
Conclusions:
Male patients with Behçet’s disease are characterized by higher genetic risk compared to female patients. This genetic difference, primarily derived from our Turkish cohort, is largely explained by risk within the HLA region. These data suggest that genetic factors might contribute to differences in disease presentation between men and women with Behçet’s disease.
Keywords: Behçet’s disease, Sex-bias, Genetics, HLA class I, GWAS, MHC
1. Introduction
Behçet’s disease is a chronic inflammatory disease characterized by recurrent oral and genital ulcers. It is a multisystemic vasculitis affecting many organs, such as the eyes, skin, blood vessels, central nervous system, and gastrointestinal tract [1,2]. Behçet’s disease is also known as the “Silk Road disease”. Although patients have been diagnosed with Behçet’s disease worldwide, it is found to be most prevalent in populations originating from the region along the ancient trading route [3]. The etiology of Behçet’s disease is not clearly understood, however, genetic factors along with environmental triggers are thought to play a role in the susceptibility to the disease. It is suggested that infectious pathogens might contribute to the onset of Behçet’s disease by triggering abnormal immune responses in genetically predisposed individuals [4]. The genetic studies performed to date in Behçet’s disease have identified multiple robust genetic susceptibility loci for the disease [5]. However, until recent years, genome-wide association studies were limited to small sample sizes due to the low prevalence of Behçet’s disease in select populations [6]. It is well established that the human leukocyte antigen (HLA) class I region is the most robust genetic susceptibility locus associated with Behçet’s disease [7]. Several independent susceptibility loci located within the HLA region have been identified, such as HLA-B/MICA, HLA-A, and HLA-C [8–10]. In addition, susceptibility loci outside the HLA region with genome-wide level of significance have also been identified, such as IL12RB2, IL10, IFNGR1, STAT4, LNCAROD/DKK1, IRF8, FUT2, and many more, providing valuable insights into the pathogenesis of the disease [5,11–18]. Behçet’s disease is known to affect both sexes, however, the disease tends to be more severe in men compared to women [19,20]. The reasons for the difference in severity of the disease between male and female patients are unknown. To better understand sex-bias in Behçet’s disease, we sought to investigate the sex-specific genetic effects in the disease by performing a case-case genetic association analysis in a large cohort of patients.
2. Methods
2.1. Study population and genotyping
A total of 2213 patients (1330 male and 883 female patients) and 1533 healthy controls (897 male and 636 female controls) of Turkish origin were included in the initial phase of this study. To confirm the results, a multi-ancestral metanalysis was performed, including 5 additional populations. These consisted of 224 patients from Spain (118 female and 106 male patients), 194 patients from Korea (96 female and 98 male patients), 126 patients from Italy (59 female and 67 male patients), 117 patients from Japan (27 female and 90 male patients), and 104 patients from Tunisia (33 female and 71 male patients). All patients included in the study fulfilled the 1990 international study group classification criteria for Behçet’s disease [21]. Genotyping was performed as previously described [6], using the Illumina Immunochip custom arrays (Infinium ImmunoArray-24 V.1.0 or V.2.0 BeadChip). Genotyping data from 654 male and 545 female Turkish patients, and 748 male and 530 female Turkish controls were obtained from dbGAP (accession number phs000272.v1.p1) [12]. The study was approved by the institutional review boards and the ethics committees at the participating institutions, and all study participants signed a written informed consent.
2.2. Data quality assessment and imputation
Genotyping data quality assessment, imputation, and subsequent filtering of genetic variants and individuals were performed as previously described [6]. Briefly, samples were filtered out if they had a genotyping call <95%, and one individual from duplicates or first-degree relatives (Pi-HAT>0.4) was excluded randomly. SNPs were removed if they had a genotyping call <98% or minor allele frequency (MAF) < 1% or deviated from Hardy-Weinberg equilibrium (HWE, P < 001). Sex chromosomes were excluded and not analyzed in this study. Principal component analysis was performed using linkage disequilibrium (LD) pruned SNPs with r2 threshold of <0.2, with Eigensoft 6.1.4 software [22]. Samples more than 6 SD from the center of the cluster were considered outliers and removed [23]. Assembly GRCh37 hg19 was used for SNP annotation [24].
Imputation was performed using genotyping data post-quality control, using the Michigan Imputation Server using Minimac3 [25]. SHAPEIT haplotype phasing software was used for haplotype reconstruction with the Haplotype Reference Consortium r1.1 as the reference population [26]. Only SNPs with high imputation accuracy values (r2 > 0.9) were used for association analyses, and imputed variants with MAF<1% or HWE P < 0.001 were also excluded.
The total number of patients and controls evaluated in this study, as listed above, is the final number of samples following the exclusion of samples based on quality control measures, and following the exclusion of samples with missing sex information or with genotyping-based imputed sex that mismatched reported sex.
2.3. Data analysis and genetic risk score calculation
Plink v. 1.9 [27] was used to conduct a case-case association analysis using a logistic regression model. Genome-wide association study (GWAS) threshold of P < 5 × 10−8 was set for evidence of significant association (sex bias).
The aggregate genetic risk was measured by calculating cumulative genetic risk scores (GRS) of individuals with Behçet’s disease. A total of 20 SNPs representing the previously reported susceptibility loci for Behçet’s disease were used to calculate GRS. Each locus was represented by one SNP, apart from Interferon Regulatory Factor 8 (IRF8), displaying 2 distinct genetic effects in this locus [5]. Only individuals with 100% genotyping success rate for these markers were included. If a particular marker was not genotyped, imputed genotype data were used for the calculation. Odds ratios (OR) used to calculate the genetic risk scores at each risk locus were those obtained from case-control association analyses for accurate representation of the impact of each locus [5]. Risk scores were calculated by multiplying the natural logarithm of the OR at each locus by the number of effect alleles: () [5,28], then Welch’s t-test was used to assess differences between male and female patients. We also implemented a non-parametric multifactor-dimensionality reduction (MDR) analysis to further validate our results [29,30].
Confirmation followed by multi-ancestral meta-analysis was performed to evaluate and confirm the sex-bias detected in rs2848712 (HLA-B/MICA) in multiple populations. Meta-analysis was performed with programming language R [31–33], using fixed-effect inverse variance method measuring the odds ratio for rs2848712 (HLA-B/MICA) comparing male and female patients for all populations. Fixed effect model was chosen in this case because both heterogeneity index (I2) and Cochran’s Q test P value suggested no evidence of heterogeneity (I2 < 0.5 and Q > 0.1) [6].
3. Results
A Case-Case genetic association analysis was performed in the Turkish population to compare male and female patients with Behçet’s disease using a logistic regression model (Fig. 1A). Our results revealed that the most significant differences between male and female patients were in the HLA region with 6 genetic variants reaching the GWAS level of significance (P < 5 × 10−8). Differences were observed in the HLA class I region (HLA-B/MICA), and the most significant sex-associated SNPs in Behçet’s disease were rs2848712, rs2596527, rs2848713 (OR = 1.46 (95% CI 1.28 to 1.66), P = 1.22 × 10−8), followed by rs77563643 (OR = 1.45 (95% CI 1.27 to 1.65), P = 2.47 × 10−8), rs3819272 (OR = 1.44 (95% CI 1.27 to 1.64), P = 3.15 × 10−8), and rs79953803 (OR = 1.43 (95% CI 1.26 to 1.63), P = 4.30 × 10−8) (Table 1, Fig. 1B). It is important to note that a similar control-control genetic association analysis including 897 male and 636 female Turkish healthy individuals revealed no significant differences across the genome (Supplementary Fig. 1).
Fig. 1.
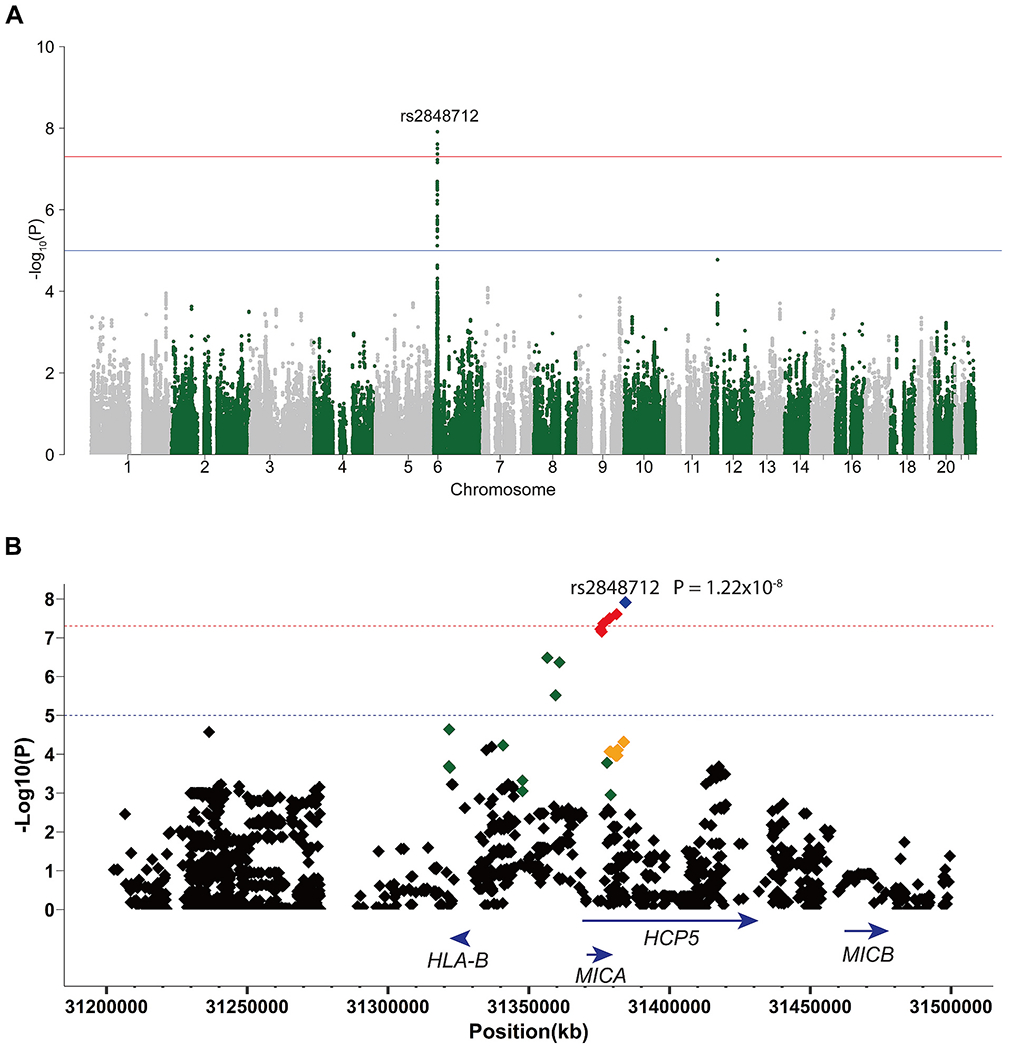
(A) A Manhattan plot showing the results of the case-case genetic association analysis of male and female patients with Behcet’s disease of Turkish origin. The −Log10(P) value for each variant is plotted against its chromosomal position. The red line represents the genome-wide level of significance (P < 5 × 10−8) and the blue line represents the suggestive level of significance (P < 1 × 10−5). (B) Regional plot displaying SNPs in LD with rs2848712 in the HLA region in the Turkish population (LD = 1 is blue, LD = 0.99–0.80 is red, LD = 0.79–0.60 is orange, LD = 0.59–0.40 is green, and LD = 0.39–0.00 is black). The −Log10(P) value for each variant is plotted against its physical position on chromosome 6. The red line represents GWAS level of significance (P < 5 × 10−8) and the blue line represents suggestive level of significance (P < 1 × 10−5). (Assembly_GRCh37/hg19 by Ensembl was used).
Table 1.
Results showing SNPs with GWAS level of significance (P < 5 × 10−8) from the genetic association analysis comparing men and women with Behçet’s disease in the Turkish cohort.
| MAF Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Locus | Chr | SNP | Male | Female | OR | 95% CI LL | 95% CI UL | P-value |
| HLA-B/MICA | 6 | rs2848712 | 0.41 | 0.33 | 1.46 | 1.28 | 1.66 | 1.22 × 10−8 |
| HLA-B/MICA | 6 | rs2596527 | 0.41 | 0.33 | 1.46 | 1.28 | 1.66 | 1.22 × 10−8 |
| HLA-B/MICA | 6 | rs2848713 | 0.41 | 0.33 | 1.46 | 1.28 | 1.66 | 1.22 × 10−8 |
| HLA-B/MICA | 6 | rs77563643 | 0.41 | 0.33 | 1.45 | 1.27 | 1.65 | 2.47 × 10−8 |
| HLA-B/MICA | 6 | rs3819272 | 0.41 | 0.33 | 1.44 | 1.27 | 1.64 | 3.15 × 10−8 |
| HLA-B/MICA | 6 | rs79953803 | 0.41 | 0.33 | 1.43 | 1.26 | 1.63 | 4.30 × 10−8 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio of male compared to female patients (case-case analysis); CI LL, confidence interval lower limit; CI UL, confidence interval upper limit; P-value, P-values from the logistic regression analysis.
To confirm the sex-bias effect detected within the HLA region in patients with Behçet’s disease, we evaluated the genetic association with male sex in a multiancestral cohort of patients with Behçet’s disease, representing 5 additional populations. A meta-analysis between all 6 cohorts confirmed that the frequency of the minor allele in the lead SNP within the HLA region, rs2848712 (HLA-B/MICA), was significantly higher in male compared to female patients with Behçet’s disease (ORmeta = 1.39 (95% CI 1.23 to 1.56), Pmeta = 1.83 × 10−8) (Fig. 2). Indeed, the direction trend of this effect was consistent in almost all populations studied (Turkish OR = 1.46 (95% CI 1.28 to 1.66) P = 1.22 × 10−8; Korean OR = 1.14 (95%CI 0.73 to 1.76), P = 0.57; Italian OR = 1.75 (95% CI 1.00 to 3.06), P = 0.048; Japanese OR = 1.15 (95% CI 0.60 to 2.17), P = 0.68; Tunisian OR 2.42 (95% CI 0.94 to 5.71), P = 0.068), with an exception being the Spanish population with OR = 0.76 (95% CI 0.47 to 1.23), P = 0.26. The Turkish cohort had the majority of the weight (76.3%) in this analysis due to the number of Turkish individuals in the study resulting in greater power.
Fig. 2.
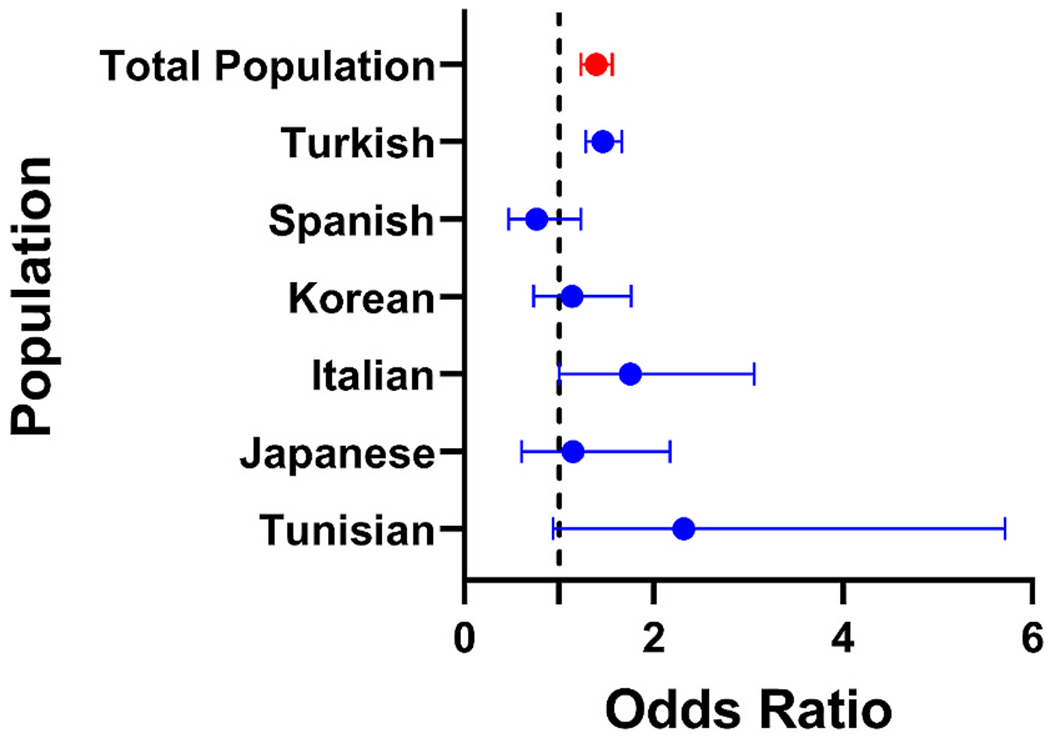
Meta-analysis forest plot of rs2848712 (HLA-B/MICA) depicting the sex-specific differences in genetic association from different populations at this locus. Individual cohorts are shown in blue and the total study population in red.
Next, we investigated sex-specific differences in overall genetic risk between men and women with Behçet’s disease in the Turkish cohort by calculating and comparing a cumulative genetic risk score (GRS). Scores were based on the odds ratios obtained from case-control association analyses [5]. Our results indicate that on average, men with Behçet’s disease have higher disease genetic risk than women (P = 3.18 × 10−8; Fig. 3A). This difference was largely driven by the genetic variants within the HLA region, and disappeared after the HLA loci were excluded (P = 0.34; Fig. 3B). No difference in GRS was detected between male and female Turkish healthy controls (P = 0.61; Fig. 3C).
Fig. 3.
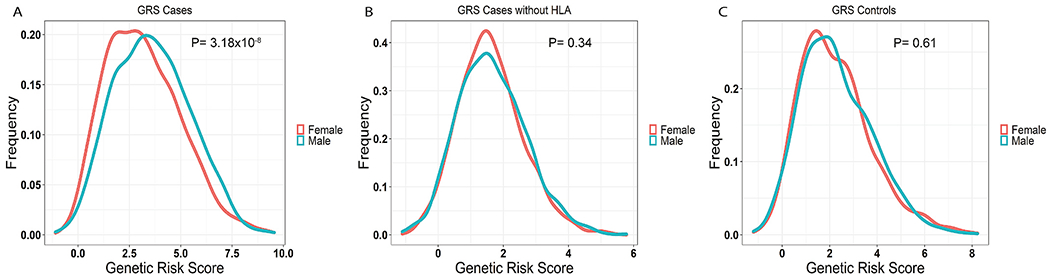
(A) Density plot of genetic risk scores (GRS) for Behçet’s disease in Turkish patients. The frequencies of individuals are plotted against their respective GRS (men in blue and women in red), showing higher genetic risk in men than women (P = 3.18 × 10−8). (B) Density plot of GRS in Turkish patients with Behçet’s disease without the SNPs in the HLA region (men in blue and women in red). The difference between men and women disappears (P = 0.34). (C) Density plot of GRS in healthy Turkish controls (men in blue and women in red) displaying no significant difference between men and women (P = 0.61).
Of the previously reported disease susceptibility loci for Behçet’s disease, the locus within HLA-B/MICA (rs116799036, OR = 1.45, P = 1.95 × 10−8) had the largest impact on the cumulative genetic risk score difference between male and female patients, followed by the locus in HLA-C (rs12525170, OR = 1.46, P = 5.66 × 10−7) (Table 2). Of note, rs116799036 was not included in the case-case GWAS analysis, as this variant was filtered out due to failing to pass the HWE test filter (HWE, P < 0.001). The susceptibility locus in KLRC4 (rs2617170, OR = 1.20 P = 0.019) also showed higher risk in male patients, though the imputation accuracy (r2 = 0.51) was low and therefore data on this locus should be investigated further for confirmation. Interestingly, the risk locus in IFNGR1 (rs4896243, OR = 0.86, P = 0.011) was associated with higher disease risk in female compared to male patients.
Table 2.
Sex-specific differences in Behçet’s disease susceptibility loci between men and women with Behçet’s disease in the Turkish cohort.
| Susceptibility Loci |
|
|
Effect Allele Frequency |
|
|
|
||
|---|---|---|---|---|---|---|---|---|
| Gene/Locus | Chr | SNP | Effect Allele | Male | Female | OR | P-value | Imputation Accuracy (r2) |
| IL12RB2 | 1 | rs924080 | A | 0.66 | 0.67 | 0.95 | 0.44 | 0.99 |
| IL10 | 1 | rs1518111 | A | 0.38 | 0.36 | 1.09 | 0.30 | 0.93 |
| IL1A/IL1B | 2 | rs3783550 | G | 0.35 | 0.36 | 1.04 | 0.55 | 0.95 |
| STAT4 | 2 | rs7574070 | A | 0.46 | 0.46 | 1.03 | 0.61 | 0.63 |
| CCR1/CCR3 | 3 | rs7616215 | T | 0.73 | 0.73 | 1.01 | 0.88 | 0.94 |
| IL12A | 3 | rs17753641 | G | 0.073 | 0.072 | 1.01 | 0.97 | 0.95 |
| ERAP1 | 5 | rs17482078 | T | 0.18 | 0.16 | 1.15 | 0.11 | 0.97 |
| IFNGR1 | 6 | rs4896243 | C | 0.45 | 0.48 | 0.86 | 0.011 | 0.99 |
| HLA-A | 6 | rs114854070 | A | 0.27 | 0.25 | 1.11 | 0.12 | 0.94 |
| HLA-C | 6 | rs12525170 | A | 0.26 | 0.19 | 1.46 | 5.66 × 10−7 | 0.90 |
| HLA-B/MICA | 6 | rs116799036 | A | 0.38 | 0.30 | 1.45 | 1.95 × 10−8 | 0.94 |
| LNCAROD/DKK1 | 10 | rs1660760 | T | 0.48 | 0.50 | 0.91 | 0.13 | 0.93 |
| ADO/EGR2 | 10 | rs224127 | A | 0.46 | 0.48 | 0.96 | 0.56 | 0.96 |
| JRKL/CNTN5 | 11 | rs2848479 | A | 0.45 | 0.48 | 1.014 | 0.83 | 0.98 |
| KLRC4 | 12 | rs2617170 | C | 0.80 | 0.77 | 1.20 | 0.019 | 0.51 |
| LACC1 | 13 | rs2121033 | G | 0.24 | 0.25 | 0.98 | 0.71 | 0.96 |
| IRF8 | 16 | rs7203487 | C | 0.16 | 0.15 | 1.08 | 0.54 | 0.84 |
| IRF8 | 16 | rs11117433 | C | 0.073 | 0.089 | 0.82 | 0.081 | 0.82 |
| FUT2 | 19 | rs681343 | T | 0.53 | 0.53 | 0.99 | 0.92 | 0.78 |
| CEBPB/PTPN1 | 20 | rs913678 | C | 0.32 | 0.34 | 0.91 | 0.17 | 0.62 |
SNP, single nucleotide polymorphism; OR, odds ratio of male compared to female patients (case-case analysis); P-value, p-value from comparing allele frequency differences between male and female patients.
We then, performed a non-parametric analysis to confirm sex-gene interactions in the specific loci that had significant allele frequency differences between male and female patients. This was applied to test sex-gene interactions in HLA-B/MICA, HLA-C, KLRC4 and IFNGR1, and confirmed our results obtained from parametric tests (Table 3).
Table 3.
Multifactor dimensionality reduction (MDR) analysis in the Turkish cohort providing confirming evidence for interaction between sex and susceptibility loci in Behçet’s disease.
|
|
|
MDR Analysis |
|||
|---|---|---|---|---|---|
| Locus | SNP | Cross-validation consistency | Balanced Accuracy | χ2 | P-value |
| HLA-B/MICA | rs116799036 | 10/10 | 0.56 | 30.76 | 2.92 × 10−8 |
| HLA-C | rs12525170 | 10/10 | 0.56 | 31.81 | 1.7 × 10−8 |
| KLRC4 | rs2617170 | 10/10 | 0.52 | 4.26 | 0.039 |
| IFNGR1 | rs4896243 | 10/10 | 0.52 | 5.23 | 0.019 |
Cross-validation consistency is a measure of the number of times the same MDR model is identified in each possible 90% of the subjects [31]; Balanced Accuracy is defined as (sensitivity + specificity/2). Sensitivity represents true positive rate (true positives/(true positives + false negatives)), and specificity represents true negative rate (true negatives/(false positives + true negatives)). Balanced accuracy is used as an estimator of true accuracy to mitigate imbalanced sampling [34].Degree of freedom (df) of 1 was used to calculate the P Values from χ2. SNP, single nucleotide polymorphism.
4. Discussion
We explored for the first time the genetic differences between male and female patients with Behçet’s disease, using a large set of patients from 6 independent populations and a genome-wide approach. The most significant sex-specific genetic differences in Behçet’s disease were observed in the HLA class I region, specifically in the HLA-B/MICA locus (tagged by rs2848712). Indeed, the difference in GRS for Behçet’s disease between male and female patients was abrogated when genetic variants within the HLA region were excluded from the analysis.
Genetic susceptibility loci in the HLA region are among the most robust contributing factors to many immune-mediated diseases, including Behçet’s disease. Several independent susceptibility loci within the HLA region have been reported in Behçet’s disease, with the most robust association localized to the HLA-B/MICA locus [8,9]. The genetic variant rs2848712, which showed the most significant difference between male and female patients with Behçet’s disease, resides in the HLA-B/MICA region. This variant is in LD with the previously reported HLA-B/MICA intergenic SNP rs116799036, which is associated with genetic risk for Behçet’s disease [8], (r2 = 0.79 in our Turkish population).
Although sexual dimorphism in Behçet’s disease is poorly understood, our data demonstrated significant genetic differences in the HLA region between male and female patients. Whether these differences in the HLA region also affect disease manifestations needs further investigation. A previous meta-analysis demonstrated that HLA-B*51/HLA-B5 was slightly but significantly present at a higher frequency in male patients (OR = 1.14), and that HLA-B*51/HLA-B5 moderately increases the risk of ocular, skin, and genital involvement [34]. In an earlier study, male-sex has been found to be the strongest determinant of disease severity in Behçet’s disease, although HLA-B*51 was not associated with a more severe disease course [35]. Several HLA-A alleles were associated with various manifestations of Behçet’s disease in Korean and Japanese populations. HLA-A*26:01 and HLA-A*30:04 were shown to have higher prevalence in patients with uveitis and vascular lesions, respectively [36].
Some studies have reported other possible explanations for sexual dimorphism in Behçet’s disease. Bang et al. reported that 18 out of 27 Korean patients with Behçet’s disease experienced worsening of disease during pregnancy, mostly in the first trimester, suggesting possible influence from hormones in the disease [37]. However, conflicting results have been reported, and in larger studies it appears that the majority of patients showed no exacerbation of disease during pregnancy [38]. Testosterone activates neutrophils, and male patients with Behçet’s disease manifest increased neutrophil oxidative burst responses compared to female patients [39]. Nonetheless, the effect of hormonal influences on the presentation and sex-bias in Behçet’s disease remains incompletely evaluated. Measurements of serum vitamin D3 levels failed to demonstrate significant differences between male and female patients with Behçet’s disease [40].
Our data suggest that the Behçet’s disease risk variant in IFNGR1 is more common in female compared to male patients. Interestingly, IFNGR1 has been also identified as a possible genetic factor associated with the development of recurrent mouth ulcers (OR = 1.08) [41]. It has been reported that in Behçet’s disease patients, oral ulcers are more prevalent in female compared to male patients [20]. IFNGR1 polymorphisms are also observed to be associated with systemic lupus erythematosus [42]. Though autoimmune clinical features and autoantibody production are not classical features of Behçet’s disease, immune responses to some autoantigens and evidence for the activation of intracellular signaling pathways such as JAK/STAT pathway have been reported [43].
KLRC4 is a member of killer cell lectin-like receptor subfamily C, and encodes a calcium dependent (C type) lectin receptor. The exact function of KLRC4 remains unknown, although it was closely associated with cytotoxic activity in natural killer cells and γδ T cells [44]. It is important to note that while the Behçet’s disease risk allele in KLRC4 (rs2617170) was more prevalent in male patients, our imputation accuracy was low at this locus (r2 = 0.51).
Our data suggest that genetic factors might contribute to differences in disease presentation between men and women with Behçet’s disease. However, a sub-phenotype analysis to examine the relationship between genetic risk and Behçet’s disease severity, or the frequency of organ threatening involvement, in men compared to women with the disease are necessary to support this conclusion. In addition, as in any genetic study, independent replication of our findings is needed to confirm our results. Our data might also be interpreted to suggest that men require a higher genetic load to develop Behçet’s disease than women. This logic would then suggest that Behçet’s disease should be more common in women than men, as the genetic threshold to develop the disease is lower in women. However, this is not consistent with epidemiologic data for Behçet’s disease [45]. Therefore, an alternative explanation would be that non-genetic factors, such as environmental or hormonal factors, play a more dominant role in the etiopathogenesis of Behçet’s disease in women. A different explanation might be that there are other genetic factors that play a more dominant role in women, and which could not be assessed by our approach, such as genetic regions not assessed in our study, gene-gene interactions, rare genetic variants, or epigenetic differences between men and women with Behçet’s disease. Indeed, epigenetic differences between patients with Behçet’s disease and healthy controls have been reported [46]. However, a comparative epigenetic analysis between men and women with Behçet’s disease would be insightful.
5. Conclusion
Our study comprehensively evaluated genetic contribution to sex-bias in Behçet’s disease for the first time. We demonstrated that the largest genetic difference between men and women with Behçet’s disease is in the HLA region, resulting in a higher overall genetic risk for the disease in male compared to female patients across multiple populations. Further studies to evaluate epigenetic differences between male and female patients, and how genetic and epigenetic factors affect specific disease manifestations in Behçet’s disease are warranted.
Supplementary Material
Funding information
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health grant number R01AR070148.
Footnotes
Authorship statement
All authors fulfilled the following criteria: Substantial contributions to the conception or design of the work, or the acquisition, analysis or interpretation of data. Drafting the work or revising it critically for important intellectual content. Final approval of the version published.
Declaration of competing interest
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaut.2022.102882.
References
- [1].Betti ol A, Prisco D, Emmi G, Behcet: the syndrome, Oxford), Rheumatology 59 (2020). iii101–iii7. [DOI] [PubMed] [Google Scholar]
- [2].Alibaz-Oner F, Sawalha AH, Direskeneli H, Management of Behcet’s disease, Curr. Opin. Rheumatol. 30 (2018) 238–242. [DOI] [PubMed] [Google Scholar]
- [3].Verity DH, Marr JE, Ohno S, Wallace GR, Stanford MR, Behcet’s disease, the Silk Road and HLA-B51: historical and geographical perspectives, Tissue Antigens 54 (1999) 213–220. [DOI] [PubMed] [Google Scholar]
- [4].Mumcu G, Direskeneli H, Triggering agents and microbiome as environmental factors on Behcet’s syndrome, Intern Emerg Med 14 (2019) 653–660. [DOI] [PubMed] [Google Scholar]
- [5].Ortiz-Fernandez L, Sawalha AH, Genetics of behcet’s disease: functional genetic analysis and estimating disease heritability, Front. Med 8 (2021), 625710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ortiz Fernandez L, Coit P, Yilmaz V, Yentur SP, Alibaz-Oner F, Aksu K, et al. , Genetic association of a gain-of-function IFNGR1 polymorphism and the intergenic region LNCAROD/DKK1 with behcet’s disease, Arthritis Rheumatol. 73 (2021) 1244–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].de Menthon M, Lavalley MP, Maldini C, Guillevin L, Mahr A, HLA-B51/B5 and the risk of Behcet’s disease: a systematic review and meta-analysis of case-control genetic association studies, Arthritis Rheum. 61 (2009) 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hughes T, Coit P, Adler A, Yilmaz V, Aksu K, Duzgun N, et al. , Identification of multiple independent susceptibility loci in the HLA region in Behcet’s disease, Nat. Genet 45 (2013) 319–324. [DOI] [PubMed] [Google Scholar]
- [9].Ombrello MJ, Kirino Y, de Bakker PI, Gul A, Kastner DL, Remmers EF, Behcet disease-associated MHC class I residues implicate antigen binding and regulation of cell-mediated cytotoxicity, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 8867–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gensterblum-Miller E, Wu W, Sawalha AH, Novel transcriptional activity and extensive allelic imbalance in the human MHC region, J. Immunol 200 (2018) 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Takeuchi M, Mizuki N, Meguro A, Ombrello MJ, Kirino Y, Satorius C, et al. , Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behcet’s disease susceptibility, Nat. Genet 49 (2017) 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, et al. , Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease, Nat. Genet 42 (2010) 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mizuki N, Meguro A, Ota M, Ohno S, Shiota T, Kawagoe T, et al. , Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet’s disease susceptibility loci, Nat. Genet 42 (2010) 703–706. [DOI] [PubMed] [Google Scholar]
- [14].Ortiz-Fernandez L, Carmona FD, Montes-Cano MA, Garcia-Lozano JR, Conde-Jaldon M, Ortego-Centeno N, et al. , Genetic analysis with the Immunochip platform in Behcet disease. Identification of residues associated in the HLA class I region and new susceptibility loci, PLoS One 11 (2016), e0161305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, et al. , Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1, Nat. Genet 45 (2013) 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xavier JM, Shahram F, Sousa I, Davatchi F, Matos M, Abdollahi BS, et al. , FUT2: filling the gap between genes and environment in Behcet’s disease? Ann. Rheum. Dis 74 (2015) 618–624. [DOI] [PubMed] [Google Scholar]
- [17].Hou S, Yang Z, Du L, Jiang Z, Shu Q, Chen Y, et al. , Identification of a susceptibility locus in STAT4 for Behcet’s disease in Han Chinese in a genome-wide association study, Arthritis Rheum. 64 (2012) 4104–4113. [DOI] [PubMed] [Google Scholar]
- [18].Kappen JH, Medina-Gomez C, van Hagen PM, Stolk L, Estrada K, Rivadeneira F, et al. , Genome-wide association study in an admixed case series reveals IL12A as a new candidate in Behcet disease, PLoS One 10 (2015), e0119085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yazici H, Tuzun Y, Pazarli H, Yurdakul S, Ozyazgan Y, Ozdogan H, et al. , Influence of age of onset and patient’s sex on the prevalence and severity of manifestations of Behcet’s syndrome, Ann. Rheum. Dis 43 (1984) 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ucar-Comlekoglu D, Fox A, Sen HN, Gender differences in behcet’s disease associated uveitis, J Ophthalmol 2014 (2014), 820710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Criteria for diagnosis of behcet’s disease. International study group for behcet’s disease, Lancet 335 (1990) 1078–1080. [PubMed] [Google Scholar]
- [22].Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D, Principal components analysis corrects for stratification in genome-wide association studies, Nat. Genet 38 (2006) 904–909. [DOI] [PubMed] [Google Scholar]
- [23].Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. , Next-generation genotype imputation service and methods, Nat. Genet 48 (2016) 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. , ENCODE data in the UCSC Genome Browser: year 5 update, Nucleic Acids Res. 41 (2013) D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. , A reference panel of 64,976 haplotypes for genotype imputation, Nat. Genet 48 (2016) 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J, Haplotype estimation using sequencing reads, Am. J. Hum. Genet 93 (2013) 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, Second-generation PLINK: rising to the challenge of larger and richer datasets, GigaScience 4 (2015) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hughes T, Adler A, Merrill JT, Kelly JA, Kaufman KM, Williams A, et al. , Analysis of autosomal genes reveals gene-sex interactions and higher total genetic risk in men with systemic lupus erythematosus, Ann. Rheum. Dis 71 (2012) 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. , Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer, Am. J. Hum. Genet 69 (2001) 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, et al. , A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction, Genet. Epidemiol 31 (2007) 306–315. [DOI] [PubMed] [Google Scholar]
- [31].R.c. Team, R: A Language and Environment for Statistical Computing, 2021. [Google Scholar]
- [32].Balduzzi S, Rücker G, Schwarzer G, How to perform a meta-analysis with R: a practical tutorial, Evid. Base Ment. Health 22 (4) (2019) 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Turner qqman S, An R package for visualizing GWAS results using Q-Q and manhattan plots, J. Open Sour. Software (2018) 731. [Google Scholar]
- [34].Maldini C, Lavalley MP, Cheminant M, de Menthon M, Mahr A, Relationships of HLA-B51 or B5 genotype with Behcet’s disease clinical characteristics: systematic review and meta-analyses of observational studies, Rheumatology 51 (2012) 887–900. [DOI] [PubMed] [Google Scholar]
- [35].Gul A, Uyar FA, Inanc M, Ocal L, Tugal-Tutkun I, Aral O, et al. , Lack of association of HLA-B*51 with a severe disease course in Behcet’s disease, Rheumatology 40 (2001) 668–672. [DOI] [PubMed] [Google Scholar]
- [36].Kang EH, Kim JY, Takeuchi F, Kim JW, Shin K, Lee EY, et al. , Associations between the HLA-A polymorphism and the clinical manifestations of Behcet’s disease, Arthritis Res. Ther 13 (2011) R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bang D, Chun YS, Haam IB, Lee ES, Lee S, The influence of pregnancy on Behcet’s disease, Yonsei Med. J 38 (1997) 437–443. [DOI] [PubMed] [Google Scholar]
- [38].Barros T, Braga A, Marinho A, Braga J. Behcet’s disease and pregnancy: a retrospective case-control study, Yale J. Biol. Med 94 (2021) 585–592. [PMC free article] [PubMed] [Google Scholar]
- [39].Yavuz S, Ozilhan G, Elbir Y, Tolunay A, Eksioglu-Demiralp E, Direskeneli H, Activation of neutrophils by testosterone in Behcet’s disease, Clin. Exp. Rheumatol 25 (2007) S46–S51. [PubMed] [Google Scholar]
- [40].Hamzaoui K, Ben Dhifallah I, Karray E, Sassi FH, Hamzaoui A, Vitamin D modulates peripheral immunity in patients with Behcet’s disease, Clin. Exp. Rheumatol 28 (2010) S50–S57. [PubMed] [Google Scholar]
- [41].Dudding T, Haworth S, Lind PA, Sathirapongsasuti JF, T. andMe Research, Tung JY, et al. , Genome wide analysis for mouth ulcers identifies associations at immune regulatory loci, Nat. Commun 10 (2019) 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nakashima H, Inoue H, Akahoshi M, Tanaka Y, Yamaoka K, Ogami E, et al. , The combination of polymorphisms within interferon-gamma receptor 1 and receptor 2 associated with the risk of systemic lupus erythematosus, FEBS Lett. 453 (1999) 187–190. [DOI] [PubMed] [Google Scholar]
- [43].Tulunay A, Dozmorov MG, Ture-Ozdemir F, Yilmaz V, Eksioglu-Demiralp E, Alibaz-Oner F, et al. , Activation of the JAK/STAT pathway in Behcet’s disease, Gene Immun. 16 (2015) 176. [DOI] [PubMed] [Google Scholar]
- [44].Hayashi T, Imai K, Morishita Y, Hayashi I, Kusunoki Y, Nakachi K, Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance, Cancer Res. 66 (2006) 563–570. [DOI] [PubMed] [Google Scholar]
- [45].Mahr A, Belarbi L, Wechsler B, Jeanneret D, Dhote R, Fain O, et al. , Population-based prevalence study of Behcet’s disease: differences by ethnic origin and low variation by age at immigration, Arthritis Rheum. 58 (2008) 3951–3959. [DOI] [PubMed] [Google Scholar]
- [46].Hughes T, Ture-Ozdemir F, Alibaz-Oner F, Coit P, Direskeneli H, Sawalha AH, Epigenome-wide scan identifies a treatment-responsive pattern of altered DNA methylation among cytoskeletal remodeling genes in monocytes and CD4+ T cells from patients with Behcet’s disease, Arthritis Rheumatol 66 (2014) 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


