Significance
Vast numbers of senescence-associated EVs (S-EVs) are released and carry different contents than EVs from proliferating cells (P-EVs), suggesting that they may harbor important factors critical for the senescence microenvironment and pathologies associated with age. We characterized the proteins, especially surface proteins, on P-EVs and S-EVs, and identified DPP4 as a specific marker on the surface of S-EVs. Although S-EVs were taken up by macrophages, they were refractory to uptake by many other cell types, including cancer cells. DPP4 on the surface of S-EVs appeared to function as a signal to prevent S-EVs internalization.
Keywords: surfaceome, extracellular vesicles, senescence
Abstract
Senescent cells are beneficial for repairing acute tissue damage, but they are harmful when they accumulate in tissues, as occurs with advancing age. Senescence-associated extracellular vesicles (S-EVs) can mediate cell-to-cell communication and export intracellular content to the microenvironment of aging tissues. Here, we studied the uptake of EVs from senescent cells (S-EVs) and proliferating cells (P-EVs) and found that P-EVs were readily taken up by proliferating cells (fibroblasts and cervical cancer cells) while S-EVs were not. We thus investigated the surface proteome (surfaceome) of P-EVs relative to S-EVs derived from cells that had reached senescence via replicative exhaustion, exposure to ionizing radiation, or treatment with etoposide. We found that relative to P-EVs, S-EVs from all senescence models were enriched in proteins DPP4, ANXA1, ANXA6, S10AB, AT1A1, and EPHB2. Among them, DPP4 was found to selectively prevent uptake by proliferating cells, as ectopic overexpression of DPP4 in HeLa cells rendered DPP4-expressing EVs that were no longer taken up by other proliferating cells. We propose that DPP4 on the surface of S-EVs makes these EVs refractory to internalization by proliferating cells, advancing our knowledge of the impact of senescent cells in aging-associated processes.
Cellular senescence arises in response to sublethal damage caused by a range of stimuli and is characterized by indefinite growth arrest. However, senescent cells remain metabolically active, express unique subsets of RNAs and proteins, and display a robust senescence-associated secretory phenotype (SASP) (1, 2). Senescent cells can be beneficial during development, as they contribute to morphogenesis and wound repair, and prevent tumorigenesis in young persons. However, their accumulation in older human tissues can exacerbate pathologies associated with aging, such as cancer, neurodegeneration, and cardiovascular disease (3–5). The SASP is a major detrimental component of senescent cells and includes the secretion of soluble proinflammatory factors, tissue-remodeling factors, and extracellular vesicles (EVs) (6).
EVs are particles carrying cytoplasmic content enclosed by a lipid bilayer and range in diameter between 30 nm and several μm. They are broadly classified by biogenesis as ectosomes arising from the plasma membrane, with a wide range of sizes, and exosomes (mostly 30 to 150 nm in diameter), arising from endosomal invaginations into multivesicular bodies (MVBs) (7). Collectively known as EVs, these particles play key roles in cell-to-cell communication through their cargo, which includes specific proteins (signaling molecules, receptors, integrins, and cytokines), bioactive lipids, and nucleic acids (mRNAs, microRNAs, long noncoding RNAs, and DNA) (8).
Proteins on the surface of EVs can help regulate the interaction of EVs with their microenvironment, influencing EV distribution in the body, time in circulation, and pharmacokinetics (9). For example, the levels of the protein programmed death ligand 1 (PD-L1) on the surface of circulating EVs are predictive of the response to anti-PD-L1 therapy in metastatic melanoma (10), and the presence of the integrin ITGB3 on the surface of EVs informs about intracellular communication in cancer metastasis (11). Moreover, EV surface proteins may be exploited to enhance retention of therapies in target areas. For example, CD47-enriched EVs loaded with chemotherapeutic nanoparticles evade phagocytosis, while a homing peptide promotes their retention by cells in tumor tissue (12).
An important link between EVs and senescence is that EV production increases in multiple models of cellular senescence (6, 13, 14). Senescence-associated EVs (S-EVs) have altered contents compared with EVs from proliferating cells (P-EVs), and hence, they may harbor essential factors that mediate the effects of senescent cells in their microenvironment and on pathologies associated with age (15, 16).
The current study was initiated to characterize the proteins differentially abundant in S-EVs relative to P-EVs, including both total EV proteins and proteins on the surface of EVs. Interestingly, while all EVs (P-EVs and S-EVs) were avidly taken up by macrophages, S-EVs were not taken up by proliferating human fibroblasts or human cervical carcinoma (HeLa) cells. To investigate why P-EVs were internalized but S-EVs were not, membrane-impermeant biotin-labeled EV surface proteins were identified by mass spectrometry. Among the proteins identified as being highly and selectively expressed on the surface of the S-EVs, dipeptidyl peptidase 4 (DPP4) present on S-EVs appeared to actively reduce their internalization, as overexpressing DPP4 in proliferating cells rendered P-EVs that were no longer taken up by proliferating cells. We propose that DPP4 is a key protein on the surface of S-EVs that prevents EV uptake by proliferating cells.
Results
EVs Are Enriched in Conditioned Medium from Senescent Compared to Proliferating Cells.
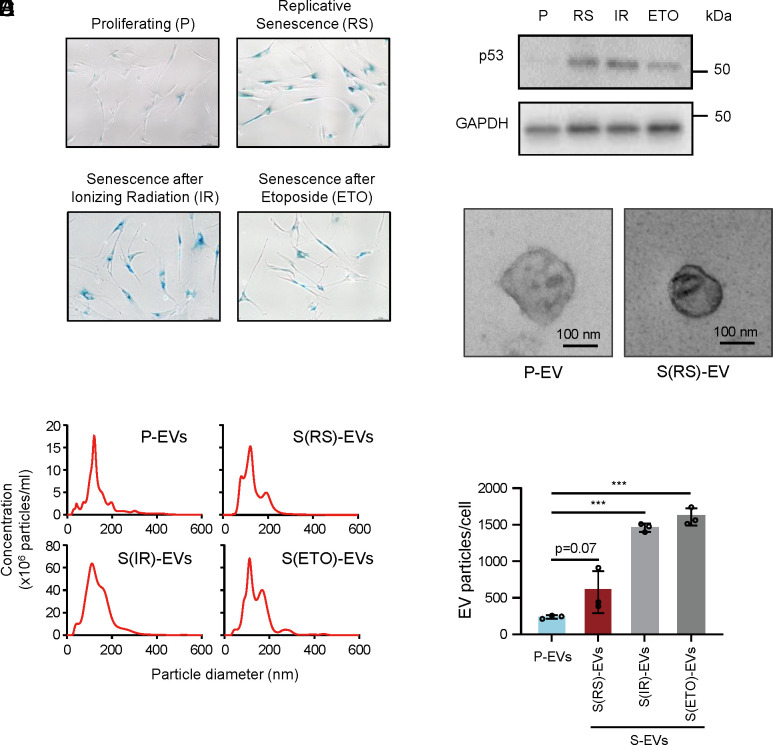
We sought to understand how senescent cells might communicate with surrounding cells by studying the extracellular vesicles (EVs) they produce. Proliferating (P) human diploid fibroblasts (WI-38) were rendered senescent using three methods, as described (17). First, WI-38 fibroblasts at the population doubling level (PDL)20 were allowed to divide repeatedly until they reached replicative senescence (RS) at approximately PDL52. Second, proliferating WI-38 cells were exposed to 10 Gray (Gy) of ionizing radiation (IR) and harvested 8 d later. Third, proliferating WI-38 cells were treated with etoposide (ETO, at 50 μM every 72 h) and harvested on day 6. Senescence was monitored by assessing senescence-associated (SA)-β-galactosidase (SA-β-gal) activity (Fig. 1A) and the levels of the senescence marker protein TP53 (p53) (Fig. 1B) in these cultures.
Fig. 1.
Levels of secreted EVs increase with cell senescence. (A) Senescence was monitored by assessing the senescence-associated (SA)-β-gal activity (blue staining) of WI-38 cells and confirmed in three different senescence paradigms, while proliferating cells (P) do not show staining. WI-38 human diploid fibroblasts were either left untreated (Proliferating, P) or were rendered senescent by exposure to 10 Gy ionizing radiation (IR) followed by incubation for 8 d, by treatment with 50 μM etoposide every 72 h and harvested at day 6 (ETO) or reached replicative senescence (RS) by proliferation to exhaustion, from population doubling number (PDL) 20 (P fibroblasts) to senescence at ~PDL52. (B) The levels of the senescence marker protein p53 (TP53) in each of the four fibroblast populations were monitored by western blot analysis; the levels of the housekeeping control protein GAPDH were assessed to monitor differences in loading across the four groups. (C) Representative transmission electron microscopy (TEM) images showing the morphology of S-EVs from fibroblasts rendered RS as described in A. See also SI Appendix, Fig. S1B. (D) Size and concentration (EVs per mL) of EVs collected from the populations described in A and measured by Nanosight NS300 Nanoparticle tracking analysis (NTA, Methods). (E) The numbers of EVs per cell were calculated based on measurements taken over 48 h in the fibroblast populations described in A. Data in E are the mean ± SEM from three biological replicates; ***P ≤ 0.001. Data were analyzed by one-way ANOVA.
We then enriched EVs from each of the four groups by ultracentrifugation (Methods and SI Appendix, Fig. S1A), evaluated them by TEM imaging (Fig. 1C and SI Appendix, Fig. S1B), and assessed their size distribution and concentration by NTA. As shown (Fig. 1D), all senescence-associated EVs (S-EVs), including EVs from the RS, IR, and ETO groups, were similar in diameter (100 to 200 nm) to those from P-EVs. However, more EVs were consistently found in senescent cultures, particularly in the ETO and IR groups, than in proliferating cultures (Fig. 1E). We then set out to study whether the protein contents of S-EVs and P-EVs differed.
S-EVs Are Rejected by Proliferating Cells but Taken up by Macrophage-Like Cells.
To investigate the uptake of P-EVs (from fibroblasts at ~PDL20) and S-EVs (from fibroblasts at ~PDL52), we measured their surface charge and labeled them with the dye PKH26. The zeta potential, a measure of surface charge and colloidal stability (18), was approximately −13 and −11 mV for P-EVs and S-EVs, respectively (SI Appendix, Fig. S1C). The efficiency of labeling with PKH26, as measured with a NanoFCM NanoAnalyzer (SI Appendix, Fig. S2A), showed ~threefold higher labeling for P-EVs (37.7%) compared to S-EVs (12.7%). We incubated proliferating (P) WI-38 fibroblasts and HeLa cells for 24 h with equal amounts of labeled EVs, taking into consideration these labeling efficiencies. Control incubations included “PBS” (PKH26 without EVs) and “no EV” to detect background fluorescence.
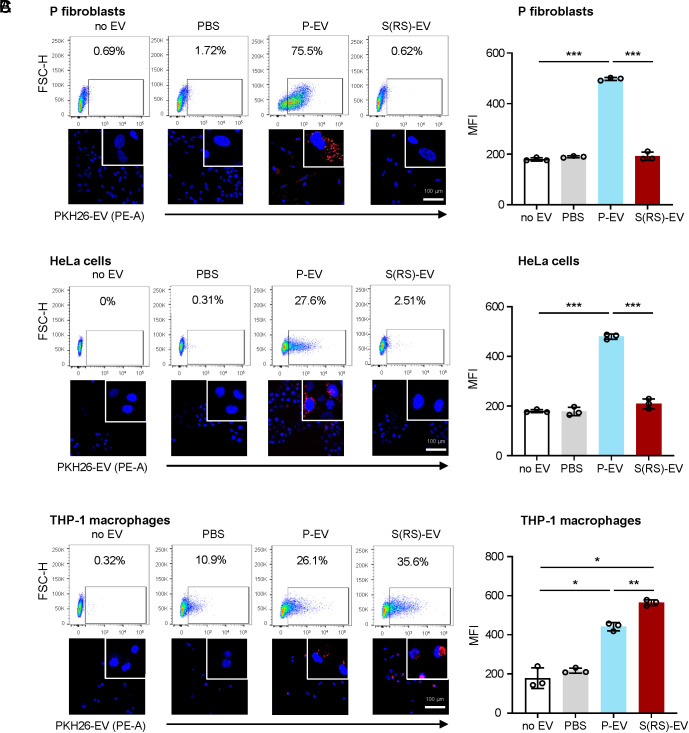
Recipient cells were analyzed by flow cytometry and confocal microscopy. Unexpectedly, the uptake of S-EVs was drastically lower than the uptake of P-EVs by P cells and HeLa cells (Fig. 2 A and B). These results were confirmed by confocal microscopy, where only incubation with P-EVs yielded strong intracellular signals in fluorescent micrographs, and by flow cytometry and quantification of mean fluorescence intensity (MFI) (Fig. 2 A and B). Similarly, P cells and HeLa cells were refractory to uptake of labeled S(ETO)-EVs and S(IR)-EVs, as determined by flow cytometry (SI Appendix, Fig. S2B). Together, these data suggest that EVs from senescent cells of all treatment conditions were not internalized efficiently by proliferating fibroblasts or HeLa cells.
Fig. 2.
Distinct uptake by different cells of P-EVs relative to S-EVs prepared from WI-38 fibroblasts. Flow cytometric analysis (Top) and representative confocal microscopy images (Bottom) of the uptake of P-EVs and S-EVs (as well as control incubations without EVs, ‘no EV’, and with PKH26 in PBS, 'PBS only') by proliferating WI-38 fibroblasts (A), HeLa cells (B), and THP-1 cells differentiated to macrophages by incubation for 48 h with PMA (C). Right, quantification of MFI in flow cytometric assays in each of the three recipient cells and the four incubation groups. Data in A–C are the mean ± SEM from three biological replicates; statistical analysis was performed by one-way ANOVA for multiple group comparison (P-EVs, S-EVs, or PBS vs. no EV group) and Student’s t test for P-EVs vs. S-EVs group. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Given that macrophages are specialized in phagocytosis, we investigated their ability to uptake S-EVs. Human monocyte leukemia-derived THP-1 cells were differentiated into macrophages by treatment with 150 nM PMA for 48 h. Strikingly, THP-1 macrophages were capable of internalizing both P-EVs and S-EVs, as determined by flow cytometry and confocal microscopy coupled with MFI quantification (Fig. 2C), and moreover, S-EVs appeared to be taken up more avidly than P-EVs. The internalization of PKH26-labeled S-EVs by THP-1 macrophages was documented by time-lapse imaging (Movie S1 and SI Appendix, Fig. S3) and revealed that EVs accumulated in the cytoplasm gradually from 1 to 24 h. Other labeling methods (using CFSE) showed similar uptake patterns by P cells and HeLa cells, namely that it was robust for P-EVs but not for S-EVs. Uptake by macrophages was comparable between EVs (SI Appendix, Fig. S2C).
In summary, proliferating fibroblasts and HeLa cells were refractory to the uptake of S-EVs, while they avidly internalized P-EVs. In contrast, THP-1 macrophages internalized S-EVs comparably to, if not better, than P-EVs. Accordingly, we hypothesize that S-EVs may have distinct surface molecules compared with P-EVs that prevent their uptake by nonphagocytic cells.
S-EVs Have Distinct Proteomes with High Levels of DPP4.
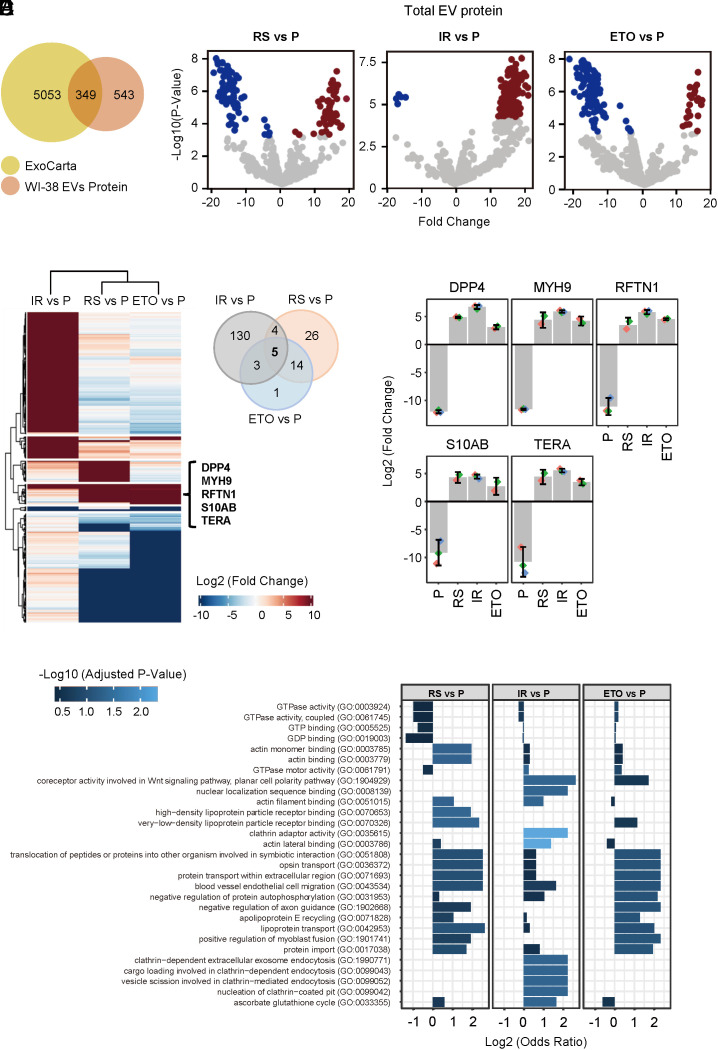
To elucidate the protein content of P-EVs and S-EVs, we performed liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) on whole EVs prepared from proliferating (~PDL20) WI-38 fibroblasts and from WI-38 fibroblasts rendered senescent by three different methods (S, IR, ETO). After further filtering proteins based on missing values (SI Appendix, Methods), a group of 892 proteins was found in all biological replicates in at least one group (P, S, IR, or ETO) (Datasets S1–S3). Among them, 349 proteins had previously been reported in EV preparations [ExoCarta (19)], while an additional 543 were found in our study (Fig. 3A).
Fig. 3.
Mass spectrometry profiling of proteomes from whole EVs from proliferating and senescent WI-38 fibroblasts. (A) Venn diagram of proteins (892 total) identified in the EV samples from all four groups combined (P, RS, IR, and ETO) compared with proteins annotated in the ExoCarta databases (5,402 proteins). (B) Volcano plots of proteins differentially abundant in S-EVs from fibroblasts rendered senescent by RS, IR, or ETO relative to proliferating (P) fibroblasts. (C) Left, heatmap of proteins differentially abundant in EVs from fibroblasts rendered senescent (S) by RS, IR, or ETO relative to EVs from proliferating (P) fibroblasts. Right, Venn diagram indicating five proteins enriched in EVs from all three senescent paradigms compared with proliferating fibroblasts. (D) Bar graphs depicting the abundance of the five specific proteins (DPP4, MYH9, RFTN1, S10AB, and TERA) in whole S-EVs [prepared from senescent (RS, IR, and ETO) cells] compared with P-EVs (prepared from proliferating cells), as measured by mass spectrometry profiling. (E) Gene Ontology enrichment analysis of the proteins preferentially abundant in EVs from S fibroblasts (RS, IR, and ETO) compared with EVs from P fibroblasts.
The proteomic profiles of EVs in each senescence group were distinct. Compared with P-EVs, different proteins were more abundant and less abundant in S(RS)-EVs, S(IR)-EVs, and S(ETO)-EVs, shown as volcano plots (Fig. 3B) and heatmaps (Fig. 3C). Not all changes in differentially abundant proteins were shared across the comparisons. For example, S(RS)-EVs and S(ETO)-EVs were markedly enriched in Keratin type II cytoskeletal protein (K2C1), while the most enriched protein in S(IR)-EVs was the synaptic vesicle membrane protein VAT1 (SI Appendix, Fig. S4A). However, compared with P-EVs, all three S-EVs showed elevated levels of five shared proteins: DPP4, MYH9, RFTN1, S10AB, and TERA (Fig. 3D), while only one protein, GPX3, was more abundant in P-EVs than in EVs from any of the three senescence models (SI Appendix, Fig. S4B).
As a class, the proteins that are enriched in S-EVs (125 proteins enriched in RS vs. P, 146 proteins in IR vs. P, and 125 proteins in ETO vs. P; Fig. 3B) are involved in binding RNA (20, 21), as well as in nucleotide binding, transport, fusion, migration, endocytosis, and other internalization-related functions (Fig. 3E). Comparison with other published datasets (6) identified S-EVs proteins DPP4, ANXA1, ANXA6, S10AB, MYH9, RFTN1, and TERA shared between the two studies.
DPP4 Is Expressed on the Surface of S-EVs.
Although proteins on the surface of EVs are involved in EV uptake by recipient cells (22, 23), for most EVs, the surface proteins have not been characterized in detail. Given the importance of internalization patterns and the unexpected observation that S-EVs are less readily taken up (Fig. 2), we studied whether some of the differentially abundant proteins in S-EVs were present on the surface and further tested whether they were functionally related to internalization efficiency.
To identify EV surface proteins, we labeled proteins specifically present on the surface of EVs with EZ-Link Sulfo-NHS-SS-Biotin, then lysed EVs and isolated biotinylated proteins for proteomic analysis using LC-MS/MS. We identified surface proteins from EVs obtained from all three senescence groups (RS, IR, and ETO) and from proliferating (P) cells. A total of 1238 proteins were identified in at least one of the four EV groups (P, RS, IR, and ETO) with one or more unique peptides (Datasets S4–S14). After we applied more stringent criteria to include the proteins with unique Uniprot identifiers that are present in all replicates of at least one condition, we identified 456 proteins on the EV surface (Dataset S15).
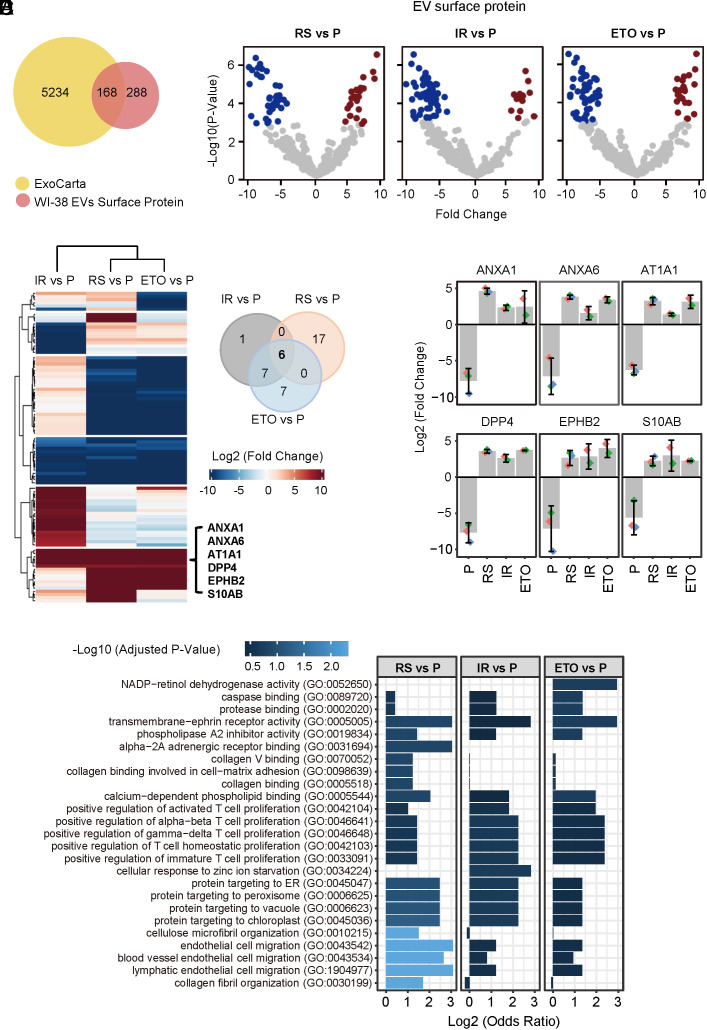
Of the 456 EV surface proteins, 168 proteins were annotated in ExoCarta as being previously identified in human EVs, while 288 proteins are reported in this study (Fig. 4A), mindful of the fact that the method for identifying surface proteins using biointylation and affinity pulldown is very sensitive. Uniprot categorized 136 of the 456 proteins (29.82%) proteins as being present in membranes, on the cell membrane, or on the cell surface; 65 of them, including CD81, CD9, and CD63, have at least one (and up to 12) transmembrane domains (Dataset S15). Gene ontology analysis identified these EV surface proteins as being present in extracellular vesicles, plasma membrane, membrane-enclosed organelles, cell junctions, and the cell surface (SI Appendix, Fig. S5A). Additionally, 73 EV surface proteins, including the highly enriched protein DPP4, were documented as components of the cell surfaceome and were classified into functional categories (SI Appendix, Fig. S5 B and C). Proteins on the surface of EVs were compared to proteins in whole EVs (SI Appendix, Fig. S5 D, Left); with senescence, the changes of the EV surfaceome and the whole-EV proteome (SI Appendix, Fig. S5 D, Right) did not correlate, although the reasons are unclear at present. Proteins on the surface of EVs from each senescence group were further compared with proteins on the surface of EVs from proliferating cells (Fig. 4B). At the overlap of the three comparison groups, the most enriched EV surface proteins from senescent populations are ANXA1, ANXA6, AT1A1, DPP4, EPHB2, and S10AB (Fig. 4 C and D). ANXA1, DPP4, EPHB2, and AT1A1 are cell membrane proteins that contain up to 10 transmembrane domains; plasma membrane proteins ANXA1, ANXA6, and chemokine S10AB can be released and localized in EVs (9, 24). In keeping with their enriched presence on the surface of S-EVs but not within S-EVs, these proteins are implicated in binding other proteins, particularly those involved in positively regulating T cell proliferation (Fig. 4E).
Fig. 4.
Mass spectrometry profiling of surfaceome of EVs from proliferating and senescent fibroblasts. (A) Venn diagram of proteins (456 total) identified in the surfaces of EV samples from all four groups combined (P, RS, IR, and ETO) compared with proteins annotated in the ExoCarta databases (5,402 proteins). (B) Volcano plot of proteins differentially abundant on the surface of EVs from proliferating (P) fibroblasts relative to the surface of EVs from fibroblasts rendered senescent (S) by RS, IR, or ETO. (C) Left, heatmap of proteins differentially abundant on the surface of EVs from proliferating (P) fibroblasts relative to the surface of EVs from fibroblasts rendered senescent (S) by RS, IR, or ETO. Right, Venn diagram indicating the six proteins enriched on the surface of EVs from all three senescent paradigms compared with EVs from proliferating fibroblasts. (D) Bar graph depicting the abundance of the 6 specific proteins (ANXA1, ANXA6, AT1A1, DPP4, EPHB2, and S10AB) on the surface of S-EVs [prepared from S (RS, IR, ETO) cells] compared with P-EVs (prepared from P cells), as measured by proteomic analysis. (E) Gene Ontology enrichment analysis of the proteins preferentially abundant on the surface of EVs from S fibroblasts (RS, IR, and ETO) compared with EVs from P fibroblasts.
Reducing DPP4 Increases, while Increasing DPP4 Reduces Internalization of S-EVs.
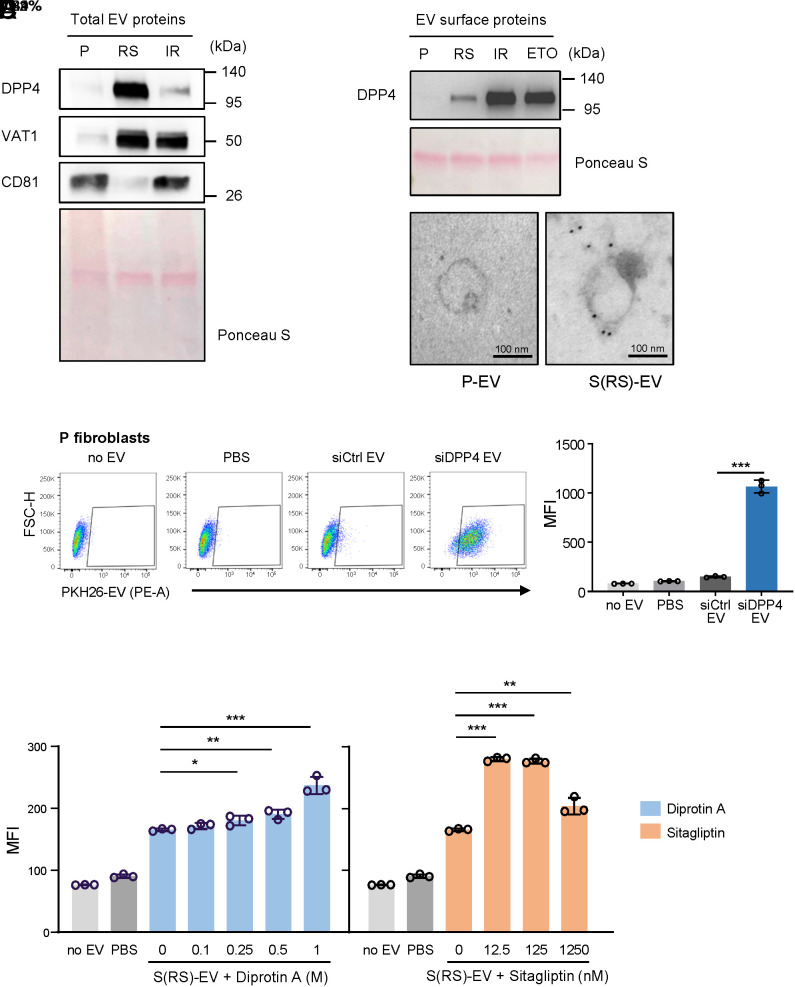
Given that the uptake of S-EVs was rejected by P fibroblasts and HeLa cells (Fig. 2C), that DPP4 was highly and preferentially expressed on the surface of S-EVs (Figs. 3D and 4D), and that DPP4 was implicated in the interaction of pulmonary endothelial cells with environmental elements (25), we sought to test whether DPP4 was directly implicated in preventing EV internalization. To study this possibility, we first assessed the presence of DPP4 in whole-protein lysates of EVs released from cells that were either proliferating (P) or rendered senescent by replicative exhaustion (RS) or exposure to IR (IR). As shown, western blot analysis revealed that levels of DPP4 (as well as VAT1) increased in EVs from RS and IR populations, consistent with EV proteomic profiles; CD81 served as a control, and the evenness of loading was monitored by Ponceau S staining (Fig. 5A). Further analysis of EV surface proteins, prepared by using EV-surface biotinylation (proteins associated with plasma membrane labeled with a reactive biotin ester, Methods) confirmed that DPP4 levels were higher on the surface of EVs released by all three senescence populations tested (RS, IR, ETO) relative to EVs from proliferating cells (Fig. 5B); some biological variability was observed, possibly because DPP4 has two forms, one surface-associated and the other soluble and possibly inside the EV. The localization of DPP4 on the surface of EVs was confirmed using immunogold electron microscopy (Fig. 5C).
Fig. 5.
Validation of DPP4 on the surface of S-EVs. (A and B) Western Blot analysis of the levels of DPP4 in total lysates (A) and on the surface (B) of EVs isolated from P fibroblasts and from S fibroblasts [S (RS), IR]; Ponceau S staining of the transfer membrane was carried out to monitor loading of the samples. (C) Representative TEM images of DPP4-specific immunogold staining (black dots) of EVs isolated from proliferating and senescent (RS) WI-38 fibroblasts. (D) Left, flow cytometric analysis of the uptake by P fibroblasts of EVs isolated from WI-38 fibroblasts (PDL45) that had been transfected with either a nontargeting siRNA control (siCtrl) or DPP4-targeting siRNA (siDPP4); control incubations with no EVs and PBS only were included as in Fig. 2. Right, quantification of MFI for each sample. (E) Quantification (MFI) of the flow cytometric analysis of P fibroblasts to measure the uptake of PKH26-labeled S(RS)-EVs that were pre-treated with DPP4 inhibitors Diprotin A or Sitagliptin; control samples with no EVs and PBS only were included. Data in D and E are the mean ± SEM from three biological replicates; statistical analysis was performed by one-way ANOVA for multiple group comparison (siCtrl EVs, siDPP4 EVs, PBS, no EV groups) and Student’s t test for siCtrl EVs vs. siDPP4 EVs group. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
We extended the analysis of DPP4 EVs to other models of senescence. We included expression of the oncogene HRASG12V in WI-38 fibroblasts by lentiviral delivery (Methods) to trigger oncogene-induced senescence (OIS) followed by analysis 8 d later, and rat H9c2 primary myoblasts and human HSkM skeletal myoblasts exposed to 10 Gy IR and analyzed 10 d later. Senescence was evaluated by SA-β-gal activity and increased cellular DPP4 activity (SI Appendix, Fig. S6 A and B). As shown, when comparing the uptake of P-EVs from WI-38 fibroblasts to the uptake of EVs prepared from WI-38 OIS fibroblasts [S(OIS)-EVs], all recipient cells tested (P WI-38 fibroblasts, human HaCaT keratinocytes, and human HMC3 microglia) showed reduced uptake of S(OIS)-EVs (SI Appendix, Fig. S7). Similarly, when comparing the uptake of P-EVs and S(IR)-EVs prepared from rat H9c2 myoblasts, uptake was greater for the P-EVs, as measured on recipient rat H9c2 myoblasts, NF8383 rat macrophages, and human HSkM myoblasts (SI Appendix, Fig. S8).
To further test whether the increased presence of DPP4 on the surface of senescent EVs was responsible for the reduced uptake, we silenced DPP4 by transfecting WI-38 cells at PDL45 with an siRNA directed at DPP4 (siDPP4) alongside a control siRNA (siCtrl). We confirmed the silencing efficiency by western blot analysis (SI Appendix, Fig. S6 B and C), collected EVs, labeled them with CFSE, and examined the uptake efficiency of EVs from the siCtrl and siDPP4 groups by proliferating cells. As shown, there was increased uptake of EVs collected from the siDPP4 group relative to EVs from the siCtrl group (Fig. 5D). In addition to silencing DPP4, we asked whether simply inhibiting the peptidase function of DPP4 in the S-EVs used for co-culture might also enable the uptake of EVs from senescent cells. Accordingly, we tested the effect of two DPP4 inhibitors, Diprotin A and Sitagliptin, each capable of specifically blocking the enzymatic activity of DPP4 by binding to its active site. Interestingly, uptake efficiency improved with either inhibitor (Fig. 5E and SI Appendix Fig. S9) at a range of concentrations as low as of 0.25 M and 12.5 nM for Diprotin A and Sitagliptin, respectively.
Finally, we asked the question from a complementary perspective, that is, whether increasing DPP4 levels reduced EV uptake. We overexpressed DPP4 by infection with a lentivirus that expressed a tagged form of DPP4 (DPP4-Myc) in HeLa cells, which otherwise express negligible levels of endogenous DPP4. After puromycin selection of the infected population (HeLa-DPP4-Myc) for 20 d, we collected EVs for uptake efficiency testing (Fig. 6A); control HeLa populations were infected with a lentivirus that expressed only the Myc antigen (HeLa-Myc). We first validated the production of DPP4-Myc fusion protein, along with loading control GAPDH and EV marker CD81, in HeLa whole-cell lysates, total EVs, and EV surface protein preparations. Compared with HeLa-Myc cells, HeLa-DPP4-Myc cells produced EVs that were positive for DPP4-Myc when assessed in whole EVs and in proteins present on the EV surface (Fig. 6B). We then labeled EVs from HeLa-Myc cells and HeLa-DPP4-Myc cells with PKH26 fluorescent dye and incubated these EVs with P WI-38 cells (2 × 108 EVs incubated with 5 × 104 cells, for an EV/cell ratio ~4,000:1). As shown (Fig. 6 C, Left), P cells showed strong uptake of PKH26-labeled EVs from HeLa-Myc cells (Myc EV), with 59.9% cells showing uptake of fluorescence. Interestingly, however, when exposed to the same number of EVs from HeLa-DPP4-Myc cells (DPP4-Myc EV), fewer P fibroblasts (26.9%) took up the fluorescent EVs. Average MFI measurements reflected this trend, with significantly lower MFI values in the DPP4-Myc EV uptake group than the Myc EV group (Fig. 6 C, Right). Similar to S-EVs, DPP4-Myc EV uptake was reduced relative to Myc EV uptake by proliferating WI-38 cells, as well as by HeLa cells, although there was no difference in uptake by THP-1 macrophages (SI Appendix, Fig. S5E). Together, these findings support the notion that the presence of DPP4 on the EV surface reduces the uptake of EVs by proliferating cells, as depicted in the schematic (Fig. 6D).
Fig. 6.
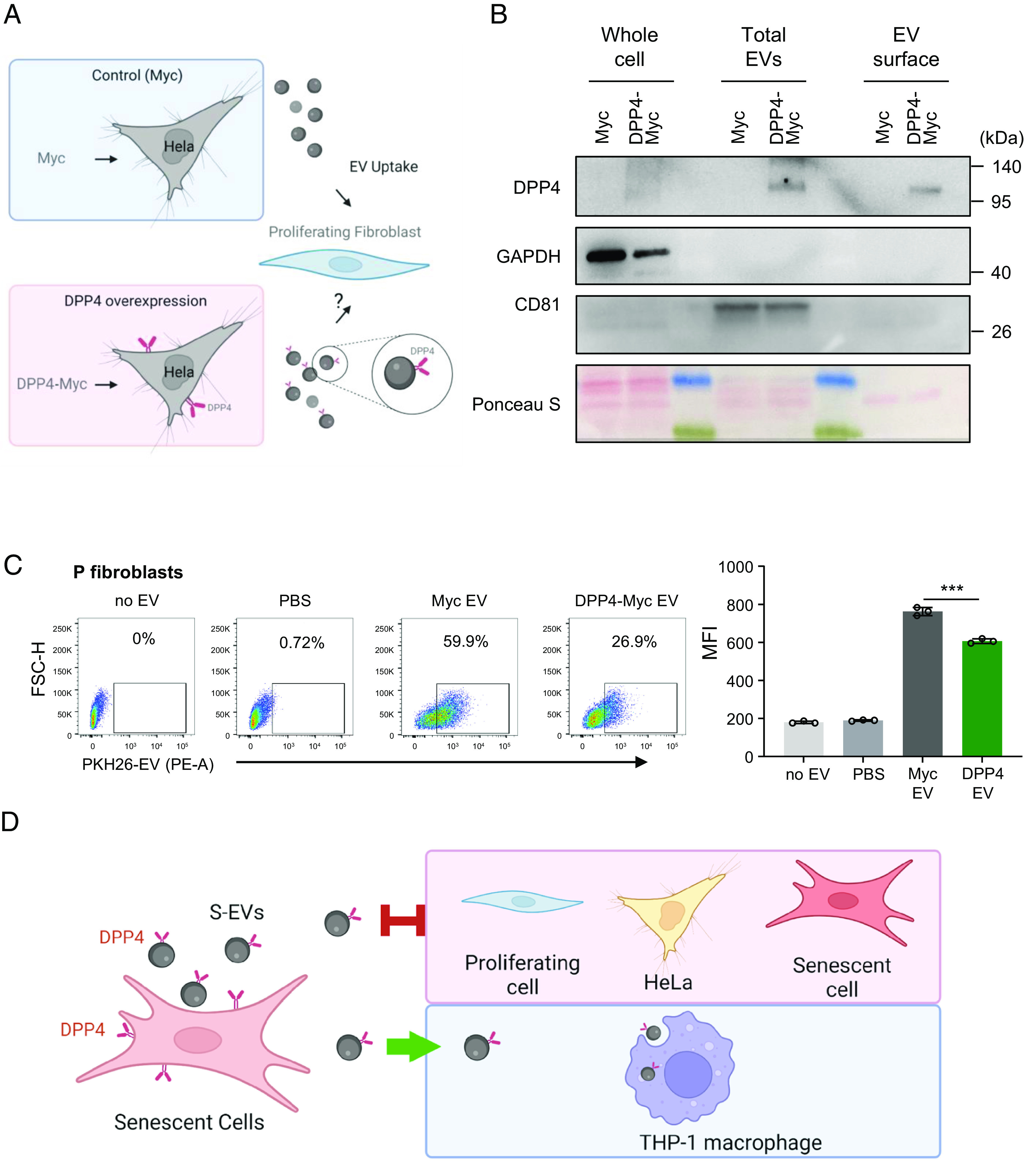
Impact of ectopic lentiviral overexpression of DPP4 on EV internalization efficiency by HeLa cells. (A) Schematic of the experiment: Hela cells were infected with a control lentivirus (Myc) or a lentivirus that overexpressed DPP4 (DPP4-Myc); after selection for 20 d, EVs were then collected for analysis. (B) Western blot analysis of DPP4 levels in HeLa cells expressing in the control group (Myc lentivirus) and the DPP4 overexpressing group (DPP4-Myc virus). Proteins were collected from whole cells, from total EVs derived from the lentivirus transfected cells, and from the surface membrane of the isolated EVs. The levels of housekeeping control protein GAPDH and the EV marker CD81 were also monitored, and overall loading and membrane transfer were monitored by Ponceau S staining. (C) Left, flow cytometric analysis showing the uptake by proliferating (P) WI-38 fibroblasts after incubation with no EVs, with only PBS, and with PKH26-labeled EVs prepared from HeLa cells that were infected with the Myc lentivirus or with the DPP4-Myc lentivirus. Right, bar graphs quantify the uptake of fluorescence (MFI) after the incubations shown on the left. Data are the means ± SEM of three biological replicates; Student’s t test is used for two groups comparison. (D) A proposed model showing that S-EVs bearing DPP4 on the surface showed reduced internalization by fibroblasts (proliferating or senescent) and by HeLa cells, while they are taken up by macrophages. ***P ≤ 0.001.
Discussion
EVs have emerged as key biological structures in cell-to-cell communication and in the elimination of cell contents. Senescent cells release many EVs that likely contribute to the overall senescence phenotype in aging and age-related disorders (14, 16, 26–28). Moreover, it is well recognized that the enhanced secretome of senescent cells (SASP) includes EVs along with many proinflammatory and matrix-remodeling factors. There is also growing evidence that there are variations in the factors and EVs that comprise the SASP depending on the cell type, the type and intensity of the senescence trigger, the time after onset of senescence, and the surrounding environment (6, 13, 29, 30).
We characterized the EVs released by cells rendered senescent by IR, etoposide, or replicative exhaustion. Collectively termed S-EVs, their size and morphology were indistinguishable from those released from proliferating cells (P-EVs), as determined by NTA and electron microscopy. However, more EVs were found in senescent cell culture media than in the culture media of proliferating cells, as reported by others (14, 31), and the protein content of S-EVs differed from that in P-EVs. These findings suggest that S-EVs may harbor important factors that can influence the tissue microenvironment, as well as the organ, system, and organism, and hence they may be relevant to pathologies associated with age.
Although various pathways have been identified for EV uptake by recipient cells, the specific mechanisms that dictate the internalization of EVs by cells remain largely unknown (32). The striking finding that S-EVs were rejected by proliferating fibroblasts and HeLa cells suggests that the composition of S-EVs surface molecules makes them refractory to uptake by other cells and thus they may accumulate in the microenvironment as well as systemically. Along these lines, we found consistently higher levels of EVs in the cultured media from senescent cells compared to proliferating cells and attributed this rise to the increased secretion of EVs by senescent cells. However, it is theoretically possible that senescent and proliferating cells secrete similar numbers of EVs, but P-EVs are readily taken up and removed from the cultured media, while S-EVs are not actively taken up and instead accumulate in the extracellular space. It will be important to elucidate whether it is increased secretion, reduced reuptake, or a combination of the two that explains the accumulation of S-EVs. Thus, characterization of S-EV proteomes, with particular attention to the EV surface proteomes, was pursued in order to shed light on their preferential cellular uptake.
Systematic characterization of the S-EV proteomes revealed the enrichment of several RNA-binding proteins (RBPs) and five other proteins (DPP4, S10AB, MYH9, RF1N1, and TERA) in all three senescence groups studied. Among these, DPP4 and S10AB were also preferentially expressed on the surface of the S-EVs as determined by surface proteome analysis; other EV surface proteins were ANXA1, ANXA6, AT1A1, and EPHB2. We focused our efforts on DPP4, not only because it was most consistently elevated across all senescence groups but also because, overall, it was the most abundant protein showing differential expression. Importantly, reducing DPP4 levels in EVs (by silencing DPP4 expression in producing cells) and suppressing DPP4 function by treatment of EVs with chemical inhibitors caused significant increases in uptake of the resulting EVs (Fig. 5). Conversely, overexpression of DPP4 levels to elevate DPP4 abundance in EVs reduced the uptake of such EVs (Fig. 6). While other proteins selectively present or absent from S-EVs may also contribute to the reduced uptake of S-EVs, DPP4 significantly contributes to this lower internalization.
DPP4 regulates glucose-dependent insulin secretion by breaking down incretins GIP (glucose-dependent insulinotropic polypeptide) and GLP-1 (glucagon-like peptide-1) (33). DPP4-containing EVs were found significantly increased in the circulation of women with gestational diabetes mellitus (DM) (34) and in the urine of persons with DM (35), suggesting links between DM and DPP4-bearing EVs. Although senescent cells accumulate in different tissues in association with diabetes (36), it is not known whether such senescent cells are responsible for the rise in DPP4-bearing EVs. DPP4-containing EVs were also found in plasma from persons with acute myeloid leukemia (AML) and in leukemia cells capable of suppressing colony formation of hematopoietic stem cells (HSCs) (37); whether the source of these EVs were senescent cells (38) is also unknown.
In order to elucidate the mechanism whereby DPP4-bearing EVs are rejected from internalization by other cells, it will be important to first establish how proliferating fibroblasts and HeLa cells take up EVs. We hypothesize that EVs are internalized by endocytosis or by fusion (39, 40). In this scenario, the presence of DPP4, a protease (41), might lead to the cleavage of membrane constituents responsible for docking and fusing the EV. The nature of such putative membrane proteins cleavable by DPP4 is unknown and is the focus of active work in our laboratory.
Interestingly, however, THP-1 macrophages (differentiated from their proliferative monocyte stage) did take up S-EVs. We hypothesize that macrophages in this paradigm engulf EVs, in keeping with their typical function, and do not rely on the presence of membrane detection proteins, as previously described (42, 43). When internalized by phagocytosis, the proteins present on the surface of EVs do not actively influence the process of internalization, so the presence or absence of DPP4 does not affect uptake. Efforts are also underway to employ methods that help us distinguish between fusion and phagocytosis of S-EVs.
Additional experiments are also underway to study the function of P-EVs and S-EVs. Whether the different cargo proteins identified in EVs (Figs. 3 and 4) have a functional impact on the recipient cells remains to be investigated. Future studies will further examine cargo proteins using intensity-based analysis, which offers improved accuracy, precision, and dynamic range (44, 45) and data-independent acquisition (DIA) approaches. Perhaps these approaches will uncover better correlations between the proteomes in whole EVs and on the surface of EVs during senescence. Future studies will also focus on the RNA content of each EV population, including mRNAs and small RNAs (such as microRNAs) that could influence metabolic processes in the recipient cell. Metabolomic analysis can also be used to identify other molecules (e.g., bioactive lipids) present in EVs that may also have biological roles. In addition to biological communication, it is possible that S-EVs represent a form of elimination of cellular components under conditions when autophagy pathways are diminished, as occurs during senescence (46).
Finally, the presence of proteins on the surface of EVs, such as DPP4 on S-EVs, could eventually serve as markers to find specific subsets of EVs in populations of mixed EVs. Techniques such as Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) could then be used to identify and study select subgroups of EVs that have specific surface markers, as has been done in cells (47). Such approaches are likely to have high diagnostic and prognostic value, as efforts escalate to intervene in disease processes exacerbated by senescent cells.
Methods
Cell Culture, Transfection, and Transduction.
The culture conditions of human and rodent cells are explained in SI Appendix, SI Text.
Senescence Assays.
For replicative senescence, WI-38 cells and EVs were collected at the population doubling level (PDL) 20, PDL32, and PDL52 (proliferating, presenescent, and senescent, respectively). For IR-induced senescence, proliferating WI-38 cells (PDL18-20) were irradiated with 10 Gy and harvested on day 8. For etoposide-induced senescence, WI-38 cells were treated with 50 μM etoposide every 72 h, and senescent cells were harvested on day 6. OIS was achieved by transducing a lentiviral vector capable of expressing HRASG12V, (48) alongside transduction of a control lentivirus for 16 h; cells were harvested on day 8. In each case, senescence was evaluated by measuring SA-β-galactosidase activity (Cell Signaling Technology) and by evaluating the levels of the senescent marker p53 by western blot analysis.
Collection and Identification of EVs.
WI-38 fibroblasts and HeLa cells were cultured in 15-cm dishes using DMEM supplemented with 10% “Exosome-Depleted FBS” (Gibco) until they reached 90% confluency. Conditioned medium was collected, and cell viability was measured using the ViaStain AO/PI Staining Solution (Nexcelom). Medium from cultures with >85% viability was used for EVs collection as described (49, 50). To determine the ratio of EVs to cells, medium was collected every 48 h from each group, and the cells and EV concentration were subsequently quantified. Intact EVs were purified from the conditioned medium by first spinning at 500 × g for 10 min at 25 °C and then at 2,000 × g for 20 min to remove debris and apoptotic bodies, and finally at 15,000 × g for 40 min on an SW32Ti Rotor (Beckman Coulter) at 4 °C. The recovered supernatants were subjected to ultracentrifugation (120,000 × g for 16 h at 4 °C) and resuspended in 4 mL of PBS (previously filtered, 0.22-µm filter, Millipore). Additional ultracentrifugation steps were done at 126,000 × g for 1 h at 4 °C, followed by washes and collection of the EV pellets for protein analysis (SI Appendix, Fig. S1A). All EV characterization methods including the Nanoparticle tracking analysis, Zeta potential measurements, TEM, and western blotting are detailed in SI Appendix.
Surface Protein Labeling.
EV surface proteins were biotinylated using the Pierce™ Cell Surface Biotinylation and Isolation Kit (Thermo Fisher Scientific, A44390) following the manufacturer’s protocol with minor modifications. Briefly, ~8 × 109 to 1010 particles were pelleted at 126,000 × g (TLA-110 Rotor, Beckman Coulter; 1 h, 4 °C). The EV-enriched pellet was resuspended and labeled with 0.25 mg/mL Sulfo-NHS-SS-Biotin for 30 min at 4 °C. EVs were centrifuged at 126,000 × g for 1 h and washed using ice-cold Tris Buffered Saline (TBS). EVs and source cell lysates were also prepared and processed by using the Pierce™ Cell Surface Biotinylation and Isolation Kit (Thermo Fisher Scientific, A44390). EZ-Link Sulfo-NHS-SS-Biotin was used to label mammalian cells and EVs. Biotin-labeled EVs were lysed on ice for 30 min, and after lysis, NeutrAvidin Agarose was used to collect the tagged proteins, and dithiothreitol (DTT) was employed in the elution to break down the disulfide bonds in the biotin label. Surface proteins on cells and EVs were isolated for mass spectrometry (MS) and western blot analysis.
Labeling and Uptake of EVs.
To monitor EV uptake by cultured cells, EVs were labeled with PKH26 (Sigma-Aldrich) following the manufacturer’s protocol with modification. PKH26 dye was diluted in 1 mL diluent C to a final concentration of 12 μM as dye solution. Particles (1010) were spun down at 55,000 rpm, gently resuspended in dye solution, incubated on ice for 5 min, and quenched by addition of 2 mL 10% BSA and 5 mL Exosome-Depleted FBS (Gibco). PKH26-labeled EVs were separated from the mixture by placing on a 0.971 M sucrose solution (1.5 mL) followed by ultracentrifugation at 190,000 × g for 2 h at 4 °C (SW41 Ti; Beckman Coulter). The pellets were gently resuspended in filtered PBS. For carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling (C34554 and C24564, Invitrogen), ~1010 EV particles in 1 mL PBS were mixed with 1 mL of 40 μM CFSE solution and incubated at 25 °C for 4 h. The unbound dye was removed by size exclusion chromatography (SEC) or by adding PBS to the Amicon filter (100 kDa) (Millipore Sigma) and centrifugation. EV concentration and labeling efficiency by PKH26 and CFSE were measured by NTA or NanoFCM (Flow NanoAnalyzer). Freshly prepared EVs were used.
To further study the efficiency of uptake of EVs by cultured cells, EVs and cells were cocultured for 48 h and then analyzed by confocal microscopy (Zeiss LSM880) and flow cytometry (Canto II flow cytometer or BD SORP FACSAria Fusion, BD Biosciences, and FlowJo software, v10.2). A PAULA lapse fluorescence microscopy Cell Imager (Leica Microsystems) was used for the time-lapse imaging of PKH26-labeled S-EVs (from PDL53 fibroblasts) internalized in THP-1 macrophages. Images were captured at 1-h intervals for 24 h.
Mass Spectrometry–Based Proteomics of EVs.
Total and surface EV proteins isolation, mass spectrometry processing, and proteomics data analysis are listed in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Dataset S12 (XLSX)
Dataset S13 (XLSX)
Dataset S14 (XLSX)
Dataset S15 (XLSX)
Movie S1.
Acknowledgments
This work was supported by the National Institute on Aging Intramural Research Program of the NIH, and by the National Cancer Institute, Center for Cancer Research, Intramural Research Program of the NIH.
Author contributions
Q.M., P.S., K.A., and M.G. designed research; Q.M., C.C., N.Y., O.G., C.S., C.A.D., M.R., J.L.M., M.D., C.H.S., J.-H.Y., K.W.W., and A.B. performed research; Q.M., N.Y., O.G., N.B., S.B., P.F.J., P.S., and K.W.W. contributed reagents/analytic tools; Q.M., C.C., N.Y., O.G., C.S., M.R., J.L.M., N.B., J.D., M.D., K.M.-M., C.H.S., K.W.W., A.B., K.A., and S.D. analyzed data; and Q.M., O.G., K.W.W., and M.G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Qiong Meng, Email: joanmeng11227@gmail.com.
Supriyo De, Email: supriyo.de@nih.gov.
Myriam Gorospe, Email: myriam-gorospe@nih.gov.
Data, Materials, and Software Availability
All raw mass spec data have been submitted to the MassIVE database (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the dataset identifier MSV000090503 (PXD037225) (51).
Supporting Information
References
- 1.Di Micco R., Krizhanovsky V., Baker D., d’Adda di Fagagna F., Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumari R., Jat P., Mechanisms of cellular senescence: Cell cycle arrest and senescence associated secretory phenotype. Front. Cell Dev. Biol. 9, 645593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacic J. C., Moreno P., Nabel E. G., Hachinski V., Fuster V., Cellular senescence, vascular disease, and aging: Part 2 of a 2-part review: Clinical vascular disease in the elderly. Circulation 123, 1900–1910 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Baar M. P., et al. , Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Cue C., Rueda N., Cellular senescence in neurodegenerative diseases. Front. Cell Neurosci. 14, 16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basisty N., et al. , A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 18, e3000599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzas E. I., Toth E. A., Sodar B. W., Szabo-Taylor K. E., Molecular interactions at the surface of extracellular vesicles. Semin Immunopathol. 40, 453–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanez-Mo M., et al. , Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rai A., Fang H., Claridge B., Simpson R. J., Greening D. W., Proteomic dissection of large extracellular vesicle surfaceome unravels interactive surface platform. J. Extracell Vesicles 10, e12164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G., et al. , Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuentes P., et al. , ITGB3-mediated uptake of small extracellular vesicles facilitates intercellular communication in breast cancer cells. Nat. Commun. 11, 4261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belhadj Z., et al. , A combined “eat me/don’t eat me” strategy based on extracellular vesicles for anticancer nanomedicine. J. Extracell Vesicles 9, 1806444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borghesan M., et al. , Small extracellular vesicles are key regulators of non-cell autonomous intercellular communication in senescence via the interferon protein IFITM3. Cell Rep. 27, 3956–3971.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallis R., Mizen H., Bishop C. L., The bright and dark side of extracellular vesicles in the senescence-associated secretory phenotype. Mech. Ageing Dev. 189, 111263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takasugi M., et al. , Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 8, 15729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melidoni A., Small extracellular vesicles combat senescence. Nat. Rev. Mol. Cell Biol. 21, 498–499 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Casella G., et al. , Transcriptome signature of cellular senescence. Nucleic Acids Res. 47, 7294–7305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frohlich E., The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomedicine 7, 5577–5591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keerthikumar S., et al. , ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 428, 688–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caudron-Herger M., Jansen R. E., Wassmer E., Diederichs S., RBP2GO: A comprehensive pan-species database on RNA-binding proteins, their interactions and functions. Nucleic Acids Res. 49, D425–D436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebauer F., Schwarzl T., Valcarcel J., Hentze M. W., RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Mulcahy L. A., Pink R. C., Carter D. R., Routes and mechanisms of extracellular vesicle uptake. J. Extracell Vesicles 3, 24641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esmaeili A., Alini M., Baghaban Eslaminejad M., Hosseini S., Engineering strategies for customizing extracellular vesicle uptake in a therapeutic context. Stem Cell Res. Ther. 13, 129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Zhu T., Miao H., Liang B., The calcium binding protein S100A11 and its roles in diseases. Front. Cell Dev. Biol. 9, 693262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y., et al. , Functional roles for CD26/DPP4 in mediating inflammatory responses of pulmonary vascular endothelial cells. Cells 10, 3508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon O. H., et al. , Senescence cell-associated extracellular vesicles serve as osteoarthritis disease and therapeutic markers. JCI Insight 4, e125019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boulestreau J., Maumus M., Rozier P., Jorgensen C., Noel D., Mesenchymal stem cell derived extracellular vesicles in aging. Front. Cell Dev. Biol. 8, 107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misawa T., Tanaka Y., Okada R., Takahashi A., Biology of extracellular vesicles secreted from senescent cells as senescence-associated secretory phenotype factors. Geriatr. Gerontol. Int. 20, 539–546 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Terlecki-Zaniewicz L., et al. , Small extracellular vesicles and their miRNA cargo are anti-apoptotic members of the senescence-associated secretory phenotype. Aging (Albany NY) 10, 1103–1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riquelme J. A., et al. , Increased production of functional small extracellular vesicles in senescent endothelial cells. J. Cell Mol. Med. 24, 4871–4876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi E. J., Kil I. S., Cho E. G., Extracellular vesicles derived from senescent fibroblasts attenuate the dermal effect on keratinocyte differentiation. Int. J. Mol. Sci. 21, 1022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathieu M., Martin-Jaular L., Lavieu G., Thery C., Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Brown D. X., Evans M., Choosing between GLP-1 receptor agonists and DPP-4 inhibitors: A pharmacological perspective. J. Nutr. Metab. 2012, 381713 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kandzija N., et al. , Placental extracellular vesicles express active dipeptidyl peptidase IV; levels are increased in gestational diabetes mellitus. J. Extracell Vesicles 8, 1617000 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun A. L., et al. , Dipeptidyl peptidase-IV is a potential molecular biomarker in diabetic kidney disease. Diab. Vasc Dis. Res. 9, 301–308 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Palmer A. K., Gustafson B., Kirkland J. L., Smith U., Cellular senescence: At the nexus between ageing and diabetes. Diabetologia 62, 1835–1841 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namburi S., Broxmeyer H. E., Hong C. S., Whiteside T. L., Boyiadzis M., DPP4(+) exosomes in AML patients’ plasma suppress proliferation of hematopoietic progenitor cells. Leukemia 35, 1925–1932 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim K. M., et al. , Identification of senescent cell surface targetable protein DPP4. Genes Dev. 31, 1529–1534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tkach M., Thery C., Communication by extracellular vesicles: Where we are and where we need to go. Cell 164, 1226–1232 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Kalluri R., LeBleu V. S., The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulvihill E. E., Drucker D. J., Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr. Rev. 35, 992–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sosale A., Saboo B., Sosale B., Saroglitazar for the treatment of hypertrig-lyceridemia in patients with type 2 diabetes: Current evidence. Diabetes Metab. Syndr. Obes. 8, 189–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidson A. D., et al. , Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 12, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox J., et al. , Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blein-Nicolas M., Zivy M., Thousand and one ways to quantify and compare protein abundances in label-free bottom-up proteomics. Biochim. Biophys. Acta 1864, 883–895 (2016). [DOI] [PubMed] [Google Scholar]
- 46.He C., Klionsky D. J., Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoeckius M., et al. , Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu S. K., et al. , A RAS-CaMKKβ-AMPKα2 pathway promotes senescence by licensing post-translational activation of C/EBPβ through a novel 3’UTR mechanism. Oncogene 37, 3528–3548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeppesen D. K., et al. , Reassessment of exosome composition. Cell 177, 428–445.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim K. M., et al. , Mitochondrial RNA in Alzheimer’s disease circulating extracellular vesicles. Front. Cell Dev. Biol. 8, 581882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng Q., et al. , Surfaceome analysis of extracellular vesicles from senescent cells uncovers uptake repressor DPP4. MassIVE. https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000090503. Deposited 10 October 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Dataset S12 (XLSX)
Dataset S13 (XLSX)
Dataset S14 (XLSX)
Dataset S15 (XLSX)
Movie S1.
Data Availability Statement
All raw mass spec data have been submitted to the MassIVE database (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the dataset identifier MSV000090503 (PXD037225) (51).