Abstract
It was demonstrated that Epstein-Barr virus (EBV)-encoded small RNAs (EBERs) were nonessential for B-lymphocyte growth transformation. We revisited this issue by producing a large quantity of EBER-deleted EBV by using an Akata cell system. Although the EBER-deleted virus efficiently infected B lymphocytes, its 50% transforming dose was approximately 100-fold less than that of the EBER-positive EBV. We then engineered the genome of EBER-deleted virus and generated a recombinant virus with the EBER genes reconstituted at their native locus. The resultant EBER-reconstituted EBV exhibited restored transforming ability. In addition, lymphoblastoid cell lines established with the EBER-deleted EBV grew significantly more slowly than those established with wild-type or EBER-reconstituted EBV, and the difference between the growth rates was especially highlighted when the cells were plated at low cell densities. These results clearly demonstrate that EBERs significantly contribute to the efficient growth transformation of B lymphocytes by enhancing the growth potential of transformed lymphocytes.
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus that establishes life-long latent infections in B lymphocytes following the primary infection (10, 18). EBV is associated with various malignancies, such as Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, gastric carcinoma, and lymphoproliferative diseases in immunosuppressed patients. In these tumor cells, the EBV genome is maintained as an ∼170-kb plasmid form and expresses a limited number of viral gene products (10, 18).
EBV readily infects human resting B cells in vitro and transforms B cells into indefinitely proliferating lymphoblastoid cell lines (LCLs). LCLs express only 11 EBV gene products, including 6 EBV nuclear antigens (EBNA-1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, and EBNA-LP), 3 latent membrane proteins (LMP-1, LMP-2A, and LMP-2B), BamHI A rightward transcripts, and 2 EBV-encoded small RNAs (EBER1 and EBER2) (10, 18). Among them, EBNA-1, EBNA-2, EBNA-3A, EBNA-3C, EBNA-LP, and LMP-1 have been reported to be essential for growth transformation, whereas EBNA-3B, LMP-2A, LMP-2B, BamHI A rightward transcripts, and EBERs are not essential (10).
EBERs are the most abundant viral transcripts observed in cells with an EBV latent infection. There are two EBERs, EBER1 and EBER2. EBER1 and EBER2 are nonpolyadenylated, untranslated RNAs of 167 and 172 nucleotides long, respectively, and transcribed by RNA polymerase III (3, 19). DNA sequence analyses of various EBV isolates have revealed that the EBER genes are structurally very highly conserved (1). The high levels of expression and sequence conservation strongly suggest that EBERs have some important biological functions.
Several reports have described growth-stimulatory roles of EBERs (12, 21, 30). EBERs are known to make complexes with several cellular proteins, such as RNA-activated protein kinase PKR (2), ribosomal protein L22 (28), and La antigen (15). Therefore, EBERs may exert various biological effects through their direct interactions with these cellular proteins. For example, the significance of the interaction between EBERs and PKR, a key mediator of the antiviral effect of alpha interferon (IFN-α), has been well studied (2, 17, 22). We have shown that, in Burkitt's lymphoma cells, EBERs confer resistance to IFN-α-induced apoptosis by directly binding to PKR and inhibiting its phosphorylation (17). Alternatively, EBERs can also induce the expression of cellular growth factors. We have also shown that EBERs induce the expression of interleukin 10 (IL-10) in B cells (11), IL-9 in T cells (31), and insulin-like growth factor 1 in epithelial cells (9), each of which acts as an autocrine growth factor. However, the mechanism by which EBERs induce the expression of such growth factors remains unclear.
Regarding the role of EBERs in the process of EBV-induced B-cell transformation, Swaminathan et al. demonstrated that EBERs were not essential for the immortalization of B lymphocytes or for the replication of the virus (26). They tried to restore the transformation defect of the EBV P3HR-1 strain, having a deletion of the essential transforming gene encoding EBNA-2, by letting it homologously recombine with an EBER-deleted [EBER(−)] EBV DNA fragment spanning the EBNA-2 locus. Their attempt resulted in the obtaining of LCLs harboring only EBER-deleted recombinant viruses, indicating that EBERs are dispensable for B-cell transformation (26). They also demonstrated that the LCLs carrying EBER-deleted EBV episomes were permissive for producing progeny viruses and that the progeny virus still transformed B lymphocytes. However, they failed to produce a large quantity of pure EBER-deleted EBV. Instead, a cocultivation method was used to passage the EBER-deleted EBV from primary LCLs to secondary LCLs. Therefore, the titer of EBER-deleted EBV to transform B lymphocytes has never been determined by using a pure recombinant virus.
We hypothesized that the reported growth-stimulatory role of EBERs could also contribute to the process of EBV-mediated B-cell transformation. Therefore, we set out to reinvestigate the role of EBERs by generating recombinant EBVs lacking the EBER genes using EBV Akata strain. It is advantageous to use the Akata cell system, as one can establish isogenic cell lines to produce large quantities of various pure recombinant viruses (24). We utilized cre-mediated site-specific recombination to keep the EBER loci of the recombinant genomes free of marker genes. We generated three isogenic Akata cell lines harboring recombinant EBVs with the EBER genes intact, the EBER genes deleted, or the EBER genes reconstituted to the EBER-deleted genome [EBER(+)]. Our data provide the first direct evidence that EBERs significantly contribute to efficient B-cell transformation.
MATERIALS AND METHODS
Cells and culture.
The EBV-negative Akata cell line (23) was grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (40 U/ml), and streptomycin (50 μg/ml) at 37°C in 5% CO2. The Akata cell line harboring NeorEBV (24) was maintained in complete culture medium containing 700 μg of G418 (Sigma-Aldrich Fine Chemicals, St. Louis, Mo.)/ml.
Plasmids.
The EBER1 and EBER2 genes are located within the BamHI C fragment of Akata EBV DNA. pUC-A/CΔPstI/EcoK contained the part of the BamHI C fragment of Akata EBV genome (corresponding to the EcoRI-PstI fragment spanning nucleotides 4163 through 9699 of EBV B95-8 strain). A hygromycin resistance marker gene driven by simian virus 40 early enhancer and promoter was cloned into the BamHI site of pBS246 (Life Technologies) to make pBS246/hyg so that the hygromycin resistance marker gene was flanked by two loxP sites. The AccI fragment of pUC-A/CΔPstI/EcoK spanning the EBER genes (corresponding to nucleotides 6612 through 7263 of EBV B95-8 strain) was replaced with the NotI fragment of pBS246/hyg to make pEBER-KO. pEBER-KOwas used as a targeting construct to generate EBER knockout EBV. The NotI fragment of pBS246/hyg was inserted into the SacI site (nucleotide 6285 of EBV B95-8 strain) of pUC-A/CΔPstI/EcoK to make pEBER-KI, which was used as a targeting construct to generate EBER knock-in EBV. Other known EBV open reading frames were not affected by this insertion. pSG-Cre was constructed by cloning the blunted XhoI-MluI fragment of pBS185 (Life Technologies), containing a cre recombinase gene, into the blunted BglII site of pSG5 (Stratagene).
Generation of homologously recombined EBVs.
Twenty micrograms of the targeting construct was digested with BstPI and EcoRV and introduced into EBV-infected Akata cells by an electroporation method as described previously (33). Transfected cells were cultured for 2 days and plated in 96-well plates at 104 cells per well in medium containing 150 μg of hygromycin B (Wako)/ml for selection. Half of the culture medium was replaced with fresh medium containing hygromycin B every 5 days. To obtain cell clones harboring the EBER knockout EBV, a total of 483 hygromycin-resistant cell clones that arose from 3,840 wells were screened for the existence of homologously recombined EBV episomes by Southern blotting. To obtain cell clones harboring the EBER knock-in EBV, a total of 507 hygromycin-resistant cell clones that arose from 2,400 wells were screened. Both screenings resulted in obtaining one clone each which harbored homologously recombined EBV episomes.
Virus infection and drug selection.
Virus production was induced by cross-linking surface immunoglobulin G (IgG) as described previously (24). Briefly, Akata cells harboring EBV episomes (2 × 106 cells) were resuspended with 1 ml of fresh medium containing 0.5% rabbit anti-human IgG (DakoCytomation, Carpiteria, Calif.) and incubated for 6 h to induce lytic replication. The culture medium was replaced with fresh medium, and 2 days later, the culture supernatant was harvested. The culture supernatant was filtered by 0.45-μm-pore-size membrane and used as a virus solution. For infection, EBV-negative Akata cells (106) were suspended in 1 ml of virus solution and incubated at 37°C for 90 min with continuous gentle mixing. After the removal of the virus solution, the infected cells were cultured for 2 days. These cells were then plated in 96-well plates at 104 cells per well in medium containing 150 μg of hygromycin B (Wako)/ml or 700 μg of G418 (Sigma)/ml for selection. Half of the culture medium was replaced with appropriate medium every 5 days. Drug-resistant clones were screened by Southern blotting to obtain cell clones harboring recombinant EBVs of interest.
Excision of the hygromycin resistance gene by cre expression.
To excise the hygromycin resistance gene from EBV genomes, Akata cells carrying episomes of either EBER(−)HygrEBV or EBER(+)HygrEBV were transfected with 20 μg of pSG-Cre by an electroporation method. At 2 days posttransfection, the cells were induced for virus production, and the culture supernatants were harvested 2 days later. EBV-negative Akata cells were then infected with the culture supernatants, and the infected cells were selected by 700 μg of G418/ml. Drug-resistant cell clones with EBV episomes with the Hygr gene deleted were screened by Southern blotting.
Southern blotting.
DNA was extracted by using the AquaPure genomic DNA isolation kit (Bio-Rad) according to the manufacturer's instructions. Extracted DNA was digested with BamHI, separated by electrophoresis in 0.8% agarose gel, and transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech). The HincII-SacI fragment (1,115 bp) of the BamHI C fragment of Akata EBV DNA (nucleotides 5176 through 6285 of EBV B95-8 strain) was used as a probe. Probe labeling was carried out using the AlkPhos direct labeling kit (Amersham), and the signals were detected with CDP-Star detection reagent (Amersham).
Northern blotting.
Total RNA was isolated using TRI reagent (Sigma) according to the manufacturer's instructions. The RNAs (7.5 μg each) prepared from each cell clone were electrophoresed in 1% agarose gel containing 0.66 M formaldehyde and transferred to a Hybond N+ nylon membrane. 18S rRNAs were visualized by ethidium bromide staining. Probes were prepared by PCR amplification using primers 5′-AGGACCTACGCTGCCCTAGA-3′ and 5′-AAAACATGCGGACCACCAGC-3′ for EBER1 (27) and primers 5′-AGGACAGCCGTTGCCCTAGTGGTTTCG-3′ and 5′-AAAAATAGCGGACAAGCCGAATACC-3′ for EBER2 (17). PCR conditions were described previously (17). The EcoRI-SacI fragment of the BamHI C fragment (nucleotides 6286 through 7315 of B95-8 strain EBV) (designated the EKS fragment; 1,045 bp [12]) was also used as a probe for detecting both of the EBERs. Probe labeling and detection procedures were the same as those for the Southern blot analysis.
Quantification of virions in the culture supernatants.
Akata cells (7.5 × 107) were induced for virus production by surface IgG cross-linking, and virus solutions were prepared as described above. Thirty milliliters of the virus solutions were ultracentrifuged by using an SW28 rotor (Beckman) at 15,000 rpm (30,000 × g) at 4°C for 2 h. The pelleted virions were resuspended in 400 μl of phosphate-buffered saline (without calcium and magnesium), and 200 μl of lysis buffer (75 mM Tris-HCl [pH 8.0], 25 mM EDTA, 3% sodium dodecyl sulfate) and 3 μl of proteinase K (20 mg/ml) were added. After the lysates were incubated at 37°C for 1 h, they were extracted twice with phenol and once with a chloroform-isoamyl alcohol (24:1) mixture. The viral DNAs were then ethanol precipitated and dissolved in 30 μl of H2O. Five microliter aliquots of each viral DNA were digested with EcoRI and analyzed by 0.8% agarose gel electrophoresis. The amounts of produced viruses were estimated by checking the band intensities of the ethidium bromide-stained DNAs.
Immunofluorescence.
EBV-negative Akata cells (106) were infected with equivalent amounts of recombinant EBVs at 37°C for 90 min with continuous gentle mixing. The medium was replaced with fresh medium, and the infected cells were cultured for 2 days. The infected cells were smeared on a slide glass and fixed with a 1:1 mixture of acetone and methanol. The expression of EBNA proteins was visualized by the anticomplement immunofluorescence method. A human serum reactive to EBNA proteins was used as a primary antibody, and fluorescein isothiocyanate-conjugated anti-human C3C antibody (DaKo) was used as a secondary antibody.
Transformation assay.
Cord blood lymphocytes were plated at a density of 1.2 × 105 to 1.3 × 105 cells per well in 96-well plates, and they were infected with serial 10-fold dilutions of each recombinant virus. The infected lymphocytes were cultured while half of the culture medium was replaced with fresh medium every 5 days. The number of wells with proliferating lymphocytes was counted at 6 weeks postinfection, and the titers for transformation (50% transforming doses [TD50/ml]) were calculated by the Reed-Muench method (4).
LCL growth assay.
Serially diluted LCLs were plated in 96-well plates, and they were cultured while half of the culture medium was replaced with fresh medium every 5 days. The number of wells with proliferating cells was counted at 4 weeks after starting cell culture.
Immunoblotting.
Immunoblot analysis was performed as previously described (32). Whole cell lysates were prepared from various cell clones (105 cells), and they were separated in sodium dodecyl sulfate-polyacrylamide (6% for EBNAs and 10% for LMP-1) gels. The expression of EBNA proteins was detected with a human serum reactive to six EBNA proteins as a primary antibody and a horseradish peroxidase-conjugated anti-human IgG (Amersham) as a secondary antibody, while the expression of LMP-1 was detected using monoclonal antibody S12 (specific for LMP-1) and peroxidase-conjugated anti-mouse IgG (Amersham).
RT-PCR.
Reverse transcription PCR (RT-PCR) analysis was carried out to investigate the expression of EBV latent genes and BamHI Q promoter-initiated mRNA as described previously (8, 11). Total cellular RNA was isolated using TRI reagent (Sigma) and then treated with DNase I (Sigma) according to the manufacturer's instructions. cDNA synthesis was performed using Molony murine leukemia virus reverse transcriptase (Invitrogen) and 100 pmol of random primer (Takara) at 37°C for 60 min, followed by heat inactivation of reverse transcriptase at 94°C for 10 min. The cDNA samples were then subjected to 30 cycles of PCR in a thermal cycler. The PCR products (7.5 μl each) were electrophoresed in 2% agarose gel, transferred to Hybond N+ (Amersham), and specifically amplified DNA was detected by the Gene Images 3′-oligolabeling module (Amersham) and ECL detection reagents (Amersham). The quality of RNA was checked by PCR amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA.
RESULTS
Generation of recombinant EBV lacking EBER genes.
We generated a recombinant EBV lacking both EBER1 and EBER2 genes to examine the roles of EBERs in B-lymphocyte growth transformation. Figure 1 shows a schematic diagram of our strategy to generate the EBER-deleted EBV. Akata cells having episomes of NeorEBV (24), a recombinant EBV carrying a neomycin resistance gene inserted into the BXLF1 open reading frame, were used for the EBER-targeting. The BXLF1 open reading frame is nonessential for infection and replication, and the phenotype of NeorEBV is indistinguishable from that of wild-type EBV (24).
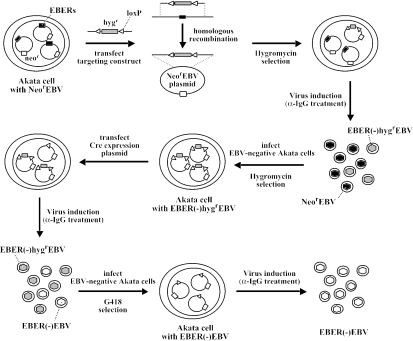
FIG. 1.
Experimental strategies for the construction of EBER knockout EBV in Akata cells. The EBER genes (black boxes), neomycin resistance gene (neor; open boxes), and hygromycin resistance gene (hygr; gray boxes), and loxP sites (white arrowheads) are indicated. Akata cells having episomes of NeorEBV were subjected to the EBER targeting by using the Hygr gene as a marker gene. The Hygr gene was then removed from the genome of the homologous recombinant by the excision reaction of cre recombinase.
First, the EBER genes of NeorEBV were replaced with a hygromycin resistance gene, which was flanked by two loxP sites, via homologous recombination in Akata cells (Fig. 2A). Akata cells carrying NeorEBV were transfected with a targeting construct containing the Hygr gene, and transfected cells were selected by hygromycin. Hygromycin-resistant cell clones were screened by Southern blotting for the presence of cell clones with homologously recombined EBV episomes. One of the cell clones proved to contain homologously recombined episomes, designated EBER(−)HygrEBV, in addition to NeorEBV episomes (Fig. 2B, lane 2). The obtained cell clone was treated with anti-human IgG for virus production, and the mixture of EBER(−)HygrEBV and NeorEBV was used to infect EBV-negative Akata cells to derive cells carrying only EBER(−)HygrEBV episomes. Hygromycin selection of the infected cells resulted in the isolation of cell clones carrying only EBER(−)HygrEBV episomes (Fig. 2B, lane 3).
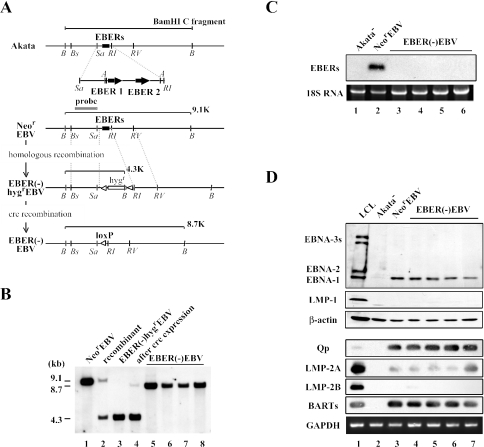
FIG. 2.
(A) Map of the genome of Akata strain EBV surrounding the EBER locus before and after the EBER targeting. The EBER genes of Neor EBV were replaced with the hygromycin resistance gene by homologous recombination. Subsequently, the Hygr gene was removed by the transient expression of cre recombinase in Akata cells. The positions of restriction enzyme recognition sequences are indicated by B (BamHI), Bs (BstPI), Sa (SacI), A (AccI), RI (EcoRI), and RV (EcoRV). A gray line indicates a probe used for Southern blotting. White arrowheads indicate loxP sites. The EBER genes (black boxes) and hygromycin resistance gene (hygr; open box) are also indicated. (B) Southern blotting of Akata cell clones harboring various recombinant EBVs. The results from a NeorEBV-infected cell clone (lane 1), a targeted cell clone (recombinant; lane 2), an EBER(−)HygrEBV-infected cell clone (lane 3), the same cell clone with transient cre expression (lane 4), and EBER(−)EBV-infected cell clones (lanes 5 through 8) are shown. (C) Northern blotting of EBERs. The results from a EBV-negative Akata cell clone (Akata−; lane 1), a NeorEBV-infected cell clone (lane 2), and EBER(−)EBV-infected cell clones (lanes 3 through 6) are shown. (D) EBV gene expression in EBER(−) EBV-infected cell clones. Immunoblotting was used to examine the expression of EBNA proteins and LMP-1 protein (top), whereas RT-PCR analysis was used to examine BamHI Q promoter (Qp) usage and the expressions of other latent genes (bottom). The results from an LCL (lane 1), an EBV-negative Akata cell clone (Akata−; lane 2), a NeorEBV-infected cell clone (lane 3), and EBER(−)EBV-infected cell clones (lanes 4 through 7) are shown.
Second, the Hygr marker gene, which was flanked by two loxP sites, was removed from the targeted locus by the excision reaction mediated by cre recombinase. This method enabled us to generate an EBER-deleted EBV without leaving a heterologous enhancer and promoter activity of the Hygr gene at the EBER locus. We could thereby minimize the artifacts affecting the expression of viral genes surrounding the EBER locus. Akata cells harboring EBER(−)HygrEBV were transiently transfected with a plasmid encoding cre recombinase. A band representing the episome that had lost the Hygr gene, designated EBER(−)EBV, appeared after the transient expression of cre recombinase (Fig. 2B, lane 4). The cre-transduced cells, having EBER(−)EBV episomes in addition to EBER(−)HygrEBV episomes, were then treated with anti-human IgG to induce virus replication. The mixture of EBER(−)EBV and EBER(−)HygrEBV was then used to infect EBV-negative Akata cells, and the infected cells were subjected to G418 selection. Cell clones having only EBER(−)EBV were obtained by screening the G418-resistant cell clones. We finally obtained cell clones harboring only EBER(−)EBV (Fig. 2B, lanes 5 through 8). The resultant EBER(−)EBV had the EBER genes (EBER1 and EBER2) replaced by one residual loxP site (and short flanking sequences), and no heterologous enhancer and promoter activity remained at the EBER locus.
We next confirmed the absence of EBER RNAs in the obtained Akata cell clones harboring EBER(−)EBV episomes by Northern blotting. The result revealed that Akata cell clones harboring EBER(−)EBV were devoid of EBER RNA, whereas an Akata cell clone harboring NeorEBV expressed abundant EBERs (Fig. 2C). We also checked the expressions of various viral genes by immunoblotting and RT-PCR analysis and found that EBER(−)EBV-infected cell clones exhibited a viral gene expression pattern identical to that of NeorEBV-infected cell clones (Fig. 2D), except for the absence of EBERs (Fig. 2C). Taken together, these results demonstrate that the EBER genes were successfully knocked out while the expression patterns of other viral genes were kept intact. It was therefore appropriate to use the obtained EBER(−)EBV for examining the effect of EBER loss on B-cell growth transformation.
Large-scale production of EBER-deleted EBV.
A previous report demonstrated that LCLs harboring EBER-deleted EBV episomes could produce progeny viruses, indicating that EBERs were dispensable for viral lytic replication and infection (26). However, the amount of the EBER-deleted virus obtained from LCLs was small, as LCLs usually exhibit poor virus-producing abilities (26). Therefore, we next employed the Akata cell system to produce EBER-deleted viruses in a large quantity. Akata cell clones harboring either EBER(−)EBV or NeorEBV were induced with anti-IgG for virus production. Immunofluorescence analyses revealed that approximately 60% of Akata cells harboring EBER(−)EBV became positive for gp110 (BALF4 gene product) expression after anti-IgG treatment (data not shown), indicating that EBER(−)EBV entered the lytic cycle of viral replication as efficiently as NeorEBV did. We then harvested the culture supernatants, extracted viral DNAs from the pelleted virions, and analyzed the EcoRI-digested DNAs by agarose gel electrophoresis. The amounts of virions were estimated based on the band intensities of the ethidium bromide-stained DNAs. The results revealed that the band intensities of EBER(−)EBV were comparable to those of NeorEBV (Fig. 3A), demonstrating the successful production of a large quantity of the EBER-deleted EBV. Furthermore, these results indicated that EBERs had no quantitative effect on viral late protein synthesis or on virion maturation.
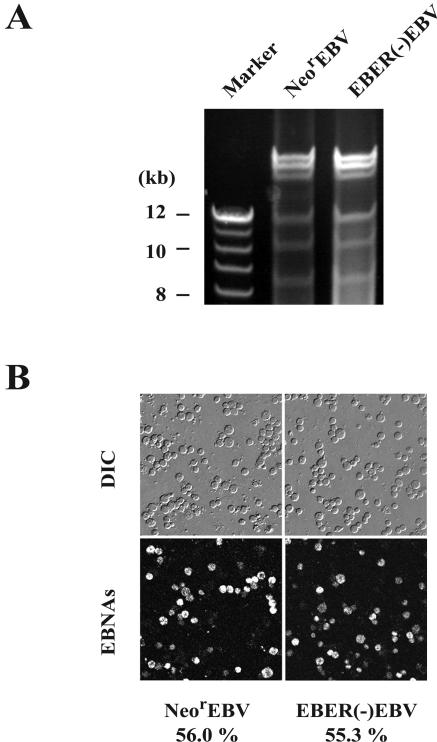
FIG. 3.
(A) Comparison of the amounts of the produced recombinant viruses. Akata cell clones harboring either NeorEBV or EBER(−)EBV were treated with anti-IgG, and the culture supernatants were harvested. Viral DNAs were extracted from the pelleted virions, digested with EcoRI, and analyzed by agarose gel electrophoresis. (B) EBV-negative Akata cells infected with either NeorEBV (left) or EBER(−)EBV (right) were processed for immunofluorescence analysis to detect the expression of EBNA proteins at 48 h postinfection (bottom). The corresponding differential interference contrast (DIC) images are shown (top). The percentages of cells that are positive for EBNA staining are also indicated.
We next examined the infectivity of EBER(−)EBV compared to that of NeorEBV. Equivalent amounts of these viruses were used for infecting EBV-negative Akata cells. The expression of EBNA proteins in the recipient cells was examined by indirect immunofluorescence analysis at 2 days postinfection. We found that more than 50% of the recipient cells became EBNA-positive, a result which was comparable to that obtained with NeorEBV (Fig. 3B). Therefore, the loss of EBERs did not affect the infectivity of the virus.
Thus, we succeeded in producing a large quantity of pure EBER-deleted recombinant viruses, and the obtained EBER-deleted viruses fully retained the infectivity of wild-type EBV.
EBER(−)EBV exhibited impaired B-cell transforming ability.
A previous report demonstrated that EBERs were nonessential for B-lymphocyte growth transformation (26). However, the roles of EBERs may have been overlooked due to the unavailability of a large quantity of pure EBER-deleted virus. The establishment of the cell clones producing large quantities of the EBER-deleted virus enabled us, for the first time, to quantitatively examine the role of EBERs in B-cell growth transformation. We produced large quantities of EBER(−)EBV and NeorEBV by treating the respective cells with anti-IgG. The amounts of the viruses in the culture supernatants were quantitated by agarose gel electrophoresis. We first confirmed that, when equivalent amounts of EBER(−)EBV and NeorEBV were used for infection, both viruses induced EBNA proteins in the recipient cells at similar efficiencies. Based on the EBNA-staining results obtained by infecting EBV-negative Akata cells with serially diluted virus solutions, we estimated that the EBNA induction titers of NeorEBV and EBER(−)EBV were 9.5 × 105 and 9.9 × 105 EBNA induction units/ml, respectively.
Cord blood lymphocytes were plated at a fixed cell density (1.2 × 105 to 1.3 × 105 cells per well) and then infected with serial 10-fold dilutions of either NeorEBV or EBER(−)EBV. The 50% transforming doses (TD50 per milliliter) of EBER(−)EBV and NeorEBV were determined by counting the numbers of wells with proliferating lymphocytes at 6 weeks postinfection. The calculated TD50/ml of EBER(−)EBV was approximately 100-fold less than of that of NeorEBV (Table 1). These results demonstrated that the loss of the EBER genes significantly impaired the ability of EBV to transform B lymphocytes.
TABLE 1.
Comparison of 50% transforming doses of NeorEBV and EBER(−)EBV
| EBV type | TD50/ml
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| Neor | 105.7 | 105.7 |
| EBER(−) | 103.7 | 103.5 |
Generation of EBER knock-in EBV.
The impaired transforming ability of the EBER(−)EBV could have been due to accidental genetic alterations acquired elsewhere on the EBV genome during the course of EBER deletion. We therefore tested whether the titer of EBER(−)EBV to transform B lymphocytes could be restored by putting the EBER genes back into the genome of EBER(−)EBV. The processes of restoring the EBER genes are referred to as “EBER knock-in processes” hereafter.
A targeting construct was designed so that the EBER genes (EBER1 and EBER2) would be inserted back into the native EBER locus of EBER(−)EBV. The marker Hygr gene was used for getting homologous recombinants, as EBER(−)EBV had only the marker Neorgene (Fig. 1 and 4A). Akata cells carrying EBER(−)EBV episomes were transfected with a targeting construct for the EBER knock-in, and transfected cells were selected by hygromycin. The results of Southern blotting revealed that one of the hygromycin-resistant cell clones contained homologously recombined episomes, designated EBER(+)Hygr EBV, in addition to EBER(−)EBV episomes (Fig. 4B, lane 2). The virus mixture of EBER(+)HygrEBV and EBER(−)EBV was produced from the cell clone, and after the infection of EBV-negative Akata cells with the mixture and hygromycin selection, cell clones harboring only EBER(+)HygrEBV episomes were obtained (Fig. 4B, lane 3).
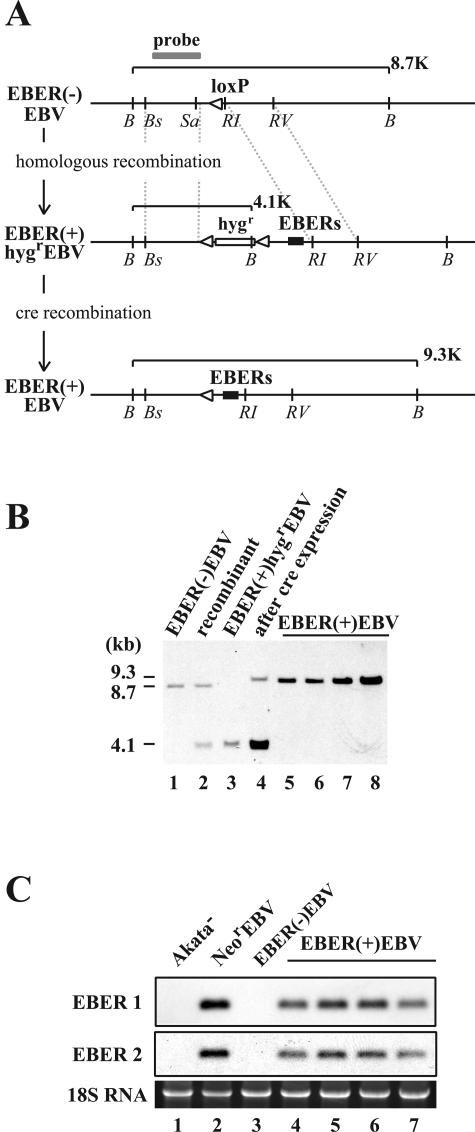
FIG. 4.
(A) Map of the genome of Akata strain EBV surrounding the EBER locus before and after the EBER knock-in. The EBER genes were reconstituted in the genome of EBER(−)EBV via homologous recombination by using the hygromycin resistance gene as a marker gene. Subsequently, the Hygr gene was removed by the transient expression of cre recombinase in Akata cells. The positions of restriction enzyme recognition sequences are indicated by B (BamHI), Bs (BstPI), Sa (SacI), RI (EcoRI), and RV (EcoRV). A gray line indicates a probe used for Southern blot analysis. White arrowheads indicate loxP sites. The EBER genes (black boxes) and the hygromycin resistance gene (hygr; open box) are also indicated. (B) Southern blot analysis of Akata cell clones harboring various recombinant EBVs.The results from an EBER(−)EBV-infected cell clone (lane 1), a targeted cell clone (recombinant; lane 2), an EBER(+)HygrEBV-infected cell clone (lane 3), the same cell clone with transient cre expression (lane 4), and EBER(+)EBV-infected cell clones (lanes 5 through 8) are shown. (C) Northern blot analysis of EBERs. The results from an EBV-negative Akata cell clone (Akata−; lane 1), a NeorEBV-infected cell clone (lane 2), a EBER(−)EBV-infected cell clone (lane 3), and EBER(+)EBV-infected cell clones (lanes 4 through 7) are shown.
Subsequently, the Hygr gene of the EBER(+)HygrEBV was removed by expressing cre recombinase in Akata cells harboring EBER(+)HygrEBV. A band representing the episome that had lost the Hygr gene, designated EBER(+)EBV, appeared in the cre-transduced cells (Fig. 4B, lane 4). A virus mixture of EBER(+)EBV and EBER(+)HygrEBV was obtained, and after EBV-negative Akata cells were infected with the virus mixture and the G418-resistant cell clones were screened, cell clones harboring only EBER(+)EBV episomes were obtained (Fig. 4B, lanes 5 through 8). The genome of EBER(+)EBV was expected to be identical to that of NeorEBV except for one residual loxP site (and short flanking sequences) inserted upstream of the reconstituted EBER genes.
We then examined whether the expression of EBERs was restored in Akata cells infected with the EBER knock-in EBV. Total RNAs prepared from Akata cells harboring NeorEBV, EBER(−)EBV, or EBER(+)EBV were examined for the expression of EBERs by Northern blotting. We found that four independent Akata cell clones harboring EBER(+)EBV expressed both EBER1 and EBER2 as expected (Fig. 4C, lanes 4 through 7), although the expression levels of EBERs were slightly lower than those in Akata cells harboring NeorEBV (Fig. 4C, compare lane 2 and lanes 4 through 7). Furthermore, we found that EBER(+)EBV-infected cell clones exhibited a viral gene expression pattern identical to that of NeorEBV- and EBER(−)EBV-infected cell clones (data not shown), except for the difference of EBER expression (Fig. 4C). Thus, we successfully restored the expression of EBERs by putting the EBER genes back into the EBER-deleted EBV, and the resultant EBER knock-in EBV exhibited normal viral gene expression, just like the wild-type counterpart.
EBER knock-in restored the ability to efficiently transform B lymphocytes.
We thus made three isogenic Akata cells lines; the first one contained the wild-type episomes (NeorEBV), the second one contained the EBER knockout episomes [EBER(−)EBV], and the third one contained EBER knock-in episomes [EBER(+)EBV]. These three cell lines were treated with anti-IgG for virus production, and NeorEBV, EBER(−)EBV, and EBER(+)EBV were produced in large quantities. Again, the amounts of virions in the culture supernatants were estimated by agarose gel electrophoresis. We then examined a set of three viruses for their EBNA induction titers, and the values were 9.9 ×105, 9.8 × 105, and 9.6 ×105 EBNA induction units/ml for NeorEBV, EBER(−)EBV, and EBER(+)EBV, respectively. Thus, we used equivalent amounts of each virus for infecting cord blood lymphocytes, and we found similar levels of EBNA induction in the recipient cells (data not shown). The results of three independent experiments, utilizing three independent preparations of the viruses, revealed that the TD50/ml of EBER(+)EBV were approximately 20-fold higher than those of EBER(−)EBV, approaching the TD50/ml of NeorEBV (Table 2). These results provided concrete evidence that the loss of EBERs was truly responsible for the impaired transforming ability of EBER(−)EBV.
TABLE 2.
Comparison of 50% transforming doses of NeorEBV, EBER(−)EBV, and EBER(+)EBV
| EBV type | TD50/ml
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| Neor | 104.7 | 105.5 | 105.5 |
| EBER(−) | 102.7 | 103.0 | 103.2 |
| EBER(+) | 104.0 | 104.3 | 104.5 |
EBERs are critical for the growth of LCLs, especially at low cell density.
A previous report demonstrated no significant difference between the growth rates of EBER-positive LCLs and EBER-negative LCLs (25, 26). The ability to produce each of the three different viruses, NeorEBV, EBER(−)EBV, and EBER(+)EBV, in large quantities enabled us to reexamine this issue. Using viruses with high EBNA induction titers for infecting lymphocytes enabled the rapid outgrowth of the LCLs, and we could eliminate the artifacts that might arise from long-term cultivation.
We successfully established two independent sets of LCLs derived from each of the three viruses. Each set of LCLs was derived from cord blood lymphocytes of different donors. LCLs belonging to each set were established in parallel, and they were maintained in culture for the same time period before they were subjected to the assays. Immunoblotting and RT-PCR analyses revealed that EBER(−)EBV-infected LCLs were indistinguishable from NeorEBV- or EBER(+)EBV-infected LCLs in their expression of viral latent proteins (Fig. 5 and data not shown).
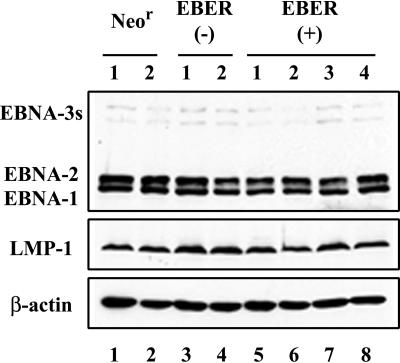
FIG. 5.
Immunoblot analysis of EBNA proteins and LMP-1 protein in the established LCLs. The results from two independent NeorEBV-infected LCLs (lanes 1 and 2), two independent EBER(−)EBV-infected LCLs (lanes 3 and 4), and four independent EBER(+)EBV-infected LCLs (lanes 5 through 8) are shown. Note that all of the LCLs express similar amounts of EBNA-1, EBNA-2, EBNA-3s, and LMP-1.
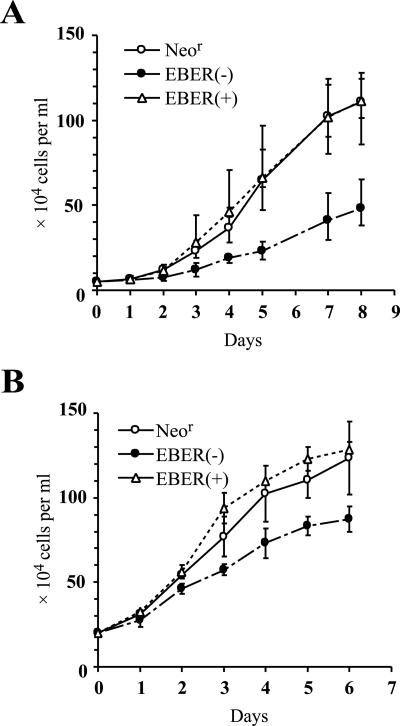
We first compared the growth rates of the established LCLs. We employed two independent sets of LCLs for this assay (Fig. 6). Using both sets of LCLs, we found that EBER(−)EBV-infected LCLs grew significantly more slowly than NeorEBV-infected LCLs. By contrast, EBER(+)EBV-infected LCLs grew as well as NeorEBV-infected LCLs (Fig. 6). When the growth rates of the established LCLs were examined shortly after infection, the results clearly demonstrated that the expression of EBERs substantially accelerated LCL growth.
FIG. 6.
Comparison of the growth rates of the established LCLs at low cell densities. Two independent sets of LCLs (panels A and B), each derived from cord blood lymphocytes of different donors, were subjected to the assay. The LCLs established by infection with NeorEBV, EBER(−)EBV, or EBER(+)EBV were plated to the wells of 24-well plates at a density of either 5 × 104 (A) or 2 × 105 (B) cells per ml, and the numbers of cells were counted daily (except for day 6). The results for NeorEBV-infected LCLs (open circles), EBER(−)EBV-infected LCLs (solid circles), and EBER(+)EBV-infected LCLs (open triangles) are shown. The growth curves represent the averaged results (means ± standard deviations) obtained with four independent LCLs (A) or two independent LCLs (B).
We next examined whether the established LCLs could grow when they were plated at low cell densities. Serially twofold-diluted LCLs were plated in 96-well plates, and the number of wells with proliferating cells was counted at 4 weeks after starting the culture. We again employed two sets of LCLs for this assay (Table 3). We found a marked difference in cell proliferation between EBER(−)EBVinfected LCLs and NeorEBV-infected or EBER(+)EBV-infected LCLs. EBER(−)EBV-infected LCLs did not proliferate at low cell densities, while NeorEBV-infected and EBER(+)EBV-infected LCLs proliferated well even when they were plated at low cell densities (Table 3). These results revealed that plating the LCLs at low cell densities could augment the effect of EBER loss on LCLs.
TABLE 3.
Numbers of wells with proliferating cells after the plating of various LCLs at low cell densities
| EBV type of LCL | No. of wells with indicated no. of plated cells per wella
|
||||
|---|---|---|---|---|---|
| 1,000 | 500 | 250 | 125 | 62.5 | |
| Expt 1 | |||||
| Neor | 16 | 16 | 16 | 15 | 12 |
| EBER(−) | 16 | 11 | 3 | 0 | 0 |
| EBER(+) | 16 | 16 | 16 | 12 | 11 |
| Expt 2 | |||||
| Neor | 16 | 16 | 16 | 13 | 11 |
| EBER(−) | 16 | 8 | 1 | 0 | 0 |
| EBER(+) | 16 | 16 | 15 | 10 | 7 |
Numbers of wells are given out of a total of 16 wells.
The overall result clearly demonstrated that EBERs significantly contributed to EBV-induced B-cell transformation by enhancing the growth potential of transformed lymphocytes.
DISCUSSION
We employed the Akata system to produce a large quantity of pure EBER-deleted virus and examined the role of EBERs in B-cell transformation. The EBER-deleted virus exhibited impaired transforming ability, and the LCLs established by the EBER-deleted virus grew significantly more slowly than those established with the wild-type counterpart (NeorEBV). Importantly, reconstituting the EBER genes at their native locus of the EBER-deleted EBV resulted in restored transformation efficiency, demonstrating that the loss of EBERs was truly responsible for the impaired transformation efficiency. Our data represent the first demonstration that EBERs play significant roles in B-cell transformation. The data also underpin our previous studies in which we demonstrated the growth-stimulatory effects of EBERs by using already dividing B cells (12, 17).
Our data fit well with a previous finding (26) that EBERs are nonessential for transforming B lymphocytes and producing progeny viruses. The discrepancy between our study and the previous study resides in the relative transformation efficiency of the EBER-deleted virus compared to that of the EBER-positive virus. In the previous study, lethally irradiated EBER-negative LCLs were cocultivated with primary lymphocytes in order to passage the transforming virus (26). The researchers found that EBER-positive and EBER-negative LCLs yielded secondary transformants with similar efficiencies, leading to the conclusion that EBERs were not significant for the entire process of B-cell transformation. However, they failed to show that similar amounts of viruses were produced from the primary LCLs and were used for generating secondary LCLs. By contrast, we quantitated the amounts of virions in the culture supernatants, checked their infectivities, and used equivalent amounts of recombinant viruses for the transformation assay. Therefore, our system is more appropriate for accurately comparing the transforming efficiencies of EBER-positive and EBER-negative viruses.
Another critical discrepancy resides in the growth rate of the established EBER-negative LCLs. While Swaminathan et al. did not see any significant difference in the growth rates of EBER-positive and EBER-negative LCLs (25, 26), we found that EBER(−)EBV-infected LCLs grew significantly more slowly than NeorEBV-infected- or EBER(+)EBV-infected LCLs (Fig. 6). We also observed significant differences between the outgrowth of EBER-negative LCLs and that of EBER-positive LCLs when they were plated at low cell densities (Table 3). We speculate that the EBER-negative LCLs used for the previous study may not represent typical EBER-negative LCLs, because establishing secondary LCLs by cocultivating lymphocytes with irradiated LCLs apparently requires long-term cultivation and it is difficult to exclude the possibility that untypical LCLs, having acquired additional growth advantages, have been selected during the period of expansion. By contrast, we produced large amounts of recombinant viruses, and they all exhibited high EBNA induction titers. As a result, many lines of LCLs were established relatively quickly, within 4 weeks at the earliest, which can eliminate artifacts that might arise from long-term cultivation.
One can argue that reconstituting the EBER genes into the EBER-deleted virus did not fully compensate for the EBER defect (Table 2). One possible explanation is that the EBER expression level of EBER(+)EBV is slightly lower than that of NeorEBV, as has been observed for infected Akata cells (Fig. 4C). It has been shown that the upstream regions of the EBER genes contain elements for efficient EBER expression (7). A slight structural change introduced to the further-upstream region of the reconstituted EBER genes (Fig. 4) may result in reduced EBER expression from EBER(+)EBV. Our previous study demonstrated that there was a good correlation between the amount of EBER transgene expression and the malignant phenotype of Akata cells (12). We speculate that EBER(+)EBV is not as transformation competent as NeorEBV, due to its slight decrease of EBER expression.
The molecular mechanism by which EBERs enhance transformation as well as growth potentials of LCLs remains unknown. One possibility is that EBERs confer resistance to apoptosis in LCLs, as has been demonstrated for Burkitt's lymphoma cells (17). However, we did not see any significant increase of apoptotic cells in EBER(−)EBV-infected LCLs (M. Yajima, unpublished result), making such a possibility unlikely. We also found that the loss of EBERs did not impair the insensitivity of the EBV-transformed LCLs to the antiproliferative effect of IFN-α, consistent with a previous report that EBERs did not modulate the effect of interferons in the established LCLs (25). The antiapoptotic effect of EBERs, however, may be masked by LMP-1 expression, as LMP-1 has been shown to upregulate the expression of antiapoptotic proteins, such as A20 (13) and bcl-2 (6, 20). Furthermore, we compared the levels of phosphorylated α subunit of translation initiation factor (eIF-2α) in the LCLs, since we previously showed that EBERs inhibit the phosphorylation of eIF-2α through their direct binding to PKR in case of Burkitt's lymphoma cells (17). However, no significant increase of the levels of phosphorylated eIF-2α in EBER(−)EBV-infected LCLs was observed (unpublished result). The inhibition of the phosphorylation of eIF-2α by EBERs may not be obvious in case of LCLs, as LMP-1 can phosphorylate eIF-2α in EBV-infected cells (14).
Another possibility is that EBERs induce the expression of growth factors in LCLs, just as they do in Burkitt's lymphoma cells (11), T cells (31), and epithelial cells (9). We therefore checked the expression levels of various growth-stimulatory cytokines (IL-1β, IL-2, IL-4, IL-6, and IL-10) in EBER-positive and EBER-negative LCLs. We did not see any differences in the expression levels of these cytokines in EBER-negative and EBER-positive LCLs (data not shown). Again, the effect of EBERs on cytokine expression might have been masked by LMP-1 expression, as LMP-1 has been demonstrated to induce the expression of various cytokines, such as IL-6 (5) and IL-10 (16, 29).
The biological significance of EBERs has been enigmatic until recently, but accumulating evidence indicates that the expression of EBERs is critical for the life cycle of EBV. EBERs may have additional functions that are critical for EBV infection in vivo. Further studies are required to clarify the molecular mechanisms by which EBERs exert their biological activities.
Acknowledgments
We thank A. Nanbo, L. Yang, K. Takasawa, and H. Yoshiyama for helpful discussions.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (T.K. and K.T.), the Akiyama Memorial Foundation (T.K.), and the Uehara Memorial Foundation (K.T.).
REFERENCES
- 1.Arrand, J. R., L. S. Young, and J. D. Tugwood. 1989. Two families of sequences in the small RNA-encoding region of Epstein-Barr virus (EBV) correlate with EBV types A and B. J. Virol. 63:983-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke, P. A., M. Schwemmle, J. Schickinger, K. Hilse, and M. J. Clemens. 1991. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 19:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemens, M. J. 1993. The small RNAs of Epstein-Barr virus. Mol. Biol. Rep. 17:81-92. [DOI] [PubMed] [Google Scholar]
- 4.Condit, R. C. 2001. Principles of virology, p. 19-51. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 5.Eliopoulos, A. G., M. Stack, C. W. Dawson, K. M. Kaye, L. Hodgkin, S. Sihota, M. Rowe, and L. S. Young. 1997. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-kappaB pathway involving TNF receptor-associated factors. Oncogene 14:2899-2916. [DOI] [PubMed] [Google Scholar]
- 6.Henderson, S., M. Rowe, C. Gregory, D. Croom-Carter, F. Wang, R. Longnecker, E. Kieff, and A. Rickinson. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107-1115. [DOI] [PubMed] [Google Scholar]
- 7.Howe, J. G., and M. D. Shu. 1993. Upstream basal promoter element important for exclusive RNA polymerase III transcription of the EBER 2 gene. Mol. Cell. Biol. 13:2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwakiri, D., Y. Eizuru, M. Tokunaga, and K. Takada. 2003. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 63:7062-7067. [PubMed] [Google Scholar]
- 10.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 11.Kitagawa, N., M. Goto, K. Kurozumi, S. Maruo, M. Fukayama, T. Naoe, M. Yasukawa, K. Hino, T. Suzuki, S. Todo, and K. Takada. 2000. Epstein-Barr virus-encoded poly(A)(−) RNA supports Burkitt's lymphoma growth through interleukin-10 induction. EMBO J. 19:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komano, J., S. Maruo, K. Kurozumi, T. Oda, and K. Takada. 1999. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J. Virol. 73:9827-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laherty, C. D., H. M. Hu, A. W. Opipari, F. Wang, and V. M. Dixit. 1992. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J. Biol. Chem. 267:24157-24160. [PubMed] [Google Scholar]
- 14.Lam, N., M. L. Sandberg, and B. Sugden. 2004. High physiological levels of LMP1 result in phosphorylation of eIF2 alpha in Epstein-Barr virus-infected cells. J. Virol. 78:1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerner, M. R., N. C. Andrews, G. Miller, and J. A. Steitz. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 78:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagomi, H., R. Dolcetti, M. T. Bejarano, P. Pisa, R. Kiessling, and M. G. Masucci. 1994. The Epstein-Barr virus latent membrane protein-1 (LMP1) induces interleukin-10 production in Burkitt lymphoma lines. Int. J. Cancer 57:240-244. [DOI] [PubMed] [Google Scholar]
- 17.Nanbo, A., K. Inoue, K. Adachi-Takasawa, and K. Takada. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 21:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 19.Rosa, M. D., E. Gottlieb, M. R. Lerner, and J. A. Steitz. 1981. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1:785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowe, M., M. Peng-Pilon, D. S. Huen, R. Hardy, D. Croom-Carter, E. Lundgren, and A. B. Rickinson. 1994. Upregulation of bcl-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-kappa B activation and to induction of cell surface markers. J. Virol. 68:5602-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruf, I. K., P. W. Rhyne, C. Yang, J. L. Cleveland, and J. T. Sample. 2000. Epstein-Barr virus small RNAs potentiate tumorigenicity of Burkitt lymphoma cells independently of an effect on apoptosis. J. Virol. 74:10223-10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp, T. V., M. Schwemmle, I. Jeffrey, K. Laing, H. Mellor, C. G. Proud, K. Hilse, and M. J. Clemens. 1993. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 21:4483-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu, N., A. Tanabe-Tochikura, Y. Kuroiwa, and K. Takada. 1994. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol. 68:6069-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, N., H. Yoshiyama, and K. Takada. 1996. Clonal propagation of Epstein-Barr virus (EBV) recombinants in EBV-negative Akata cells. J. Virol. 70:7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaminathan, S., B. S. Huneycutt, C. S. Reiss, and E. Kieff. 1992. Epstein-Barr virus-encoded small RNAs (EBERs) do not modulate interferon effects in infected lymphocytes. J. Virol. 66:5133-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swaminathan, S., B. Tomkinson, and E. Kieff. 1991. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc. Natl. Acad. Sci. USA 88:1546-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tierney, R. J., N. Steven, L. S. Young, and A. B. Rickinson. 1994. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J. Virol. 68:7374-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toczyski, D. P., and J. A. Steitz. 1991. EAP, a highly conserved cellular protein associated with Epstein-Barr virus small RNAs (EBERs). EMBO J. 10:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vockerodt, M., B. Haier, P. Buttgereit, H. Tesch, and D. Kube. 2001. The Epstein-Barr virus latent membrane protein 1 induces interleukin-10 in Burkitt's lymphoma cells but not in Hodgkin's cells involving the p38/SAPK2 pathway. Virology 280:183-198. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, N., T. Takizawa, Y. Iwanaga, and N. Shimizu. 2000. Malignant transformation of B lymphoma cell line BJAB by Epstein-Barr virus-encoded small RNAs. FEBS Lett. 484:153-158. [DOI] [PubMed] [Google Scholar]
- 31.Yang, L., K. Aozasa, K. Oshimi, and K. Takada. 2004. Epstein-Barr virus (EBV)-encoded RNA (EBER) promotes growth of EBV-infected T-cells through interleukin-9 induction. Cancer Res. 64:5332-5337. [DOI] [PubMed] [Google Scholar]
- 32.Yang, L., S. Maruo, and K. Takada. 2000. CD21-mediated entry and stable infection by Epstein-Barr virus in canine and rat cells. J. Virol. 74:10745-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshiyama, H., N. Shimizu, and K. Takada. 1995. Persistent Epstein-Barr virus infection in a human T-cell line: unique program of latent virus expression. EMBO J. 14:3706-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]