Significance
The TGF-beta family members Nodal and Vg1 are the major inducers of mesendoderm formation during vertebrate embryogenesis. We previously established that the Vg1 proprotein is retained in the endoplasmic reticulum (ER) and that Nodal and Vg1 form heterodimers to pattern the early embryo. However, the mechanisms underlying the retention, processing, secretion, and signaling of Vg1 have been unclear. We found two mechanisms that embryos use to efficiently generate active Nodal-Vg1 heterodimers: 1) Vg1 employs its chaperone-binding motifs to ensure its retention as a ready-to-heterodimerize monomer in the ER, and 2) using a Synthetic Processing (SynPro) System, we found that Vg1 must be processed for signaling to occur, but its processing location is flexible.
Keywords: Vg1, Nodal, retention, processing, zebrafish
Abstract
The TGF-beta signals Vg1 (Dvr1/Gdf3) and Nodal form heterodimers to induce vertebrate mesendoderm. The Vg1 proprotein is a monomer retained in the endoplasmic reticulum (ER) and is processed and secreted upon heterodimerization with Nodal, but the mechanisms underlying Vg1 biogenesis are largely elusive. Here, we clarify the mechanisms underlying Vg1 retention, processing, secretion, and signaling and introduce a Synthetic Processing (SynPro) system that enables the programmed cleavage of ER-resident and extracellular proteins. First, we find that Vg1 can be processed by intra- or extracellular proteases. Second, Vg1 can be processed without Nodal but requires Nodal for secretion and signaling. Third, Vg1-Nodal signaling activity requires Vg1 processing, whereas Nodal can remain unprocessed. Fourth, Vg1 employs exposed cysteines, glycosylated asparagines, and BiP chaperone-binding motifs for monomer retention in the ER. These observations suggest two mechanisms for rapid mesendoderm induction: Chaperone-binding motifs help store Vg1 as an inactive but ready-to-heterodimerize monomer in the ER, and the flexibility of Vg1 processing location allows efficient generation of active heterodimers both intra- and extracellularly. These results establish SynPro as an in vivo processing system and define molecular mechanisms and motifs that facilitate the generation of active TGF-beta heterodimers.
The TGF-beta signals Nodal and Vg1 (Dvr1/Gdf3) play crucial roles in vertebrate development (1, 2), including the induction of mesendoderm and the generation of left-right asymmetry (3–17). For example, secreted Vg1-Nodal heterodimers induce a gradient of signaling that patterns the embryonic mesendoderm in zebrafish (10). Vg1-Nodal heterodimers exert their effects as ligands for a receptor complex that comprises Activin serine-threonine kinase receptors and an essential coreceptor called Oep (Tdgf1/CRIPTO) (18–20). Activated ligand-receptor complexes catalyze phosphorylation of Smad2 (pSmad2), which accumulates in the nucleus to induce the expression of mesendodermal genes (21).
We previously proposed a 4-step model for how Vg1-Nodal heterodimers pattern the mesendoderm of zebrafish embryos (10): 1) Maternally ubiquitous Vg1 proprotein is retained as a monomer in the endoplasmic reticulum (ER) of embryonic cells. 2) Expression of zebrafish Nodal genes, cyclops (cyc) and squint (sqt), initiates at the yolk margin at ~3 h postfertilization (hpf). 3) Nodal forms heterodimers with preexisting Vg1. 4) Vg1-Nodal heterodimers are processed and secreted to activate signaling. This model explains how Nodal and Vg1 interact during early embryogenesis, but the molecular mechanisms that regulate Vg1 retention, processing, secretion, and signaling have remained unclear.
Previous studies of growth factor processing and ER retention provide potential mechanisms for how Vg1 localization, processing, and activity might be regulated. TGF-beta ligands are synthesized as preproproteins that comprise an amino-terminal signal sequence, a long prodomain, and a shorter bioactive mature domain. A conserved cysteine in the mature domain is primarily responsible for dimer formation via an intermolecular disulfide bond (22). TGF-beta prodomain processing can occur in the Golgi apparatus and on the cell surface, where proprotein convertases are found (23–26), but it is unclear whether Vg1 needs to be processed intra- or extracellularly or whether Vg1 processing depends on Nodal. It is also unclear how Vg1 is retained in the ER. Vg1 has an intact prodomain when it is retained in the ER (10), raising the possibility that Vg1 cannot be secreted because the Vg1 prodomain cannot be processed (3, 4, 27). Alternatively, a suite of protein-folding chaperones could potentially interact with specific motifs on the Vg1 proprotein to block its release from the ER and subsequent processing. For example, protein disulfide isomerases (PDIs) that bind to exposed cysteines facilitate the folding of nascent proproteins and promote the retention of unassembled protein complexes in the ER (28, 29). Additionally, the lectin chaperones, calnexin and calreticulin, bind to asparagine-linked glycosyl groups on nascent secreted proteins (30). Glycosylated proteins are released from the ER after N-linked sugar moieties are trimmed off (30, 31). Moreover, the BiP chaperone (also known as Hspa5/Grp78) aids in protein folding by binding to predominantly hydrophobic heptapeptide sequences and can also retain proteins in the ER (32–34). Vg1 has exposed cysteines and asparagines that can be glycosylated (35), but it is unclear whether Vg1 or other TGF-beta signals use these chaperone-binding motifs to ensure retention in the ER.
In this study, we investigate the molecular mechanisms that regulate Vg1 retention, processing, secretion, and signaling. To control prodomain processing, we created a Synthetic Processing (SynPro) system that enables programmed cleavage of ER-resident and extracellular proteins. Using SynPro, we find that Vg1 can be cleaved and activated either cell-autonomously or non-cell-autonomously. Vg1 can be processed without Nodal but requires Nodal for secretion and signaling. We further show that Vg1, but not Nodal, must be processed for signaling activity. Finally, we identify several chaperone-binding motifs in the prodomain and mature domains of Vg1 that function in ER retention. These molecular mechanisms and sequence motifs control Vg1 biogenesis and contribute to the temporal and spatial specificity of mesendoderm formation.
Results
Creation of a Synthetic Processing System (SynPro).
The activity of Vg1 depends on its dimerization with Nodal (10), but where processing must occur relative to dimerization and secretion has remained unclear. For example, Vg1 might be processed before or after secretion, and processing might occur by proteases expressed in Vg1-secreting cells or in neighboring cells. To address these questions, we first set out to control processing orthogonally by creating SynPro. SynPro employs the ability of a synthetic protease to cleave a specific peptide cleavage site (Fig. 1 and SI Appendix, Fig. S1). We screened several proteases for their ability to cleave short peptide sequences and to be produced in zebrafish embryos without deleterious effects. These included proteases from the tobacco etch virus (TEVp), tobacco vein mottling virus (TVMVp), human rhinovirus 3C, and enterokinase (36). We found that most commercially available proteases are toxic when produced from mRNAs injected into zebrafish embryos (SI Appendix, Fig. S1A). Thus, we synthesized zebrafish-codon-optimized mRNAs of TEVp, TVMVp, and 13 additional proteases of the Potyviridae family (37). All 15 proteases were nontoxic when expressed in zebrafish embryos (SI Appendix, Fig. S1B).
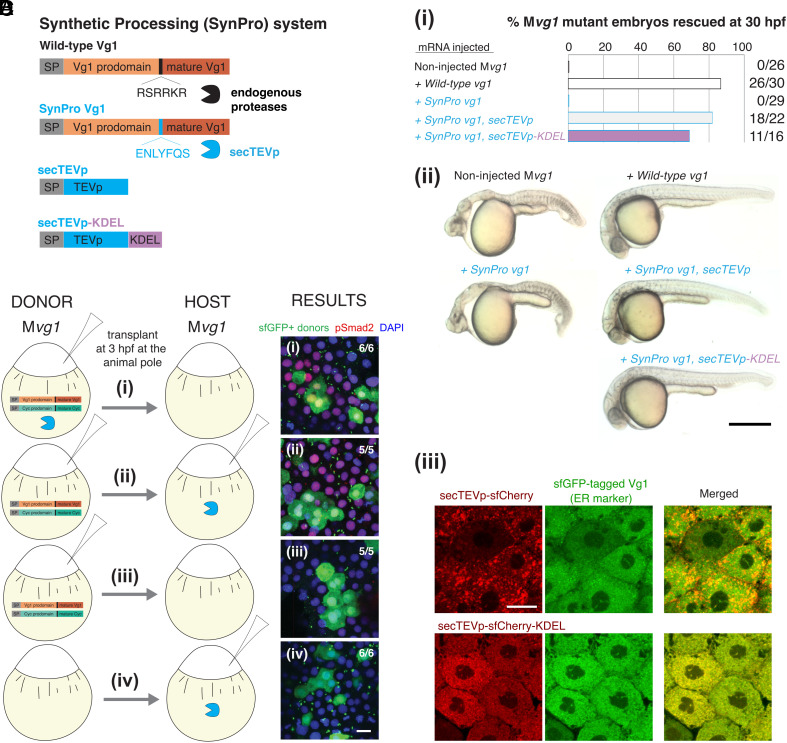
Fig. 1.
The Synthetic Processing (SynPro) system shows that prodomain cleavage can be non-cell-autonomous. (A) The SynPro system comprises an orthogonal secreted protease derived from tobacco etch virus (secTEVp) and a cognate sequence that replaces the endogenous cleavage site of Vg1 (SynPro Vg1, RSRRKR → ENLYFQS). (B) (i) Rescue percentage after 30 hpf of Mvg1 embryos injected with 50 pg of vg1, SynPro vg1, SynPro vg1 and secTEVp, or SynPro vg1 and secTEVp-KDEL mRNAs. (ii) Representative images of 30 hpf Mvg1 embryos for the indicated injection condition. Minor brain and tail defects are noted in embryos transiently rescued with mRNAs of the SynPro system. (Scale bar, 0.5 mm.) (iii) Fluorescence images of Mvg1 embryos at 50 to 60% epiboly that were injected with mRNAs for sfGFP-tagged wild-type Vg1 and secTEVp-sfCherry (Top) or secTEVp-sfCherry-KDEL (Bottom). (Scale bar, 20 μm.) (C) Schematic of transplantation assay. Mvg1 embryos were injected with 50 pg mRNA each of: (i) DONOR: cyc, SynPro vg1, and secTEVp; HOST: none; (ii) DONOR: cyc and SynPro vg1; HOST: secTEVp; (iii) DONOR: cyc and SynPro vg1; HOST: none; (iv) DONOR: none; HOST: secTEVp. All Mvg1 donor embryos were marked by also injecting 50 pg sfGFP mRNA. At high stage, before the onset of Nodal signaling, sfGFP-marked DONOR cells were transplanted to the animal pole of HOST Mvg1 embryos. (D) At 50 to 60% epiboly, chimeric embryos were fixed and immunostained for sfGFP and pSmad2. DAPI, nuclei. (Scale bar, 20 μm.)
To test whether the synthetic proteases are functional, we designed a fluorescent reporter of proteolytic cleavage (SI Appendix, Fig. S1C; see Materials and Methods for details). Briefly, a protease-cleavable substrate is initially localized in the cytoplasm. Protease-catalyzed cleavage results in the release into the nucleus and reconstitution of a split fluorescent protein, mNeonGreen2 (38). Coexpression of the cleavable substrate and the nuclear reporter in zebrafish embryos did not lead to reconstitution of nuclear mNeonGreen2 fluorescence (SI Appendix, Fig. S1 D, i). By contrast, nuclear mNeonGreen2 fluorescence was observed when TEVp was coexpressed with its cognate substrate and the nuclear reporter (SI Appendix, Fig. S1 D, ii). We observed protease-induced fluorescence reconstitution for 6 out of 15 proteases tested (SI Appendix, Fig. S1E), indicating that the synthetic proteases can cleave their cognate sequences in zebrafish embryos.
Synthetically Processed Vg1 Rescues vg1 Mutants.
While the TEV protease has been shown to be active in the cytoplasm, none have been shown to be active in the secretory system in vivo. To generate a secreted protease for the SynPro system, we added an amino-terminal signal sequence to TEVp to produce a secreted variant, secTEVp (see Materials and Methods for details). We also introduced five amino acid substitutions in secTEVp that are known to promote solubility (39) and prevent oxidation in the secretory compartments (40). To determine whether secTEVp is functional and can replace endogenous convertases that cleave the Vg1 prodomain, we generated SynPro Vg1. In this Vg1 derivative, the native cleavage sequence, “RSRRKR”, was replaced with the cognate cleavage sequence of secTEVp, “ENLYFQS” (Fig. 1A). Remarkably, the coexpression of SynPro vg1 and secTEVp mRNAs rescued mesendoderm formation in maternal vg1 mutant (Mvg1) embryos, whereas the expression of SynPro vg1 mRNA alone failed to rescue Mvg1 mutants (Fig. 1B). Notably, coproduction of SynPro Vg1 and an ER-localized secTEVp-KDEL (Fig. 1 B, iii) also rescued Mvg1 embryos, revealing that intracellular processing of Vg1 can result in normal Vg1 activity. These results show that endogenous enzymes that process Vg1 can be replaced by an orthogonal protease and that SynPro provides a tool to control processing and protein activity in the secretory system.
Vg1 Processing Is Not Sufficient for Secretion.
The coexpression of SynPro vg1 and secTEVp rescued Mvg1 mutants but did not induce abnormal overexpression phenotypes. This result suggests that Vg1 signaling activity was restricted to domains of coexpression with endogenous Nodal, raising two hypotheses: (1) Vg1 cleavage by SynPro depends on Nodal or (2) Vg1 cleavage by SynPro is independent of Nodal, but its secretion requires Nodal. To test these models, we coexpressed SynPro vg1 and ER-resident secTEVp-KDEL and assessed the location and processing of SynPro Vg1 with or without the zebrafish Nodal cyc (SI Appendix, Fig. S2). We found that SynPro Vg1 was processed but not secreted in the absence of the Nodal signal Cyclops. These results support hypothesis (2): Synthetic processing of Vg1 does not require Nodal, but Nodal is necessary for Vg1 secretion.
Vg1 Processing Can Be Non-Cell-Autonomous.
The results above show that intracellular processing in the secretory compartments is sufficient to produce functional Vg1. We next determined whether processing of the Vg1 proprotein in the extracellular milieu might also be sufficient to generate active Vg1. By leveraging the versatility of the SynPro system, we generated scenarios where SynPro Vg1 and secTEVp were expressed in the same or in different cells. We performed a series of transplant experiments in Mvg1 embryos that were injected with mRNAs expressing components of the SynPro system (Fig. 1 C and D). First, donor Mvg1 cells that coexpress SynPro vg1, secTEVp, and cyc mRNAs were transplanted into host Mvg1 embryos (Fig. 1C). In this scenario, ligands and protease are coexpressed in the same cells. As expected, we observed Nodal signaling activity based on immunostaining of nuclear pSmad2 in both donor and neighboring host cells (Fig. 1 D, i). This result indicates that donor cells secreted active secTEVp-processed SynPro Vg1 and Cyc. Second, donor cells coexpressing SynPro vg1 and cyc were transplanted into host Mvg1 embryos expressing secTEVp. In this scenario, ligands and protease are not coexpressed in the same cells. Strikingly, we observed nuclear pSmad2 accumulation in both donor and surrounding host cells (Fig. 1 D, ii). In control experiments, nuclear pSmad2 was never observed when secTEVp was absent, nor when donor embryos did not express SynPro Vg1 and Cyc (Fig. 1 D, iii and iv). Thus, host-secreted secTEVp was able and required to cleave and activate the donor-secreted SynPro Vg1 and Cyc. This application of the SynPro system indicates that Vg1 can be processed and activated non-cell-autonomously.
Processing Is Not Required for Secretion of Vg1 and Nodal.
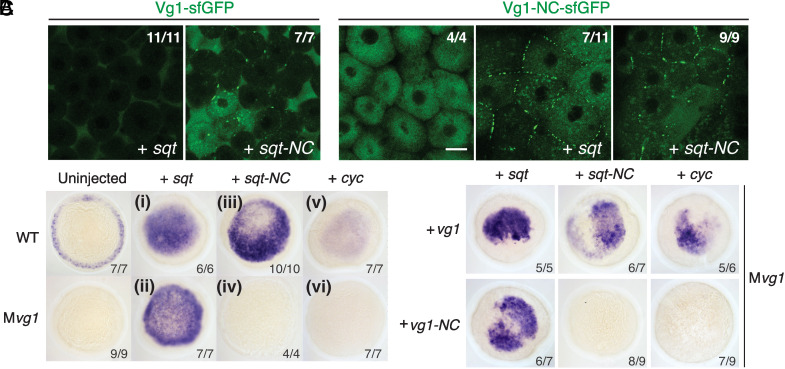
The observation that extracellular secTEVp was sufficient to generate active signaling around cells producing SynPro Vg1 and Nodal suggested that an unprocessed Vg1 proprotein can be secreted in the presence of Nodal. To further test this idea, we generated noncleavable mutants of Vg1 and Nodal. We inactivated the cleavage site of Vg1 and of the zebrafish Nodal Squint (Sqt) and inserted a superfolder green fluorescent protein (sfGFP) to generate the noncleavable variants, vg1-NC-sfGFP and sqt-NC-sfGFP. We could not generate a noncleavable variant of Cyc due to additional cryptic cleavage sites (SI Appendix, Fig. S3). We found that both sfGFP-tagged Vg1 (vg1-sfGFP) or noncleavable Vg1 (vg1-NC-sfGFP) were secreted when coproduced with Sqt or noncleavable Sqt (sqt-NC) in Mvg1 embryos (Fig. 2A). Notably, endogenous levels of Cyc-Vg1 and Sqt-Vg1 heterodimers are so low that secretion of Vg1-sfGFP cannot be detected without ectopic addition of Sqt (10). Secretion was incomplete under conditions in which at least one signal was noncleavable, as evidenced by intracellular and cell membrane localization. These results indicate that Vg1 and Nodal can be secreted in the absence of processing.
Fig. 2.
Prodomain cleavage affects Vg1-Nodal signaling but not secretion. (A) Live fluorescence imaging of Mvg1 coinjected with 50 pg of vg1-sfGFP or noncleavable vg1-sfGFP (vg1-NC-sfGFP, RSRRKR → SQNTSN) mRNA and 50 pg of sqt or sqt-NC (RRHRR → SQNTS) mRNA. (Scale bar, 17 μm.) (B) Nodal target gene (lefty1) expression at 50% epiboly in WT and Mvg1 embryos injected with 50 pg of sqt, sqt-NC, or cyc mRNA. (C) lefty1 expression in Mvg1 embryos coinjected with 50 pg sqt, sqt-NC, or cyc; and vg1 or vg1-NC mRNA.
Processing Is Not Required for Nodal Activity in the Presence of Processed Vg1.
To test whether noncleavable Vg1 (Vg1-NC) and Nodal are physiologically active, we injected mRNAs of the noncleavable constructs into wild-type and Mvg1 embryos and assessed the induction of Nodal target genes. Remarkably, noncleavable Sqt was able to induce target gene expression in embryos expressing wild-type Vg1 (Fig. 2B, iii). By contrast, noncleavable Sqt was inactive in the absence of Vg1 (Mvg1 mutants) (Fig. 2B, iv) or when coexpressed with Vg1-NC (Fig. 2C). Vg1-NC was also inactive when coexpressed with Cyclops (Fig. 2C). These results indicate that the Vg1 prodomain—but not the Nodal prodomain—must be cleaved for active heterodimer signaling.
Cysteine Thiol and N-glycosylation Sites Retain the Vg1 Prodomain in the ER.
Previous studies have shown that replacement of the Vg1 prodomain with other TGF-beta prodomains resulted in Vg1 processing and mesoderm-inducing activity (3, 4, 27, 41, 42). Similarly, we found that a chimeric Vg1 protein that fused the Nodal prodomain to the Vg1 mature domain resulted in a secreted and active Vg1 (SI Appendix, Fig. S4A). Conversely, replacing the prodomain of zebrafish Nodals with the prodomain of Vg1 inhibited the secretion and activity of these chimeric TGF-betas (SI Appendix, Fig. S4B). These results suggest that the Vg1 prodomain has features that block secretion and promote ER retention.
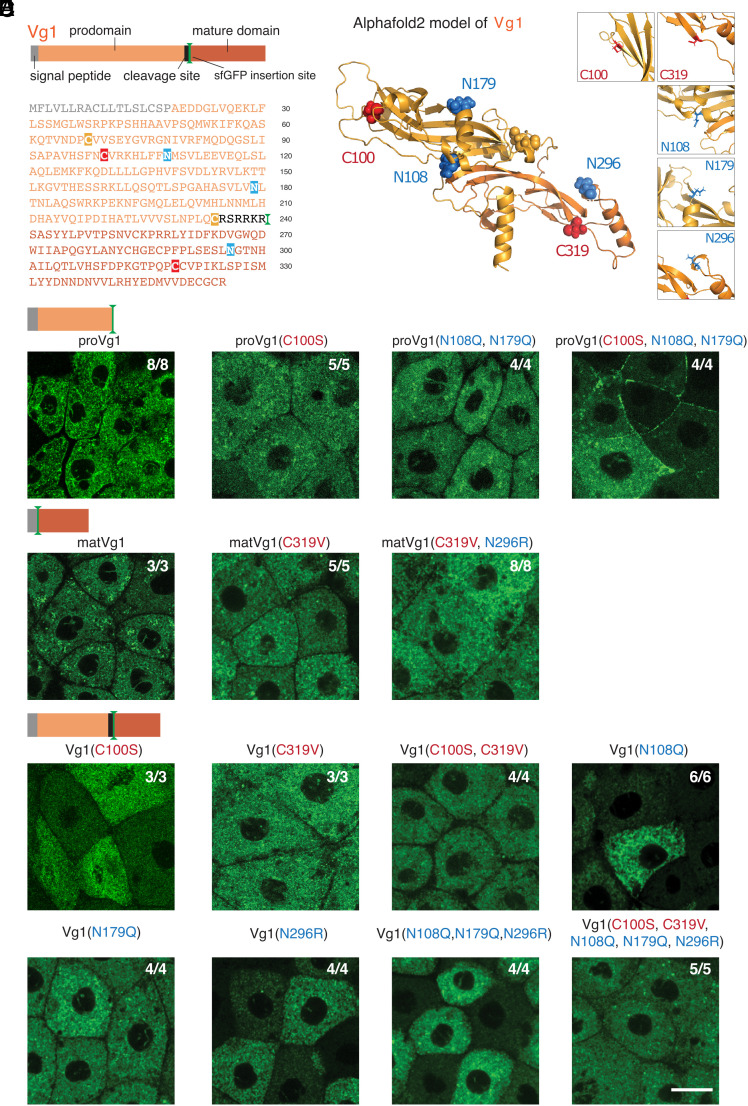
To determine how the Vg1 prodomain might mediate ER retention, we searched for putative sequence motifs that might mediate retention. We did not find any KDEL sequences in Vg1; these motifs are found at the C termini of ER-resident proteins and are recognized by KDEL receptors that trigger Golgi-to-ER retrograde transport (43). However, we found putative sequence motifs in Vg1 that bind to ER-resident chaperones, including exposed cysteines that bind to PDIs (28, 29) and glycosylated asparagines in NX[S/T] motifs that bind to calnexin and calreticulin (30). In particular, the prodomain of Vg1 (proVg1) has one exposed cysteine (C100) and two potential N-glycosylation sites (N108, N179). Notably, the mature domain of Vg1 (matVg1) also has a free cysteine (C319) and a potential N-glycosylation site (N296), raising the possibility that the mature domain also plays a role in ER retention (Fig. 3A). To determine whether these residues are accessible at the surface, we used AlphaFold2 (44) (there is no biophysically determined Vg1 structure). The top model for Vg1 in AlphaFold2 shows that the cysteine and asparagine residues are indeed exposed at the surface (Fig. 3B), suggesting that ER-resident chaperones can potentially access them.
Fig. 3.
Cysteine and N-linked glycosylation sites retain the Vg1 prodomain in the ER. (A) Schematic and primary amino acid sequence of zebrafish Vg1 preproprotein. Cysteines (red, yellow) and asparagines (blue) are highlighted. (B) Alphafold2 model for Vg1 is shown in cartoon representation, whereas the cysteine (red, yellow) and asparagine (blue) residues are shown in spheres (Insets show zoomed-in views of the residues mutated in this study). We make a minor note here that two cysteine residues, C68 and C234 (yellow), are predicted to form a disulfide bond and thus were not further studied. (C–E) Fluorescence images of fixed Mvg1 embryos injected with 50 pg mRNA of sfGFP-tagged vg1 prodomain (proVg1) (C), vg1 mature domain (matVg1) (D), and full-length vg1 (E), with or without the indicated cysteines and asparagines mutated. sfGFP was inserted into vg1 downstream of the predicted basic cleavage site in all constructs. (Scale bar, 20 μm.)
To elucidate whether these residues promote Vg1 retention in the ER, we systematically mutated the cysteine and N-glycosylation residues in sfGFP-tagged Vg1, proVg1, and matVg1 constructs and visualized their localization. We injected Mvg1 embryos with sfGFP-tagged vg1, proVg1, or matVg1 mRNAs and performed fluorescence microscopy to determine the subcellular localization of the resulting proteins. Similar to full-length Vg1, we observed ER localization of sfGFP-tagged proVg1 and matVg1 (Fig. 3 C and D). Mutants for the prodomain cysteine (C100S) or glycosylation sites (N108Q and N179Q) were retained in the ER, but the triple mutant proVg1(C100S, N108Q, and N179Q) was secreted to the extracellular space (Fig. 3C). By contrast, the matVg1 cysteine and glycosylation mutants (C319S and N296R) as well as the quintuple mutant Vg1(C100S, N108Q, N179Q, C319S, and N296R) were still retained in the ER (Fig. 3 D and E). We verified the secretion or localization of these constructs in the ER of Mvg1 embryos by coexpressing them with a fluorescent ER marker (sfCherry-KDEL) or with a nucleocytoplasmic marker (sfCherry-Smad2) (SI Appendix, Fig. S4C). We also found that the loss of N-glycosylation sites was more deleterious to signaling activity than the loss of cysteines (SI Appendix, Fig. S5). Taken together, our mutagenesis results show that N-glycosylation sites and an exposed cysteine thiol retain the Vg1 prodomain in the ER.
BiP-Binding Motifs Retain the Vg1 Mature Domain in the ER.
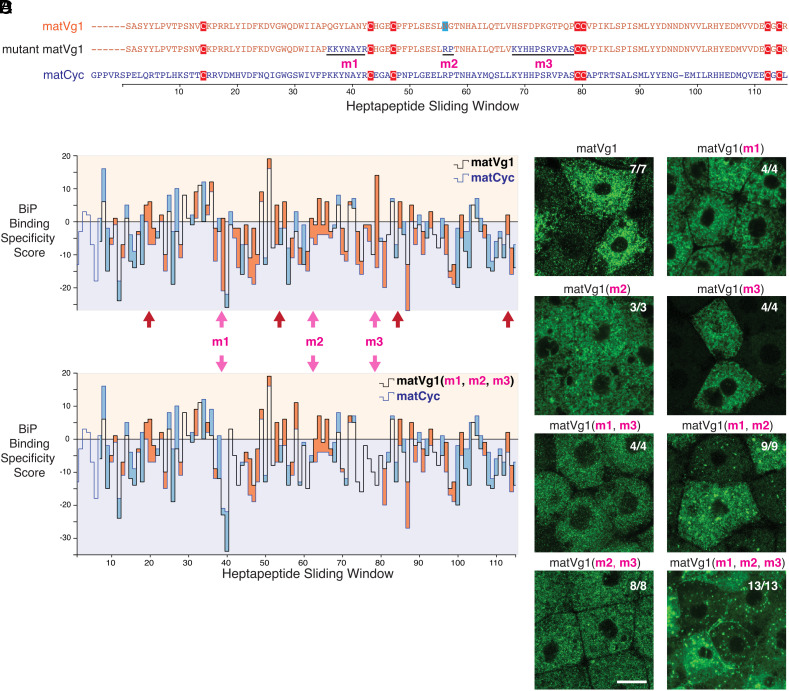
The observation that the free cysteine and N-glycosylation mutants of matVg1 and full-length Vg1 are still retained in the ER suggests that matVg1 possesses additional ER-retention motifs. We hypothesized that a third chaperone, BiP (Hspa5/Grp78), might promote ER retention. To elucidate which sequence features of matVg1 may bind to BiP, we utilized the Gething-Sambrook scoring system (34). Since mature Nodals (matSqt and matCyc) are secreted (SI Appendix, Fig. S6), we compared the sequences of matVg1 and matCyc and scored for all possible BiP-binding heptapeptides (Fig. 4 A and B and SI Appendix, Table S1; see Materials and Methods for details). To test whether the high-scoring heptapeptides of matVg1 promote ER retention, we systematically mutated three high-scoring matVg1 sequences to their corresponding low-scoring matCyc sequences (labeled as m1, m2, and m3 in Fig. 4 A and C). While all single mutants and double mutants were found in the ER, the triple mutant matVg1(m1, m2, and m3) exhibited extracellular localization (Fig. 4D and SI Appendix, Fig. S4C). Conversely, a mutant matCyc possessing the high-scoring features of matVg1 was retained in the ER (SI Appendix, Table S1 and Fig. S7). These results indicate that BiP-binding sequences retain the Vg1 mature domain in the ER.
Fig. 4.
Binding motifs for BiP promote ER retention of the Vg1 mature domain. (A) Amino acid sequence alignment of the mature domains of Vg1 (matVg1) and Cyc (matCyc) and mutant matVg1. Cysteines (red) and a potentially glycosylated asparagine (blue) are highlighted. (B and C) Difference charts of the BiP binding specificity scores (34) between two protein sequences along a sliding window of seven amino acids. Orange fills indicate the matVg1 or matVg1(m1, m2, m3) score (black line) > matCyc score (blue line). Conversely, blue fills indicate matCyc score > matVg1 or matVg1(m1, m2, m3) score. Arrows denote regions where matVg1 score > 0 and matCyc score < 0. Red arrows specifically denote regions that contain cysteines involved in cystine-knot formation. (B) Differences in BiP binding specificity scores between matVg1 (black line) and matCyc (blue line). (C) Differences in BiP binding specificity scores between mutant matVg1(m1, m2, m3) (black line) and matCyc (blue line). Note the loss of orange fills in m1, m2, and m3 regions (pink arrows) when compared to (B). (D) Fluorescence images of fixed Mvg1 embryos injected with 50 pg mRNA of sfGFP-tagged vg1 mature domain variants. sfGFP was inserted upstream of the vg1 mature domain in all constructs. (Scale bar, 20 µm.)
Discussion
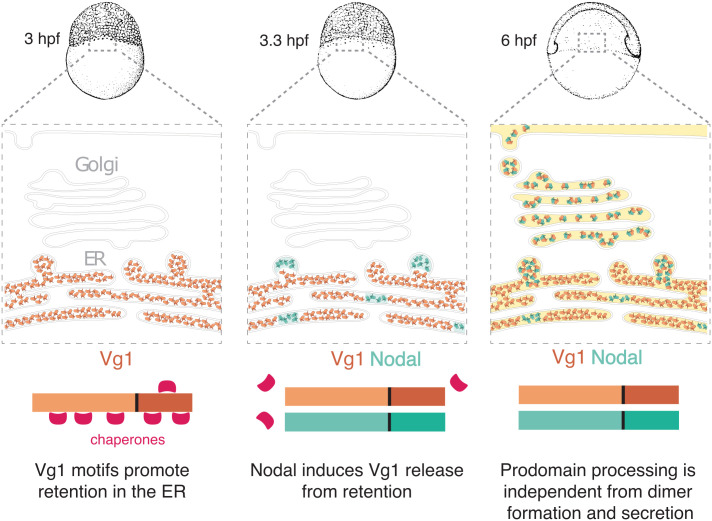
This study reveals features of Vg1 that regulate its retention, processing, secretion, and signaling during early zebrafish embryogenesis (Fig. 5): 1) ER retention of Vg1 is mediated by exposed cysteines, glycosylated asparagines, and BiP chaperone-binding motifs. 2) Vg1 can be processed without Nodal but requires Nodal for secretion and signaling. 3) Vg1 can be processed cell-autonomously or non-cell-autonomously. 4) Vg1-Nodal signaling activity requires processing of Vg1 but not of Nodal. These conclusions unify and extend several previous observations about Vg1-Nodal signaling:
Fig. 5.
Model of Vg1-Nodal heterodimer formation and processing. Maternal Vg1 is retained in the ER via chaperone-binding motifs; during maternal-zygotic transition, Nodal is produced and induces release of Vg1 from the ER via heterodimer formation; processing of the Vg1 prodomain is required for the activity of Vg1-Nodal heterodimers but can be independent from dimer formation and secretion.
First, our study defines Vg1 sequence motifs that have the hallmarks of binding sites for ER-resident chaperones. Our mutational analyses indicate that these motifs help retain monomeric Vg1 in the ER until Nodal is expressed and heterodimerizes with Vg1. We find that the Vg1 prodomain is specifically retained in the ER via its N-glycosylation sites and exposed cysteine. Thus, Vg1 is a member of a class of secreted proteins that are transiently retained in the ER until their cysteines form intermolecular disulfide bonds (45–48). In addition, Vg1 employs BiP-binding regions as chaperone motifs for ER retention. Mutating the BiP-binding regions (m1, m2, m3) of the Vg1 mature domain leads to secretion. Interestingly, the m3 region is juxtaposed to cysteine C319, which mediates heterodimerization with Nodal. It is conceivable that BiP binding blocks the formation of mature Vg1 homodimers. We note that Vg1 mutant constructs that remain in the ER may have signaled to ER chaperones that they are misfolded, preventing us from determining whether additional retention motifs exist or whether mutations in the construct led to misfolding.
We found no evidence for KDEL-mediated ER retention of Vg1. This mode of ER retention differs from chaperone-mediated retention in that it mediates retrograde transport from the Golgi apparatus to the ER lumen (43). By contrast, chaperone-mediated retention prevents the escape of proteins from the ER. We speculate that the Vg1 system has evolved to trap Vg1 in the ER until Nodal is imported and displaces the chaperones as a heteromeric partner.
Second, our study reveals shared features of the Vg1-Nodal system with the heavy- and light-chain immunoglobulin (Ig) assembly system (46, 49–60). Both Nodal and free light chains are readily secreted (57, 58), whereas both Vg1 and heavy chains are retained in the ER until they are covalently bonded to Nodal and light chains, respectively. Mutating the exposed cysteine in the heavy chain leads to premature secretion of monomers (59, 60). Our results suggest a similar role for the exposed cysteine in Vg1, but Vg1 additionally employs N-glycosylation sites and BiP-binding regions as chaperone motifs for ER retention. During antibody assembly, the BiP chaperone can stably bind to unstructured heavy chains (55, 59) and cooperates with PDI and calnexin to properly assemble light and heavy chains (52, 61). A similar mechanism might control Vg1-Nodal heterodimer assembly. In this scenario, the Vg1 monomer remains in an unstructured, immature state through interaction with various chaperones in the ER and might fold only upon interaction with Nodal. How the chaperone network hands over the Vg1 monomer to Nodal remains to be elucidated.
Third, our study clarifies the roles of Vg1 processing in secretion. Early studies suggested that Vg1 cannot be secreted because its prodomain cannot be processed (3, 4, 27). However, our application of the SynPro system shows that prodomain processing in the ER is not sufficient to induce the secretion of Vg1 (SI Appendix, Fig. S2). In fact, unprocessed Vg1 can be secreted upon dimerizing with Nodal (Fig. 2). These results are consistent with the observations that mouse Nodal and its proprotein convertases are not coexpressed in the same cell (24, 25, 62, 63). In this case, prodomain processing of Gdf1/3 (the orthologs of Vg1) and Nodal dimers occurs after they are released in the extracellular space or bound to the target cell surface and in early endosomes. Using the SynPro system in zebrafish embryos, we show that Vg1 can also be cleaved after its Nodal-induced release in the extracellular space (Fig. 1). These findings establish that prodomain processing of Vg1 can be location-independent and separable from Nodal-induced secretion.
Fourth, our results elucidate the roles of Nodal and Vg1 processing in signaling. We find that Vg1-Nodal heterodimers with noncleavable prodomains are secreted but cannot signal, whereas unprocessed Nodal in combination with processed Vg1 remains partially active. This result demonstrates that Nodal processing is not required for secretion or partial activity, consistent with previous studies that found that unprocessed mouse Nodal still promotes mesoderm formation but cannot position or maintain the primitive streak (64). Our finding that the Vg1 prodomain must be processed for signaling suggests the possibility that the activity of unprocessed mouse Nodal also relies on processed Vg1 (Gdf1/3).
Fifth, our study extends the previous suggestion that the Vg1-Nodal heterodimeric system allows for the rapid activation of the Nodal signaling pathway (10). Instead of a time delay caused by the accumulation of sufficient levels of Nodal needed to form homodimers, the preformed pool of Vg1 allows for rapid heterodimer formation as soon as Nodal is synthesized. Our study suggests that two features of Vg1 facilitate this strategy of rapid signaling: chaperone-mediated monomer retention in the ER stores Vg1 in an inactive but ready-to-heterodimerize form, and location-independent processing allows efficient generation of active heterodimers both intra- and extracellularly.
Last, the SynPro system introduced here has broad applications in the targeted processing and regulation of secreted proteins. Our results show that endogenous Vg1 can be replaced by a Vg1 derivative containing an engineered cleavage sequence in combination with the corresponding SynPro protease. The Mvg1 rescue experiments with the exogenous SynPro system exhibited minor morphological defects, which might be more fully recovered when the SynPro system is stably integrated into the genome. The SynPro system might be used for the orthogonal regulation of TGF-beta and other secreted signals. Endogenous proprotein convertases cleave the same polybasic motif in multiple secreted proteins. By contrast, each SynPro protease recognizes a unique cleavage sequence. Different SynPro proteases could be combined to separately control the activity of multiple secreted signals. For example, different TGF-beta signals could be processed independently to reveal distinct spatial and temporal requirements. Beyond TGF-beta proteins, the SynPro system could be used to independently process several bioactive peptides from hormone or neuropeptide precursors. By releasing individual peptides one at a time from a polyprotein, the effects of each peptide could be analyzed. Thus, the SynPro system has the potential to accelerate the functional assignment of bioactive peptides generated by secreted polyproteins.
Materials and Methods
Genotyping of vg1 Mutants.
Genomic DNA was isolated via the HotSHOT method from either excised adult caudal fin tissue or individual fixed embryos (65). Genotyping was carried out via PCR using standard conditions followed by 2% gel electrophoresis. Mutant vg1 fish have the vg1a165allele, which contains a 29-bp deletion in the first exon of vg1 and was detected as described (10). Allele designation was omitted for brevity in the rest of the text.
Zebrafish Husbandry and Fish Lines.
Fish were maintained per standard laboratory conditions (66). Embryos were raised at 28.5 °C in embryo medium (250 mg/L Instant Ocean salt and 1 mg/L methylene blue in reverse osmosis water adjusted to pH 7 with sodium bicarbonate) and staged according to a standard staging series (67). Wild-type fish and embryos represent the TLAB strain. The vg1 mutant fish line was maintained as previously described (10). Mvg1 embryos were generated by crossing zygotic homozygous vg1 female fish to TLAB wild-type male fish.
Cloning of Expression Constructs and the Synthetic Processing (SynPro) System.
Standard molecular cloning techniques, such as PCR, Gibson Assembly (68), and site-directed mutagenesis, were performed to assemble constructs used in this study.
The coding sequences (CDS) of vg1 (10), sqt, cyc (69), and their variants tagged with superfolder GFP (sfGFP) (70) were previously assembled into the pCS2(+) vector that contains a β-globin 5′ UTR and an SV40 late polyA signal at the 3′ UTR. For sfGFP-tagged Vg1 prodomain or mature domain, site-directed mutagenesis (Q5 Kit, New England Biolabs) of pCS2(+)-vg1-sfGFP was performed to truncate the full-length construct into respective domains (as depicted in Fig. 3). All point mutants, indels, and epitope tags were subsequently generated using site-directed mutagenesis. For noncleavable vg1-NC, vg1-NC-sfGFP, and proVg1-sfGFP, the cleavage site “RSRRKR” was replaced with “SQNTSN.” For noncleavable sqt-NC and sqt-NC-sfGFP, the cleavage site “RRHRR” was replaced with “SQNTS.” For chimeras of Squint/Cyclops prodomain and sfGFP-tagged Vg1 mature domain (sfGFP-matVg1), the Squint and Cyclops prodomains (including their respective cleavage sites) were PCR-amplified from pCS2(+)-squint and pCS2(+)-cyclops, and sfGFP-matVg1 was PCR-amplified from pCS2(+)-vg1-sfGFP.
Commercially available sequences for tobacco etch virus protease (TEVp, Addgene Plasmid #8835), tobacco vein mottling virus protease (TVMVp; Addgene Plasmid #8832), and human rhinovirus 3C protease (HRV 3Cp; Addgene Plasmid #78571) were gifts from David Waugh (National Cancer Institute, Frederick, MD) (71–73). Commercially available sequence for bovine enterokinase (Addgene Plasmid #49048) was a gift from Hans Brandstetter (University of Salzburg, Austria) (74). These sequences were PCR-amplified and subcloned into the pCS2(+) vector for downstream applications in vivo.
Zebrafish codon-optimized sequences of 15 Potyviral proteases listed in SI Appendix, Fig. S1 were synthesized by Integrated DNA Technologies and subcloned into the pCS2(+) vector. For the protease cleavage reporter assay described in SI Appendix, Fig. S1, Component 1 is a cleavable substrate for Potyviral proteases and was constructed via Gibson assembly of zebrafish beta-Arrestin (arrb2a gene PCR-amplified from high-stage cDNA library) and mNeonGreen2(1–10) (a gift from Bo Huang; University of California, San Francisco, CA) (38) into the pCS2(+) vector. The cognate cleavage site inserted between arrb2a and mNeonGreen2(1–10) was encoded in the primer overhangs. Component 2 is the codon-optimized Potyviral protease described above. Component 3 was constructed via Gibson assembly of four tandem copies of mNeonGreen2(11) (a gift from Bo Huang) (38) and mCherry-tagged Histone 2B (a gift from Jeffrey Farrell; National Institute of Child Health and Human Development, Bethesda, MD) into the pCS2(+) vector.
For the SynPro system, the codon-optimized TEV protease was further modified using site-directed mutagenesis to generate secTEVp. The secTEVp construct contains a secretion signal sequence derived from zebrafish Toddler (translated signal peptide: MRFFHPLYLLLLLLTVLVLISA) and 5 point mutations (N23Q, C130S, T173G, L56V, and S135G). The secTEVp-sfCherry-KDEL construct additionally contains a C-terminal fusion of sfCherry3C (38) sequence and an ER-targeting motif “(GSGS)EEKDEL.” SynPro Vg1 was derived from pCS2(+)-vg1-sfGFP using site-directed mutagenesis to replace the “RSRRKR” cleavage site with “(GSGS)ENLYFQS(GS).” The ER marker sfCherry-KDEL was generated using site-directed mutagenesis of the sfCherry3C sequence to insert the secretion signal sequence from Toddler at the N terminus and the C-terminal sequence “EEKDEL.” To fluorescently mark the cytoplasm and nucleus, sfCherry-Smad2 was cloned via Gibson assembly, where sfCherry3C is N-terminal to zebrafish Smad2 via a “GSGSGS” linker. To verify whether the zebrafish proteome contains sequences identical to the cleavage motif of secTEVp, a BLASTP alignment search showed that a secreted protein RgmA contains the best sequence match of “EDLYFQS,” which is predicted by Alphafold2 to be not surface-accessible (https://alphafold.ebi.ac.uk/entry/Q1LVM8) and should also not be recognized by secTEVp.
Determination of BiP-Binding Scores.
To focus our mutagenesis efforts, we ignored corresponding matVg1 and matCyc heptapeptides that both scored positive in the Gething-Sambrook scoring system because BiP is predicted to bind to both. We also ignored corresponding regions where both heptapeptides scored zero or negative because BiP is predicted to not bind at all. We focused on seven regions wherein matVg1 heptapeptides scored positive and the corresponding matCyc sequences scored negative (Fig. 4B, arrows). We ignored four out of the seven regions because they included six cysteines that participate in cystine-knot formation (Fig. 4B, red arrows). BiP-binding scores were visualized using the D3.js chart library (https://d3js.org/), and the code used is available on GitHub (https://github.com/davedingal/BiP_binding_score).
mRNA Synthesis and Microinjection into Embryos.
All pCS2(+) plasmids were linearized with NotI and subsequently purified with the E.Z.N.A. Cycle Pure Kit (Omega). Capped mRNAs were synthesized with the Sp6 mMessage mMachine kit (Invitrogen) using the purified linearized plasmids as templates. Capped mRNAs were then purified with the E.Z.N.A. Total RNA Kit I (Omega). Capped mRNA concentrations were measured using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). All kits were used according to the respective manufacturer’s protocols. If not mentioned otherwise, all mRNAs were injected at 50 pg into embryos at the one-cell stage using standard methods (66).
Transplantation.
For transplantation experiments, donor and host Mvg1 embryos at one-cell stage were injected with 50 pg of each mRNA relevant to the experiment and then grown to 1,000-cell stage (3 hpf). At 1,000-cell stage, cells were transplanted from donor embryos to host embryos and were grown to shield stage (6 hpf) before fixation for immunostaining.
Live Imaging and Immunofluorescence Imaging.
Embryos were injected with 50 pg of each mRNA, grown to sphere stage, and then embedded in 1% low melting temperature agarose (Aquapor) on glass-bottomed dishes (MatTek). Imaging was performed on a Zeiss LSM 880 inverted confocal microscope.
In Situ Hybridization.
Embryos were grown to 50% epiboly and then fixed in 4% formaldehyde overnight at 4 °C. Whole-mount in situ hybridizations were performed according to standard protocols (75). A DIG-labeled antisense RNA probe targeting lefty1 was synthesized using a DIG Probe Synthesis Kit (Roche). NBT/BCIP/Alkaline phosphatase-stained embryos were dehydrated in methanol before clearing and imaging in 2:1 benzyl benzoate:benzyl alcohol (BBBA) using a Zeiss Axio Imager.Z1 microscope.
Immunoblotting.
Embryos were injected at the 1-cell stage with 50 pg of mRNA and then allowed to develop to 50% epiboly. Eight embryos per sample were manually deyolked with forceps and frozen in liquid nitrogen. Samples were boiled for 5 min at 95 °C with 2× SDS loading buffer and DTT (150 mM final concentration) and then loaded onto Any kD protein gels (Bio-Rad). Samples were transferred to PVDF membranes (GE Healthcare), blocked in 5% nonfat milk (Bio-Rad) in TBST, and incubated in primary antibodies (1:5,000 rabbit anti-GFP, ThermoFisher A11122, RRID:AB_221569) at 4 °C overnight. Proteins were detected using HRP-coupled secondary antibody (1:15,000 goat anti-rabbit, Jackson ImmunoResearch Labs 111-035-144, RRID:AB_2307391). Chemiluminescence was detected using Amersham ECL reagent (GE Healthcare).
Immunostaining.
A previous protocol (76) was modified to improve signal-to-noise ratio. Embryos were fixed in 4% paraformaldehyde overnight at 4 °C in PBSTw (1× phosphate-buffered saline + 0.1% v/v Tween 20), washed three times in PBSTw for 10 min each, dehydrated in a MeOH/PBSTw mixture series (25%, 50%, 75%, and 100% methanol) at 5 min per wash at room temperature (RT), and stored in 100% MeOH at –20 °C for at least 2 h. Embryos were rehydrated in a MeOH/PBSTr (1× PBS + 1% Triton X-100) mixture series (75%, 50%, 25% MeOH) for 5 min each, washed three times in PBSTr for 10 min each at RT, and manually deyolked. Embryos were then incubated in antibody binding buffer (PBSTr + 1% v/v dimethyl sulfoxide) for 2 h at RT and subsequently stored overnight at 4 °C in antibody binding buffer containing relevant primary antibodies. After primary antibody incubation, embryos were washed six times with PBSTr for 10 min each, before a 30-min incubation in antibody binding buffer at RT. Embryos were then incubated for 2 h at RT in antibody binding buffer containing appropriate fluorescent secondary antibodies. Embryos were then washed six times with PBSTr. To label DNA in cell nuclei, embryos were incubated with 1 μg/mL DAPI in PBSTw for 30 min at RT. Last, embryos were washed three times in PBSTw for 10 min each at RT and then mounted for microscopy.
Primary antibodies were used against GFP (1:1,000 chicken IgY, Aves Lab, RRID:AB_2307313) and phosphorylated Smad2 (1:1,000 rabbit IgG, Cell Signaling, RRID:AB_2798798). Fluorescent secondary antibodies used were goat anti-chicken Alexa 488 conjugate (1:2,000, Thermo Fisher, RRID:AB_2534096) and goat anti-rabbit Alexa 647 conjugate (1:2,000, Thermo Fisher, RRID:AB_2633282).
Image Processing.
All images were processed in Fiji/ImageJ (77). Brightness, contrast, and color balance were uniformly applied to images.
Ethics.
Animal experimentation.
All vertebrate animal works were performed at the facilities of Harvard University, Faculty of Arts & Science (HU/FAS). The HU/FAS animal care and use program maintains full AAALAC accreditation, is assured with OLAW (A3593-01), and is currently registered with the USDA. This study was approved by the HU/FAS Standing Committee on the Use of Animals in Research & Teaching under Protocol No. 25-08.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Richard Losick, Nathan Lord, Maxwell Shafer, Madalena Madeira Reimão Pinto, Jan Christian, and two reviewers for helpful comments on the manuscript, Yiqun Wang for the figure schematics, and Stephen D. Hansen for programming and data visualization. We thank the members of the Schier lab for advice, expertise, insights, and discussions. We also thank the Harvard Center for Biological Imaging and Jacopo Ferruzzi for microscopy infrastructure and support. This project was supported by NIH DP1-HD094764, the Allen Discovery Center for Cell Lineage Tracing, and a UT Dallas Startup Fund to P.C.D.P.D.
Author contributions
P.C.D.P.D., A.N.C., T.G.M., and A.F.S. designed research; P.C.D.P.D., A.N.C., T.G.M., and M.B.L.S. performed research; P.C.D.P.D. and A.F.S. analyzed data; and P.C.D.P.D. and A.F.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: R.D.B., Princeton University; and S.D., University of Georgia.
Contributor Information
P. C. Dave P. Dingal, Email: davedingal@utdallas.edu.
Alexander F. Schier, Email: alex.schier@unibas.ch.
Data, Materials, and Software Availability
Code data have been deposited in Github (https://zenodo.org/badge/latestdoi/317948804) (78).
Supporting Information
References
- 1.Schier A. F., Nodal morphogens. Cold Spring Harb. Perspect. Biol. 1, a003459 (2009), 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers K. W., Muller P., Nodal and BMP dispersal during early zebrafish development. Dev. Biol. 447, 14–23 (2019), 10.1016/j.ydbio.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Dale L., Matthews G., Colman A., Secretion and mesoderm-inducing activity of the TGF-beta-related domain of Xenopus Vg1. EMBO J. 12, 4471–4480 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomsen G. H., Melton D. A., Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell 74, 433–441 (1993), 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X., Sasaki H., Lowe L., Hogan B. L., Kuehn M. R., Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature 361, 543–547 (1993), 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- 6.Conlon F. L., et al. , A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120, 1919–1928 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Feldman B., et al. , Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 395, 181–185 (1998), 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 8.Birsoy B., Kofron M., Schaible K., Wylie C., Heasman J., Vg 1 is an essential signaling molecule in Xenopus development. Development 133, 15–20 (2006), 10.1242/dev.02144. [DOI] [PubMed] [Google Scholar]
- 9.Bisgrove B. W., Su Y. C., Yost H. J., Maternal Gdf3 is an obligatory cofactor in Nodal signaling for embryonic axis formation in zebrafish. Elife 6, e28534 (2017), 10.7554/eLife.28534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montague T. G., Schier A. F., Vg1-Nodal heterodimers are the endogenous inducers of mesendoderm. Elife 6, e28183 (2017), 10.7554/eLife.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelliccia J. L., Jindal G. A., Burdine R. D., Gdf3 is required for robust Nodal signaling during germ layer formation and left-right patterning. Elife 6, e28635 (2017), 10.7554/eLife.28635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin M., Johnson R. L., Stern C. D., Kuehn M., Tabin C., A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 82, 803–814 (1995), 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- 13.Collignon J., Varlet I., Robertson E. J., Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381, 155–158 (1996), 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 14.Lowe L. A., et al. , Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381, 158–161 (1996), 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- 15.Pagan-Westphal S. M., Tabin C. J., The transfer of left-right positional information during chick embryogenesis. Cell. 93, 25–35 (1998), 10.1016/s0092-8674(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 16.Long S., Ahmad N., Rebagliati M., The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130, 2303–2316 (2003), 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 17.Montague T. G., Gagnon J. A., Schier A. F., Conserved regulation of Nodal-mediated left-right patterning in zebrafish and mouse. Development 145, dev171090 (2018), 10.1242/dev.171090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gritsman K., et al. , The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97, 121–132 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Yeo C., Whitman M., Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell. 7, 949–957 (2001), 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S. K., Olale F., Bennett J. T., Brivanlou A. H., Schier A. F., EGF-CFC proteins are essential coreceptors for the TGF-beta signals Vg1 and GDF1. Genes. Dev. 17, 31–36 (2003), 10.1101/gad.1041203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker J. C., Harland R. M., A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes. Dev. 10, 1880–1889 (1996), 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 22.Hinck A. P., Mueller T. D., Springer T. A., Structural biology and evolution of the TGF-beta family. Cold Spring Harb. Perspect. Biol. 8, a022103 (2016), 10.1101/cshperspect.a022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazono K., Olofsson A., Colosetti P., Heldin C. H., A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 10, 1091–1101 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck S., et al. , Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 4, 981–985 (2002), 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- 25.Blanchet M. H., et al. , Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J. 27, 2580–2591 (2008), 10.1038/emboj.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidah N. G., Prat A., The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug. Discov. 11, 367–383 (2012), 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 27.Dohrmann C. E., Kessler D. S., Melton D. A., Induction of axial mesoderm by zDVR-1, the zebrafish orthologue of Xenopus Vg1. Dev. Biol. 175, 108–117 (1996), 10.1006/dbio.1996.0099. [DOI] [PubMed] [Google Scholar]
- 28.Reddy P. S., Corley R. B., Assembly, sorting, and exit of oligomeric proteins from the endoplasmic reticulum. Bioessays 20, 546–554 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson B., Gilbert H. F., Protein disulfide isomerase. Biochim. Biophys. Acta 1699, 35–44 (2004), 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 30.Helenius A., Aebi M., Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004), 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 31.Hammond C., Braakman I., Helenius A., Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. U.S.A. 91, 913–917 (1994), 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn G. C., Chappell T. G., Rothman J. E., Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 (1989), 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 33.Flynn G. C., Pohl J., Flocco M. T., Rothman J. E., Peptide-binding specificity of the molecular chaperone BiP. Nature 353, 726–730 (1991), 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 34.Blond-Elguindi S., et al. , Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 (1993), 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 35.Tannahill D., Melton D. A., Localized synthesis of the Vg1 protein during early Xenopus development. Development 106, 775–785 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Waugh D. S., An overview of enzymatic reagents for the removal of affinity tags. Protein Expr. Purif. 80, 283–293 (2011), 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wylie S. J., et al. , ICTV virus taxonomy profile: Potyviridae. J. Gen. Virol. 98, 352–354 (2017), 10.1099/jgv.0.000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng S., et al. , Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8, 370 (2017), 10.1038/s41467-017-00494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabrita L. D., et al. , Enhancing the stability and solubility of TEV protease using in silico design. Protein Sci. 16, 2360–2367 (2007), 10.1110/ps.072822507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cesaratto F., Lopez-Requena A., Burrone O. R., Petris G., Engineered tobacco etch virus (TEV) protease active in the secretory pathway of mammalian cells. J. Biotechnol. 212, 159–166 (2015), 10.1016/j.jbiotec.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 41.Dale L., Matthews G., Tabe L., Colman A., Developmental expression of the protein product of Vg1, a localized maternal mRNA in the frog Xenopus laevis. EMBO J. 8, 1057–1065 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C., et al. , The Vg1-related protein Gdf3 acts in a Nodal signaling pathway in the pre-gastrulation mouse embryo. Development 133, 319–329 (2006), 10.1242/dev.02210. [DOI] [PubMed] [Google Scholar]
- 43.Munro S., Pelham H. R., A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 (1987), 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 44.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021), 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas I. G., Wabl M., Immunoglobulin heavy chain binding protein. Nature 306, 387–389 (1983), 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 46.Bole D. G., Hendershot L. M., Kearney J. F., Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 102, 1558–1566 (1986), 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu L., Sun T. T., Kreibich G., Specific heterodimer formation is a prerequisite for uroplakins to exit from the endoplasmic reticulum. Mol. Biol. Cell 13, 4221–4230 (2002), 10.1091/mbc.e02-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu C. C., et al. , Assembly of a membrane receptor complex: Roles of the uroplakin II prosequence in regulating uroplakin bacterial receptor oligomerization. Biochem. J. 414, 195–203 (2008), 10.1042/BJ20080550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sitia R., et al. , Developmental regulation of IgM secretion: The role of the carboxy-terminal cysteine. Cell 60, 781–790 (1990), 10.1016/0092-8674(90)90092-s. [DOI] [PubMed] [Google Scholar]
- 50.Alberini C. M., Bet P., Milstein C., Sitia R., Secretion of immunoglobulin M assembly intermediates in the presence of reducing agents. Nature 347, 485–487 (1990), 10.1038/347485a0. [DOI] [PubMed] [Google Scholar]
- 51.Melnick J., Dul J. L., Argon Y., Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature 370, 373–375 (1994), 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 52.Mayer M., Kies U., Kammermeier R., Buchner J., BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J. Biol. Chem. 275, 29421–29425 (2000), 10.1074/jbc.M002655200. [DOI] [PubMed] [Google Scholar]
- 53.Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M., A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 13, 4456–4469 (2002), 10.1091/mbc.e02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y., Hendershot L. M., ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol. Biol. Cell. 16, 40–50 (2005), 10.1091/mbc.e04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feige M. J., et al. , An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell. 34, 569–579 (2009), 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcinowski M., et al. , Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 (2011), 10.1038/nsmb.1970. [DOI] [PubMed] [Google Scholar]
- 57.Haas I. G., BiP (GRP78), an essential hsp70 resident protein in the endoplasmic reticulum. Experientia 50, 1012–1020 (1994), 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- 58.Hendershot L., et al. , Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc. Natl. Acad. Sci. U.S.A. 93, 5269–5274 (1996), 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanhove M., Usherwood Y. K., Hendershot L. M., Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity 15, 105–114 (2001), 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- 60.Hendershot L., Bole D., Köhler G., Kearney J. F., Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J. Cell Biol. 104, 761–767 (1987), 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shusta E. V., Raines R. T., Pluckthun A., Wittrup K. D., Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16, 773–777 (1998), 10.1038/nbt0898-773. [DOI] [PubMed] [Google Scholar]
- 62.Blanchet M. H., et al. , Cripto localizes Nodal at the limiting membrane of early endosomes. Sci. Signal. 1, ra13 (2008), 10.1126/scisignal.1165027. [DOI] [PubMed] [Google Scholar]
- 63.Tessadori F., et al. , Nodal signaling range is regulated by proprotein convertase-mediated maturation. Dev. Cell. 32, 631–639 (2015), 10.1016/j.devcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 64.Ben-Haim N., et al. , The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell 11, 313–323 (2006), 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Meeker N. D., Hutchinson S. A., Ho L., Trede N. S., Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 43, 610, 612, 614 (2007), 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 66.Westerfield M., The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) (University of Oregon Press, Eugene, ed. 4, 2000). [Google Scholar]
- 67.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev Dyn. 203, 253–310 (1995), 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 68.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6, 343–345 (2009), 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 69.Muller P., et al. , Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science 336, 721–724 (2012), 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pedelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006), 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 71.Kapust R. B., et al. , Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001), 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 72.Nallamsetty S., et al. , Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 38, 108–115 (2004), 10.1016/j.pep.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 73.Raran-Kurussi S., Waugh D. S., A dual protease approach for expression and affinity purification of recombinant proteins. Anal. Biochem. 504, 30–37 (2016), 10.1016/j.ab.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skala W., Goettig P., Brandstetter H., Do-it-yourself histidine-tagged bovine enterokinase: A handy member of the protein engineer’s toolbox. J. Biotechnol. 168, 421–425 (2013), 10.1016/j.jbiotec.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008), 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 76.Rogers K. W., et al. , Nodal patterning without Lefty inhibitory feedback is functional but fragile. Elife 6, e28785 (2017), 10.7554/eLife.28785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012), 10.1038/nmeth.2019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dave P. C., Dingal P., davedingal/BiP_binding_score: BiP binding score v1.0 (2023). Zenodo. 10.5281/zenodo.7879730. Deposited 30 April 2023. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Code data have been deposited in Github (https://zenodo.org/badge/latestdoi/317948804) (78).