Abstract
The fusel alcohols 3-methyl-1-butanol, 2-methyl-1-butanol, and 2-methyl-propanol are important flavor compounds in yeast-derived food products and beverages. The formation of these compounds from branched-chain amino acids is generally assumed to occur via the Ehrlich pathway, which involves the concerted action of a branched-chain transaminase, a decarboxylase, and an alcohol dehydrogenase. Partially purified preparations of pyruvate decarboxylase (EC 4.1.1.1) have been reported to catalyze the decarboxylation of the branched-chain 2-oxo acids formed upon transamination of leucine, isoleucine, and valine. Indeed, in a coupled enzymatic assay with horse liver alcohol dehydrogenase, cell extracts of a wild-type Saccharomyces cerevisiae strain exhibited significant decarboxylation rates with these branched-chain 2-oxo acids. Decarboxylation of branched-chain 2-oxo acids was not detectable in cell extracts of an isogenic strain in which all three PDC genes had been disrupted. Experiments with cell extracts from S. cerevisiae mutants expressing a single PDC gene demonstrated that both PDC1- and PDC5-encoded isoenzymes can decarboxylate branched-chain 2-oxo acids. To investigate whether pyruvate decarboxylase is essential for fusel alcohol production by whole cells, wild-type S. cerevisiae and an isogenic pyruvate decarboxylase-negative strain were grown on ethanol with a mixture of leucine, isoleucine, and valine as the nitrogen source. Surprisingly, the three corresponding fusel alcohols were produced in both strains. This result proves that decarboxylation of branched-chain 2-oxo acids via pyruvate decarboxylase is not an essential step in fusel alcohol production.
Saccharomyces cerevisiae has been used for centuries in the production of bread and alcoholic beverages. Along with ethanol and carbon dioxide, fermenting cultures of this yeast produce a variety of low-molecular-weight flavor compounds (including alcohols, diacetyl, esters, organic acids, organic sulfides, and carbonyl compounds). The compounds 3-methyl-1-butanol, 2-methyl-1-butanol, and 2-methyl-1-propanol, commonly known as fusel alcohols, and their esters make an important contribution to the flavor of alcoholic beverages and bread (1, 14).
A metabolic pathway for production of fusel alcohols by yeast was first proposed by Ehrlich (6). The Ehrlich pathway starts with the enzyme-catalyzed decarboxylation of branched-chain 2-oxo acids to the corresponding aldehydes. Subsequently, the aldehyde is reduced to the corresponding fusel alcohol by an alcohol dehydrogenase (11, 16, 24). The branched-chain 2-oxo acid substrates for the Ehrlich pathway can be produced by the deamination of l-leucine, l-isoleucine, or l-valine. Growth of S. cerevisiae with any of these three amino acids as the nitrogen source results in the accumulation of the corresponding fusel alcohol (2, 3, 21). Alternatively, branched-chain 2-oxo acids may be synthesized de novo from carbohydrates as intermediates of branched-chain amino acid synthesis (13).
The conversion of branched-chain oxo acids into their respective aldehydes and alcohols via the Ehrlich pathway resembles the fermentative metabolism of pyruvate, which yields ethanol and carbon dioxide. In both cases, the decarboxylation of a 2-oxo acid is followed by the reduction of the resulting aldehyde. Partially purified preparations of yeast pyruvate decarboxylase have been shown to catalyze the decarboxylation of various 2-oxo acids, including the putative intermediates of the Ehrlich pathway (8, 12, 16, 21). However, it has not been conclusively proven that pyruvate decarboxylase is essential for or even involved in fusel alcohol production by S. cerevisiae.
Dickinson and Dawes (4) have reported that, at least under some conditions, oxidative decarboxylation by a mitochondrial branched-chain oxo acid dehydrogenase complex (17) is involved in the catabolism of branched-chain 2-oxo acids. Mutants that did not express the lipoamide dehydrogenase subunit of this enzyme complex accumulated branched-chain oxo acids in batch cultures grown on media containing leucine, isoleucine, or valine (4), thus casting some doubt on the exclusive role of pyruvate decarboxylase in the decarboxylation of branched-chain oxo acids.
The aim of this study was to reinvestigate the role of pyruvate decarboxylase in the production of fusel alcohols by S. cerevisiae. The S. cerevisiae genome harbors three structural genes (PDC1, PDC5, and PDC6) that can each encode an active pyruvate decarboxylase (9). In wild-type yeast strains, PDC6 expression is either very low or absent (7, 9). However, revertants of pdc1-pdc5 double mutants, in which a recombination event has caused a fusion of the PDC1 promoter and the PDC6 open reading frame, express a functional enzyme (10). Therefore, studies on the physiological effects of pyruvate decarboxylase deficiency are most easily interpreted when they are performed with strains in which all three PDC genes are disrupted.
In the present study, the decarboxylation of branched-chain 2-oxo acids was studied in cell extracts of wild-type S. cerevisiae and in extracts of an isogenic pyruvate decarboxylase-negative mutant. Furthermore, conversion of branched-chain amino acids to the corresponding fusel alcohols by intact cells was analyzed in ethanol-grown cultures of a wild-type S. cerevisiae strain and in those of the Pdc− mutant.
MATERIALS AND METHODS
Strains and growth conditions.
The yeast strains used in this study are listed in Table 1. S. cerevisiae T2-3D and the isogenic, prototrophic pyruvate decarboxylase-negative strain GG570 were grown in aerobic carbon-limited chemostat cultures (dilution rate, [D] = 0.10 h) on a mineral medium containing 7.125 g of glucose/liter. To meet the requirement of the Pdc− mutant for cytosolic acetyl-coenzyme A, 0.375 g of acetate/liter was also added to the reservoir medium (for further details on growth conditions, see reference 7). Strains GG567 and GG569 were grown overnight at 30°C in shake flask cultures on complex medium (Difco yeast extract [10 g/liter], Difco peptone [20 g/liter], glucose [20 g/liter]) prior to preparation of cell extracts.
TABLE 1.
S. cerevisiae strains used in the present study
| Strain | Genotype | Reference |
|---|---|---|
| T2-3D | HO/HO PDC1/PDC1 PDC5/PDC5 PDC6/PDC6 | 22 |
| GG 570 | HO/HO pdc1::Tn5ble/pdc1::Tn5ble pdc5::Tn5ble/pdc5::Tn5ble pdc6::APT1/pdc6::APT1 | 7 |
| GG 567 | HO/HO PDC1/PDC1 pdc5::Tn5ble/pdc5::Tn5ble pdc6::APT1/pdc6::APT1 | 7 |
| GG 569 | HO/HO pdc1::Tn5ble/pdc1::Tn5ble PDC5/PDC5 pdc6::APT1/pdc6::APT1 | 7 |
Analysis of fusel alcohol production by growing cells.
Precultures were grown at 30°C in shake flask cultures containing mineral medium with vitamins (20) supplemented with 1% (vol/vol) ethanol, with an initial pH of 6.0. The ammonium sulfate concentration in the mineral medium was decreased to 0.5 g/liter to obtain a nitrogen-depleted inoculum culture. After 24 h of incubation, the cells were centrifuged at 20,000 × g for 5 min and aseptically transferred to a 1,000-ml shake flask containing 400 ml of mineral medium supplemented with 1% (vol/vol) ethanol, with an initial pH of 6.0. The mineral medium lacked ammonium sulfate and contained instead a mixture of leucine, isoleucine, and valine (15 mM each) as a nitrogen source. The flasks were shaken (200 rpm) at 30°C. Samples were taken at appropriate time intervals and analyzed for optical density at 660 nm (23). Culture supernatants, obtained by centrifugation at 20,000 × g for 5 min, were analyzed for amino acids and fusel alcohols.
Analytical procedures.
Biomass concentrations were determined as described previously (7). Concentrations of 3-methyl-butanol, 2-methyl-butanol, and 2-methyl-propanol were analyzed by gas chromatography on a Perkin-Elmer 5800 gas chromatograph fitted with a Chrompack CP-SIL5CB column (length, 50 m; internal diameter, 0.32 mm; film thickness, 1.2 μm). The identity of the peaks was confirmed by high-pressure liquid chromatography (HPLC) analysis of the same samples on a Rezex ROA organic acid column at 60°C. The HPLC column was eluted with 0.5 g of H2SO4/liter; detection was by means of an ERMA ERC-7515A refractive index detector coupled with a Hewlett-Packard 3390A integrator. Concentrations of leucine, isoleucine, and valine were analyzed after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) with the Waters AccQ.Fluor kit. Derivatized samples were then analyzed by HPLC on a Waters Nova-Pak C18 column. The two mobile phases were (i) 60 mM ammonium acetate (pH 5.00) and (ii) 50% of mobile phase A plus 50% (vol/vol) acetonitrile. Gradient conditions were as follows: 27 min linearly from 97% of mobile phase A to 89% of mobile phase A and thereafter in 22 min linearly to 54% mobile phase A. Column temperature was 25°C; flow rate was 1 ml/min.
Preparation of cell extracts and enzyme assays.
Cell extracts were prepared as described previously (7). Pyruvate decarboxylase was assayed spectrophotometrically at 30°C. The assay mixture consisted of 40 mM imidazole-HCl buffer (pH 6.5), 0.2 mM thiamine pyrophosphate, 5 mM MgCl2, 150 μM NADH, yeast alcohol dehydrogenase (88 U/ml; Boehringer Mannheim) 0.05% Triton X-100, and cell extract. The reaction was started by the addition of 10 mM pyruvate. Essentially the same coupled assay was used to measure decarboxylation of α-ketoisocaproate, α-keto-β-methyl valerate, or α-keto isovalerate by cell extracts. However, to measure decarboxylation of these substrates, horse liver alcohol dehydrogenase (2 U/ml in assay mixture; Sigma) was used instead of yeast alcohol dehydrogenase (see Results). Alcohol dehydrogenase (EC 1.1.1.1) was assayed in a reaction mixture (1 ml) containing glycine-KOH buffer (pH 9.0), 50 mM; NAD (lithium salt), 1 mM; and cell extract. The reaction was started by the addition of 10 mM of either ethanol, 2-methylbutanol, 3-methylbutanol, or 2-methylpropanol.
Protein determination.
Protein concentrations in cell-free extracts were determined by the Lowry method (12a). Bovine serum albumin (fatty acid free; Sigma Chemical Co.) was used as a standard.
Biochemicals.
Partially purified preparations of pyruvate decarboxylase, yeast alcohol dehydrogenase, and horse liver alcohol dehydrogenase were obtained from Sigma.
RESULTS
Decarboxylation of 2-oxo acids by cell extracts of wild-type S. cerevisiae.
Cell extracts of the wild-type strain S. cerevisiae T2-3D, pregrown in aerobic carbon-limited chemostat cultures, exhibited high activities of pyruvate decarboxylase (Table 2) in a coupled spectrophotometric assay with yeast NAD-dependent alcohol dehydrogenase. Although the extracts exhibited some activity when pyruvate was replaced by the branched-chain 2-oxo acid α-keto-β-methyl valerate, α-keto isovalerate, or α-ketoisocaproate, only very low activities were observed (ca. 1% of those found with pyruvate). Furthermore, these low rates were not constant over time and not always linearly proportional to the amount of cell extract added to the assays. Control experiments showed that, even at an alcohol concentration of 10 mM, the activity of the coupling enzyme yeast alcohol dehydrogenase with fusel alcohols was much lower than when ethanol was used as the substrate (Table 3).
TABLE 2.
Decarboxylation of pyruvate and branched-chain 2-oxo acids by cell extracts of wild-type S. cerevisiae and isogenic strains affected in the expression of one or more PDC genes
| Strain (genotype) | 2-Oxo acid decarboxylation by cell extracts (U/mg of protein)b
|
|||
|---|---|---|---|---|
| Pyruvate | α-Keto-isovalerate | α-Keto-β-methyl valerate | α-Keto isocaproate | |
| T2-3Da (wild type) | 0.70 ± 0.05 | 0.082 (12%) | 0.027 (4%) | 0.039 (6%) |
| GG 570a (Δpdc1 Δpdc5 Δpdc6) | <0.003 | <0.003 | <0.003 | <0.003 |
| T2-3Dc (wild type) | 1.08 ± 0.03 | 0.129 (12%) | 0.070 (7%) | 0.064 (6%) |
| GG 567c (PDC1 Δpdc5 Δpdc6) | 0.81 ± 0.03 | 0.122 (15%) | 0.046 (6%) | 0.051 (6%) |
| GG 569c (Δpdc1 PDC5 Δpdc6) | 0.36 ± 0.02 | 0.064 (18%) | 0.026 (7%) | 0.029 (8%) |
Cells from carbon-limited chemostat cultures.
Data are the averages of at least two independent assays; standard deviations were less than 20% for all assays. Percentages in parentheses indicate the activity with branched-chain substrates relative to the rate of pyruvate decarboxylation.
Cells from exponential-phase shake flask cultures (see Materials and Methods).
TABLE 3.
Relative activities of commercial preparations of NAD-dependent alcohol dehydrogenases from yeast and equine liver with ethanol and fusel alcohols
| Substrate | Alcohol dehydrogenase activity (%)a
|
|
|---|---|---|
| Yeast | Horse liver | |
| Ethanol | 100 ± 4.4 | 100 ± 0.8 |
| 2-Methyl butanol | 9.0 ± 0.8 | 88 ± 2.3 |
| 3-Methyl butanol | 3.8 ± 0.8 | 41 ± 1.8 |
| 2-Methyl propanol | 8.2 ± 0.3 | 86 ± 6.5 |
Alcohol dehydrogenase activities were assayed at a substrate concentration of 25 mM as indicated in Materials and Methods; activity with ethanol was set at 100%.
According to the literature, horse liver alcohol dehydrogenase has a much broader substrate specificity than the yeast enzyme (19). Indeed, activities of the horse liver enzyme with the three fusel alcohols were of the same order of magnitude as those observed when ethanol was the substrate. With horse liver enzyme as the coupling enzyme, significant rates of conversion of the branched-chain 2-oxo acids could be measured (Table 2). At a substrate concentration of 10 mM, the activities with the branched-chain 2-oxo acids were 4 to 12% of those observed with pyruvate (Table 2).
The commercial preparation of horse liver alcohol dehydrogenase used in this study could not be used as a coupling enzyme in the pyruvate decarboxylase assay, as it was contaminated with lactate dehydrogenase. This contaminating activity did not catalyze NADH-dependent reduction of branched-chain 2-oxo acids (data not shown) and therefore did not interfere with the branched-chain 2-oxo acid decarboxylation assays.
Pyruvate decarboxylase is involved in decarboxylation of branched-chain 2-oxo acids by cell extracts.
To determine the role of pyruvate decarboxylase in the decarboxylation of branched-chain 2-oxo acids by cell extracts, assays were performed with extracts of the isogenic Pdc− strain S. cerevisiae GG570 (7). Extracts of this strain had no activity when pyruvate or the branched-chain 2-oxo acids were used as a substrate (Table 2). The addition of commercial, partially purified pyruvate decarboxylase to reaction mixtures containing Pdc− cell extracts restored decarboxylation activities (data not shown). This result demonstrated that pyruvate decarboxylase was the sole component lacking for the conversion of α-oxo acids to aldehydes by cell extracts.
The question of whether PDC1- and PDC5-encoded pyruvate decarboxylases are both able to catalyze the conversion of branched-chain 2-oxo acids was addressed by experiments with cell extracts of strains GG564 and GG762. In these strains, PDC1 and PDC5, respectively, are the only functional PDC genes (Table 1). Extracts of both strains catalyzed the conversion of pyruvate, α-ketoisocaproate, α-keto-β-methyl valerate, and α-keto isovalerate (Table 2), thus demonstrating that the gene products of these two PDC genes are both able to perform the conversion of branched-chain 2-oxo acids in vitro.
Production of fusel alcohols by cell suspensions does not require an active pyruvate decarboxylase.
We examined the role of pyruvate decarboxylase during the production of fusel alcohols from three branched-chain amino acids (leucine, isoleucine, and valine) by growing wild-type S. cerevisiae T2-3D and its isogenic Pdc− strain GG570 with a mixture of these three amino acids as the nitrogen source. Ethanol was used as the carbon source to circumvent the inability of the Pdc− strain to grow on glucose in batch cultures (7).
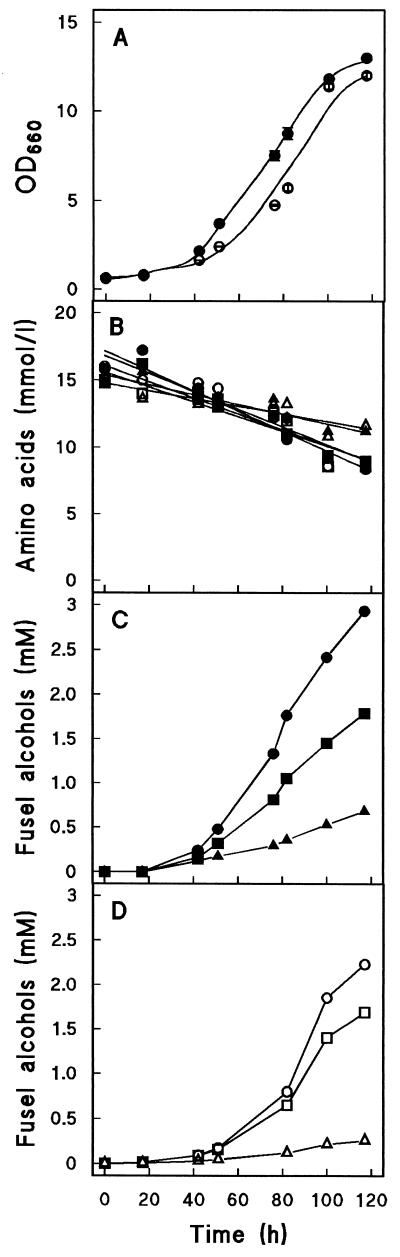
Growth rates of the two strains in the ethanol-grown shake flask cultures were similar, although the lag phase of the Pdc− strain appeared to be slightly longer (Fig. 1A). All three amino acids were consumed during growth, although the amount of valine that was utilized was about twofold less than the overall consumption of leucine and isoleucine (Fig. 1B). This result was reflected in the production of 2-methylpropanol, which in wild-type cultures was produced in lower amounts than the fusel alcohols derived from leucine and isoleucine (Fig. 1C).
FIG. 1.
Production of fusel alcohols by whole cells of S. cerevisiae T2-3D (wild type) and the isogenic pyruvate decarboxylase-negative S. cerevisiae GG570. Cells were grown on ethanol with a mixture of leucine, isoleucine, and valine (15 mM each) as the nitrogen source. In an independent duplicate experiment, optical density at 660 nm (OD660) and concentrations of fusel alcohols differed from those shown in the figure by less than 20%. Closed symbols represent wild-type cultures, and open symbols represent the Pdc− strain. (A) Optical density at 660 nm. (B) Concentrations of amino acids (• and ○, leucine; ▪ and □, isoleucine; ▴ and ▵, valine). (C) Concentrations of fusel alcohols in wild-type culture (•, 3-methylbutanol; ▪, 2-methylbutanol; ▴, 2-methylpropanol). (D) Concentrations of fusel alcohols in Pdc− culture (○, 3-methylbutanol; □, 2-methylbutanol; ▵, 2-methylpropanol).
Surprisingly, all three fusel alcohols were also produced by the pyruvate decarboxylase-negative mutant (Fig. 1C and D). Only in the case of 2-methylpropanol was the final product concentration substantially lower (over 50%) in mutant cultures than in wild-type cultures.
To rule out the possibility that the production of fusel alcohols by the Pdc− strain was due to any unintended presence of pyruvate decarboxylase (e.g., due to a reversion or to a contamination with wild-type cells), several control experiments were performed. Assays of pyruvate decarboxylase confirmed its absence in cell extracts of the Pdc− strain. Moreover, when samples from the ethanol-grown cultures were transferred to a medium containing glucose as the sole carbon source, no growth was observed, consistent with a Pdc− phenotype (7).
DISCUSSION
Our results demonstrate that, although pyruvate decarboxylase is able to catalyze the decarboxylation of branched-chain 2-oxo acids, this enzyme is not essential for the production of fusel alcohols by S. cerevisiae. Consequently, an alternative mechanism for fusel alcohol production must exist in this yeast. Our data do not exclude involvement of the Ehrlich pathway (including pyruvate decarboxylase) in fusel alcohol production by wild-type S. cerevisiae. Indeed, the rate of fusel alcohol production in the Pdc− strain was lower than that in the wild type, particularly in the case of 2-methylpropanol (Fig. 1C). The very poor activity of yeast NAD-dependent alcohol dehydrogenase with the putative intermediates of the Ehrlich pathway (Table 3) identifies the alcohol dehydrogenase reaction as an interesting target for attempts to improve productivity of fusel alcohols via this pathway. Expression of the horse liver enzyme in S. cerevisiae is an interesting option to test the viability of such an approach.
Discussion of the nature of the pathway for fusel alcohol production by the Pdc− strain is speculative. One possibility is that a decarboxylase other than pyruvate decarboxylase is present in S. cerevisiae and is not detected by the coupled enzyme assay used in this study. Dickinson and Dawes (4) have reported the involvement of a mitochondrial branched-chain 2-oxo acid dehydrogenase complex in the catabolism of branched-chain 2-oxo acids. To act as an intermediate in the Ehrlich pathway, the coenzyme A derivative formed by this enzyme complex (25) must be reduced to the corresponding aldehyde. So far, no enzyme activities have been identified in S. cerevisiae that catalyze this conversion. The only subunit of the branched-chain 2-oxo-acid dehydrogenase complex whose structural gene has been cloned is the lipoamide-dehydrogenase subunit (encoded by the LPD1 gene [15]). This subunit is also an essential part of the pyruvate-dehydrogenase and α-ketoglutarate–dehydrogenase complexes (15, 25) and of glycine decarboxylase (18), thereby complicating physiological studies of gene disruption mutants. For example, it is impossible to study the effect of an lpd1 null mutation in a Pdc− strain, since the resulting double mutant is not viable (respiratory growth requires LPD1, and fermentative growth requires a functional PDC gene). Identification of the structural gene encoding the E1 subunit of the branched-chain 2-oxo acid dehydrogenase complex, which is probably specific for this complex (4), seems to be a prerequisite for elucidating its possible role in fusel alcohol production.
Biochemical research to elucidate the pyruvate decarboxylase-independent formation of fusel alcohol production is not only of fundamental scientific interest; identification of the enzymes and genes involved may ultimately enable the optimization of fusel alcohol formation independent of the fermentative production of ethanol. Such a development might be applicable in processes such as the production of low-alcohol beers and high-gravity brewing.
ACKNOWLEDGMENTS
We thank Jaap Jongejan for advice on the substrate specificity of alcohol dehydrogenases, Max Zomerdijk and Toine van den Broek for gas chromatography, and Corrie Erkelens for amino acid analysis.
Yeast research in our groups is sponsored by the European Community (Framework IV program project “From Gene to Product in Yeast: a Quantitative Approach”). M.T.F. acknowledges a grant from The Netherlands Ministry of Economic Affairs (ABON program on Metabolic Engineering of Yeasts and Filamentous Fungi).
ADDENDUM
While the manuscript was under review, Dickinson and coworkers (5) published a study in which they convincingly demonstrated by 13C-nuclear magnetic resonance analysis of leucine metabolism in wild-type and mutant S. cerevisiae strains that neither pyruvate decarboxylase nor the branched-chain 2-oxo acid dehydrogenase complex is essential for the formation of 2-methyl butanol from leucine. Open reading frame YDL080c, which exhibits strong homology with the structural PDC genes, was proposed to encode a major decarboxylase involved in this process, although small amounts of 2-methyl butanol were still formed by null mutants (5). These observations emphasize the necessity for further research on the enzymology of fusel alcohol production and particularly on the relative importance of the various proposed pathways and enzymes as functions of environmental conditions.
REFERENCES
- 1.Berry D R, Watson D C. Production of organoleptic compounds. In: Berry D R, Russell I, Stewart G G, editors. Yeast biotechnology. London, England: Allen and Unwin; 1987. pp. 345–368. [Google Scholar]
- 2.Bigels R, Weir P D, Jones R R M, Umbarg H E. Exogenous valine reduces conversion of leucine to 3-methyl-1-butanol in Saccharomyces cerevisiae. Appl Environ Microbiol. 1983;45:658–664. doi: 10.1128/aem.45.2.658-664.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrick S, Large P J. Activities of the enzymes of the Ehrlich pathway and the formation of branched-chain alcohols in Saccharomyces cerevisiae and Candida utilis grown in continuous culture on valine or ammonium as sole nitrogen source. J Gen Microbiol. 1993;139:2783–2792. doi: 10.1099/00221287-139-11-2783. [DOI] [PubMed] [Google Scholar]
- 4.Dickinson J R, Dawes I W. The catabolism of branched chain amino acids occurs via 2-oxoacid dehydrogenase in Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2029–2033. doi: 10.1099/00221287-138-10-2029. [DOI] [PubMed] [Google Scholar]
- 5.Dickinson J R, Lanterman M M, Danner D J, Pearson B M, Sanz P, Harrison S J, Hewlins M J E. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 1997;272:26871–26878. doi: 10.1074/jbc.272.43.26871. [DOI] [PubMed] [Google Scholar]
- 6.Ehrlich F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweissaufbau der Hefe. Berichte der Deutschen Chemischen Gesellschaft. 1907;40:1027–1047. [Google Scholar]
- 7.Flikweert M T, van der Zanden L, Janssen W M T M, Steensma H Y, van Dijken J P, Pronk J T. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C247::AID-YEA911%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 8.Green D E, Herbert D, Subrahmanyan V. Carboxylase. J Biol Chem. 1941;138:327–339. [Google Scholar]
- 9.Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–7969. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohmann S. PDC6, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when fused spontaneously under the control of the PDC1 promoter. Curr Genet. 1991;20:373–378. doi: 10.1007/BF00317064. [DOI] [PubMed] [Google Scholar]
- 11.Large P J. Degradation of organic nitrogen compounds by yeasts. Yeast. 1986;2:1–34. [Google Scholar]
- 12.Lehmann H, Fischer G, Heuber G, Kohnert K-D, Schellenberger A. The influence of steric and electronic parameters on the substrate behaviour of α-oxo acids to yeast pyruvate decarboxylase. Eur J Biochem. 1973;32:83–87. doi: 10.1111/j.1432-1033.1973.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 12a.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:266–275. [PubMed] [Google Scholar]
- 13.Ouchi K, Yamamoto Y, Takagishi M, Akiyama H. Regulation of isoamyl alcohol formation via Ehrlich pathway in Saccharomyces cerevisiae. J Ferment Technol. 1980;58:301–309. [Google Scholar]
- 14.Reed G, Nagodawithana T W. Yeast technology. 2nd ed. New York, N.Y: Van Nostrand Reinhold; 1991. [Google Scholar]
- 15.Roy D J, Dawes I W. Cloning and characterization of the gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae. J Gen Microbiol. 1987;133:925–933. doi: 10.1099/00221287-133-4-925. [DOI] [PubMed] [Google Scholar]
- 16.Sentheshanmuganathan S. The mechanism of the formation of higher alcohols from amino acids by Saccharomyces cerevisiae. Biochem J. 1960;74:568–576. doi: 10.1042/bj0740568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinclair D A, Dawes I W, Dickinson J R. Purification and characterization of branched chain α-ketoacid dehydrogenase complex from Saccharomyces cerevisiae. Biochem Mol Biol Int. 1993;31:911–922. [PubMed] [Google Scholar]
- 18.Sinclair D A, Dawes I W. Genetics of the synthesis of serine from glycine and the utilization of glycine as sole nitrogen source by Saccharomyces cerevisiae. Genetics. 1995;140:1213–1222. doi: 10.1093/genetics/140.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sund H, Theorell H. Alcohol dehydrogenases. In: Boyer P, Lardy H, Myrbäck K, editors. The enzymes. New York, N.Y: Academic Press; 1963. pp. 25–83. [Google Scholar]
- 20.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 21.Watson T G, Hough J S. Conversion of α-keto-isocaproic acid to iso-amyl alcohol by yeast pyruvate decarboxylase and alcohol dehydrogenase. J Inst Brew. 1969;75:359–363. [Google Scholar]
- 22.Wenzel T J, van den Berg M A, Visser W, van den Berg J A, Steensma H Y. Characterization of Saccharomyces cerevisiae mutants lacking the E1a subunit of the pyruvate dehydrogenase complex. Eur J Biochem. 1992;209:697–705. doi: 10.1111/j.1432-1033.1992.tb17338.x. [DOI] [PubMed] [Google Scholar]
- 23.Weusthuis R A, Luttik M A H, Scheffers W A, van Dijken J P, Scheffers W A. Is the Kluyver effect in yeasts caused by product inhibition? Microbiology. 1994;140:1723–1729. doi: 10.1099/13500872-140-7-1723. [DOI] [PubMed] [Google Scholar]
- 24.Woodward J R, Cirillo V P. Amino acid transport and metabolism in nitrogen-starved cells of Saccharomyces cerevisiae. J Bacteriol. 1977;130:714–723. doi: 10.1128/jb.130.2.714-723.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeaman S J. The 2-oxo acid dehydrogenase complexes: recent advances. Biochem J. 1989;257:625–632. doi: 10.1042/bj2570625. [DOI] [PMC free article] [PubMed] [Google Scholar]