Abstract
Phytohormones play indispensable roles in plant growth and development. However, the molecular mechanisms underlying phytohormone-mediated regulation of fiber secondary cell wall (SCW) formation in cotton (Gossypium hirsutum) remain largely underexplored. Here, we provide mechanistic evidence for functional interplay between the APETALA2/ethylene response factor (AP2/ERF) transcription factor GhERF108 and auxin response factors GhARF7-1 and GhARF7-2 in dictating the ethylene–auxin signaling crosstalk that regulates fiber SCW biosynthesis. Specifically, in vitro cotton ovule culture revealed that ethylene and auxin promote fiber SCW deposition. GhERF108 RNA interference (RNAi) cotton displayed remarkably reduced cell wall thickness compared with controls. GhERF108 interacted with GhARF7-1 and GhARF7-2 to enhance the activation of the MYB transcription factor gene GhMYBL1 (MYB domain-like protein 1) in fibers. GhARF7-1 and GhARF7-2 respond to auxin signals that promote fiber SCW thickening. GhMYBL1 RNAi and GhARF7-1 and GhARF7-2 virus-induced gene silencing (VIGS) cotton displayed similar defects in fiber SCW formation as GhERF108 RNAi cotton. Moreover, the ethylene and auxin responses were reduced in GhMYBL1 RNAi plants. GhMYBL1 directly binds to the promoters of GhCesA4-1, GhCesA4-2, and GhCesA8-1 and activates their expression to promote cellulose biosynthesis, thereby boosting fiber SCW formation. Collectively, our findings demonstrate that the collaboration between GhERF108 and GhARF7-1 or GhARF7-2 establishes ethylene–auxin signaling crosstalk to activate GhMYBL1, ultimately leading to the activation of fiber SCW biosynthesis.
Transcription factors GhERF108 and GhARF7 interact to establish ethylene–auxin crosstalk, which activates downstream secondary cell wall (SCW)–related genes to facilitate fiber SCW formation in cotton.
IN A NUTSHELL.
Background: Cotton (Gossypium hirsutum) produces natural fibers (unicellular trichomes) on the seed, making it a valuable crop for the worldwide textile industry. Cotton fibers possess a unique secondary cell wall (SCW) that contains more than 90% cellulose but almost no hemicellulose or lignin. The phytohormones ethylene and auxin play indispensable roles in plant growth and development, but the molecular mechanisms for ethylene–auxin-mediated regulation of fiber SCW formation in cotton remain largely underexplored.
Question: We wanted to know if and how ethylene and auxin are involved in regulating cotton fiber SCW development.
Findings: We tested the role of these phytohormones in cotton fiber development using an in vitro cotton ovule culture assay with ethylene and auxin treatments. We also silenced GhERF108, which encodes an AP2/ethylene response factor (ERF) transcription factor from the ERF family, which showed that GhERF108 positively regulates fiber SCW development. Moreover, both ethylene and auxin responses were reduced in GhMYBL1-silenced plants. GhERF108 interacts with 2 auxin response factors (ARF) (GhARF7-1 and GhARF7-2), which respond to auxin signals to promote fiber SCW thickening. GhERF108 and the GhARF7s enhance the activation of GhMYBL1 in fibers where the SCW is thickening. GhMYBL1 directly binds to the promoters of cellulose synthase genes to activate their expression, promoting cellulose biosynthesis and boosting fiber SCW formation. Collectively, our findings demonstrate that the collaboration between GhERF108 and GhARF7s establishes crosstalk in ethylene–auxin signaling to activate GhMYBL1, ultimately leading to the activation of fiber SCW biosynthesis.
Next steps: Scientists aim to improve cotton fiber quality by genetic manipulation. Our work demonstrates that ethylene–auxin signal crosstalk may play a crucial role in improving fiber quality by regulating the expression of the response genes (such as ERF and ARF transcription factors). We hope our work provides the theoretical basis for cotton breeding.
Introduction
The plant cell wall contributes to key characteristics distinguishing plant cells from animal cells by generating a complex extracellular matrix that encompasses every cell inside every plant. At the early active growth stage, the plant cell mainly forms a thin and flexible primary cell wall (PCW), allowing cellular expansion and anisotropic growth. Many specialized plant cells including tracheary and fibrous cells, however, can create secondary cell walls (SCWs) that can not only provide strong mechanical support and transport water needed for plant growth but can also act as protective barriers safeguarding the plant against various external biotic and abiotic stresses. The main components of plant SCW are cellulose, hemicellulose, and lignin, which may vary in composition among different specialized plant cell types (Kumar et al. 2016).
Upland cotton (Gossypium hirsutum), a valuable crop for the worldwide textile industry, produces natural fibers with excellent qualities, owing to the unique characteristics of the highly specialized cotton fiber cells. Cotton fibers are the unicellular trichomes on the seed and possess a unique SCW structure that contains more than 90% cellulose with almost no hemicellulose and lignin, contributing to its unique economic value for the textile industry (Haigler et al. 2012; Han et al. 2013). Moreover, cotton fiber can serve as an excellent single-cell model for investigating the molecular mechanisms behind cell polar growth and cell wall biosynthesis in plants.
Previous studies have revealed that the master switches NAC (from NAM, ATAF1/2, and CUC2) and MYB (such as R2R3-MYB) transcription factors (TFs) mediated transcriptional network governs the SCW biosynthetic program in Arabidopsis (Arabidopsis thaliana) and a portion of other vascular plants (Zhong et al. 2008; Nakano et al. 2015). Recently, studies also demonstrated the roles of NAC and MYB TFs in the SCW biosynthesis of cotton fibers (Zhang et al. 2018; Huang et al. 2019; Fang et al. 2020). For example, the NAC TF GhFSN1 (fiber SCW–related NAC1) acts as a positive regulator in controlling fiber SCW formation by activating its downstream SCW-related genes including GhDUF231L1, GhKNL1, GhMYBL1, GhGUT1, and GhIRX12 in cotton (Zhang et al. 2018). Furthermore, the Class II KNOX TF GhKNL1 could also directly bind to the promoters of GhCesA4-2, GhCesA4-4, GhCesA8-2, and GhMYB46 for modulating cellulose biosynthesis during fiber SCW development in cotton (Gong et al. 2014; Wang et al. 2022). However, whether and how APETALA2/ethylene response factor (AP2/ERF) TFs participate in regulating fiber SCW formation in cotton remains largely underexplored.
As one of the largest TF families in plants, AP2/ERF family members contain at least 1 conserved AP2 domain that is composed of about 60 to 70 amino acids and can be classified according to the number of domains and recognition sequences into 5 groups (AP2, ERF, dehydration-responsive element-binding [DREB] proteins, related to ABI3/VP1 proteins [RAV], and Soloist) (Sakuma et al. 2002; Zhang et al. 2019). In model plants Arabidopsis and tobacco (Nicotiana tabacum), several studies have suggested AP2/ERF TFs to be involved in plant cell wall formation (Seyfferth et al. 2018; Saelim et al. 2019). In Arabidopsis, the AP2/ERF proteins (ERF035 and ERF041) may activate the expression of genes encoding PCW-type cellulose synthase (CesA) subunits and other related genes for enhancing PCW formation (Sakamoto et al. 2018; Nakata et al. 2021). Furthermore, poplar (Populus L.) ERF139 plays a key role not only in suppressing the radial expansion of vessel cells but also in stimulating the accumulation of guaiacyl-type lignin and xylan (Wessels et al. 2019). Similarly, overexpression of a poplar AP2/ERF gene PsnSHN2 in tobacco could significantly alter the SCW formation, leading to a thickened SCW with altered cell wall compositions (Liu et al. 2017). However, the precise molecular mechanisms of how these AP2/ERF TFs regulate SCW development remain unknown and need to be further investigated.
Plant hormones, as key signaling molecules, orchestrate the network of regulatory mechanisms coordinating the SCW development in plants (Didi et al. 2015). Jasmonic acid (JA) induces protoxylem differentiation in Arabidopsis roots (Ghuge et al. 2015). Also, prolonged gibberellin (GA) signaling promotes the thickening of SCW and the lignification of xylem parenchyma by restoring KNAT1's transcriptional regulatory activity in Arabidopsis (Wang et al. 2017; Felipo-Benavent et al. 2018). Furthermore, continuous brassinosteroid (BR) signaling enhances the thickening of SCWs of Casparian strip cells to safeguard against ion toxicity and maintain efficient water transport, effectively alleviating the oxidative stress in cucumber (Cucumis sativus L.) (An et al. 2018).
A well-defined auxin signaling action module known as AUXIN RESPONSE FACTOR7 (ARF7)/ARF19-LATERAL ORGAN BOUNDARIES DOMAIN 29 (LBD29)-NAC SECONDARY WALL THICKENING PROMOTING FACTORs (NSTs) has been shown to participate in SCW deposition (Johnsson et al. 2019; Lee, Du, et al. 2019). The auxin indole-3-acetic acid (IAA) promotes cambium cell division, induces xylem tracheid differentiation, and is involved in controlling PCW expansion and SCW thickening through position effects (Uggla et al. 1996). In cotton, IAA is enriched in elongating fibers and its level further rises at the SCW deposition stage, while higher IAA catabolism has been detected in the SCW development stage than in the fiber elongation stage (Jasdanwala et al. 1977; Nayyar et al. 1989; Gokani and Thaker 2002). Further study revealed that IAA could induce proGhRAC13 activity, which regulates reactive oxygen species (ROS)-triggered cellulose synthesis to promote SCW deposition in cotton fibers (Zhang, Cao, et al. 2020).
Ethylene is mainly associated with plant cell elongation and fleshy fruit ripening but has also been shown to play a key role in SCW development (Carrari and Fernie 2006; Shi et al. 2006; Seyfferth et al. 2018). Exogenous ethylene or its precursor 1-aminocyclopropane-1-carboxylic acid (ACC) can stimulate cambial activity, xylem development, and SCW deposition (Seyfferth et al. 2018). Previous studies have also revealed the potential connections between AP2/ERFs, ETHYLENE INSENSITIVE 3/ETHYLENE INSENSITIVE3-LIKE1 (EIN3/EIL1) TFs and SCW formation (Liu et al. 2017; Felten et al. 2018; Seyfferth et al. 2018; Wessels et al. 2019). Nevertheless, the detailed molecular mechanisms for ethylene-mediated SCW thickening in plants, especially cotton, remain largely underexplored.
In the current study, we provide detailed mechanistic evidence that the AP2/ERF protein GhERF108, as an ERF, physically interacts with the ARFs GhARF7-1 and GhARF7-2 to coregulate the expression of SCW-related key regulator GhMYBL1 to control fiber SCW synthesis in cotton. Thus, these findings provide insights into how AP2/ERF and ARF TFs collaborate to establish ethylene and auxin signaling crosstalk to dictate and fine-tune fiber SCW development in cotton.
Results
Ethylene promotes fiber SCW thickening in cotton
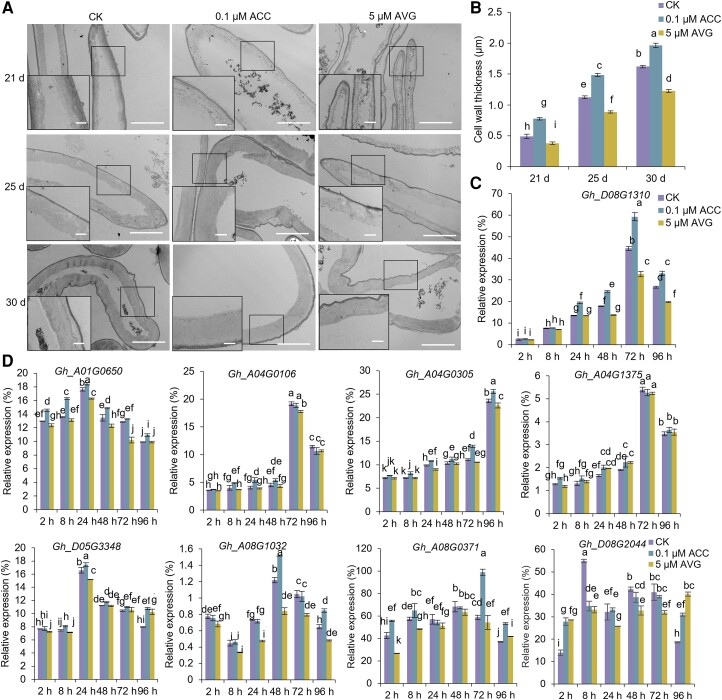
In order to explore the effect of ethylene on SCW formation in cotton (G. hirsutum), we performed the in vitro cotton ovule culture with treatments using the ethylene precursor ACC. After wild-type (WT) ovules were cultured in liquid Beasley and Ting (BT) medium for 15 d, ACC (0.1 µm) was applied in fresh BT medium for further culturing to 21, 25, and 30 d, and the status of fiber SCWs was observed and analyzed by microscopy (see Materials and methods). As shown in Fig. 1, A and B, the cell wall thickness of fibers was significantly increased by ACC treatment, whereas the cell wall thickness of fibers was significantly decreased when treated with 5 µm ethylene inhibitor aminoethoxyvinylglycine (AVG), compared with the fibers without ACC or AVG treatment (CK). These results indicated that ethylene plays a positive role in SCW thickening of cotton fibers.
Figure 1.
Effects of ethylene on cell wall formation of fibers on in vitro cultured ovules of cotton. A) Ultrathin cross-sections of WT fibers by TEM. The WT ovules with fibers collected at 1 DPA were in vitro cultured with 0.1 µm ACC, 5 µm ethylene inhibitor AVG, and without ACC or AVG (CK) for 21, 25, and 30 d, respectively, and then fibers were sectioned and photographed by TEM. Scale bars = 2.0 µm. The rectangle marked at a comparable and magnified position in each fiber cell. Scale bars = 0.5 µm. B) Measurement and statistical analysis of cell wall thickness of fibers on the cultured ovules in A) (n ≥ 50, and at least 30 ovules of 20 bolls per group). C) RT-qPCR analysis of expression of GhERF108 (Gh_D08G13101) in fibers on the cultured ovules applied with ACC or AVG for 2, 8, 24, 48, 72, and 96 h, respectively. D) RT-qPCR analysis of expressions of 8 AP2/ERF genes in fibers on the cultured ovules applied with ACC or AVG for 2, 8, 24, 48, 72, and 96 h, respectively. The mean value and Sd were calculated from 3 biological replicates. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. CK, ACC or AVG free treatment; ACC, treated with 0.1 µm ethylene synthesis precursor ACC; AVG, treated with 5 µm ethylene synthesis inhibitor AVG.
ERF TFs are a class of the AP2/ERF family that can respond to ethylene signals in plants. To investigate whether ethylene affects fiber SCW formation through the ERF genes in cotton, we analyzed the expression of 9 GhERF genes that are highly expressed in fibers at 18-d postanthesis (DPA), the stage of SCW synthesis (Supplemental Fig. S1A; Zafar et al. 2022). As shown in Fig. 1, C and D, when 18 DPA cotton ovules were treated with ACC, expressions of 6 GhERFs (Gh_A04G0305, Gh_D05G3348, Gh_A01G0650, Gh_A08G1032, Gh_A08G0371, and Gh_D08G2044) were significantly altered, but the trend of ACC treatment was not well negatively associated with that of AVG treatment, while 2 genes (Gh_A04G1375 and Gh_A04G0106) were slightly influenced by ethylene. Interestingly, we found that the expression of Gh_D08G1310 was significantly upregulated after 24-h treatment with ACC, reaching a peak at 72-h treatment, and then gradually decreasing in the treated fibers. When AVG was applied, on the contrary, its expression in fibers was decreased with increasing ACC concentrations (Fig. 1C).
Therefore, we focused on Gh_D08G1310, which may play an important role in fiber SCW formation of cotton through ethylene signaling pathway. By analyzing its amino acid sequence, we found this protein contains only 1 AP2 domain and belongs to the ERF subfamily of AP2/ERF TFs and designated it as GhERF108 (Supplemental Fig. S1B). Further experiments revealed that GhERF108 protein is mainly localized in the cell nucleus (Supplemental Fig. S1C) and has transcriptional activation activity (Supplemental Fig. S1D), indicating that GhERF108 is a typical ERF TF. Moreover, GhERF108 is expressed throughout the development of fibers and reaches its peak transcript level in 21 DPA fibers of cotton (Supplemental Fig. S1E), suggesting that it might play an important role in fiber SCW thickening.
GhERF108 functions in fiber SCW formation through the ethylene signaling pathway
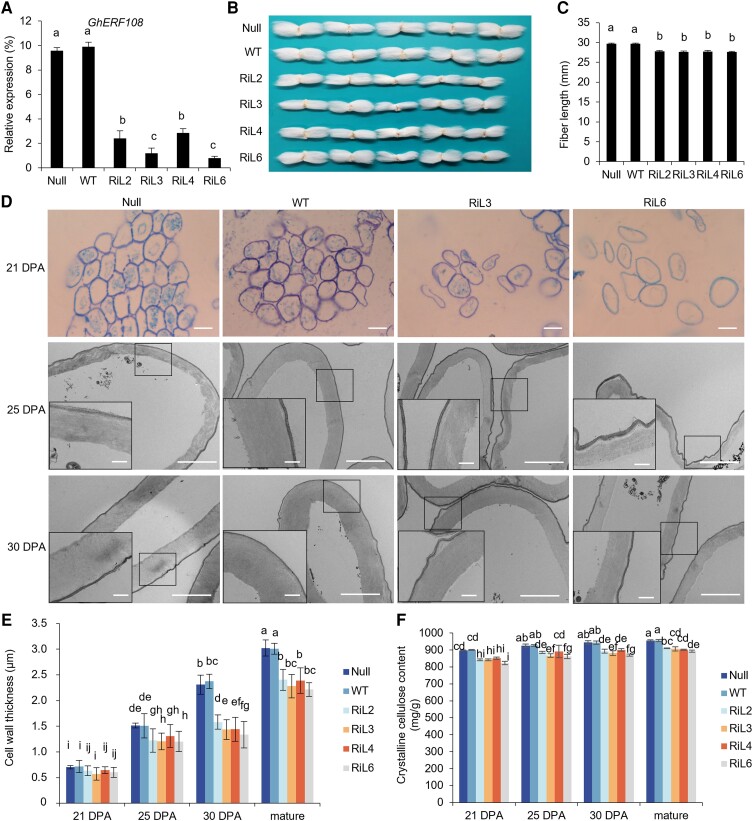
To clarify the role of GhERF108 in fiber development of cotton, we generated the GhERF108 RNA interference (RNAi) transgenic cotton plants under the control of CaMV 35S promoter. We analyzed 4 RNAi transgenic cotton lines (RiL2, RiL3, RiL4, and RiL6) with different levels of GhERF108 expression downregulation (T0 to T4 generations) (Fig. 2A). Among them, DNA gel blot analysis demonstrated that RiL2, RiL3, and RiL6 were independent single-insertion (single-copy) lines (Supplemental Fig. S2A). These GhERF108 RNAi plants exhibited reduced fiber length compared with the controls (transgenic null line [Null] and WT) (Fig. 2, B and C), whereas no variation was found in vegetative growth, seed size, and germination of the transgenic plants relative to the controls (Supplemental Fig. S2, B to E). Further study revealed that the GhERF108 RNAi cotton plants also displayed reduced cell wall thickness in the fibers (Fig. 2, D and E) and decreased crystalline cellulose content in fiber SCWs (T2 generation) (Fig. 2F). However, the content of lignin, another main component of the SCW, was not changed in fiber cell walls, compared with the controls (Supplemental Fig. S2F). Moreover, the phenotypes of the GhERF108 RNAi transgenic lines could be stably inherited in their progeny (T3 and T4 generations) (Supplemental Fig. S2, G to L). These results revealed that GhERF108 plays a vital role in fiber SCW formation of cotton.
Figure 2.
Phenotypic analysis of GhERF108 RNAi transgenic cotton. A) RT-qPCR analysis of GhERF108 expression in 21 DPA fibers of the GhERF108 RNAi transgenic lines (T2 generation) and controls (Null and WT). GhUBI1 was used as an internal control for normalization. B) Comparison of mature fiber length of the GhERF108 RNAi transgenic lines and controls (Null and WT). C) Measurement and statistical analysis of mature fiber length of the independent GhERF108 RNAi transgenic lines and controls (Null and WT) (n ≥ 50, and at least 20 bolls from 15 plants for each independent line). D) Cross-sections of fibers. (Upper) Cross half-thin sections of 21 DPA fibers of the GhERF108 RNAi transgenic lines (RiL3 and RiL6) and controls (Null and WT) by light microscopy. Scale bars = 50 µm. (Bottom) Ultrathin sections of 25 and 30 DPA fibers of the GhERF108 RNAi transgenic lines (RiL3 and RiL6) and controls (Null and WT) by TEM. Scale bars = 2.0 µm. The rectangle marked at a comparable and magnified position in each fiber cell. Scale bars = 0.5 µm. E) Measurement and statistical analysis of fiber cell wall thickness of the GhERF108 RNAi transgenic lines and controls (Null and WT) (n ≥ 50, and at least 50 seeds from 15 plants for each line). F) Measurement and statistical analysis of crystalline cellulose content in fibers of the GhERF108 RNAi transgenic lines and controls (Null and WT). The bolls in the bough located between the third and fourth internodes of cotton plants were randomly collected for fiber phenotypic analysis, and the mean value and Sd were calculated from 3 biological replicates. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. RiL, GhERF108 RNAi transgenic cotton lines.
To further determine whether GhERF108 contributes to divergence in fiber properties, mature fibers from the GhERF108 RNAi cotton plants (T2 and T3 generations) and controls (Null and WT) were collected for fiber quality assay by high-volume instrument (HVI) and advance fiber information system (AFIS). As shown in Table 1, both HVI and AFIS assays indicated that the GhERF108 RNAi transgenic plants had shorter and thinner fibers with lower breaking strength than those of controls (Null and WT). These results further demonstrated that GhERF108 functions in fiber SCW development of cotton.
Table 1.
Comparison of fiber quality parameters between GhERF108 RNAi transgenic cotton and controls (Null and WT) by HVI and AFIS
| Line no.a | HVI | Line no.a | AFIS | ||||
|---|---|---|---|---|---|---|---|
| Fiber length (mm)b | Micronaire value | Fiber breaking strength (g.tex−1)b | Upper quartile fiber length (mm)b | Short fiber rate <12.7 mm/%b | Fineness (m.tex)b | ||
| T2 (year 2020) | |||||||
| Null | 30.24 ± 0.55 | 4.08 ± 0.77 | 30.41 ± 0.75 | Null | 32.30 ± 0.26 | 4.90 ± 1.05 | 187.0 ± 2.00 |
| WT | 30.27 ± 0.57 | 4.12 ± 1.12 | 30.26 ± 1.04 | WT | 32.87 ± 0.40 | 4.17 ± 0.72 | 185.0 ± 2.00 |
| RiL2 | 29.48 ± 0.66c | 4.15 ± 1.03 | 30.14 ± 0.89 | RiL2 | 30.77 ± 0.91c | 3.87 ± 0.32 | 180.3 ± 3.79c |
| RiL3 | 28.48 ± 1.02c | 3.81 ± 0.73 | 29.30 ± 1.0c | RiL3 | 30.03 ± 1.01c | 2.73 ± 0.12c | 172.0 ± 4.36c |
| RiL4 | 29.36 ± 1.15c | 3.81 ± 1.06 | 28.84 ± 1.21c | RiL4 | 30.10 ± 1.14c | 3.67 ± 1.11c | 180.0 ± 4.58c |
| RiL6 | 26.72 ± 1.26c | 3.76 ± 0.79c | 26.93 ± 0.78c | RiL6 | 29.97 ± 0.97c | 3.30 ± 0.56c | 170.7 ± 4.04c |
| T3 (year 2021) | |||||||
| Null | 30. 04 ± 0.56 | 4.09 ± 0.78 | 32.1 ± 1.11 | Null | 32.13 ± 0.68 | 5.03 ± 0.84 | 184.0 ± 2.65 |
| WT | 29.87 ± 1.14 | 3.97 ± 0.99 | 31.9 ± 1.11 | WT | 32.07 ± 0.57 | 5.20 ± 0.72 | 184.7 ± 1.53 |
| RiL2 | 28.86 ± 0.77c | 3.95 ± 1.11 | 30.7 ± 1.05c | RiL2 | 30.57 ± 0.35c | 3.10 ± 0.26c | 178.0 ± 2.65c |
| RiL3 | 27.94 ± 1.12c | 3.90 ± 0.82 | 30.1 ± 0.96c | RiL3 | 30.10 ± 0.44c | 2.73 ± 0.21c | 180.0 ± 1.00c |
| RiL4 | 28.88 ± 1.23c | 4.01 ± 1.11 | 31.0 ± 0.78c | RiL4 | 30.83 ± 0.32c | 3.83 ± 0.85c | 177.7 ± 1.53c |
| RiL6 | 26.99 ± 0.77c | 3.71 ± 1.21c | 30.3 ± 1.04c | RiL6 | 29.97 ± 0.97c | 3.00 ± 0.26c | 171.7 ± 3.79c |
Fibers were handpicked from bolls in the bough located in the third and fourth branches on cotton plants. The transgenic cotton progeny plants (T2 and T3 generations) and controls (Null and WT) were cultivated in the field on the campus of Central China Normal University, Wuhan, China. g.tex−1 means g per tex, n > 100 cotton seeds (at least 12-g fibers per sample for HVI analysis and at least 6-g fibers per sample for AFIS analysis). Null, GhERF108 RNAi transgenic null line; RiL, GhERF108 RNAi lines.
Mean ± Sd. The mean value and Sd were calculated from 3 biological replicates.
There was significant difference (P < 0.05) in fiber length, micronaire value, fiber breaking strength, short fiber rate by weight, and fineness between GhERF108 RNAi transgenic lines and controls (Null and WT).
As GhERF108 is an ERF, we hypothesized that ethylene signaling affects fiber SCW development through GhERF108 protein. To test this hypothesis, we performed in vitro ovule culture experiment again using GhERF108 RNAi transgenic cotton ovules. When the ovules were cultured in vitro in the liquid BT medium with ACC for 25 d, fiber cell wall thickness of the GhERF108 RNAi lines (RiL3 and RiL6) was slightly increased (by 31.9% to 35.5% relative to the control), while the cell wall thickness was slightly reduced (by 31.6% to 34.7% relative to the control) when AVG was applied, compared with the same treatment in WT (Supplemental Fig. S3, A and B). Meanwhile, the expression level of GhERF108 was upregulated in the GhERF108 RNAi fibers after being treated with ACC, but the extent of its upregulation in RNAi fibers was significantly lower than that in WT with the same ACC treatment (Supplemental Fig. S3C). The above data suggested that ACC treatment may partly restore the thinner SCW phenotype of GhERF108 RNAi cotton fibers by promoting the expression of GhERF108, indicating that GhERF108 participates in ethylene signaling for promoting fiber SCW thickening in cotton.
GhERF108 interacts with GhARF7-1 and GhARF7-2 in fibers
To investigate how GhERF108 functions in fiber SCW development and participates in the ethylene signaling pathway, we examined the expression of some genes that may be associated with fiber SCW biosynthesis in the GhERF108 RNAi cotton. As shown in Supplemental Fig. S4, expression of GhCesAs, GhLBDs, GhMYB46, and GhMYBL1 were significantly lower in 21 DPA fibers of the GhERF108 RNAi cotton, compared with those in WT. Therefore, we hypothesized that GhERF108 influences fiber SCW development possibly by regulating these genes. Unfortunately, our experimental results indicated that GhERF108 does not directly activate these genes (Supplemental Fig. S5). Thus, we hypothesized that GhERF108 may form heterodimers to perform its function in cotton.
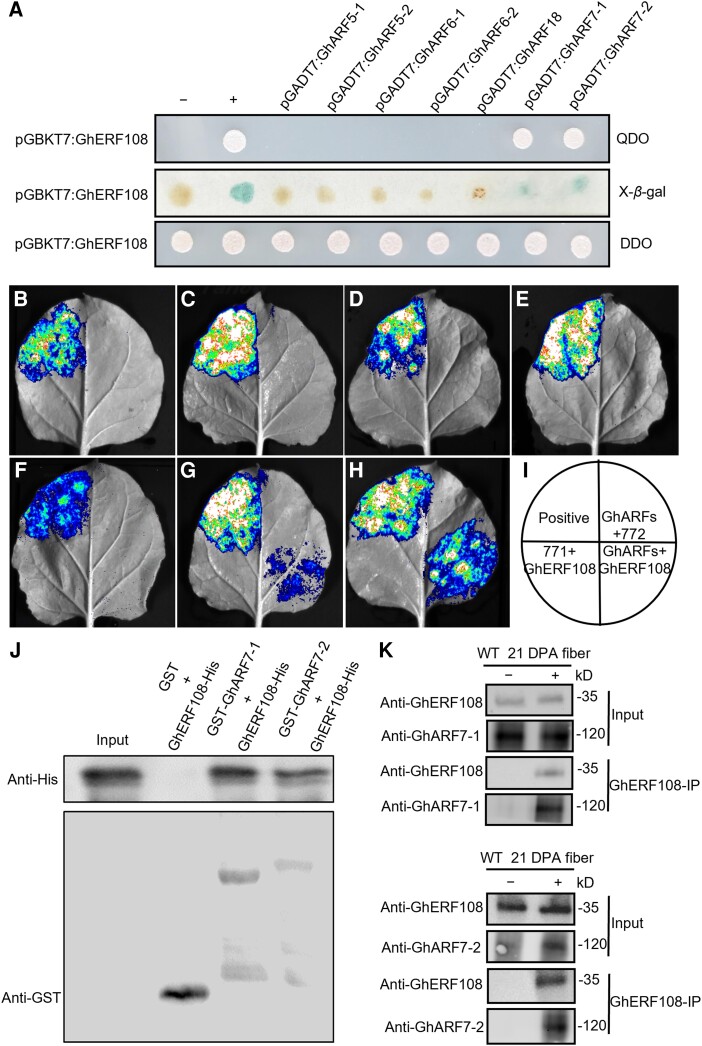
To isolate proteins that interact with GhERF108, we conducted a yeast 2-hybrid (Y2H) assay, employing the truncated GhERF108 protein in which the region with transcriptional activation activity was deleted as bait to screen the library of 21 DPA fiber cDNAs of cotton (Supplemental Fig. S6). The Y2H identified 2 ARFs GhARF7-1 (Gh_A05G0264) and GhARF7-2 (Gh_D07G0132) in the library. In order to further test the accuracy and comprehensiveness of the Y2H sieve library analysis results, we selected 7 GhARFs (GhARF5-1, GhARF5-2, GhARF6-1, GhARF6-2, GhARF18, GhARF7-1, and GhARF7-2) with high expression levels in fibers at the SCW synthesis stage (Supplemental Fig. S7A). Reverse transcription quantitative PCR (RT-qPCR) analysis also confirmed that these 7 GhARF genes are expressed at high levels in fibers undergoing SCW thickening (Supplemental Fig. S7B).
Then, the Y2H assay was employed for detecting the interaction of GhERF108 and these GhARFs. As shown in Fig. 3A, GhERF108 could only interact with GhARF7-1 and GhARF7-2 but did not interact with the others. Likewise, luciferase (LUC) complementation imaging (LCI) assay further verified the interactions of GhERF108 protein with GhARF7-1/7-2 in vivo. As shown in Fig. 3, B to I, strong LUC luminescence was observed in the leaves of Nicotiana benthamiana coexpressing GhERF108-nLUC with GhARF7-1-cLUC or GhARF7-2-cLUC, but no signal was detected in the leaves coexpressing GhERF108 and the other GhARFs. Furthermore, pull-down assays also demonstrated that glutathione S-transferase (GST)-tagged GhARF7-1 and GhARF7-2 could bind specifically to His-tagged GhERF108 in vitro (Fig. 3J). On the basis of an antibody specificity test showing that the GhERF108, GhARF7-1, and GhARF7-2 polyclonal antibodies specifically recognize GhERF108, GhARF7-1, and GhARF7-2 proteins in cotton plants, respectively (Supplemental Fig. S8, A to C), we also examined the interactions of GhERF108 and GhARF7-1 or GhARF7-2 in cotton fibers at the SCW development stage. As shown in Fig. 3K, GhERF108 could interact with GhARF7-1 and GhARF7-2 in cotton fibers. Thus, we can conclude that GhERF108 forms heterodimers with GhARF7-1 and GhARF7-2 during fiber SCW formation of cotton.
Figure 3.
GhERF108 interacts with GhARF7-1 and GhARF7-2. A) Y2H assay of GhERF108 protein interacting with GhARFs, using pGADT7 + pGBKT7-lam as a negative control and pGADT7-T + pGBKT7-53 a as positive control. (Middle) Flash-freezing filter assay of the β-galactosidase activity. (Upper and bottom) Yeast transformants streaked on QDO medium (SD/-Trp/-Leu/-His/-Ade) and DDO medium (SD/-Trp/-Leu), respectively. B to H) LCI assay of GhERF108 protein interacting with GhARF5-1 B), GhARF5-2 C), GhARF6-1 D), GhARF6-2 E), GhARF18 F), GhARF7-1 G), and GhARF7-2 H). LUC luminescence intensities represent their binding activities. I) Schematic diagram of B-H in LCI assay. J) Pull-down assay of GhERF108 protein interacting with GhARF7-1 and GhARF7-2 protein in vitro, respectively. GhERF108-His protein was incubated with GST-GhARF7-1 or GST-GhARF7-2 protein in vitro, using GST protein as a control. The precipitated proteins were analyzed by immunoblotting with anti-His or anti-GST antibody. K) Coimmunoprecipitation (Co-IP) assay of GhERF108 with GhARF7-1 or GhARF7-2 in 21 DPA fibers of WT cotton. Soluble proteins were extracted in 21 DPA fibers of WT before (input) or after (IP) immunoprecipitation with anti-GhERF108 antibody and then detected by immunoblot with anti-GhERF108 (Cys-DGESPTQNGVAPQDS), anti-GhARF7-1 and anti-GhARF7-2 antibodies, respectively.
GhERF108 acts as a coactivator of GhARF7-1/7-2 to regulate GhMYBL1 expression in fibers
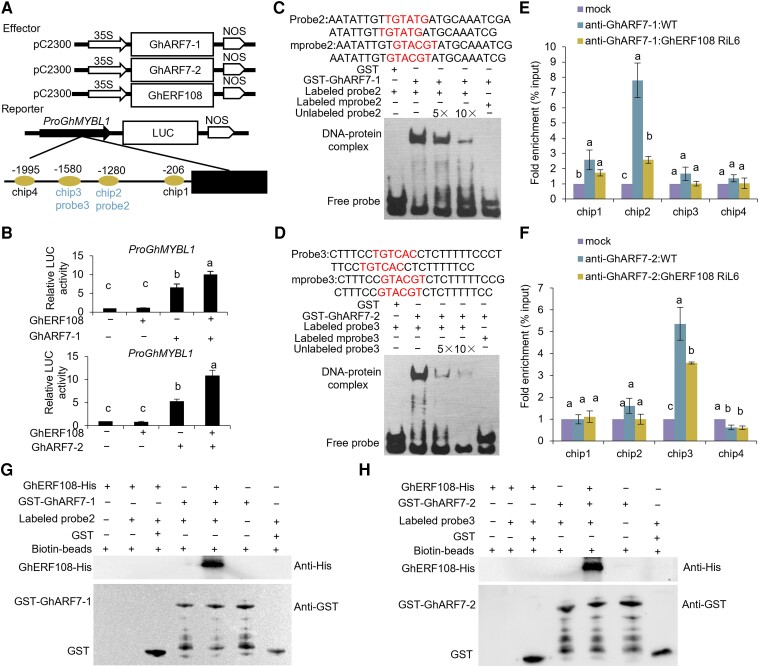
ARFs bind to the auxin response elements (AuxREs) in the promoter regions of early auxin response genes to activate or repress their transcription (Guilfoyle and Hagen 2001). Through bioinformatics analysis and dual-LUC transcriptional activation assay, we screened some genes, which are reported to be involved in fiber SCW formation in cotton, as potential target genes of GhARF7-1 and GhARF7-2. As shown in Supplemental Fig. S9A, the promoter sequences of these candidate genes contain multiple AuxRE cis-elements. GhARF7-1 and GhARF7-2 could activate expression of GhMYBL1 and GhLBD30 (Fig. 4, A and B; Supplemental Fig. S9B). GhMYBL1 is specifically expressed in SCW thickening fibers, and GhLBD30 is preferentially expressed in fibers at SCW developing stage (Supplemental Fig. S10). Therefore, we chose GhMYBL1 as the top candidate target gene of GhARF7-1 and GhARF7-2 for further studies.
Figure 4.
GhERF108 acts as a coactivator of GhARF7-1 and GhARF7-2 to coregulate GhMYBL1 expression. A) Diagram of vectors and AuxREs in GhMYBL1 promoter for Dual-LUC assay, ChIP-qPCR analysis, and EMSA. (Bottom) Horizontal line represents the GhMYBL1 promoter, the ovals represent AuxREs in the promoter, the chip1 to chip4 indicate the fragments detected by ChIP-qPCR, and the probe2 and probe3 indicate the fragments used in EMSA. The numbers along the gene model are relative to the ATG. B) Dual-LUC assay of GhERF108 and GhARF7-1/7-2 on GhMYBL1 promoter activity. The relative LUC activities were normalized to the reference REN LUC. The corresponding effector (+) and empty vector (−) were cofiltrated. The mean value and Sd are calculated from 3 biological replicates. The different letters indicate significant difference (P < 0.05) between the 2 groups according to the Tukey HSD test. C, D) EMSA of GhARF7-1 C) and GhARF7-2 D) binding to the AuxREs in the GhMYBL1 promoter. Biotin-labeled probes were incubated with GST-GhARF7-1 or GST-GhARF7-2 in vitro. Unlabeled probes were used for competition, and biotin-labeled mutated AuxREs were used as negative controls. Normal (up) or mutated AuxREs (down) are shown in C and D. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. E, F) ChIP-qPCR analysis of GhARF7-1 E) and GhARF7-2 F) bind to the GhMYBL1 promoter. Equal amounts of anti-GhARF7-1/7-2 antibodies were used in 21 DPA fibers of both WT and GhERF108 RiL6 transgenic cotton. The ChIP signal is expressed as the percentage of immunoprecipitated DNA in the total input DNA. mock, ChIP without anti-GhARF7-1 and anti-GhARF7-2 but added lgG antibody as negative control; RiL6, the GhERF108 RNAi line. The mean value and Sd are calculated from 3 biological replicates. Tukey HSD test demonstrated that there were significant differences (P < 0.05) between the mock and ChIP groups. G, H) DNA–protein pull-down assay of GhERF108 acting as a coactivator of GhARF7-1 G) and GhARF7-2 H) to bind to the GhMYBL1 promoter. GhERF108-His protein was incubated with GST-GhARF7-1 protein and labeled probe2 or GST-GhARF7-2 protein and labeled probe3 in vitro, using empty GST protein and biotin beads as a negative control. The precipitated proteins were analyzed by immunoblotting with anti-His or anti-GST antibody.
Then, we employed electrophoretic mobility shift assay (EMSA) to analyze whether GhARF7-1 and GhARF7-2 directly bind to GhMYBL1 promoter in vitro. As shown in Fig. 4C, strong signals were detected in the lane with GhARF7-1 protein and the biotin-labeled probe (p2 fragment of the GhMYBL1 promoter region, probe2). Similarly, strong signals were observed in the lane with GhARF7-2 protein and the biotin-labeled probe (p3 fragment of the GhMYBL1 promoter region, probe3) (Fig. 4D). Furthermore, the intensity of the shifted bands was gradually reduced or disappeared, with the increasing concentration of the unlabeled probe2 and probe3. When the AuxREs in probe2 and probe3 were mutated, the shifted bands disappeared (Fig. 4, C and D), indicating that GhARF7-1 and GhARF7-2 could directly bind to the AuxREs in the promoter of GhMYBL1 in vitro. Additionally, we employed chromatin immunoprecipitation (ChIP)-qPCR to detect whether GhARF7-1/7-2 binds to the promoter of GhMYBL1 in vivo. As shown in Fig. 4, E and F, the GhMYBL1 promoter chip2 and chip3 fragments were significantly enriched in ChIP-qPCR assays of GhARF7-1 and GhARF7-2, respectively. These results indicated that GhARF7-1 and GhARF7-2 directly bind to the promoter of GhMYBL1 to activate its expression in cotton fibers.
The expression level of GhMYBL1 was substantially reduced in fibers of GhERF108 RNAi cotton, compared with that in WT (Supplemental Fig. S4). However, GhERF108 could not directly activate the expression of GhMYBL1 (Fig. 4B; Supplemental Fig. S5B). These results prompted us to investigate whether GhERF108 acts as a coactivator of GhARFs for the transcriptional activation of GhMYBL1. To elucidate the biological significance of GhERF108 and GhARF7-1 or GhARF7-2 interactions, we employed the dual-LUC reporter system for the analysis and found that coexpression of GhARF7-1 or GhARF7-2 with ProGhMYBL1:LUC in N. benthamiana leaves led to significantly increased LUC activity, compared with the control. When GhERF108 was additionally coexpressed with GhARF7-1 or GhARF7-2 and ProGhMYBL1:LUC, the GhARF7-1- and GhARF7-2-dependent activation of LUC activity was further enhanced (Fig. 4B). Meanwhile, ChIP-qPCR assay revealed that the enriched chip2 and chip3 fragments of the GhMYBL1 promoter were significantly reduced in fibers of the GhERF108 RNAi transgenic cotton when detected with the same amount of GhARF7-1 or GhARF7-2 antibody, compared with those in WT (Fig. 4, E and F).
To further investigate whether GhERF108 interacts with GhARF7-1 and GhARF7-2 to promote its binding to GhMYBL1 promoter, we performed the in vitro DNA–protein pull-down assay. Specifically, the purified GST-tagged GhARF7-1/7-2 and His-tagged GhERF108 proteins were mixed with magnetic bead-bound, biotin-labeled probe2 or probe3 of GhMYBL1 promoter and then subjected to magnetic pull down of DNA–protein complexes by anti-His antibody. As shown in Fig. 4, G and H, only when both GhERF108 protein and GhARF7-1 or GhARF7-2 proteins were mixed with probe2 or probe3, the His-tagged GhERF108 protein could be detected, suggesting that GhERF108 protein can interact with GhARF7-1 and GhARF7-2 to specifically promote the binding with GhMYBL1 promoter. Collectively, the above data substantiated the idea that GhERF108 acts as a coactivator of GhARF7-1 and GhARF7-2 to regulate GhMYBL1 expression to dictate SCW thickening in fibers of cotton.
Additionally, to further explore the role of GhARF7-1 and GhARF7-2 in fiber development, we employed virus-induced gene silencing (VIGS) to generate GhARF7-1- and GhARF7-2-silenced cotton plants (Supplemental Fig. S11A). We extracted total RNA from 21 DPA fibers of TRV:GhARF7-1 and TRV:GhARF7-2 cotton plants and determined GhARF7-1 and GhARF7-2 transcript levels in fibers of these plants. As shown in Supplemental Fig. S11, B and C, expression of GhARF7-1 and GhARF7-2 was significantly downregulated in fibers of the TRV:GhARF7-1 and TRV:GhARF7-2 plants, respectively, compared with the controls (TRV:00). The suppression of GhARF7-1 and GhARF7-2 expressions in cotton impeded fiber elongation, and mature fiber length of the TRV:GhARF7-1 and TRV:GhARF7-2 plants was shorter than that of the TRV:00 control plants (Supplemental Fig. S11, D and E). Moreover, the cell wall thickness of mature fibers was distinctly less than that of the controls (Supplemental Fig. S11, F and G). We also measured the expression of GhMYBL1 and GhERF108 in fibers of the TRV:GhARF7-1 and TRV:GhARF7-2 cotton and found that GhMYBL1 expression was significantly repressed, while GhERF108 expression was not changed in these plants (Supplemental Fig. S11, H and I). Taken together, the above data indicated that GhARF7-1 and GhARF7-2 play positive roles in fiber development of cotton via activating GhMYBL1 expression.
To further confirm that GhERF108 acts as the coactivator of GhARF7-1 and GhARF7-2 in vivo, we performed ethylene response assays on the GhARF7-1- and GhARF7-2-silenced cotton plants. Compared with the same treatment in TRV:00 control plants, fiber cell wall thickness was only slightly increased (59.4% to 61.3% relative to the control) in the TRV:GhARF7-1 and TRV:GhARF7-2 cotton plants with ACC treatment, while the cell wall thickness was only slightly decreased (60.5% relative to the control) in the TRV:GhARF7-1 and TRV:GhARF7-2 plants treated with AVG (Supplemental Fig. S12, A and B). Notably, the expression fold change of the downstream effectors GhMYBL1 and GhLBD30 was also significantly reduced in the GhARF7-1- and GhARF7-2-silenced plants after ACC treatment, relative to the control TRV:00 plants (Supplemental Fig. S12, C and D). Thus, the above data substantiated the idea that GhERF108 acts as a coactivator of GhARF7-1 and GhARF7-2 to regulate GhMYBL1 expression during SCW thickening in fibers of cotton.
GhMYBL1 is essential for SCW thickening of cotton fibers
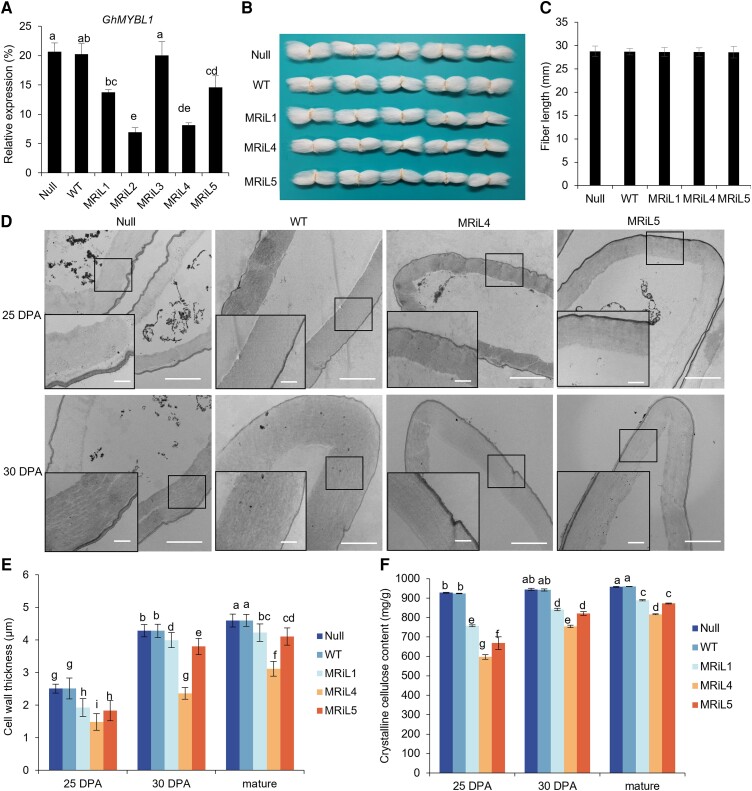
To identify the function of GhMYBL1 in regulating fiber SCW development in cotton, we generated the GhMYBL1 RNAi transgenic cotton under the control of its native promoter. More than 50 transgenic plants from 5 independent transformation lines (T0 generation) were obtained for further studies. We extracted total RNAs from 21 DPA fibers of GhMYBL1 RNAi cotton lines (T1 to T3 generations) and analyzed GhMYBL1 transcript levels in fibers of the transgenic lines by RT-qPCR. As shown in Fig. 5A, the expression of GhMYBL1 was greatly reduced in fibers of the 4 GhMYBL1 RNAi lines, except MRiL3, relative to the controls (Null and WT). On the basis of DNA gel blot analysis results (Supplemental Fig. S13A), 3 independent transgenic lines (MRiL1, MRiL4, and MRiL5) with different GhMYBL1 expression levels were selected for further analysis.
Figure 5.
Phenotypic analysis of GhMYBL1 RNAi transgenic cotton. A) RT-qPCR analysis of GhMYBL1 expression in 18 DPA fibers of the GhMYBL1 RNAi transgenic lines (T2 generation) and controls (Null and WT). GhUBI1 was used as an internal control for normalization. B) Comparison of mature fiber length of the GhMYBL1 RNAi transgenic lines and controls (Null and WT). C) Measurement and statistical analysis of mature fiber length of the independent GhMYBL1 RNAi transgenic lines and controls (Null and WT) (n ≥ 50, and at least 20 bolls from 15 plants for each independent line). D) Ultrathin sections of 25 and 30 DPA fibers of the GhMYBL1 RNAi transgenic lines (MRiL4 and MRiL5) and controls (Null and WT) by TEM. Scale bars = 2.0 µm. The rectangle marked at a comparable and magnified position in each fiber cell. Scale bars = 1.0 µm. E) Measurement and statistical analysis of fiber cell wall thickness of the GhMYBL1 RNAi transgenic lines and WT (n ≥ 50, and at least 50 seeds from 15 plants for each line). F) Measurement and statistical analysis of crystalline cellulose content in fibers of the GhMYBL1 RNAi transgenic lines and controls (Null and WT). The bolls in the bough located between the third and fourth internodes of cotton plants were randomly collected for fiber phenotypic analysis, and the mean value and Sd were calculated from 3 biological replicates. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. MRiL, GhMYBL1 RNAi transgenic cotton lines.
Phenotypic analysis showed that there was no difference in vegetative growth, seed size, seed germination, and fiber length between the RNAi plants and controls (Null and WT) (Fig. 5, B and C; Supplemental Fig. S13, B to E). We further inspected the effects of GhMYBL1 on fiber SCW biosynthesis and found fiber cell wall thickness was remarkably reduced in the GhMYBL1 RNAi lines, compared with that in controls (Null and WT) (Fig. 5, D and E). Moreover, the content of crystalline cellulose was significantly decreased in fiber cell walls of the GhMYBL1 RNAi lines, relative to controls (Null and WT) (Fig. 5F). The above phenotypes of GhMYBL1 RNAi cotton lines could be stably inherited in their progenies (Supplemental Fig. S13, F to J). These data revealed that GhMYBL1 plays a vital role in fiber SCW formation of cotton. Additionally, mature fibers from the GhMYBL1 RNAi cotton plants (T2 and T3 generations) and controls (Null and WT) were collected to determine fiber quality by both HVI and AFIS (see Materials and methods). As shown in Table 2, the GhMYBL1 RNAi transgenic cotton showed lower breaking strength and thinner fibers than those of controls (Null and WT). These results demonstrated that GhMYBL1 functions in fiber SCW development of cotton.
Table 2.
Comparison of fiber quality parameters between GhMYBL1 RNAi transgenic cotton and controls (Null and WT) by HVI and AFIS
| Line no.a | HVI | Line no.a | AFIS | ||||
|---|---|---|---|---|---|---|---|
| Fiber length (mm)b | Micronaire value | Fiber breaking strength (g.tex−1)b | Upper quartile fiber length (mm)b | Short fiber rate <12.7 mm/%b | Fineness (m.tex)b | ||
| T2 (year 2021) | |||||||
| Null | 30.63 ± 0.86 | 5.13 ± 0.12 | 32.37 ± 0.50 | Null | 32.40 ± 0.10 | 4.90 ± 1.05 | 187.0 ± 2.0 |
| WT | 30.27 ± 1.21 | 4.93 ± 0.06 | 31.87 ± 0.15 | WT | 32.90 ± 0.44 | 4.17 ± 0.72 | 185.0 ± 2.0 |
| MRiL1 | 28.73 ± 0.76 | 4.57 ± 0.29 | 30.47 ± 0.51c | MRiL1 | 32.23 ± 0.76 | 4.37 ± 1.27 | 174.0 ± 3.0c |
| MRiL4 | 29.33 ± 0.29 | 4.43 ± 0.15c | 30.63 ± 1.07c | MRiL4 | 32.03 ± 0.31 | 4.17 ± 1.25 | 172.0 ± 6.0c |
| MRiL5 | 30.00 ± 0.62 | 4.37 ± 0.06c | 30.93 ± 0.31c | MRiL5 | 32.47 ± 1.02 | 4.07 ± 0.47 | 160.7 ± 2.5c |
| T3 (year 2022) | |||||||
| Null | 30.97 ± 0.31 | 4.96 ± 0.15 | 32.00 ± 0.36 | Null | 32.16 ± 0.64 | 5.03 ± 0.84 | 184.0 ± 2.6 |
| WT | 31.23 ± 0.27 | 4.93 ± 0.25 | 32.33 ± 0.61 | WT | 32.73 ± 0.85 | 4.93 ± 0.31 | 184.7 ± 1.5 |
| MRiL1 | 30.17 ± 0.54 | 4.26 ± 0.15c | 29.93 ± 0.59c | MRiL1 | 32.57 ± 0.06 | 4.83 ± 0.23 | 169.7 ± 5.1c |
| MRiL4 | 30.47 ± 1.34 | 4.30 ± 0.10c | 29.63 ± 0.83c | MRiL4 | 32.90 ± 0.70 | 4.90 ± 0.96 | 167.7 ± 9.3c |
| MRiL5 | 30.80 ± 0.15 | 4.37 ± 0.02c | 30.13 ± 1.08c | MRiL5 | 32.17 ± 0.51 | 4.37 ± 0.97 | 160.7 ± 3.8c |
Fibers were handpicked from bolls in the bough located in the third and fourth branches on cotton plants. The transgenic cotton progeny plants (T2 and T3 generations) and controls (Null and WT) were cultivated in the field on the campus of Central China Normal University, Wuhan, China. g.tex−1 means g per tex, n > 100 cotton seeds (at least 12-g fibers per sample for HVI analysis and at least 6-g fibers per sample for AFIS analysis). Null, GhMYBL1 RNAi transgenic null line; MRiL, GhMYBL1 RNAi lines.
Mean ± Sd. The mean value and Sd were calculated from 3 biological replicates.
There was significant difference (P < 0.05) in fiber length, micronaire value, fiber breaking strength, short fiber rate by weight, and fineness between GhMYBL1 RNAi transgenic lines and controls (Null and WT).
GhMYBL1 directly binds to the promoters of GhCesA4-1, GhCesA4-2, and GhCesA8-1 to activate their transcription in fibers
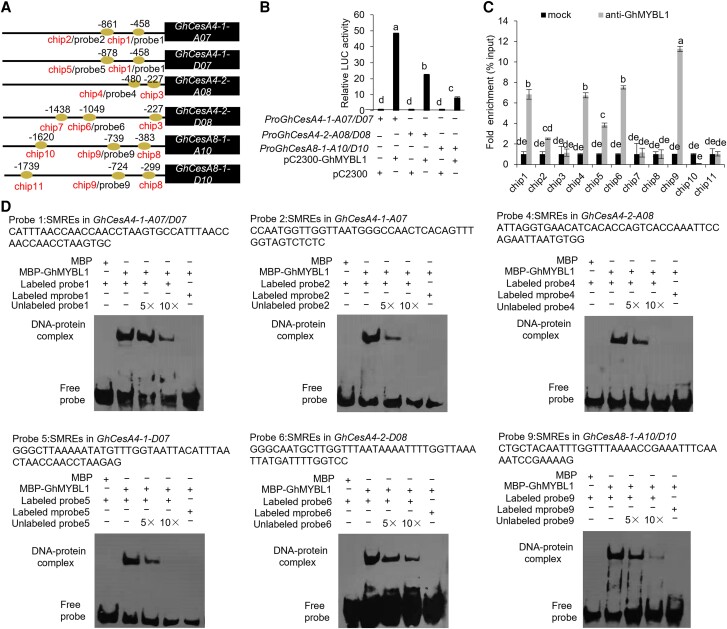
Previous study demonstrated that the R2R3-MYB TF may affect fiber SCW development by regulating cellulose biosynthesis (Sun et al. 2015). To investigate whether GhMYBL1 directly regulates cellulose biosynthesis to affect SCW development in cotton fibers, we analyzed the expression of various SCW-related GhCesA genes (GhCesA4-1-A07, GhCesA4-1-D07, GhCesA4-2-A08, GhCesA4-2-D08, GhCesA7-1-A05, GhCesA7-1-D05, GhCesA7-2-A07, GhCesA7-2-D07, GhCesA8-1-A10, GhCesA8-1-D10, and GhCesA8-2-D05) in developing fibers of the GhMYBL1 RNAi transgenic cotton and WT. As shown in Supplemental Fig. S14, expression level of GhMYBL1 was remarkably downregulated in the GhMYBL1 RNAi fibers. Likewise, expression of GhCesAs significantly declined in the GhMYBL1 RNAi cotton compared with those in WT. To work out how GhMYBL1 regulates the expression of these GhCesA genes, we examined the promoter sequences of these GhCesAs and found several possible secondary wall MYB-responsive elements (SMREs) in GhCesA4-1-A07, GhCesA4-1-D07, GhCesA4-2-A08, GhCesA4-2-D08, GhCesA8-1-A10, and GhCesA8-1-D10 promoters (Fig. 6A). Therefore, we hypothesized that these SCW-related GhCesAs might be the candidate target genes of GhMYBL1.
Figure 6.
GhMYBL1 directly binds to the promoters of SCW-related GhCesAs in cotton. A) A schematic diagram depicting SMREs in the promoters of GhCesAs. Horizontal lines represent the promoters, the ovals represent SMREs, the chip1 to chip11 indicate the fragments detected by ChIP-qPCR analysis, and the probe1 to probe9 indicate the fragments detected by EMSA. B) Dual-LUC assay of GhMYBL1 on the promoter activities of GhCesAs (GhCesA4-1-A07/D07, GhCesA4-2-A08/D08, and GhCesA8-1-A10/D10). The relative LUC activities were normalized to the reference REN LUC, and the corresponding effector (+) and empty vector (−) were cofiltrated. The different letters indicate significant difference (P < 0.05) between the 2 groups according to the Tukey HSD test. C) ChIP-qPCR analysis of GhMYBL1's binding to the promoters of GhCesAs. An anti-GhMYBL1 polyclonal antibody was used for ChIP, followed by qPCR analysis of the bound chromatin from 21 DPA cotton fiber cells. The ChIP signal is expressed as the percentage of immunoprecipitated DNA in the total input DNA. Mock, ChIP with control lgG antibody as a negative control. The mean value and Sd were calculated from 3 biological replicates. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. D) EMSA of GhMYBL1's binding to the SMREs in the promoters of GhCesAs. Biotin-labeled probes were incubated with MBP-GhMYBL1 in vitro. Unlabeled probes were used for competition, and biotin-labeled mutated SMREs were used as negative controls.
To test this hypothesis, we performed dual-LUC assay to verify the function of GhMYBL1 in activating these GhCesA promoters. As shown in Fig. 6B, LUC activity controlled by GhCesA4-1-A07/D07 promoter was elevated remarkably when GhMYBL1 was expressed. Similarly, LUC activities driven by GhCesA4-2-A08/D08 and GhCesA8-1-A10/D10 promoters were significantly enhanced when GhMYBL1 was expressed. Thus, we concluded that GhMYBL1 could directly activate the expression of GhCesA4-1, GhCesA4-2, and GhCesA8-1. Furthermore, we investigated the binding activities of GhMYBL1 to these GhCesA promoters using ChIP-qPCR assay, based on the specificity of GhMYBL1 antibody (Supplemental Fig. S8D). The experimental results showed that GhMYBL1 could directly bind to the promoters of GhCesA4-1-A07, GhCesA4-1-D07, GhCesA4-2-A08, GhCesA4-2-D08, GhCesA8-1-A10, and GhCesA8-1-D10 (Fig. 6C). In addition, EMSA was also used to verify the interactions between GhMYBL1 and these GhCesA promoters in vitro. As shown in Fig. 6D, strong signals were observed in the lane with the GhMYBL1 protein and the biotin-labeled cis-elements. However, the intensity of the shifted bands was gradually reduced with the increasing concentrations of the unlabeled cis-element competitors. When the conserved binding sequences SMREs of the GhCesA promoters were mutated (mprobes), GhMYBL1 protein could no longer bind to the target probes. Taken together, the above data indicated that GhMYBL1 is capable of binding to the promoters of GhCesA4-1-A07/D07, GhCesA4-2-A08/D08, and GhCesA8-1-A10/D10 to activate these genes for promoting cellulose biosynthesis and SCW formation of cotton fibers.
GhMYBL1 integrates ethylene and auxin signaling to upregulate fiber SCW synthesis in cotton
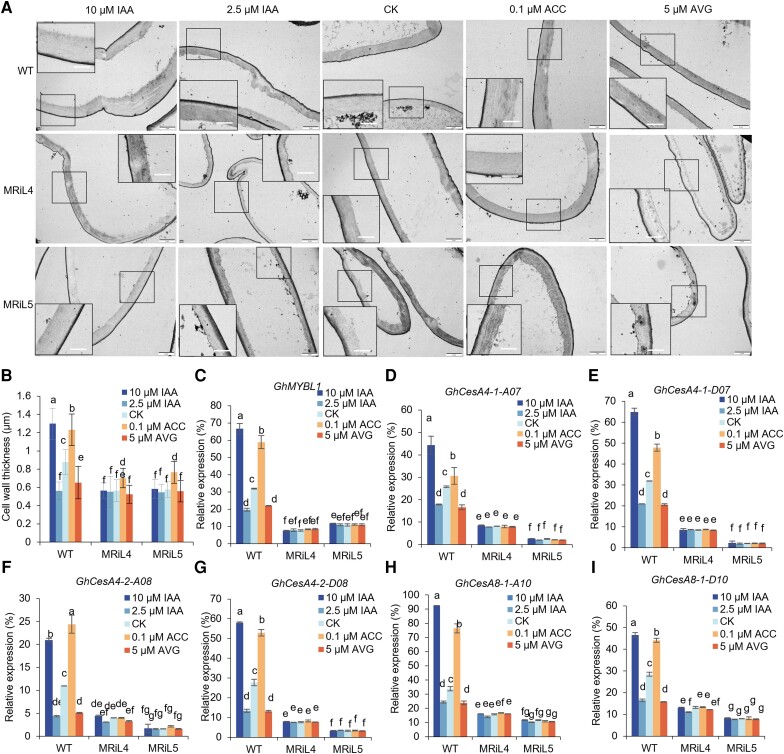
GhARF7-1 and GhARF7-2 belong to the ARF family. To verify whether they regulate fiber SCW synthesis through auxin signaling pathway, we firstly in vitro cultured 1 DPA ovules from the WT cotton in BT medium normally for 15 d and then further cultured in BT medium with 2.5, 5, and 10 µm IAA for 25 d, respectively. The thickness of fiber cells, which were treated with the reduced IAA (2.5 µm), significantly lagged behind that in normal ovule culture condition (CK, 5 µm IAA), whereas the fiber cell wall thickness was remarkably increased when treated with a higher IAA concentration (10 µm IAA) (Fig. 7, A and B). These data indicated that auxin plays a positive role in fiber cell wall thickening of cotton.
Figure 7.
Effects of auxin and ethylene on cell wall formation of fibers on in vitro cultured ovules of GhMYBL1 RNAi cotton. A) Ultrathin cross-sections of fibers. The ovules with fibers (at 1 DPA) collected from the GhMYBL1 RNAi transgenic plants (MRiL4 and MRiL5) and WT were in vitro cultured in BT medium with 2.5 or 10 µm IAA and 0.1 µm ACC or 5 µm AVG for 25 d, and then fibers were sectioned and photographed by TEM. Scale bars = 2.0 µm. The ovules cultured in BT medium with 5 µm IAA were used as control (CK). The rectangle marked at a comparable and magnified position in each fiber cell. Scale bars = 1.0 µm. B) Measurement and statistical analysis of cell wall thickness of fibers on the cultured ovules in A (n ≥ 50, and at least 30 ovules from 20 bolls per group). C) RT-qPCR analysis of GhMYBL1 expression in fibers on the GhMYBL1 RNAi ovules cultured in vitro in A). D to I) RT-qPCR analysis of GhCesAs expressions in fibers on ovules of the GhMYBL1 RNAi lines cultured in vitro in A). The mean value and Sd were calculated from 3 biological replicates. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. MRiL, GhMYBL1 RNAi transgenic cotton lines.
In order to further explore the role of GhMYBL1 in auxin-promoting fiber cell wall thickening, we analyzed the effects of auxin on fiber SCW development of the GhMYBL1 RNAi cotton by in vitro ovule culture similarly as mentioned above. The results confirmed that no matter a lower concentration of IAA (2.5 µm) or a higher concentration of IAA (10 µm) was applied, there was no significant change in fiber cell wall thickness of the GhMYBL1 RNAi lines, compared with that in WT plants under the same IAA concentration treatments (Fig. 7, A and B). Furthermore, the expression of GhMYBL1 was significantly inhibited in fibers treated with a lower IAA concentration (2.5 µm), while the expression of GhMYBL1 was significantly elevated in fibers treated with a higher IAA concentration (10 µm), compared with that in fibers treated with a normal IAA concentration (5 µm) in WT. However, no matter a lower concentration of IAA (2.5 µm) or a higher concentration of IAA (10 µm) was applied, there was no significant change of GhMYBL1 expression in the GhMYBL1 RNAi lines (Fig. 7C). The changes in expression levels of GhCesAs were similar to that of GhMYBL1 in in vitro ovule culture assays (Fig. 7, D to I). Collectively, the above results suggested that GhMYBL1 may play a central role in auxin-promoting fiber SCW thickening process in cotton.
Additionally, we also detected auxin responses in the GhERF108 RNAi plants. The results showed that the fiber cell wall thickness was slightly increased in the GhERF108 RNAi lines when 10 µm IAA was applied, while it was mildly reduced in the GhERF108 RNAi cotton when 2.5 µm IAA was applied, compared with that in WT plants under the same IAA treatment conditions (Supplemental Fig. S3, A and B). Furthermore, the expression of GhERF108 was not significantly changed in the GhERF108 RNAi fibers after being treated with no matter a lower or a higher concentration of IAA (Supplemental Fig. S3C). We also detected the expression of GhARF7-1, GhARF7-2, GhMYBL1, and GhLBD30 in fibers on the in vitro cultured ovules of GhERF108 RNAi cotton. The results showed that expression levels of GhARF7-1 and GhARF7-2 were not changed (Supplemental Fig. S3, D and E), but transcripts of GhMYBL1 and GhLBD30 were significantly downregulated in fibers of the GhERF108 RNAi cotton lines, compared with those in WT, no matter under ethylene or auxin treatments (Supplemental Fig. S3, F and G). Thus, the above data suggested that GhMYBL1 may be also involved in GhERF108-mediated ethylene signaling pathway for fiber SCW thickening of cotton.
To further verify whether GhMYBL1 integrates ethylene and auxin signaling to promote fiber SCW synthesis in cotton, we also detected ethylene response in the GhMYBL1 RNAi plants. As shown in Fig. 7, A and B, when treated with ACC or AVG, the fiber cell wall thickness was slightly changed in GhMYBL1 RNAi plants, compared with that in WT under the same treatment conditions. The expression levels of GhMYBL1 and its target genes GhCesAs also remained almost the same under both auxin and ethylene treatments in GhMYBL1 RNAi lines, compared with these in WT plants under the same treatment conditions (Fig. 7, C to I). On the contrary, expression of GhMYBL1 and GhCesAs was remarkably increased in WT fibers with ACC treatments and significantly decreased in WT fibers treated with AVG, compared with that in fibers without ACC or AVG treatments (CK) (Fig. 7, C to I). Moreover, the expression of GhERF108, GhARF7-1, and GhARF7-2 remained almost the same in fibers of the GhMYBL1 RNAi cotton plants under auxin treatments, compared with those in WT under the same treatment conditions (Supplemental Fig. S15). Taken together, our data revealed that GhMYBL1 integrates ethylene and auxin signaling pathways to positively regulate fiber SCW synthesis in cotton.
Discussion
The biosynthesis of SCW in plants involves the highly coordinated expression of SCW-related genes spatiotemporally regulated by a cascade of TFs (TFs) (Zhong et al. 2008; Du and Groover 2010; Grant et al. 2010; Zhang et al. 2018). To investigate the molecular mechanisms of fiber cell differentiation and development important for improving fiber productivity and quality, it is necessary to identify key regulators or switches whose changes can boost or suppress low hierarchical TFs and downstream cellulose biosynthesis pathway-related genes, as these key molecules can serve as targets of manipulations to increase the economic value of cotton fibers produced for the worldwide textile industry (Walford et al. 2011; Xiao et al. 2018; Tian et al. 2020; Zhang, Cao, et al. 2020).
AP2/ERF TFs regulate plant secondary growth and environmental responses and may have pleiotropic effects on multiple biological processes, especially may potentially serve as high hierarchical TFs in SCW development (Liu, Chen, et al. 2018; Zhang et al. 2019; Feng et al. 2020). For example, Populus ERF139 has been reported to play a role not only in suppressing the radial expansion of vessel elements but also in stimulating the accumulation of guaiacyl-type lignin and xylan (Wessels et al. 2019).
In this study, we have identified AP2/ERF TF GhERF108 as a key regulator of SCW formation in cotton (G. hirsutum). GhERF108 RNAi transgenic cotton plants show shorter fibers with thinner SCWs, compared with controls (Null and WT). Both HVI and AFIS fiber quality analyses have indicated that GhERF108 enhances fiber length, breaking strength and fiber fineness, suggesting GhERF108 as a key factor required for high-quality fibers. Subsequent molecular mechanistic evidence has confirmed that GhERF108 can physically collaborate with GhARF7-1 and GhARF7-2 to activate the downstream target gene GhMYBL1 for fiber development.
GhMYBL1 is a key TF in the NAC-MYB multistage regulatory network that controls SCW development. The transgenic cotton plants with downregulated GhMYBL1 expression show the same phenotype on fiber cell wall thickness as GhERF108 RNAi cotton plants with thinner fiber SCWs and decreased cellulose content. Our results show that GhMYBL1 affects SCW synthesis by directly regulating the expression of GhCesA4-1-A07/D07, GhCesA4-2-A08/D08, and GhCesA8-1-A10/D10 that is associated with cellulose biosynthesis in fibers of cotton. Unlike GhERF108 RNAi transgenic cotton, however, GhMYBL1 RNAi transgenic cotton has not shown the “shorter fiber” phenotype. To assess why they have different phenotypes in fiber length, we have compared their expression patterns in cotton fibers and have found that GhERF108 is expressed throughout the development of fibers (Supplemental Fig. S1), while GhMYBL1 mRNA is specifically accumulated in fibers at the stage of SCW biosynthesis (Supplemental Fig. S10). Therefore, GhERF108 influences both fiber elongation and SCW formation, but GhMYBL1 may function only in fiber SCW synthesis in cotton.
Phytohormones, including auxin, GAs, JA, and BRs, play vital roles in regulating cotton fiber initiation and development (Xiao et al. 2010; Tan et al. 2012; Zhou et al. 2015; Xia et al. 2018). Ethylene also plays an essential role in promoting fiber elongation possibly by upregulating the expression levels of genes encoding sucrose synthase, tubulin, and expansin or by generating hydrogen peroxide, an active ROS, which significantly promotes the elongation of fiber cells in vitro (Shi et al. 2006; Qin et al. 2008). Bioactive GAs may promote fiber SCW deposition by enhancing sucrose synthase expression in cotton (Bai et al. 2014). Auxin regulates the time of initiation of ROS production by directly upregulating expression of GhRAC13, which regulates ROS-triggered cellulose synthesis to affect fiber SCW deposition (Zhang, Cao, et al. 2020). In our study, we have revealed that fiber SCWs in WT were thickened when exogenous IAA was applied at a higher concentration (10 µm), while fiber SCWs in WT were thinner when the concentration of IAA applied was reduced to 2.5 µm, compared with the control (5 µm IAA). However, no matter a lower or higher concentration of IAA was applied, there was no significant change in fiber cell wall thickness of the GhMYBL1 RNAi lines, compared with that in WT under the same treatment conditions (Fig. 7), suggesting that GhMYBL1 may play a crucial role in auxin-promoting fiber SCW thickening of cotton.
Ethylene signaling pathway has been shown to affect SCW deposition in certain woody plants (Liu et al. 2017; Felten et al. 2018; Seyfferth et al. 2018; Wessels et al. 2019). However, its effect on fiber SCW development remains elusive in cotton so far. In this study, we have found that exogenous application of ACC results in cotton fiber SCW thickening, suggesting ethylene as a positive regulator of fiber SCW deposition in cotton. The expression level of GhERF108 (as an ERF) significantly increases in fibers treated with ACC, while AVG inhibits the expression of GhERF108 in fibers (Fig. 1). Similarly, the GhERF108 RNAi transgenic plants display a reduction in fiber cell wall thickness compared with controls (Null and WT), which is corroborated by the evidence of a decreased crystalline cellulose content in the fiber SCWs of these transgenic cotton plants (Fig. 2). Furthermore, we have found that the increase or decrease in fiber cell wall thickness of the GhERF108 RNAi lines is much less than that in WT when ACC or AVG is applied (Supplemental Fig. S3). These data indicate that GhERF108 regulates fiber SCW synthesis through ethylene signaling pathway in cotton.
The relationship between ethylene and auxin signaling pathways during cotton fiber SCW deposition was unexplored, though the individual contributions of ethylene or auxin to plant development have been examined previously. It has been shown that ethylene's effects on root growth are mediated by stimulating auxin synthesis and transport in Arabidopsis. Earlier studies indicate that ethylene stimulates the production and transport of auxin toward root elongation zone where auxin sensitizes cells to the growth inhibitory effects of ethylene (Ruzicka et al. 2007; Stepanova et al. 2007; Swarup et al. 2007). A latter study reveals that hypocotyl cell elongation is regulated by a network involving ethylene and auxin signaling, which is mediated by interactions between ERF72 and ARF6 (Liu, Li, et al. 2018). Furthermore, ERF1 mediates crosstalk between ethylene and auxin biosynthesis during primary root elongation by regulating ANTHRANILATE SYNTHASE α1 (ASA1) expression in Arabidopsis (Mao et al. 2016). RhERF1 and RhERF4 integrate and coordinate ethylene and auxin signals to modulate pectin metabolism in Rosa hybrida (Gao et al. 2019). MAP kinase14 (MPK14)-mediated auxin signaling modulates lateral root development via ERF13-regulated very-long-chain fatty acid (VLCFA) biosynthesis (Lv et al. 2021). In cotton, researchers also reveal the individual roles of ethylene or auxin in fiber development (Kim and Triplett 2004; Ahmed et al. 2018; Xiao et al. 2019).
However, the scientific question regarding a potential crosstalk between ethylene and auxin signaling to coregulate fiber development, especially SCW synthesis, has remained unaddressed. Our data reveal an additive effect of ethylene and auxin on fiber SCW deposition of cotton. GhERF108 expression can be significantly upregulated by ethylene. Although expression of GhARFs remained almost the same in fibers when ACC or IAA was applied, GhARF7-1 and GhARF7-2 link ethylene and auxin signaling pathways via their interactions with GhERF108 to transcriptionally activate GhMYBL1 expression, promoting fiber SCW biosynthesis and deposition in cotton. GhMYBL1 is crucial in auxin-promoting fiber SCW thickening and also involved in GhERF108-mediated ethylene signaling pathway in cotton (Fig. 7; Supplemental Fig. S3). Collectively, we conclude that GhMYBL1 integrates GhERF108-GhARF7-1 or GhERF108-GhARF7-2 complex–mediated ethylene and auxin signaling to promote fiber SCW synthesis in cotton.
Previous studies have shown that the development of lateral roots is strongly dependent on ARF7 and ARF19, which activates the expression of LBD TFs (LBD16, LBD18, and LBD29) in Arabidopsis (Okushima et al. 2007; Lee, Cho, et al. 2019; Zhang, Tao, et al. 2020). Furthermore, the expression of downstream homolog of PR-1 (PRH1) gene depends on the regulation of ARF7 and LBDs. In prh1 mutant, certain α-expansins (EXPAs) genes related to cell elongation are downregulated, suggesting that PRH1 may regulate lateral root elongation by affecting expression of EXPs with cell wall loosening (Zhang, Tao, et al. 2020). In this study, we provide evidence that GhERF108 knockdown in cotton leads to a significantly shorter fiber length as well as a remarkably thinner fiber cell wall thickness. Meanwhile, GhARF7-1 and GhARF7-2 could directly activate the expression of GhLBD30 (Supplemental Fig. S9), while certain GhEXPAs (GhEXPA2D/4D/13A) genes with expression specificity to the elongation stage of cotton fibers are significantly downregulated in fibers of GhERF108 RNAi cotton, compared with WT (Supplemental Fig. S16). Therefore, it is plausible that suppression of GhERF108 expression in cotton may cause the downregulation of EXPA genes, leading to the shortened fiber length of these GhERF108 RNAi cotton. However, the molecular mechanism of GhERF108-regulated fiber elongation in cotton still remains to be explored in details.
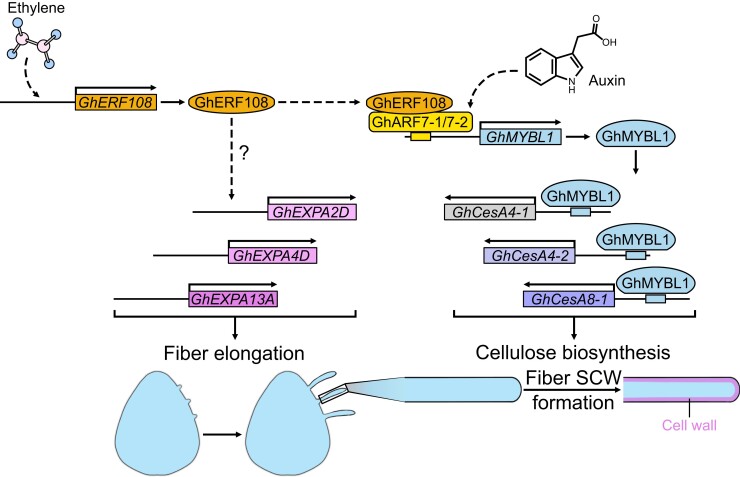
In brief, our data reveal an ethylene and auxin signaling–mediated regulatory mechanism for promoting fiber SCW formation in cotton (Fig. 8). At the stage of early fiber cell development, GhERF108 may participate in regulating fiber cell elongation potentially by influencing the expression of GhEXPA-related genes. During the fiber SCW developmental stage, GhERF108, as an ERF, can be significantly upregulated by ethylene and positively regulate the fiber SCW formation through ethylene signaling pathway. Furthermore, GhERF108 as a transcriptional coactivator can interact with GhARF7-1 and GhARF7-2 to bind to the promoter of GhMYBL1 and enhance the transcriptional activation of GhMYBL1. This cascade of biological intermolecular binding events subsequently promotes enhanced activation signals for low hierarchical regulatory networks associated with cellulose biosynthesis, resulting in the augmented fiber SCW thickening in cotton. On the other hand, GhARF7-1 and GhARF7-2, as ARFs, can respond to auxin signals, activating the expression of GhMYBL1 for fiber SCW thickening.
Figure 8.
A schematic model depicting the detailed molecular mechanism of GhERF108 regulating fiber SCW formation in cotton. GhERF108 could respond to ethylene and GhARFs could respond to auxin. GhERF108 interacts with GhARF7-1 or GhARF7-2 to dictate the crosstalk between ethylene and auxin signaling pathways, leading to the activation of GhMYBL1-mediated cellulose biosynthesis to regulate fiber SCW formation in cotton. Otherwhile, GhERF108 may also influence the expression of GhEXPAs to regulate fiber elongation. Arrows indicate facilitation.
Mechanistically, GhMYBL1 integrates the biological responses of ethylene and auxin signaling pathways and then binds to the SMRE cis-acting elements in the promoters of downstream target genes GhCesA4-1, GhCesA4-2, and GhCesA8-1 to transcriptionally activate their expression for cellulose biosynthesis, thereby promoting fiber SCW formation of cotton. Consistently, all GhERF108 RNAi and GhMYBL1 RNAi cotton plants and TRV:GhARF7-1 and TRV:GhARF7-2 VIGS cotton plants display the reduced fiber cell wall thickness phenotype compared with controls (Null and WT). These comprehensive in vivo data suggest that GhERF108-GhARF7-1 or GhERF108-GhARF7-2 complex–mediated ethylene–auxin signaling crosstalk positively regulates fiber SCW synthesis through activating the downstream GhMYBL1, which promotes GhCesA4-1-, GhCesA4-2-, and GhCesA8-1-controlled cellulose biosynthesis in cotton. Collectively, this study not only provides mechanistic evidence with comprehensive in vivo data on the biological significance of GhERF108-GhARF7-1 or GhERF108-GhARF7-2 complex established ethylene and auxin signaling crosstalk in regulating fiber SCW development of cotton but also identifies GhERF108, GhARF7-1, and GhARF7-2 to be the key regulators of this crosstalk that may have potentials as targets for manipulations to improve cotton fiber quality for worldwide textile industry.
Materials and methods
Plant materials, growth conditions, and cotton transformation
Cotton (G. hirsutum, cv. Coker 312) seeds were surface sterilized with 75% (v/v) ethanol for 1 min and 10% (v/v) H2O2 for 1 to 2 h, followed by washing with distilled water. Then, the sterilized seeds germinated on half-strength MS (½MS) medium under 16-h light (provided by 12-W LED light bulb; 5,000 lux light intensity [100 µE/m2/s])/8-h dark cycles at 27 °C for 5 to 6 d. The transgenic seedlings were transplanted into the field on the campus of Central China Normal University in Wuhan, China for 3 to 4 consecutive years (3 replicates of each line for each year). The seedlings were sown in rows that were 5 m in length with 25 plants, and the rows were spaced 0.8 m apart. Each transgenic line was planted at intervals with the controls (Null and WT). The average planting density was about 45,000/ha, and at least 15 plants of each line were used in the analysis. Additionally, hypocotyls as explants were cut from the 5-d sterile cotton seedlings for Agrobacterium tumefaciens-mediated cotton transformation as described previously (Li et al. 2002).
N. benthamiana seeds were surface-sterilized and sown on ½MS medium in a growth chamber under 16-h light (provided by 12-W LED light bulb; 5,000 lux light intensity [100 µE/m2/s])/8-h dark cycles at 25 °C for 7 d and then transplanted into the soil for 4 to 6 wk for agrobacteria (A. tumefaciens) infiltration.
Construction of vectors
For GhERF108 RNAi vector construction, a 250-bp specific sequence of the GhERF108 gene was cloned into pBlue-script SK vector to create an inverted repeat transgene and then cloned into pMD vector (New England Biolabs, Ipswich, MA, USA) at Sal I and Xba I sites under the control of CaMV 35S promoter (pMD-Pro35S:GhERF108-RNAi). Likewise, approximately 250-bp specific sequence of GhMYBL1 was also used to construct GhMYBL1-RNAi vector under the control of GhMYBL1 promoter, and the fragments (300 bp) in coding sequence (CDS) regions of GhARF7-1 and GhARF7-2 were inserted into TRV2 vector (Testobio, TSPLA10888, Zhejiang, China) to generate TRV:GhARF7-1 and TRV:GhARF7-2 constructs, respectively. Furthermore, protein prokaryotic expression vectors of GhERF108, GhARF7-1, GhARF7-2, and GhMYBL1 were constructed. The CDSs of GhERF108 and GhMYBL1 were separately cloned into pET-32a and pMAL-c2X (New England Biolabs, Ipswich, MA, USA), the part of the CDS of GhARF7-1 or GhARF7-2 containing DNA-binding domain were cloned into pGEX4T vector (GE Healthcare, Pittsburgh, PA, USA), respectively. To explore the regulatory relationship between GhARFs and GhMYBL1, a 1,995-bp promoter sequence of GhMYBL1 was cloned into pGreen II 0800-LUC vector (Serve Life Science, MF3732, Shanghai, China), and the CDSs of GhARFs were cloned into pC2300-C-eGFP vectors (BioVector NTCC, Beijing, China). All primers used to construct the vectors are listed in Supplemental Data Set S1.
In vitro cotton ovule culture
At least 100 cotton (cv. Coker 312) ovules at 1 DPA were randomly collected from at least 20 bolls in 15 plants of each line and cultured in the dark in liquid BT medium supplemented with 0.5 µm gibberellic acid (GA3) and 5 µm IAA at 30 °C for 15 d (Beasley and Ting 1973). Then, the old BT medium was removed from plates, and the ovules were continued culturing to 21, 25, and 30 d in fresh BT medium with extra 0.1 µm ethylene precursor ACC, 5 µm ethylene synthesis inhibitor AVG, 2.5 µm IAA, or 10 µm IAA. The ovules cultured in BT medium with 0.5 µm GA3 and 5 µm IAA were used as controls (CK). After further cultured, the fibers were used to extract RNA (at 21 d) or were cross-sectioned into slices for transmission electron microscopy (TEM) (at 21 to 30 d). The number of ovules counted in each line was at least 50, and each experiment was repeated at least 3 times. Tukey honestly significant difference (HSD) test of 1-way ANOVA was used for statistical analysis (significant difference: P < 0.05).
RNA extraction and RT-qPCR
Total RNA was isolated from 21 DPA fibers of the GhERF108 and GhMYBL1 RNAi transgenic cotton, TRV:GhARF7-1 and TRV:GhARF7-2 VIGS cotton, and WT. In brief, fibers were quickly separated from cotton ovules and frozen and grounded in liquid nitrogen. Then, total RNA was extracted from the fiber samples with the RNAprep Pure Polysaccharide Polyphenol Plant Total RNA Extraction Kit (TIANGEN, Beijing, China) according to the manufacturer's instructions. About 1 to 2 µg of total RNA was reversely transcribed into cDNA using Hiscript III Reverse Transcriptase (Vazyme, Nanjing, China) according to the manufacturer's instructions. The cDNAs were used as templates in qPCR reactions with gene-specific primers, and a cotton polyubiquitin gene GhUBI1 (EU604080) was used as reference. The detailed experimental method refers to Wang et al. (2022). Each RT-qPCR reaction was performed in triplicates, and gene-specific primers used in this analysis are listed in Supplemental Data Set S1.
Test of fiber quality characteristics by HVI and AFIS
Mature fiber samples (at least 12 g of each sample for HVI and at least 6 g of each sample for AFIS) randomly collected from at least 20 bolls in the bough located between the third and fourth internodes in at least 15 GhERF108 RNAi transgenic cotton plants (T2 and T3 generations), 15 GhMYBL1 RNAi transgenic cotton plants (T2 and T3 generations), and controls (Null and WT) were prepared under field planting environment before measurements. Fiber quality characteristics (including fiber length, fiber strength, and micronaire) were determined with a high-volume fiber test system (HVI) (Premier HFT 9000, Premier Evolvics Pvt. Ltd, Coimbatore, India) by the Institute of Agricultural Quality Standards and Testing Technology Research, Hubei Academy of Agricultural Sciences (Wuhan, China). Fiber quality characteristics (including upper quartile fiber length, short fiber rate by weight [<12.7 mm/%], and fiber fineness) were also detected by AFIS in Institute of Cotton Research, Chinese Academy of Agricultural Sciences (Anyang, China). The ginning type of MY98 was adopted for all of the mature fiber samples prior to automated fiber quality analysis by AFIS. Three biological replicates were performed for fiber sample of each line, and the Tukey HSD test of 1-way ANOVA was used for statistical analysis (significant difference: P < 0.05).
Transactivation activity and Y2H assay
The CDS of GhERF108 was cloned into the Y2H vectors pGBKT7 (bait vector) and pGADT7 (prey vector), respectively. pGBKT7-GhERF108 construct was introduced into yeast (Saccharomyces cerevisiae) strain Y187, and pGADT7-GhERF108 construct was transferred into yeast strain AH109. Both transactivation activity assay and mating reaction were performed according to the method described in the previous study (Liu et al. 2020). A Y2H library of 21 DPA cotton fiber cDNAs was constructed using the BD Matchmaker Library Construction and Screening Kit (Clontech) according to the manufacturer's instruction (Zhang et al. 2010). After removing the transactivation region of GhERF108, we screened the target proteins for interaction with GhERF108 and verified the interactions between the screened proteins and GhERF108 by the previous method (Liu et al. 2020). Sequences were analyzed using BLASTP on the tetraploid cotton genome databases (https://www.cottonfgd.org/sequenceserver/). The corresponding gene sequences were determined in the cotton genome. All the primers used in the above methods are listed in Supplemental Data Set S1.
LCI assay
The CDS of GhERF108 was constructed into JW771 vector (BioVector NTCC, Beijing, China) and fused to the amino terminus of LUC (nLUC), while the CDSs of GhARFs were constructed into JW772 vector (BioVector NTCC, Beijing, China) and fused to the carboxy terminus of LUC (cLUC). The GhERF108-nLUC and GhARF-cLUC were coexpressed in N. benthamiana leaves by Agrobacterium-mediated transfer (contain the helper pSoup-P19 plasmid) according to the method described earlier (Gou et al. 2011). Empty vectors JW771 and JW772 were used as negative controls. The LUC luminescence in leaf cells of N. benthamiana was observed under the chemiluminescence imaging system (tanon-5,200 multi) with spraying 0.8 mm fluorescein on the transformed leaves after 48 to 72 h.
In vitro pull-down assay
GhERF108 was fused with His protein tag, and GhARF7-1 and GhARF7-2 were fused to GST tags. GhERF108-His protein was produced in Escherichia coli strain BL21 (WeiDi, Shanghai, China) with the addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37 °C and then purified using Ni-NTA resin. The purified GhERF108-His protein was incubated with glutathione sepharose (GE Healthcare) that was combined with different GST fusion proteins (GST-GhARF7-1 and GST-GhARF7-2) for 2 to 3 h at 4 °C, respectively. Then, the beads were washed 3 times with 1× phosphate buffer saline (PBS). Proteins were eluted from the beads with 100-µL elution buffer (1 × PBS + 10 mm glutathione + 50 mm Tris-Cl, pH 8.0), and loaded onto the SDS–PAGE gel. Gel blots were analyzed using an anti-His antibody (working dilution 1:2,000; Abcam, UK) and anti-GST antibody (working dilution 1:2,000; Abcam, UK), and the empty GST tag was used as the control.
ChIP-qPCR analysis
ChIP-qPCR assay was performed following the protocol described earlier (Liu et al. 2020). Same quality fibers (at 21 DPA) from WT and GhERF108 RNAi transgenic cotton were collected and quickly frozen in liquid nitrogen. After grinding, the powders were transferred to nuclear isolation buffer (10 mm HEPES pH 7.6, 1 m sucrose, 5 mm KCl, 5 mm MgCl2, 5 mm EDTA, 1% [w/v] PVP, 14 mm β-mercaptoethanol, 0.6% [v/v] Triton X-100, and 1× protease inhibitor cocktail) containing formaldehyde at room temperature for 10-min cross-linking, and then 2 m glycine was added to stop the cross-linking. After isolation and sonication, chromatin complexes in the fiber samples were incubated with same amount of purified anti-GhARF7-1, anti-GhARF7-2, or anti-GhMYBL1 polyclonal antibody (20 µg), and specific primer sets were used to analyze the interactions of GhARFs and GhMYBL1 promoter or the interactions of GhMYBL1 and GhCesAs promoters. At the same time, the sample added the same amount of lgG antibody was used as negative control (mock). The above protein antibodies used for ChIP assays were prepared by GL Biochem Ltd (Shanghai, China). In brief, a specific amino acid sequence in each protein (GhARF7-1: Cys-AMRSNSGLIDGDAPPS; GhARF7-2: Cys-SPSSFLSRSQQVP; and GhMYBL1: Cys-QGVLLQDVNVRGEV) was selected to synthesize peptides. Then, the synthetic peptides were injected into rabbits, and the antiserum was collected for peptide affinity purification. The purified antibodies were detected by ELISA at >1:32,000. Three biological replicates were performed for each test, and the Tukey HSD test of 1-way ANOVA was used for statistical analysis (significant difference: P < 0.05). All of the primers used are listed in Supplemental Data Set S2.
EMSA
Complementary oligonucleotides were synthesized and annealed to double-stranded DNAs (95 °C, 5 min, and then slowly cool down to RT and then transfer to 4 °C), and GhMYBL1 and GhCesAs DNA probes were respectively incubated with 300-ng purified GhARFs proteins or GhMYBL1 protein in binding buffer (50 mm KCl, 1 mm DTT, 5 mm MgCl2, 2.5% [v/v] glycerol, and 10 mm Tris-HCl, pH 7.5) for 30 min at 4 °C. DNA–protein complexes were separated by 4% (w/v) nondenaturing PAGE on ice. The biotin-labeled EMSA was performed according to the manufacturer's instruction for the Chemiluminescent Nucleic Acid Detection Module Kit (0020158; Thermo Scientific, Rockford, IL, the USA). For the competition assay, 5-fold and 10-fold amounts of unlabeled DNA fragments (as competitors) or the mutated conserved binding sequences (mprobes) were added to the reactions. All oligonucleotides used in EMSA are listed in Supplemental Data Set S2.
Dual-LUC assay
1.5- to 2-kb sequences of upstream of the ORFs of the candidate target genes (GhLBD18/19/30/38/39, GhCesA4-1/4-2/8-1, GhMYBL1, GhMYB46, GhFSN1, and GhKNL1) were amplified from cotton genomic DNAs using the primer listed in Supplemental Data Set S1. The reporter constructs were produced by inserting promoters of these genes into pGreen II 0800-LUC vector, using the Renilla (REN) LUC driven by the CaMV 35S promoter on the same vector as a reference to calibrate transfection efficiency (Hellens et al. 2005). The CDSs of GhERF108, GhMYBL1, GhARF7-1, and GhARF7-2 were cloned into modified pC2300 vector downstream of CaMV 35S promoter to make effector vectors. The constructs were transformed into A. tumefaciens (strain GV3101) with the helper plasmid pSoup-P19, which prevents gene silencing. Agrobacterium-mediated transformation of N. benthamiana leaves was performed, and promoter activity was detected as described previously (Huang et al. 2019). For each promoter-driven dual-LUC activity assay, 3 independent experiments were performed (at least 6 leaves from 3 N. benthamiana plants for each independent experiment), and the mean and Sd were calculated. The Tukey HSD test of 1-way ANOVA was used for statistical analysis (significant difference: P < 0.05).
TEM
Cotton bolls in the bough located between third and fourth internodes were randomly collected from GhERF108 and GhMYBL1 RNAi transgenic cotton plants and controls (Null and WT). Twenty-one, 25, and 30 DPA and mature fibers from these bolls were embedded by resin and cross-sectioned into slices for TEM analysis as described previously (Li et al. 2011; Qin et al. 2017). In addition, in vitro ovule culture up to 21, 25, and 30 d of fiber materials were also used by the same method to observe their cell wall phenotypes. The mean values and Sd were calculated from 3 biological replicates, and at least 50 fiber cells were counted in each sample. The ImageJ software was used to measure and analyze the data, and the Tukey HSD test of ANOVA was used for statistical analysis (significant difference: P < 0.05).
Assay of crystalline cellulose content in cotton fibers
Bolls in the bough located between third and fourth internodes were randomly collected from GhERF108 and GhMYBL1 RNAi transgenic cotton plants and controls (Null and WT). Fiber samples at different developmental stages (21, 25, and 30 DPA and mature fibers) were used for determination of the crystalline cellulose content as described previously (Li et al. 2018). In brief, 10-mg alcohol-insoluble residue (AIR) per sample was hydrolyzed in 2 m trifluoroacetic acid (TFA) at 121 °C for 90 min and then centrifuged to collect the precipitates. The crystalline cellulose content was detected by hydrolyzing the remains of TFA treatment with Updegraff reagent (acetic acid:nitric acid:water = 8:1:2, v/v/v) at 100 °C for 30 min. After treating the pellets with 60% sulfuric acid for 1 h until completely dissolved, the content was quantified via an anthrone assay (Li et al. 2018). Three biological replicates were performed for each experiment, and the Tukey HSD test of ANOVA was used for statistical analysis (significant difference: P < 0.05).
The statistical analyses results are shown in Supplemental Data Set S3.
Accession numbers
Sequence data from this article can be found in the CottonFGD data libraries under accession numbers_GhERF108 (Gh_D08G13101); GhARF5-1 (Gh_A01G0908); GhARF5-2 (Gh_D03G1293); GhARF6-1 (Gh_D09G0071); GhARF6-2 (Gh_A12G0813); GhARF7-1 (Gh_D07G0132); GhARF7-2 (Gh_A05G0264); GhARF18 (Gh_D09G1405); GhCesA4-1-A07 (Gh_A07G1871); GhCesA4-2-A08 (Gh_A08G0421); GhCesA4-1-D07 (Gh_D07G2083); GhCesA4-2-D08 (Gh_D08G0509); GhCesA7-1-A05 (Gh_A05G3965); GhCesA7-2-A07 (Gh_A07G0322); GhCesA7-1-D05 (Gh_D05G0079); GhCesA7-2-D07 (Gh_D07G0380); GhCesA8-1-A10 (Gh_A10G0327); GhCesA8-2-D05 (Gh_D05G1460); GhCesA8-1-D10 (Gh_D10G0333); GhLBD18 (Gh_D11G0861); GhLBD19 (Gh_A11G0741); GhLBD30 (Gh_D08G1484); GhLBD38 (Gh_A11G1963); GhLBD39 (Gh_A12G0696); GhMYB46 (Gh_A13G1873); GhMYBL1 (Gh_A03G0166); GhFSN1 (Gh_A12G1049); GhKNL1 (Gh_D08G1910); GhEXPA2D (Gh_D10G1238); GhEXPA4A (Gh_A10G1496); GhEXPA4D (Gh_D09G1663); GhEXPA8A (Gh_A13G2631); GhEXPA13A (Gh_A07G079500); and GhEXPA13D (Gh_D07G1058).
Supplementary Material
Contributor Information
Yao Wang, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Yang Li, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Shao-Ping He, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Shang-Wei Xu, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Li Li, College of Biomedicine and Health, Huazhong Agricultural University, Wuhan 430070, China; College of Life Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Yong Zheng, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Xue-Bao Li, Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, China.
Author contributions
X.-B.L. conceived and designed the research; Y.W., Y.L., S.-P.H., and S.-W.X. performed the experiments; Y.W., Y.L., L.L., Y.Z., and X.-B.L. analyzed data; and Y.W., Y.L., L.L., and X.-B.L. wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Characterization of GhERF108.
Supplemental Figure S2 . Phenotypic analysis of GhERF108 RNAi transgenic cotton (T3 generation).
Supplemental Figure S3 . Effects of ethylene and auxin on cell wall formation of fibers on in vitro cultured ovules of GhERF108 RNAi cotton.
Supplemental Figure S4 . RT-qPCR analysis of expression of the SCW-related genes in fibers of GhERF108 RNAi cotton.
Supplemental Figure S5 . Dual-LUC assay of transcriptional activation of the target genes by GhERF108.
Supplemental Figure S6 . Examination of transactivation activity of the truncated GhERF108 protein.
Supplemental Figure S7 . Analysis of expression patterns of GhARF genes in cotton.
Supplemental Figure S8 . Analysis of the antibody specificity of GhERF108, GhARF7-1, GhARF7-2, and GhMYBL1 proteins in cotton.
Supplemental Figure S9 . Dual-LUC assay of transcriptional activation of the target genes by GhARF7-1 and GhARF7-2.
Supplemental Figure S10 . RT-qPCR analysis of expression patterns of GhLBD30 and GhMYBL1 genes in cotton tissues.
Supplemental Figure S11 . Phenotypic analysis of TRV:GhARF7-1 and TRV:GhARF7-2 VIGS cotton.
Supplemental Figure S12 . Effects of ethylene on cell wall formation of fibers on in vitro cultured ovules of TRV:GhARF2-1 and TRV:GhARF7-2 cotton.
Supplemental Figure S13 . Phenotypic analysis of GhMYBL1 RNAi transgenic cotton (T3 generation).
Supplemental Figure S14 . RT-qPCR analysis of expression of SCW-related GhCesA genes in fibers of GhMYBL1 RNAi cotton.
Supplemental Figure S15 . RT-qPCR analysis of expression of GhERF108,GhARF7-1, and GhARF7-2 in fibers on the cultured ovules of GhMYBL1 RNAi cotton.
Supplemental Figure S16 . RT-qPCR analysis of expression of GhEXPAs in fibers of GhERF108 RNAi cotton.
Supplemental Data Set S1 . Primers used in the experiments.
Supplemental Data Set S2 . ChIP-qPCR primers and EMSA probes used in the experiments.
Supplemental Data Set S3 . Summary of statistical analyses.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31871667 and 31471542).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Ahmed M, Shahid AA, Din SU, Akhtar S, Ahad A, Rao AQ. An overview of genetic and hormonal control of cotton fiber development. Pak J Bot. 2018:50(1): 433–443 [Google Scholar]
- An Y-H, Zhou H, Yuan YH, Li L, Sun J, Shu S, Guo S-R. 24-Epibrassinolide-induced alterations in the root cell walls of Cucumis sativus L. under Ca(NO3)2 stress. Protoplasma 2018:255(3):841–850. 10.1007/s00709-017-1187-8 [DOI] [PubMed] [Google Scholar]
- Bai W-Q, Xiao Y-H, Zhao J, Song S-Q, Hu L, Zeng J-Y, Li X-B, Hou L, Luo M, Li D-M, et al. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS One 2014:9(5):e96537. 10.1371/journal.pone.0096537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley CA, Ting IP. Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am J Bot. 1973:60(2):130–139. 10.1002/j.1537-2197.1973.tb10209.x [DOI] [Google Scholar]
- Carrari F, Fernie AR. Metabolic regulation underlying tomato fruit development. J Exp Bot. 2006:57(9):1883–1897. 10.1093/jxb/erj020 [DOI] [PubMed] [Google Scholar]
- Didi V, Jackson P, Hejátko J. Hormonal regulation of secondary cell wall formation. J Exp Bot. 2015:66(16):5015–5027. 10.1093/jxb/erv222 [DOI] [PubMed] [Google Scholar]
- Du J, Groover A. Transcriptional regulation of secondary growth and wood formation. J Integr Plant Biol. 2010:52(1):17–27. 10.1111/j.1744-7909.2010.00901.x [DOI] [PubMed] [Google Scholar]
- Fang S, Shang XG, Yao Y, Li WX, Guo WZ. NST- and SND-subgroup NAC proteins coordinately act to regulate secondary cell wall formation in cotton. Plant Sci. 2020:301:110657. 10.1016/j.plantsci.2020.110657 [DOI] [PubMed] [Google Scholar]
- Felipo-Benavent A, Úrbez C, Blanco-Touriñán N, Serrano-Mislata A, Baumberger N, Achard P, Agustí J, Blázquez MA, Alabadí D. Regulation of xylem fiber differentiation by gibberellins through DELLA-KNAT1 interaction. Development. 2018:145(23):dev164962. 10.1242/dev.164962 [DOI] [PubMed] [Google Scholar]
- Felten J, Vahala J, Love J, Gorzsás A, Rüggeberg M, Delhomme N, Leśniewska J, Kangasjärvi J, Hvidsten TR, Mellerowicz EJ, et al. Ethylene signaling induces gelatinous layers with typical features of tension wood in hybrid aspen. New Phytol. 2018:218(3):999–1014. 10.1111/nph.15078 [DOI] [PubMed] [Google Scholar]
- Feng K, Hou X-L, Xing G-M, Liu J-X, Duan A-Q, Xu Z-S, Li M-Y, Zhuang J, Xiong A-S. Advances in AP2/ERF super-family transcription factors in plant. Crit Rev Biotech. 2020:40(6):750–776. 10.1080/07388551.2020.1768509 [DOI] [PubMed] [Google Scholar]
- Gao YR, Liu Y, Liang Y, Lu JY, Jiang CY, Fei ZJ, Jiang C-Z, Ma C, Gao JP. Rosa hybrida RhERF1 and RhERF4 mediate ethylene- and auxin-regulated petal abscission by influencing pectin degradation. Plant J. 2019:99(6):1159–1171. 10.1111/tpj.14412 [DOI] [PubMed] [Google Scholar]
- Ghuge SA, Carucci A, Rodrigues-Pousada RA, Tisi A, Franchi S, Tavladoraki P, Angelini R, Cona A. The apoplastic copper AMINE OXIDASE1 mediates jasmonic acid-induced protoxylem differentiation in Arabidopsis roots. Plant Physiol. 2015:168(2):690–707. 10.1104/pp.15.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokani SJ, Thaker VS. Physiological and biochemical changes associated with cotton fiber development: IX. Role of IAA and PAA. Field Crops Res. 2002:77(2-3):127–136. 10.1016/S0378-4290(02)00062-X [DOI] [Google Scholar]
- Gong S-Y, Huang G-Q, Sun X, Qin L-X, Li Y, Zhou L, Li X-B. (2014). Cotton KNL1, encoding a Class II KNOX transcription factor, is involved in regulation of fibre development. J Exp Bot. 65(15):4133– 4147. 10.1093/jxb/eru182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J-Y, Felippes FF, Liu C-J, Weigel D, Wang J-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011:23(4):1512–1522. 10.1105/tpc.111.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant EH, Fujino T, Beers EP, Brunner AM. Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 2010:232(2):337–352. 10.1007/s00425-010-1181-2 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Plant Growth Regul. 2001:20(3):281–291. 10.1007/s003440010026 [DOI] [Google Scholar]
- Haigler CH, Betancur L, Stiff MR, Tuttle JR. Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci. 2012:3:104. 10.3389/fpls.2012.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L-B, Li Y-B, Wang H-Y, Wu X-M, Li C-L, Luo M, Wu S-J, Kong Z-S, Pei Y, Jiao G-L, et al. The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 2013:25(11):4421–4438. 10.1105/tpc.113.116970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005:1(1):13. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JF, Guo YJ, Sun QW, Zeng W, Li J, Li XB, Xu WL. Genome-wide identification of R2R3-MYB transcription factors regulating secondary cell wall thickening in cotton fiber development. Plant Cell Physiol. 2019:60(3):687–701. 10.1093/pcp/pcy238 [DOI] [PubMed] [Google Scholar]
- Jasdanwala RT, Singh YD, Chinoy JJ. Auxin metabolism in developing cotton hairs. J Exp Bot. 1977:28(5):1111–1116. 10.1093/jxb/28.5.1111 [DOI] [Google Scholar]
- Johnsson C, Jin X, Xue W, Dubreuil C, Lezhneva L, Fischer U. The plant hormone auxin directs timing of xylem development by inhibition of secondary cell wall deposition through repression of secondary wall NAC-domain transcription factors. Physiol Plant. 2019:165(4):673–689. 10.1111/ppl.12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. Characterization of GhRac1 and GTPase expressed in developing cotton (Gossypium hirsutum L.) fibers. Biochim Biophys Acta. 2004:1679(3):214–221. 10.1016/j.bbaexp.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Kumar M, Campbell L, Turner S. Secondary cell walls: biosynthesis and manipulation. J Exp Bot. 2016:67(2):515–531. 10.1093/jxb/erv533 [DOI] [PubMed] [Google Scholar]
- Lee HW, Cho C, Pandey SK, Park Y, Kim MJ, Kim J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019:19(1):46. 10.1186/s12870-019-1659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Du Q, Zhuo CL, Qi LY, Wang HZ. LBD29-involved auxin signaling represses NAC master regulators and fiber wall biosynthesis. Plant Physiol. 2019:181(2):595–608. 10.1104/pp.19.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Wang SC, Liu YY, Chen J-G, Douglas CJ. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 2011:67(2):328–341. 10.1111/j.1365-313X.2011.04595.x [DOI] [PubMed] [Google Scholar]
- Li X-B, Cai L, Cheng N-H, Liu J-W. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol. 2002:130(2):666–674. 10.1104/pp.005538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang N-N, Wang Y, Liu D, Gao Y, Li L, Li X-B. The cotton XLIM protein (GhXLIM6) is required for fiber development via maintaining dynamic F-actin cytoskeleton and modulating cellulose biosynthesis. Plant J. 2018:96(6):1269–1282. 10.1111/tpj.14108 [DOI] [PubMed] [Google Scholar]
- Liu K, Li YH, Chen XN, Li LJ, Liu K, Zhao HP, Wang YD, Han SC. ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J Exp Bot. 2018:69(16):3933–3947. 10.1093/jxb/ery220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MC, Chen Y, Chen Y, Shin J-H, Mila I, Audran C, Zouine M, Pirrello J, Bouzayen M. The tomato ethylene response factor Sl-ERF.B3 integrates ethylene and auxin signaling via direct regulation of Sl-Aux/IAA27. New Phytol. 2018: 219(2):631–640. 10.1111/nph.15165 [DOI] [PubMed] [Google Scholar]
- Liu YY, Wei MJ, Hou C, Lu TT, Liu LL, Wei HR, Cheng YX, Wei Z. Functional characterization of Populus PsnSHN2 in coordinated regulation of secondary wall components in tobacco. Sci Rep. 2017:7(1):42. 10.1038/s41598-017-00093-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-H, Chen Y, Wang N-N, Chen Y-H, Wei N, Lu R, Li Y, Li X-B. A basic helix-loop-helix protein (GhFP1) promotes fibre elongation of cotton (Gossypium hirsutum) by modulating brassinosteroid biosynthesis and signalling. New Phytol. 2020:225(6):2439–2452. 10.1111/nph.16301 [DOI] [PubMed] [Google Scholar]
- Lv BS, Wei KJ, Hu KQ, Tian T, Zhang F, Yu ZP, Zhang DJ, Su YH, Sang YL, Zhang XS, et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol Plant. 2021:14(.2 ):285-–297.. 10.1016/j.molp.2020.11.011 [DOI] [PubMed] [Google Scholar]
- Mao J-L, Miao Z-Q, Wang Z, Yu L-H, Cai X-T, Xiang C-B. Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 2016:12(1):e1005760. 10.1371/journal.pgen.1005760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata MT, Sakamoto S, Nuoendagula KS, Mitsuda N. Fiber cell-specific expression of the VP16-fused ethylene response factor 41 protein increases biomass yield and alters lignin composition. Front Plant Sci. 2021:12:654655. 10.3389/fpls.2021.654655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci. 2015:6(288):288. 10.3389/fpls.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayyar H, Kaur K, Basra AS, Malik CP. Hormonal regulation of cotton fibre elongation in Gossypium arboreum L. in vitro and in vivo. Biochem Physiol Pflanz. 1989:185(5-6):415–421. 10.1016/S0015-3796(89)80065-4 [DOI] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007:19(1):118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L-X, Chen Y, Zeng W, Li Y, Gao L, Li D-D, Bacic A, Xu W-L, Li X-B. The cotton β-galactosyltransferase 1 (GalT1) that galactosylates arabinogalactan proteins participates in controlling fiber development. Plant J. 2017:89(5):957–971. 10.1111/tpj.13434 [DOI] [PubMed] [Google Scholar]
- Qin Y-M, Hu C-Y, Zhu Y-X. The ascorbate peroxidase regulated by H2O2 and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant Signal Behav. 2008:3(3):194–196. 10.4161/psb.3.3.5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007:19(7):2197–2212. 10.1105/tpc.107.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelim L, Akiyoshi N, Tan TT, Ihara A, Yamaguchi M, Hirano K, Matsuoka M, Demura T, Ohtani M. Arabidopsis group IIId ERF proteins positively regulate primary cell wall-type CESA genes. J Plant Res. 2019:132(1):117–129. 10.1007/s10265-018-1074-1 [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Somssich M, Nakata MT, Unda F, Atsuzawa K, Kaneko Y, Wang T, Bagman AM, Gaudinier A, Yoshida K, et al. Complete substitution of a secondary cell wall with a primary cell wall in Arabidopsis. Nat Plants. 2018:4(10):777–783. 10.1038/s41477-018-0260-4 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002:290(3):998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Seyfferth C, Wessels B, Jokipii-Lukkari S, Sundberg B, Delhomme N, Felten J, Tuominen H. Ethylene-related gene expression networks in wood formation. Front Plant Sci. 2018:9:272. 10.3389/fpls.2018.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y-H, Zhu S-W, Mao X-Z, Feng J-X, Qin Y-M, Zhang L, Cheng J, Wei L-P, Wang Z-Y, Zhu Y-X. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006:18(3):651–664. 10.1105/tpc.105.040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 2007:19(7):2169–2185. 10.1105/tpc.107.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Gong S-Y, Nie X-Y, Li Y, Li W, Huang G-Q, Li X-B. A R2R3-MYB transcription factor that is specifically expressed in cotton (Gossypium hirsutum) fibers affects secondary cell wall biosynthesis and deposition in transgenic Arabidopsis. Physiol Plant. 2015:154(3):420–432. 10.1111/ppl.12317 [DOI] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 2007:19(7):2186–2196. 10.1105/tpc.107.052100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JF, Tu LL, Deng FL, Wu R, Zhang XL. Exogenous jasmonic acid inhibits cotton fiber elongation. J Plant Growth Regulat. 2012:31(4):599–605. 10.1007/s00344-012-9260-1 [DOI] [Google Scholar]
- Tian Y, Du JJ, Wu HT, Guan XY, Chen WH, Hu Y, Ding LY, Li ML, Yang DF, Yang QL, et al. The transcription factor GhMML4_D12 regulates fiber development in cotton through interplay with the WD40-repeat protein WDR in cotton. J Exp Bot. 2020:71(12):3499–3511. 10.1093/jxb/eraa104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C, Moritz T, Sandberg G, Sundberg B. Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci U S A. 1996:93(17):9282–9286. 10.1073/pnas.93.17.9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford SA, Wu Y, Llewellyn DJ, Dennis ES. GhMYB25-like: a key factor in early cotton fibre development. Plant J. 2011:65(5):785–797. 10.1111/j.1365-313X.2010.04464.x [DOI] [PubMed] [Google Scholar]
- Wang GL, Que F, Xu Z-S, Wang F, Xiong A-S. Exogenous gibberellin enhances secondary xylem development and lignification in carrot taproot. Protoplasma 2017:254(2):839–848. 10.1007/s00709-016-0995-6 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Y, Gong S-Y, Qin L-X, Nie X-Y, Liu D, Zheng Y, Li X-B. GhKNL1 controls fiber elongation and secondary cell wall synthesis by repressing its downstream genes in cotton (Gossypium hirsutum). J Integr Plant Biol. 2022:64(1):39–55. 10.1111/jipb.13192 [DOI] [PubMed] [Google Scholar]
- Wessels B, Seyfferth C, Escamez S, Vain T, Antos K, Vahala J, Delhomme N, Kangasjärvi J, Eder M, Felten J, et al. An AP2/ERF transcription factor ERF 139 coordinates xylem cell expansion and secondary cell wall deposition. New Phytol. 2019:224(4):1585–1599. 10.1111/nph.15960 [DOI] [PubMed] [Google Scholar]
- Xiao G, Zhao P, Zhang Y. A pivotal role of hormones in regulating cotton fiber development. Front Plant Sci. 2019:10:87. 10.3389/fpls.2019.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GH, He P, Zhao P, Liu H, Zhang L, Pang CY, Yu JN. Genome-wide identification of GhARF gene family reveals GhARF2 and GhARF18 are involved in cotton fibre cells initiation. J Exp Bot. 2018:69(18):4323–4337. 10.1093/jxb/ery219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y-H, Li D-M, Yin M-H, Li X-B, Zhang M, Wang Y-J, Dong J, Zhao J, Luo M, Luo X-Y, et al. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol. 2010:167(10):829–837. 10.1016/j.jplph.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Xia X-C, Hu Q-Q, Li W, Chen Y, Han L-H, Tao M, Wu W-Y, Li X-B, Huang G-Q. Cotton (Gossypium hirsutum) JAZ3 and SLR1 function in jasmonate and gibberellin mediated epidermal cell differentiation and elongation. Plant Cell Tiss Org. 2018:133(2):249–262. 10.1007/s11240-018-1378-9 [DOI] [Google Scholar]
- Zafar MM, Rehman A, Razzaq A, Parvaiz A, Mustafa G, Sharif F, Mo H, Youlu Y, Shakeel A, Ren M. Genome-wide characterization and expression analysis of Erf gene family in cotton. BMC Plant Biol. 2022:22(1):134. 10.1186/s12870-022-03521-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Tao WQ, Sun RQ, Wang JX, Li CL, Kong XP, Tian HY, Ding ZJ. PRH1 mediates ARF7-LBD dependent auxin signaling to regulate lateral root development in Arabidopsis thaliana. PLoS Genet. 2020:16(2):e1008044. 10.1371/journal.pgen.1008044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang G-Q, Zou D, Yan J-Q, Li Y, Hu S, Li X-B. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018:217(2):625–640. 10.1111/nph.14864 [DOI] [PubMed] [Google Scholar]
- Zhang M, Cao HZ, Xi J, Zeng JY, Huang J, Li BX, Song SQ, Zhao J, Pei Y. Auxin directly up regulates GhRAC13 expression to promote the onset of secondary cell wall deposition in cotton fibers. Front Plant Sci. 2020:11:581983. 10.3389/fpls.2020.581983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ji A, Xu Z, Luo H, Song J. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol Biol. 2019:100(1-2):83–93. 10.1007/s11103-019-00845-7 [DOI] [PubMed] [Google Scholar]
- Zhang Z-T, Zhou Y, Li Y, Shao S-Q, Li B-Y, Shi H-Y, Li X-B. Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J Exp Bot. 2010:61(12):3331–3344. 10.1093/jxb/erq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008:20(10):2763–2782. 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang Z-T, Li M, Wei X-Z, Li X-J, Li B-Y, Li X-B. Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotech J. 2015:13(2):269–280. 10.1111/pbi.12275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.