Abstract
The relationship between the detection of mRNA and cellular viability in Escherichia coli was investigated in cells killed by heat or ethanol. Reverse transcription-PCR (RT-PCR) methods were developed for detecting mRNA from rpoH, groEL, and tufA genes. mRNA from all three genes was detected immediately after the cells had been killed by heat or ethanol but gradually disappeared with time when dead cells were held at room temperature. In heat-killed cells, some mRNA targets became undetectable after 2 to 16 h, whereas after ethanol treatment, mRNA was still detected after 16 h. In contrast, 16S rRNA was detected by RT-PCR in all samples containing dead cells and did not disappear during a subsequent incubation of 16 h at room temperature. Of the different types of nucleic acid, mRNA is the most promising candidate for an indicator of viability in bacteria, but its persistence in dead cells depends on the inactivating treatment and subsequent holding conditions.
Conventional methods for detecting bacterial pathogens typically involve culturing the organisms in selective media and identifying isolates according to their morphological, biochemical, and/or immunological characteristics. These methods are sensitive and can allow small numbers of organisms (<103 cells/ml) to be detected in complex environments such as foods and certain clinical samples. However, the methods also require days from initiation to readout, and identification schemes based on phenotypic properties are notoriously difficult to interpret.
Gene probe methods of detection and identification are inherently more specific and can also be extremely sensitive when coupled, for example, with PCR (26). One disadvantage of DNA-based methods is that they do not distinguish between living and dead organisms (22), which limits their use for monitoring purposes. Josephson et al. (18) performed PCR amplification of DNA from cells killed by boiling or exposure to UV light, while Masters et al. (25) showed that there was no relationship between viability and PCR detection of DNA targets in Listeria monocytogenes or Escherichia coli that had been exposed to heat, acid, hydrogen peroxide, drying, or starvation. In the latter study, even autoclaved cells provided a positive PCR signal.
These disadvantages of DNA-based methods may be partly overcome by including a preenrichment step that allows organisms to multiply before gene probe tests are applied. This procedure increases sensitivity and helps to distinguish between samples that contain living cells and those that do not; however, it also increases the time taken for analysis. There is also a possibility that samples which initially contained large numbers of dead cells would still give positive results with sensitive gene probe methods even though the number of target organisms had not increased during preenrichment.
Nucleic acid-based methods that could be applied directly to samples to give an indication of the viability of any microbes present would be of enormous significance for food, industrial, environmental, and medical applications. mRNA is turned over rapidly in living bacterial cells, with most mRNA species having a half-life of only a few minutes (2, 7). Detection of mRNA might therefore be a good indicator of living cells or those only recently dead at the time of sampling. Detection of mRNA by Northern blot hybridization has been used as an indicator of microbial metabolic activity in aquatic and soil samples (17, 28, 35), and detection of mRNA by reverse transcription-PCR (RT-PCR) was used to monitor gene expression in activated sludge (33). While these studies undoubtedly reflected the activities of viable cells in natural environments, their main purpose was not to distinguish between living and dead cells. Few studies have specifically investigated the relationship between detection of microbial mRNA and viability. Bej et al. (5, 6) used RT-PCR to examine Legionella pneumophila and Vibrio cholerae exposed to heat or starvation, respectively, and detected specific mRNA only in samples that contained viable cells detected by culturing. Similarly, Patel et al. (27) successfully assessed the viability of heat-killed Mycobacterium leprae, detecting a heat shock protein mRNA in living cells. Recently, Klein and Juneja (20) described a method for the specific detection of viable L. monocytogenes cells based on RT-PCR. Despite their potential advantages, mRNA-based approaches have proved difficult to exploit because of the complexity of the methods, the practical problems of extracting detectable levels of intact mRNA from small numbers of bacteria, and a lack of basic information about the significance of detecting mRNA in stressed cells. The aim of this work was to develop methods for detecting specific mRNA from E. coli and to examine the relationship between viability and the presence of mRNA. Since any relationship between mRNA and viability may depend on the method used to inactivate cells or the type of mRNA sought, we exposed the cells to two different stress treatments (heat and ethanol) and assayed mRNA from three different genes (rpoH, groEL, and tufA). These genes were chosen as representing a gene encoding an abundant cellular housekeeping protein (tufA) (8, 36) and genes associated with a stress response regulon (rpoH and groEL) (3, 12, 14).
MATERIALS AND METHODS
Bacterial strain and growth conditions.
E. coli (type strain NCFB 1984) was grown in Luria-Bertani (LB) broth to exponential phase (optical density at 680 nm, 0.2; approximately 2 × 108 cells/ml).
Viable counts.
To obtain quantitative estimates of viable numbers in suspensions subjected to heat or ethanol treatment and in experiments to determine the limits of detection by RT-PCR, samples were serially diluted in maximum-recovery diluent (Oxoid, Basingstoke, United Kingdom) and plated onto LB agar. Colonies were counted after incubation for 2 days at 37°C.
To monitor for the presence of viable cells in ethanol- or heat-killed suspensions used for RT-PCR, 0.1-ml samples were removed at the same time that samples were removed for RNA extraction and diluted 1:100 in LB broth. The broth was incubated aerobically at 37°C for 2 days and then examined for visible turbidity. Any survival, recovery, or multiplication of cells exposed to an inactivation treatment and then held at room temperature would thus have been detected. A broth was used as recovery medium to provide the maximum opportunity for resuscitation of injured cells (23).
Inactivation treatments.
Volumes (1 ml) of exponential-phase culture (ca. 107 cells) were treated in five different ways to kill the bacteria. They were killed by boiling at 100°C for 5 min, heating at 80°C for 10 min or at 60°C for 20 min, or exposure to 50 or 67% ethanol for 7 min. The bacteria treated with ethanol were pelleted by being centrifuged in a bench centrifuge at 10,000 × g for 5 min, washed twice, and resuspended in 100 μl of LB broth. All the treated bacteria were left in their broth at room temperature for 0, 15, or 30 min or 1, 2, 3, or 16 h after treatment before RNA isolation.
RNA isolation.
The chaotropic solution used for isolating total E. coli RNA consisted of 2% (vol/vol) DivoLab cationic detergent (Diversey Ltd., Northampton, United Kingdom), 50 mM sodium acetate, and 1% (vol/vol) β-mercaptoethanol (pH 4.5) and was prepared immediately prior to use. Chaotropic solution (500 μl) was added to the bacterial suspension (100 μl), and the mixture was transferred to a blue-capped Ribolyser tube (Hybaid Ltd., Teddington, United Kingdom) containing silica beads and 600 μl of phenol-chloroform (5:1, pH 4.7). The tubes were shaken in the Ribolyser at speed setting 6 for 20 s and cooled on ice before being centrifuged at 10,000 × g for 10 min. The aqueous layer was removed and washed twice with chloroform-isoamyl alcohol (24:1) by vortexing for 30 s and centrifuging at 10,000 × g for 10 min between each wash. RNA was precipitated with 2 equal volumes of isopropanol containing 0.01 M polyinositol and left at −20°C for at least 10 min before the nucleic acids were pelleted at 10,000 × g for 30 min. The nucleic acid pellet was washed twice with 75% (vol/vol) ice-cold ethanol, air dried, resuspended in 40 μl of sterile double-distilled diethylpyrocarbonate-treated water (Sigma, Poole, United Kingdom), and stored at −20°C. An aliquot (5 μl) of RNA was examined by gel electrophoresis on a 1.2% (wt/vol) agarose gel running in 1× TAE (0.04 M Tris acetate, 0.001 M EDTA) buffer and then stained with ethidium bromide (25 μg/ml) and examined with a UV transilluminator at 254 nm.
Elimination of contaminating DNA.
To remove any contaminating DNA, 10 μl (ca. 1 μg) of RNA was incubated at 37°C for 30 min with 30 U of RNase-free DNase (either Boehringer Mannheim, Lewes, United Kingdom, or Life Technologies Ltd., Paisley, United Kingdom) and 1 U of RNasin RNase inhibitor (Promega, Southampton, United Kingdom) in a 50-μl volume. Residual DNase was either heat inactivated at 80°C for 5 min (Life Technologies) or removed by extraction with phenol-chloroform (5:1; pH 4.7) (Boehringer Mannheim), and RNA isolation continued according to the protocol. A PCR was performed to check for any contaminating DNA by using rTth DNA polymerase with 1× EZ buffer (Perkin-Elmer, Warrington, United Kingdom) and 16S rDNA primers. DNase-treated RNA (ca. 100 ng) was amplified using with 10 μM forward primer, and 10 μM reverse primer, 5 U of rTth DNA polymerase, 1× EZ buffer (50 mM bicine, 115 mM potassium acetate, 8% [wt/vol] glycerol [pH 8.2]), 300 μM each deoxynucleoside triphosphate, and 2.5 mM manganese acetate in a reaction volume of 50 μl. The reaction mixtures were overlaid with mineral oil before being subjected to amplification for 35 cycles of 95°C for 30 s, 60°C for 45 s, and 72°C for 1 min with a final cycle of 72°C for 10 min. The PCR products (15 μl) were run on a 1.2% (wt/vol) agarose gel, stained with ethidium bromide (25 μg/ml), and photographed with 35-mm film.
Southern blotting.
To confirm the identity of PCR products, gels were Southern blotted onto Hybond N+ (Amersham International, Amersham, United Kingdom) nylon (32). PCR products from 16S rDNA, tufA, or rpoH genes were randomly fluorescein labelled as specified by the manufacturer (Tropix Inc., Bedford, Mass.) and hybridized to the blot at 65°C for 16 h in hybridization buffer (1 mM EDTA, 7% sodium dodecyl sulfate [SDS], 0.25M disodium phosphate [pH 7.2]). The blots were washed twice for 5 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 25°C, twice for 15 min with 1× SSC–0.1% SDS at 65°C, and twice for 5 min with 0.1× SSC–0.1% SDS at 25°C. The hybridized probe was detected with the Southern-Star (Tropix) nucleic acid detection system as specified by the manufacturer and exposed to BioMax X-ray film (Sigma) before being developed.
One-step RT-PCR with rTth DNA polymerase.
DNase-treated RNA samples containing any contaminating DNA (as identified by PCR) were not analyzed further. The presence of three target mRNAs, groEL, tufA, and rpoH, was analyzed by a one-step RT-PCR method in which rTth DNA polymerase (Perkin-Elmer) is used to synthesize cDNA from RNA and also to amplify the product in the subsequent PCR. DNase-treated RNA (ca. 100 ng) was amplified with 5 U of rTth DNA polymerase, 1× EZ buffer 10 μM each forward and reverse primers, 300 μM each deoxynucleoside triphosphate, and 2.5 mM manganese acetate made up to 50 μl with sterile double-distilled water. The RT-PCR samples were overlaid with 50 μl of mineral oil and amplified in a two-step thermal program: cDNA was synthesized at 60°C for 30 min and then amplified in a two-step PCR for 40 cycles (denaturing at 95°C for 45 s and annealing at 50°C for 45 s) with a final extension at 60°C for 10 min. The RT-PCR samples (15 μl) were electrophoresed on a 1% (wt/vol) agarose gel in 1× TAE buffer. As a positive control for each sample, RT-PCR was performed on the same extract with primers specific for 16S rRNA. This target was chosen as a control because it is very stable and is present in much greater quantities than is mRNA.
Two-step RT-PCR with rTth DNA polymerase.
The one-step RT-PCR system with EZ buffer was designed for use as a screening or detection tool and allows a certain degree of enzymatic transcriptional and synthetic infidelity. A change in reaction buffering conditions in a two-step RT-PCR with rTth DNA polymerase ensures high enzymatic fidelity; therefore, to compare the limits of mRNA detection, both one- and two-step RT-PCR methods were used. In the first step, DNase-treated RNA (ca. 100 ng) was subjected to RT with 2.5 U of rTth DNA polymerase–1× RT buffer (10 mM Tris-HCl, 90 mM KCl [pH 8.3]), 300 μM each dNTP–10 mM MnCl2–10 μM reverse primer in a total volume of 20 μl (made up to volume with sterile double-distilled water) at 60°C for 30 min. To this mixture, 80 μl of PCR components was added and a PCR was carried out with 40 cycles of 95°C for 45 s, 50°C for 1 min, and 72°C for 1 min, finishing with an extension at 60°C for 10 min. The PCR buffer contained 2 mM MgCl2, 10 mM forward primer, 1× chelating buffer (10 mM Tris-HCl, 0.1 M KCl, 0.05% [wt/vol] Tween 20, 0.75 mM EGTA [pH 8.3]), and sterile double-distilled water. RT-PCR products (15 μl) were electrophoresed on a 1% (wt/vol) agarose gel as described above.
Two-step RT-PCR with Superscript II and Taq DNA polymerase.
DNase-treated RNA was subjected to RT into cDNA with Moloney murine leukemia virus reverse transcriptase (Superscript RNase H− reverse transcriptase [Life Technologies Ltd.]), as specified by the manufacturer, in the presence of 1 U of RNasin. cDNA (3 μl) was then PCR amplified with Taq DNA polymerase (Applied Biosystems) under the thermal conditions and by the procedure detailed for the one-step RT-PCR system.
Oligonucleotide primer sequence design.
Oligonucleotide primers (Table 1) were designed by using the PRIMERSELECT program from a genetic analysis computer package (DNASTAR Inc., Madison, Wis.), from DNA sequences submitted to the EMBL/GenBank databases. The primers amplify between 40 and 50% of the transcribed gene.
TABLE 1.
Oligonucleotide primer sequences used for RT-PCR analysis
| Accession no.a | Target gene | Primer | Sequence (5′-3′) | Size (positions) of productb |
|---|---|---|---|---|
| J01695 | 16S rRNA | ICM16SF | CAGCGGGGAGGAAGGGAGTAAAGT | 405 (732–1137) |
| ICM16SR | ACCACCGCCCGTCACACCATG | |||
| X07850 | groEL | IMGROEF | CCGTGGCTACCTGTCTCCTTACTT | 653 (1058–1711) |
| IMGROER | CCAGCAACCACGCCTTCTTCTACC | |||
| M20668 | rpoH | IMRPOHF | CCACAGGCGGATTTGATTC | 450 (351–801) |
| IMRPOHR | GGTTTGCCGCCTGCTCTTC | |||
| J01690 | tufA | IMTUFAF | ACTTCCCGGGCGACGACACTC | 578 (624–1202) |
| IMTUFAR | CGCCCGGCATTACCATCTCTAC |
EMBL/GenBank accession numbers.
Sizes are in base pairs; positions are nucleotides.
RESULTS
Development of the RT-PCR protocol.
Several methods of extracting RNA, involving hot- and cold-phenol extractions and the use of different chaotropic agents and lytic enzymes, were tested (1, 11, 13, 21, 30). The most reproducible results were obtained when cells were rapidly disrupted with silicon beads in a chaotropic buffer and phenol-chloroform mix with the Hybaid Ribolyser system.
The choice of detergent used as a chaotrophic agent was specific to the type of bacteria being investigated (24). From a comparison of different concentrations of a range of detergents (e.g., Tween 80, SDS, Teepol), 2% Divo Lab detergent was found to be optimal for E. coli cells. Moreover, the time and speed at which the Ribolyser tubes were shaken were critical, since low speeds and short shaking times favoured the isolation of DNA from E. coli while high speeds and long times favored the isolation of RNA. The quality of total cellular RNA extracted was monitored by gel electrophoresis, but, surprisingly, the efficiency of extraction of total RNA was not a reliable guide to the success of subsequent RT-PCR of mRNA.
We observed no significant difference between the two different reverse transcriptase enzymes and three different protocols used for detecting mRNA from initial cell concentrations of 107 E. coli cells/ml. However, the method involving rTth DNA polymerase with EZ buffer was the most suitable for our purposes because it was the quickest method and required the fewest manipulations. In the one-tube method, all the reverse-transcribed RNA (cDNA) is available for amplification, whereas in the two-tube method, only a subsample is amplified. By using this RT-PCR regimen, it was possible to detect rpoH mRNA from suspensions containing 103 cells/ml, and Southern blot analysis confirmed the amplification fidelity of this enzyme (data not shown).
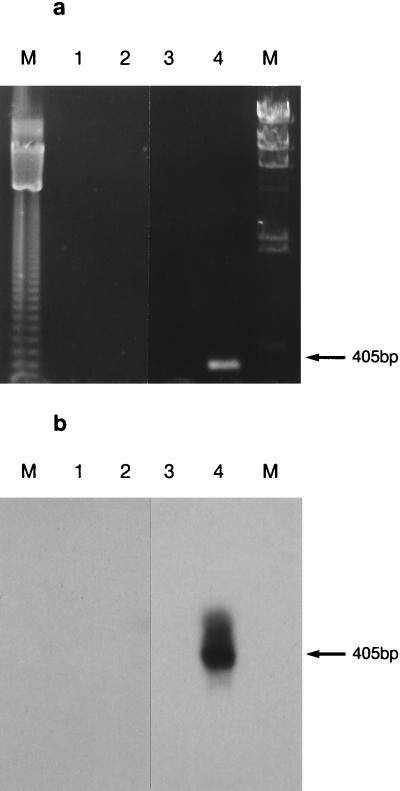
The method is not valid without an assurance of the absence of DNA. This is done quite simply by performing a control PCR on the DNase-treated RNA sample, which should give negative results. This was done for all our RNA samples. 16S rDNA was the target chosen for detecting contaminating DNA, and samples were analyzed by using rTth DNA polymerase with EZ buffer (Fig. 1a), since this was the enzyme system used to analyze mRNA in killed cells. Blotting and probing PCR agarose gels with a 16S rDNA fragment (Fig. 1b) confirmed that no contaminating DNA was present in DNase-treated RNA samples, eliminating the chance of a false-positive result in the subsequent RT-PCR analysis.
FIG. 1.
(a) PCR test to demonstrate the absence of residual contaminating 16S rDNA in DNase-treated mRNA samples. E. coli cells (107 cells/ml) were heat inactivated at 100°C for 5 min and then left at room temperature for 16 h (lane 2) after the killing treatment. Lane 1 contains an unheated control sample, lane 3 is a negative control containing sterile water in place of nucleic acid, and lane 4 is a positive control containing E. coli DNA. Lanes M contain molecular size standards. (b) Southern blot analysis of PCR products from amplification of residual contaminating 16S rDNA in DNase-treated mRNA samples. The gel in panel a was blotted and probed with a fluorescein-labelled 16S rDNA fragment.
Detection of mRNA in heat-killed cells.
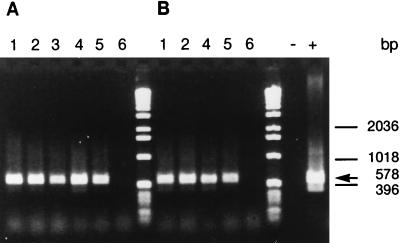
Detection of tufA mRNA in heat-killed cells is shown in Fig. 2. mRNA from groEL, rpoH, and tufA genes was detected by RT-PCR in all the cell suspensions immediately after heating at 60, 80, or 100°C (Table 2). However, during subsequent incubation at room temperature, these mRNA species became undetectable and were absent at 16 h. No viable cells were detected in samples immediately after treatment or during subsequent incubation at room temperature. The time at which mRNA became undetectable varied slightly and depended on the target. Generally, target mRNA from all three genes was detectable for up to 2 h but disappeared after 16 h (Table 2). However, groEL mRNA was undetectable at 2 h after heating at 60°C for 20 min. By contrast, 16S rRNA was detected immediately after heating and also at 16 h in all the samples.
FIG. 2.
RT-PCR detection of tufA mRNA from E. coli cells (107 cells/ml) killed by heat treatment at 100°C for 5 min (A) or 80°C for 10 min (B) and left at room temperature for 0 min (lanes 2), 30 min (lanes 3), 60 min (lanes 4), 120 min (lanes 5), or 16 h (lanes 6). Lanes 1 contain control unheated E. coli samples, and the RT-PCR-positive (lane +) and -negative (lane −) controls contain E. coli DNA and sterile water, respectively.
TABLE 2.
mRNA species detected by RT-PCR after incubation of heat-killed E. coli at room temperature
| Treatment | Target | Detection aftera:
|
|||||
|---|---|---|---|---|---|---|---|
| Untreated | 0 min | 30 min | 1 h | 2 h | 16 h | ||
| 100°C for 5 min | 16S rRNA | Y | Y | NP | Y | Y | Y |
| groEL | Y | Y | Y | Y | Y | N | |
| rpoH | Y | Y | NP | Y | Y | N | |
| tufA | Y | Y | Y | Y | Y | N | |
| 80°C for 10 min | 16S rRNA | Y | Y | Y | Y | Y | Y |
| groEL | Y | Y | Y | Y | Y | N | |
| rpoH | Y | Y | Y | Y | Y | N | |
| tufA | Y | Y | Y | Y | Y | N | |
| 60°C for 20 min | 16S rRNA | Y | Y | Y | Y | Y | Y |
| groEL | Y | Y | N | Y | N | N | |
| tufA | Y | Y | Y | Y | Y | N | |
Y, positive RT-PCR amplification; N, negative RT-PCR amplification; NP, not performed.
Detection of mRNA in ethanol-treated cells.
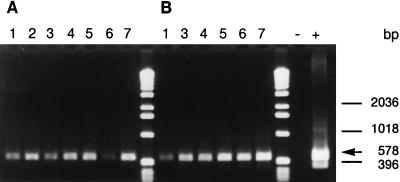
As with heat treatment, mRNA from the groEL, rpoH, and tufA genes was detected immediately after the cells had been killed (Table 3). However, these mRNA species were more stable in ethanol-treated cells than in heat-killed cells, since rpoH and tufA mRNAs were still detectable 16 h after treatment with 50% (vol/vol) ethanol and tufA was also detected 16 h after treatment with 67% (vol/vol) ethanol (Fig. 3). mRNA from groEL became undetectable before that from rpoH or tufA, disappearing by 16 and 2 h in cells killed with 50 and 67% (vol/vol) ethanol, respectively. As with the heat-killed cells, 16S rRNA was present in all ethanol-treated samples.
TABLE 3.
mRNA species detected by RT-PCR after incubation of ethanol-killed E. coli at room temperature
| Treatment | Target | Detection aftera:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Untreated | 0 min | 10 min | 30 min | 1 h | 2 h | 16 h | ||
| 50% for 7 min | 16S rRNA | Y | Y | Y | Y | Y | Y | Y |
| groEL | Y | Y | Y | Y | Y | Y | N | |
| rpoH | Y | Y | Y | Y | Y | Y | Y | |
| tufA | Y | Y | Y | Y | Y | Y | Y | |
| 67% for 7 min | 16S rRNA | Y | Y | Y | Y | Y | Y | Y |
| groEL | Y | Y | Y | Y | Y | N | N | |
| rpoH | Y | Y | Y | Y | Y | Y | N | |
| tufA | Y | Y | Y | Y | Y | Y | Y | |
Y, positive RT-PCR amplification; N, negative RT-PCR amplification.
FIG. 3.
RT-PCR detection of tufA mRNA from E. coli cells (107 cells/ml) killed with 50% (A) or 67% (B) ethanol for 7 min and left at room temperature for 0 min (lanes 2), 10 min (lanes 3), 30 min (lanes 4), 60 min (lanes 5), 120 min (lanes 6), or 16 h (lanes 7). Lanes 1 contain control untreated E. coli sample, and the RT-PCR positive (lane +) and -negative (lane −) controls contain E. coli DNA and sterile water, respectively.
DISCUSSION
The terms “life” and “living” have proved difficult to define because there is no unique property common to all things normally regarded as living. Hence, definitions tend to consist of a collection of attributes, none of which is shown by all organisms and some of which also apply to inanimate objects (29). Microbiologists tend to adopt a pragmatic approach and usually define viable organisms as those that can multiply to form colonies on agar plates or visible turbidity in broth. This definition may need qualification to accommodate the so-called viable but nonculturable organisms (31) and those that can multiply in liquid media but only to low maximum cell densities (9). We propose that a reasonable working definition of a viable bacterial cell is one that has the potential to multiply under suitable conditions.
Since replication involves the coordinated activity of homeostatic, biosynthetic, and energy-conserving systems, there is no a priori reason to suppose that any single indicator of metabolic activity will correlate with viability, so defined, under all circumstances. Nevertheless, metabolic indicators such as membrane potential, the ability to generate reducing power, or the ability to undertake DNA synthesis have proved useful indicators of the viability of cells in the natural environment (4, 19). Nucleic acids as viability indicators would have the added advantage of specificity.
In principle, the presence of one or another type of nucleic acid (DNA, rRNA, mRNA or tRNA) in bacterial cells might be a useful indicator of viability if (i) it is present only in viable cells, (ii) the kinetics of its disappearance is related to loss of viability, or (iii) it disappears from cells soon after death.
In this work, we have developed RT-PCR methods for detecting specific mRNA from E. coli and have shown that mRNA was initially present in cells killed by heat or ethanol but subsequently disappeared at a rate depending on the inactivating treatment. Because mRNA was detected in dead E. coli cells by RT-PCR, it was not an absolute indicator of viability according to criterion (i) given above. Our findings are different from those of Bej et al. (6), who detected no mRNA in cells of Vibrio cholerae that were killed by heat or starvation. Similarly, Patel et al. (27) did not detect a heat shock protein mRNA from Mycobacterium leprae killed by heat treatment, probably because the sample was left at room temperature for 5 h before analysis, allowing for any surviving mRNA to become degraded. An RT-PCR method for the specific detection of viable L. monocytogenes was recently developed (20) based on the detection of mRNA from iap, hly, or prfA genes. The greatest sensitivity was achieved with iap mRNA as the target, which allowed the detection of 10 to 15 viable cells/ml after a 1-h enrichment in broth. Cells killed by autoclaving were not detected, but the method was not tested with cells killed by other treatments.
Because of the presence of the 2′-hydroxyl group of ribose, the phosphodiester bonds of RNA are more susceptible to hydrolysis than those of DNA, particularly in the presence of divalent cations. RNA is therefore more labile than DNA and more susceptible to degradation caused directly by deleterious treatments, such as heating or acidification. However, the extent of that degradation will vary according to the type and severity of treatment, and mRNA would be expected to survive in some circumstances. Indeed, because cells have to be killed before their nucleic acid can be extracted, there can be no absolute correlation between the presence of mRNA and viability.
The three different mRNA species detected in this study were selected to represent two genes involved in stress responses (groEL and rpoH) and a gene encoding an abundant cellular housekeeping protein (tufA). Of these mRNAs, tufA and rpoH mRNAs were detected for a longer time after treatment with both heat and ethanol than was groEL. This may be correlated with the stability of the different mRNA sequences targeted by the oligonucleotide primers designed here or to the relative abundance of each mRNA type in a given cell. Either way, it appears that the type of mRNA selected for detection of viable bacteria will be important.
Detectable mRNA disappeared more quickly from heat-killed cells than from ethanol-killed cells (based on the time at which RT-PCR gave a negative result). Since the incubation conditions after heat or ethanol treatment were identical, the difference in detection times after treatment must be related to the different effects of heat and ethanol on the capacity for RNA breakdown in dead cells. In living cells, most mRNA turns over rapidly, reflecting a balance between the synthesis of mRNA and its degradation by RNases (2, 7). In dead cells, mRNA synthesis (if any) is likely to be slow and nuclease activity will continue to degrade any mRNA present. The factors controlling mRNA longevity in dead cells are not understood, but presumably mRNA would disappear most rapidly from cells killed by treatments that do not inactivate the degradative RNase enzymes. Conversely, mRNA may remain intact for longer periods in cells killed by treatments that also inactivate RNases or render the RNA resistant to attack. The relatively rapid disappearance of mRNA from dead cells shown here means that it is likely to be a much better indicator of viability than DNA according to criterion (iii) above. The factors that influence the longevity of mRNA in dead cells require clarification before its usefulness as a general indicator of viability can be judged. In particular, the effects of different inactivating treatments and holding conditions must be investigated, along with any differences in abundance or rates of disappearance of different target mRNAs.
rRNA has been suggested as an indicator of viability in Mycobacterium smegmatis (37). In cells exposed to rifampin and ofloxacin, the presence of 16S rRNA, as detected by the nucleic acid sequence-based amplification (NASBA) method, corresponded well to numbers of viable cells and a lack of signal coincided with loss of viability. A correlation between viability and cellular rRNA content was also observed in E. coli cells undergoing starvation (10) but did not occur in starved Azotobacter agilis cells (34). In heat-killed bacterial cells, the breakdown of ribosomes and rRNA is highly dependent on the medium in which cells are heated and is not necessarily related to loss of viability (16). In our study, 16S rRNA from heat- or ethanol-killed E. coli cells did not disappear after 16 h of incubation in broth at room temperature. It was not, therefore, a useful indicator of viability for the time frame over which our experiments were conducted (i.e., 16 h).
There is very little information about the persistence of tRNA in injured or dead cells. Davis et al. (10) found that tRNA survived much longer than rRNA in cells of E. coli that had died of starvation, and it is therefore unlikely to be a good indicator of viability.
This work has demonstrated that of the four species of nucleic acid, mRNA is the most promising candidate as an indicator of viability in bacteria. Klein and Juneja (20) showed a good correlation between the presence of mRNA and the viability of L. monocytogenes when comparing growing cells with those killed by autoclaving. However, in cells killed by milder treatments, the correlation is not absolute and in some cases the mRNA can persist for a while. The detection of mRNA therefore indicates either that a cell is alive or has died “fairly recently.” The length of time that mRNA persists and hence the time envelope described by the phrase “fairly recently” will depend on both the method by which cells were killed and the postmortem holding conditions. We have established that mRNA can persist for at least 16 h, but further work is required to characterize the decay rates of mRNA in dead cells under a range of conditions before the limitations of the method are fully defined. However, it should be possible to identify conditions under which mRNA disappears very rapidly and those where it does not. In this way, RT-PCR methods may be developed for particular applications that will provide a good indication of the presence of viable cells in food, clinical, or environmental samples. Methods now exist for detecting mRNA in single bacterial cells (15), and further improvements in technology are likely to increase the application of these methods in microbial monitoring and ecology.
ACKNOWLEDGMENTS
We are grateful to the Department of Health, London, United Kingdom, for supporting this work (grant DH 195).
We thank Joe Mangan for helpful discussions on methods of RNA extraction.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence of two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Alifano P, Bruni C B, Carlomagno M S. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94:157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- 3.Bakau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 4.Barer M R, Gribbon L T, Harwood C R, Nwoguh C E. The viable but nonculturable hypothesis and medical bacteriology. Rev Med Microbiol. 1993;4:183–191. [Google Scholar]
- 5.Bej A K, Mahbubani M H, Atlas R M. Detection of viable Legionella pneumophila in water by polymerase chain reaction and gene probe methods. Appl Environ Microbiol. 1991;57:597–600. doi: 10.1128/aem.57.2.597-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bej A K, Ng W-Y, Morgan S, Jones D, Mahbubani M H. Detection of viable Vibrio cholerae by reverse-transcriptase polymerase chain reaction (RT-PCR) Mol Biotechnol. 1996;5:1–10. doi: 10.1007/BF02762407. [DOI] [PubMed] [Google Scholar]
- 7.Belasco J. mRNA degradation in prokaryotic cells: an overview. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 3–12. [Google Scholar]
- 8.Bosch L, Kraal B, Van der Meide P H, Duisterwinkel F J, Van Noort J M. The elongation factor EF-Tu and its encoding genes. Prog Nucleic Acid Res Mol Biol. 1983;30:91–126. doi: 10.1016/s0079-6603(08)60684-4. [DOI] [PubMed] [Google Scholar]
- 9.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engleberg N C, Carter C, Weber D R, Cianciotto N P, Eisenstein B I. DNA sequence of mip a Legionella pneumophila gene associated with macrophage activity. Infect Immun. 1992;57:1236–1270. doi: 10.1128/iai.57.4.1263-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson J W, Vaughn V, Walter W A, Neidhart F C, Gross C A. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat-shock regulatory gene. Genes Dev. 1987;1:419–432. doi: 10.1101/gad.1.5.419. [DOI] [PubMed] [Google Scholar]
- 13.Fung M-C, Fung Y-M. PCR amplification of mRNA directly from a crude cell lysate prepared by thermophilic protease digestion. Nucleic Acids Res. 1991;19:4300. doi: 10.1093/nar/19.15.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrick J P, Hartl F-U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 15.Hodson R E, Dustman W A, Garg R P, Moran M A. In situ PCR for visualization of microscale distribution of specific genes and gene products in prokaryotic communities. Appl Environ Microbiol. 1995;6:4074–4082. doi: 10.1128/aem.61.11.4074-4082.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst A. Revival of vegetative bacteria after sublethal heating. In: Andrew M H E, Russell A D, editors. The revival of injured microbes. London, United Kingdom: Academic Press, Ltd.; 1993. pp. 77–103. [Google Scholar]
- 17.Jeffrey W H, Nazaret S, Von Haven R. Improved method for recovery of mRNA from aquatic samples and its application to detection of mer expression. Appl Environ Microbiol. 1994;60:1814–1821. doi: 10.1128/aem.60.6.1814-1821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josephson K L, Gerba C P, Pepper I L. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol. 1993;59:3513–3515. doi: 10.1128/aem.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaprelyants A S, Gottschal J C, Kell D B. Dormancy in non-sporulating bacteria. FEMS Microbiol Rev. 1993;104:271–286. doi: 10.1111/j.1574-6968.1993.tb05871.x. [DOI] [PubMed] [Google Scholar]
- 20.Klein P G, Juneja V K. Sensitive detection of viable Listeria monocytogenes by reverse-transcriptase PCR. Appl Environ Microbiol. 1997;63:4441–4448. doi: 10.1128/aem.63.11.4441-4448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladin B F, Accuroso E M, Mielenz J R, Wilson C R. A rapid procedure for isolating RNA from small scale Bacillus cultures. BioTechniques. 1992;12:672–675. [PubMed] [Google Scholar]
- 22.Lindahl T. Instability and decay of the primary structure of DNA. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 23.Mackey B M, Derrick C M. A comparison of solid and liquid media for measuring the sensitivity of heat-injured Salmonella typhimurium to selenite and tetrathionate media, and the time needed to recover resistance. J Appl Bacteriol. 1982;53:233–242. doi: 10.1111/j.1365-2672.1982.tb04682.x. [DOI] [PubMed] [Google Scholar]
- 24.Mangan J A, Sole K M, Mitchison D A, Butcher P D. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacterium. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masters C I, Shallcross J A, Mackey B M. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J Appl Bacteriol. 1994;77:73–79. doi: 10.1111/j.1365-2672.1994.tb03047.x. [DOI] [PubMed] [Google Scholar]
- 26.Olsen J E, Aabo S, Hill W, Notermans S, Werners K, Granum P E, Popovic T, Rasmussen H N, Olvic O. Probes and polymerase chain reaction for detection of foodborne bacterial pathogens. Int J Food Microbiol. 1995;28:1–17. doi: 10.1016/0168-1605(94)00159-4. [DOI] [PubMed] [Google Scholar]
- 27.Patel B K R, Banjerjee D K, Butcher P D. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat shock protein mRNA. J Infect Dis. 1993;168:799–800. doi: 10.1093/infdis/168.3.799. [DOI] [PubMed] [Google Scholar]
- 28.Pichard S L, Paul J H. Gene expression per gene dose, a specific measure of gene expression in aquatic micro-organisms. Appl Environ Microbiol. 1993;59:451–457. doi: 10.1128/aem.59.2.451-457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirie N W. The meaninglessness of the terms life and living. In: Needham J, Green D E, editors. Perspectives in Biochemistry. Cambridge, United Kingdom: Cambridge University Press; 1937. pp. 11–22. [Google Scholar]
- 30.Qiagen Ltd. Qiagen RNeasy handbook. Dorking, United Kingdom: Qiagen Ltd.; 1994. [Google Scholar]
- 31.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Selvaratnam S, Schoedel B A, McFarland B L, Kulpa C F. Application of reverse transcriptase PCR for monitoring expression of the catabolic dmpN gene in a phenol-degrading sequencing batch reactor. Appl Environ Microbiol. 1995;61:3981–3985. doi: 10.1128/aem.61.11.3981-3985.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobek J M, Charba J F, Foust W N. Endogenous metabolism of Azotobacter agilis. J Bacteriol. 1966;92:687–690. doi: 10.1128/jb.92.3.687-695.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai Y-L, Olson B H. Effects of Hg2+, CH3-Hg+, and temperature on the expression of mercury resistance genes in environmental bacteria. Appl Environ Microbiol. 1990;56:3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Meide P H, Vijgenboom E, Talens A, Bosch L. The role of EF-Tu in the expression of tufA and tufB genes. Eur J Biochem. 1983;130:397–407. doi: 10.1111/j.1432-1033.1983.tb07166.x. [DOI] [PubMed] [Google Scholar]
- 37.Van der Vliet G M E, Schepers P, Schukkink R A F, Van Gemen B, Klatser P R. Assessment of mycobacterial viability by RNA amplification. Antimicrob Agents Chemother. 1994;38:1959–1965. doi: 10.1128/aac.38.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]