Abstract
Two genes coding for isozymes of alcohol dehydrogenase (ADH); designated PsADH1 and PsADH2, have been identified and isolated from Pichia stipitis CBS 6054 genomic DNA by Southern hybridization to Saccharomyces cerevisiae ADH genes, and their physiological roles have been characterized through disruption. The amino acid sequences of the PsADH1 and PsADH2 isozymes are 80.5% identical to one another and are 71.9 and 74.7% identical to the S. cerevisiae ADH1 protein. They also show a high level identity with the group I ADH proteins from Kluyveromyces lactis. The PsADH isozymes are presumably localized in the cytoplasm, as they do not possess the amino-terminal extension of mitochondrion-targeted ADHs. Gene disruption studies suggest that PsADH1 plays a major role in xylose fermentation because PsADH1 disruption results in a lower growth rate and profoundly greater accumulation of xylitol. Disruption of PsADH2 does not significantly affect ethanol production or aerobic growth on ethanol as long as PsADH1 is present. The PsADH1 and PsADH2 isozymes appear to be equivalent in the ability to convert ethanol to acetaldehyde, and either is sufficient to allow cell growth on ethanol. However, disruption of both genes blocks growth on ethanol. P. stipitis strains disrupted in either PsADH1 or PsADH2 still accumulate ethanol, although in different amounts, when grown on xylose under oxygen-limited conditions. The PsADH double disruptant, which is unable to grow on ethanol, still produces ethanol from xylose at about 13% of the rate seen in the parental strain. Thus, deletion of both PsADH1 and PsADH2 blocks ethanol respiration but not production, implying a separate path for fermentation.
Reduction of acetaldehyde to ethanol is the last step in ethanol production. Conversely, ethanol oxidation is the first reaction of ethanol respiration. The enzymes catalyzing these reactions and their structural genes have been well studied in Saccharomyces cerevisiae. Alcohol dehydrogenase (EC 1.1.1.1), ADH1, the classical fermentative isozyme, is responsible for the last step in the yeast glycolytic pathway, the reduction of acetaldehyde to ethanol (4, 8, 11). ADH2, the oxidative isozyme, is highly repressed by fermentative conditions (12) and is derepressed in the absence of a fermentable sugar such as glucose. The function of ADH2 in the cell is to oxidize ethanol, formed during fermentation, to acetaldehyde, which can then be metabolized via the tricarboxylic acid cycle in the mitochondria and also serve as an intermediate in gluconeogenesis (30). S. cerevisiae has two other ADH genes: ADH3, coding for a mitochondrion-targeted enzyme presumably involved in ethanol oxidation (48), and ADH4, which displays no significant similarity to any other characterized yeast ADH gene (46). Four such ADHs have been characterized in the galactose-fermenting yeast Kluyveromyces lactis (23, 32, 33, 38). These genes, named KlADH1 through KlADH4, encode two cytoplasmic (KlADH1 and KlADH2) and two mitochondrial (KlADH3 and KlADH4) activities. KlADH1 and KlADH2 are preferentially expressed in the presence of glucose (33, 38). By contrast, KlADH4 is induced at the transcriptional level in the presence of ethanol (23, 38). Three of the ADH isozymes from S. cerevisiae and the four isozymes of K. lactis all belong to the group I zinc-dependent enzymes because of their sequence identity and functional similarities (27). The ADH enzymes of xylose-fermenting yeasts have not been examined previously.
The xylose-fermenting yeast Pichia stipitis is among the few organisms that use both xylose and glucose and exhibit a regulatory transition between respiratory and fermentative processes (19). The fundamental mechanisms by which fermentation is regulated differ profoundly in the glucose-fermenting yeast S. cerevisiae and the xylose-fermenting yeast P. stipitis (16, 19, 45). In S. cerevisiae, the availability of oxygen is irrelevant to fermentative metabolism. Glucose induces high levels of glycolytic enzymes and represses respiration, leading to ethanol production (4). In the Crabtree-negative yeast P. stipitis, oxygen limitation, rather than the presence of either glucose or xylose, induces fermentation (25, 45).
P. stipitis is capable of producing ethanol from xylose under anaerobic conditions because it has a single xylose reductase with dual cofactor (NADPH and NADH) specificity (1). In most yeasts and fungi, xylose reductase uses only NADPH. Xylose metabolism in yeasts proceeds via xylose reductase (XR), which catalyzes the reduction of xylose with NAD(P)H to form xylitol, and xylitol dehydrogenase (XDH), which catalyzes the oxidation of xylitol by NAD to form xylulose. Thus, a cofactor imbalance can arise under anaerobic conditions if XR uses only NADPH, XDH uses only NAD, and no mechanism exists to reduce NADP with NADH. In P. stipitis, XR can accept either NADPH or NADH, so a cofactor imbalance does not block xylose uptake under anaerobic conditions (7).
P. stipitis requires small amounts of oxygen for maximal conversion of xylose to ethanol (41). Under strictly anaerobic conditions, little ethanol is formed (19). The explanation for this oxygen effect could reside in the higher activity of its XR with NADPH. Its XDH is specific for NAD, so partial accumulation of NADP and NADH probably still occurs, despite NADH-linked XR activity in P. stipitis (20).
Two key fermentative enzyme activities, pyruvate decarboxylase (PDC) and ADH, are induced as P. stipitis becomes oxygen limited and ethanol production increases (25). However, little is known about this process in P. stipitis, at the molecular genetic and physiological levels. A better understanding of metabolic regulation of the genes involved in xylose fermentative metabolism is essential to further advance this field. A recent paper from our laboratory described two P. stipitis genes for PDC (21). The next step toward understanding the roles of fermentation and respiration in xylose metabolism is to clone and disrupt P. stipitis ADH genes.
The objective of the present study was to isolate and characterize the ADH genes necessary for ethanol production in P. stipitis. The research identified two distinct genes for ADH in P. stipitis. We determined the physiological roles for each and showed that one (PsADH1) is critically important for ethanol production.
MATERIALS AND METHODS
Microbial strains.
The P. stipitis strains used in this research are summarized in Table 1. P. stipitis CBS 6054 was the source of all derived strains and of all sequenced DNA. Escherichia coli DH5α (Gibco BRL, Gaithersburg, Md.) (F− recA1 endA1 hsdR17 [rk− mk+] supE44 thi-1 gyrA relA1) was used for all recombinant DNA experiments that required a bacterial host.
TABLE 1.
P. stipitis strains used in this study
| Strain | Genotype | Origin | Reference |
|---|---|---|---|
| CBS 6054 (= NRRL Y-11545 = ATCC 58785) | Wild type | CBSa | 20 |
| FPL-PSU-1 | Psura3-2/Psura3-2 | CBS 6054 | 47 |
| FPL-PLU20 | Psura3-3/Psura3-3 Psleu2Δ-1/Psleu2Δ-1 | FPL-DX26 (= NRRL Y-21304) | 22 |
| FPL-PSU218 | Psura3-2/Psura3-2 Psadh1::PsURA3/Psadh1::PsURA3 | FPL-PSU1 (= NRRL Y-21446) | This study |
| FPL-PLU123 | Psura3-3/Psura3-3 Psleu2Δ-1/Psleu2Δ-1 Psadh2::PsLEU2/Psadh2::PsLEU2 | FPL-PLU20 | This study |
| FPL-PLU1209 | Psura3-3/Psura3-3 Psleu2Δ-1/Psleu2Δ-1 Psadh1::PsURA3/Psadh1::PsURA3 Psadh2::PsLEU2/Psadh2::PsLEU2 | FPL-PLU123 | This study |
CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
Media and culture conditions.
Yeasts were grown in yeast-peptone-dextrose medium consisting of yeast extract at 10 g/liter, peptone at 20 g/liter, and glucose at 20 g/liter. For cultivation of ura3 and leu2 auxotrophs, media were supplemented with 100 mg of uridine per liter and 100 mg of leucine per liter, respectively. E. coli was grown in Luria-Bertani medium (34) with 50 μg of ampicillin per ml in liquid medium or 100 μg of ampicillin per ml in solid medium. Fermentation studies were done with 1.7 g of yeast nitrogen base per liter without ammonium sulfate or amino acids (YNB; Difco, Detroit, Mich.) supplemented with Bacto Peptone at 6.56 g/liter, urea at 2.27 g/liter (2× nitrogen), and 80 g of d-xylose per liter plus leucine or uridine as needed. Cells were cultivated at 25°C in 50 ml of fermentation medium shaken in 125-ml Erlenmeyer flasks at 100 rpm.
Plasmid constructions.
The PsURA3 (47) and PsLEU2 (22) selectable markers originated from P. stipitis CBS 6054. Plasmid Bluescript KSII+ was obtained from Stratagene (La Jolla, Calif.). pJY102 (PsADH1 disruption cassette) was built in three steps: (i) the EcoRI site in pUC19 was destroyed by treatment with T4 DNA polymerase (New England Biolabs, Beverly, Mass.) and ligation reactions to form pJY19; (ii) a 2.0-kbp SacI-SacI fragment of the PsADH1 gene bearing a unique EcoRI site was inserted into the SacI site of pJY19 to form pJY101; and (iii) a plasmid containing the URA3 gene of P. stipitis, pVY3 (47), was cut with PstI (New England Biolabs). The EcoRI linker (New England Biolabs) was inserted at the remaining PstI site. A 1.2-kbp EcoRI-EcoRI fragment carrying the P. stipitis URA3 gene was inserted into a unique EcoRI site within the PsADH1 gene of pJY101 to form pJY102. The resulting plasmid, pJY102, contained the 3.2-kbp SacI-SacI Psadh1::PsURA3 fragment and was digested with SacI prior to transformation. pJY202 (PsADH2 disruption cassette) was built in three steps: (i) a portion (3.5 kbp) of the 5′-flanking region was deleted from PsADH2 to leave a unique PstI site in its coding region; (ii) this site was destroyed by T4 DNA polymerase, and a 2.1-kbp blunt-ended EcoRI fragment bearing PsLEU2 gene was ligated into the middle of Psadh2 to form pJY201; and (iii) a 4.5-kbp SacI-BamHI fragment of pJY201 bearing the Psadh2::PsLEU2 construct was inserted into the corresponding sites of pUC18 to form pJY202. The resulting plasmid, pJY202, was digested with SacI and BamHI prior to transformation.
Yeast transformation.
Lithium acetate transformation of P. stipitis PSU1 (ura3-2/ura3-2) and PLU20 (ura3-3/ura3-3 leu2Δ-1/leu2Δ-1) (22) was performed as described by Ito et al. (18). Yeast transformants were selected on YNB plus 20 g of glucose per liter without uracil or leucine when URA3 and LEU2 were used as selectable markers, respectively. For solid media, 20 g of agar per liter was added.
DNA isolation.
Plasmid DNA was isolated and purified by the alkaline extraction method of Birnboim and Doly (5) or with the Qiagen Plasmid Prep kit (Qiagen Corp., Chatsworth, Calif.). Yeast genomic DNA was isolated and purified as described by Specht et al. (43) or Rose et al. (28).
Cloning of P. stipitis ADH genes.
Genomic DNA from P. stipitis CBS 6054 was cut with BamHI and SalI and electrophoresed in 0.8% agarose. DNA corresponding to ca. 6.6 kbp was isolated from the gel slice by using Gelase (Epicentre Corp., Middleton, Wis.). The purified DNA fragment was ligated into the corresponding sites of pBluescript KSII+. The DNA library was amplified in E. coli DH5α, followed by electroporation using the Pulse Controller together with the Gene Pulser from Bio-Rad (Richmond, Calif.). Inserts containing ADH sequences were identified by colony hybridization to the PCR-amplified S. cerevisiae ADH1 coding sequences, and colonies that were positive with the probe were tested further. The second genomic library was constructed from BamHI- and XbaI-digested CBS 6054 genomic DNA as described above for the isolation of P. stipitis ADH1. E. coli transformants were screened for the presence of S. cerevisiae ADH2 homologous sequences by using the colony hybridization technique (34).
Blot hybridizations.
Genomic DNA digests were electrophoresed in 0.8% agarose, blotted into Nytran filters (Schleicher & Schuell, Keene, N.H.) by standard procedures (42), and hybridized with the PCR-amplified P. stipitis PsADH2 or S. cerevisiae ADH1 or ADH2 coding sequences labeled with digoxigenin-11-dUTP by using a Genius 1 kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Hybridizations were done at 37°C under the manufacturer’s (Boehringer Mannheim) specified conditions. The colony hybridizations were performed under standard conditions (34) by using a Genius 1 kit (Boehringer Mannheim).
DNA sequencing.
The dideoxy method of Sanger et al. (35) was used for DNA sequencing (Sequenase version 2.0 DNA sequencing kit; United States Biochemical, Cleveland, Ohio). Primers were synthesized at the University of Wisconsin—Madison Biotechnology Center and Ransom Hill Bioscience, Inc. (Ramona, Calif.). Nested deletions of pJY158, a plasmid containing the PsADH2 gene, were sequenced from universal primers located adjacent to the multiple cloning site. The sequence was confirmed by sequencing other deletions with overlapping endpoints. The sequence of the coding region and 5′- and 3′-flanking regions is available under GenBank accession no. AF008244. The SpeI-XbaI fragment of pJY268, a plasmid containing PsADH1 (see Fig. 2A), was deleted, religated, and subjected to sequence analysis by primer walking from the universal primers located adjacent to the multiple cloning site. The sequence of the coding region and the 5′- and 3′-flanking regions is available under GenBank accession no. AF008245. The Genetics Computer Group programs (13) developed at the University of Wisconsin—Madison were used to evaluate DNA and derived amino acid sequences.
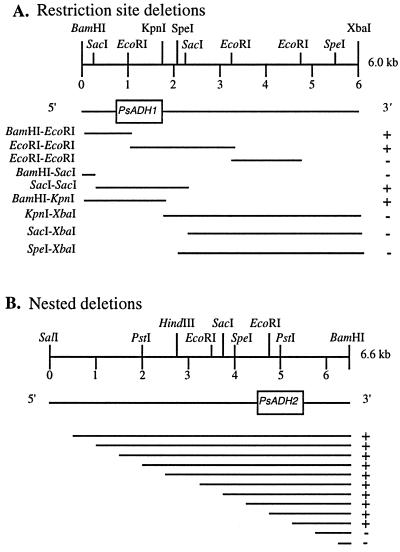
FIG. 2.
Southern hybridization to identify genomic regions on clones of PsADH1 and PsADH2. (A) Approximate location of the PsADH1 coding region on the 6.0-kbp BamHI-XbaI PsADH1 DNA fragment as determined by restriction mapping and cross-hybridizations of the resulting restriction fragments to ScADH1. +, positive hybridization; −, no hybridization. Bold lines represent the fragment containing 5′- and 3′-flanking regions of PsADH genes. (B) Approximate location of the PsADH2 coding region on the 6.6-kbp BamHI-SalI PsADH2 DNA fragment as determined by nested deletions from the 5′ end and cross-hybridizations of the resulting fragments to ScADH1. +, positive hybridization; −, no hybridization. The boxes indicate the PsADH1 (A) and PsADH2 (B) coding regions.
PCR screening of PsADH disruptants.
Amplification reactions were performed in 50 μl containing 100 ng of genomic template DNA, 200 pmol of each primer, 0.2 mM each deoxyribonucleoside triphosphate (A, T, C, and G), and 5 U of Pfu DNA polymerase (Stratagene) in 10 mM KCl–10 mM (NH4)2SO4–20 mM Tris-Cl (pH 8.8)–2 mM MgSO4–0.1% Triton X-100–100-μg/ml bovine serum albumin (the 10× reaction buffer provided with the enzyme). Times and temperatures were as follows: 5 min at 94°C, 1 min at the primer annealing temperature, and 6 min at 75°C. The reaction was completed by a 10-min incubation at 75°C. Amplification was performed in a Coy Temp Cycler II (Coy Corp., Grass Lake, Mich.). The PCR products were checked by gel electrophoresis. Primers, supplied by Genosys Biotechnologies, Inc. (The Woodlands, Tex.), were as follows (sequences are 5′ to 3′): no. 50 (20-mer near the 5′ end of the LEU2 insertion at PsADH2), TGT-CTG-TCA-CAC-CGA-CTT-GC; no. 34 (20-mer near the 3′ end of the LEU2 insertion at PsADH2), TGG-CTT-CAG-AGT-CAG-CT; no. 53 (18-mer near the 5′ end of the URA3 insertion at PsADH1), GCT-CTA-CAA-GGA-CAT-TCC; no. 35 (18-mer near the 3′ end of the URA3 insertion at PsADH1), CCT-GGT-TGG-ATT-TGA-GCG.
Analytical methods.
Determination of ethanol, xylose, and xylitol was done as described previously (9). Cell growth was determined daily by monitoring optical density at 600 nm (OD600), and cell dry weight was estimated by diluting a suspension to OD600s between 0.15 and 0.5. Under these conditions, an OD600 of 1.0 equals 0.24 g of cells/liter.
RESULTS
Identification of PsADH genes.
P. stipitis genomic DNA was digested with BamHI and SalI, and the fragments containing ADH genes were identified by Southern hybridization using S. cerevisiae ADH1 (ScADH1) as a probe. S. cerevisiae genomic DNA was used as a control. The P. stipitis genome showed two bands hybridizing to ScADH1. As expected, the S. cerevisiae genome showed at least three restriction fragments hybridizing to the probe.
Cloning of PsADH2.
One fragment of P. stipitis DNA, 6.6 kbp in length, strongly cross-hybridized to the ScADH1 coding sequences (Fig. 1A). A second fragment, about 20 kbp in length, hybridized to a lesser extent. The BamHI-SalI genomic fragments corresponding to 6.6 kbp were isolated from the gel to construct a DNA library pool for screening of the ADH gene of P. stipitis with ScADH1. Of 300 colonies screened by colony hybridization, two positive clones contained a 6.6-kbp BamHI-SalI fragment which hybridized to the ScADH1 probe following Southern blot analysis. These clones were further characterized by restriction mapping and Southern hybridizations. These DNA fragments, designated PsADH2, strongly hybridized to the ScADH1 probe and showed the same restriction patterns. Staggered deletions of about 500 bp each were prepared by opening the plasmid at one end of the insert, followed by timed digestions with exonuclease III (34). The deletion fragments were hybridized to the probe to localize and sequence the PsADH2 coding regions (Fig. 2B). The coding region of PsADH2 was tentatively located on a fragment with 4.1 and 1.5 kbp of the 5′- and 3′-flanking sequences, respectively. It was subsequently sequenced (see below).
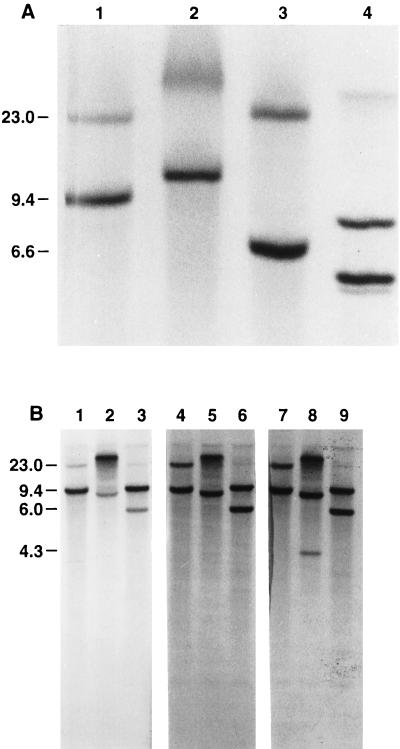
FIG. 1.
Identification of PsADH DNA fragments by Southern blot analysis. P. stipitis CBS 6054 genomic DNA was digested with the restriction enzymes indicated below and hybridized with coding sequences of the S. cerevisiae ADH1 gene (A, lanes 1 to 4; B, lanes 4 to 6), the S. cerevisiae ADH2 gene (B, lanes 7 to 9), and the P. stipitis ADH2 gene (B, lanes 1 to 3). S. cerevisiae (A, lane 4) and P. stipitis genomic DNAs (20 μg of each) were digested with BamHI (A, lanes 1 and 4; B, lanes 1, 4, and 7), SalI (A, lane 2), BamHI-SalI (A, lane 3), XbaI (B, lanes 2, 5, and 8), or BamHI-XbaI (B, lanes 3, 6, and 9). The P. stipitis ADH1 (BamHI-XbaI) and ADH2 (BamHI-SalI) DNA fragments were 6.0 and 6.6 kbp long, respectively. These fragments were isolated from the gel slice. The values on the left are molecular sizes in kilobase pairs.
Cloning of PsADH1.
PsADH1 was identified by hybridizing BamHI and XbaI digests of P. stipitis genomic DNA to the coding sequences of PsADH2, ScADH1, and ScADH2 (Fig. 1B). Two fragments, 9.4 and 6.0 kbp in length, strongly cross-hybridized to ScADH1 and ScADH2 (Fig. 1B, lanes 6 and 9). The hybridization of ScADH2 to the XbaI genomic fragments showed that a third fragment, about 4.3 kbp in length, cross-hybridized to a lesser extent to this probe (Fig. 1B, lane 8). In addition, PsADH2 cross-hybridized more to the 9.4-kbp band than to the 6.0-kbp band (Fig. 1B, lane 3). The putative ADH1 gene of P. stipitis hybridized more strongly to ScADH1 and ScADH2 than to PsADH2.
To isolate PsADH1, a DNA library was constructed as described previously and screened by colony hybridization with ScADH2 as the probe, and positive colonies were tested further. The restriction map of this fragment differed from that of PsADH2 in both the coding and flanking regions. The PsADH1 gene was localized and oriented on the 6.0-kbp fragment by hybridization to ScADH2 and sequence comparison with other ADH genes (Fig. 2A).
DNA sequence analysis.
Analysis of the sequence of PsADH1 revealed an open reading frame (ORF) of 1,047 nucleotides which codes for a polypeptide of 349 amino acids with a calculated molecular weight of 36,496. Potential promoter elements containing a TATA-like sequence were also found upstream from the putative ORF at position −177 to position −183 (Fig. 3A). However, very little identity can be observed between the upstream sequences of the ADH genes from P. stipitis and S. cerevisiae. PsADH2 contains an ORF of 1,047 nucleotides coding for a polypeptide of 349 residues and a calculated molecular weight of 36,541. The upstream sequences of PsADH2 show a putative TATA element (consensus sequence, TATAT/AAT/A), upstream of the RNA initiation site, at position −110 to position −117 (Fig. 3B).
FIG. 3.
Upstream sequences of PsADH genes and localization of their coding regions. (A) PSADH1 upstream region showing a putative TATA element (TATATATAA, bold and underlined), hairpins (bold), and repeats (underlined). (B) PSADH2 upstream region showing a putative TATA element (TATAAATA, bold and underlined) and hairpins (bold).
A comparison of the polypeptides encoded by these genes with the ADH polypeptides of S. cerevisiae and K. lactis showed considerable identity with the other yeast ADHs (Fig. 4). The amino acid sequences of the PsADH1 and PsADH2 isozymes are 80.5% identical to one another and are 71.9 and 74.7% identical to S. cerevisiae ADH1. The P. stipitis ADH proteins are presumed to be localized in the cytoplasm, as they do not possess the amino-terminal extension of mitochondrion-targeted ADHs (26, 44, 48).
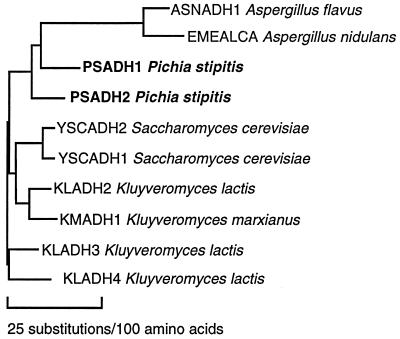
FIG. 4.
Phylogenetic tree showing similarity among yeast ADH genes. Sequences of 10 different ADH genes from the GenBank and EMBL data banks were compared by using the Genetics Computer Group (12) PileUp, Distances, and Grow Tree programs. Source names are shown.
Disruption of PsADH genes and analysis of disruptant strains.
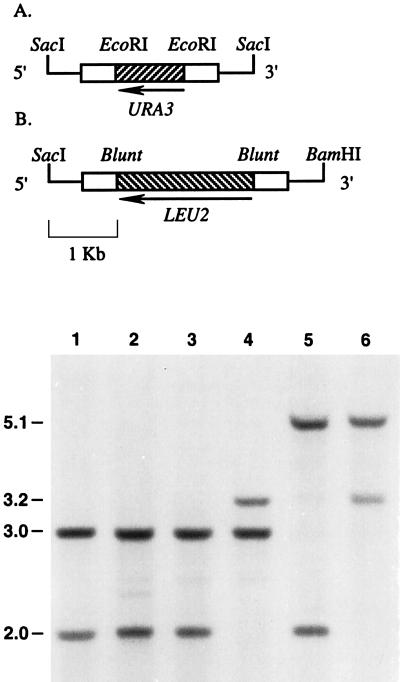
To examine the physiological role of the two P. stipitis ADH isozymes known, we created Psadh disruptant strains of P. stipitis by the one-step gene replacement technique (29). P. stipitis ura3 auxotroph PSU1 was transformed with the linear construct carrying the disrupted copy of Psadh1, and transformants were selected on glucose medium lacking uracil. The Ura+ transformants were picked randomly and screened by Southern blot analysis (Fig. 5). Genomic DNAs derived from these transformants, parental strain PSU1, and wild-type strain CBS 6054 were digested with SacI. Southern blotting of CBS 6054 and PSU1 genomic DNAs with a PsADH2 probe revealed 2.0- and 3.0-kbp bands for the PsADH1 and PsADH2 fragments, respectively (Fig. 5, lanes 1 and 2). Southern blotting of a Ura+ transformant probed with PsADH2 coding sequences revealed the pattern of fragments expected for a simple gene replacement at PsADH1 with the Psadh1::PsURA3 fragment: 3.2- and 3.0-kbp bands were detected for the SacI digests (Fig. 5, lane 4). The resulting Psadh1::PsURA3/Psadh1::PsURA3 homozygous disruptant was named PSU-218, and its fermentative behavior was tested further.
FIG. 5.
Construction and Southern blot analysis confirming replacement of PsADH genes. Linear fragments carrying disrupted copies of the PsADH1 (pJY102) (A) and PsADH2 (pJY202) (B) genes were constructed and introduced into the genome as described in the text. These disruption constructs contain approximately 1.0 kbp of 5′- and 3′-flanking PsADH1 (A) and PsADH2 (B) homology. Bold lines represent fragments containing 5′- and 3′-flanking regions used for gene replacement. White boxes indicate PsADH1 (A) and PsADH2 (B) coding regions, and striped boxes represent functional LEU2 (A) and URA3 (B) genes from P. stipitis. Southern hybridization results are shown at the bottom. Strain CBS 6054 (lane 1) is the P. stipitis wild type. P. stipitis PSU1 (lane 2) and PLU20 (lane 3) are parental strains used for PsADH1 and PsADH2 gene replacements, respectively. PSU218 (lane 4) and PLU123 (lane 5) are strains disrupted in PsADH1 and PsADH2, respectively. PLU1209 (lane 6) is disrupted in both PsADH1 and PsADH2.
In the case of the PsADH2 disruption, PsLEU2 was inserted into the PsADH2 coding region (see Materials and Methods). A 4.5-kbp fragment of pJY202 containing the disrupted copy of Psadh2 was generated by digestion with restriction enzymes BamHI and SacI and then transformed into the P. stipitis ura3 Δleu2 auxotroph PLU20 in the presence of restriction enzymes BamHI and SacI. The resulting transformants were selected for growth in the absence of leucine and screened by PCR by using primers specific for the point at which the PsLEU2 DNA fragment was inserted into the PsADH2 gene. One of the transformants selected for growth in the absence of leucine, named PLU-123, showed a 2.1-kbp DNA fragment amplified by PCR, as expected for correct integration (data not shown). Correct homozygous integration of the fragment carrying the disrupted copy of Psadh2 (Psadh2::PsLEU2) into the corresponding genomic locus showed that 3.0-kbp fragments were replaced in the Southern blot analysis by new fragments of increased length because of the insertion of the 2.1-kbp fragments of PsLEU2 (Fig. 5, lane 5).
We then constructed a strain lacking both PsADH genes. PLU-123, a derivative of PLU20 carrying disrupted copies of Psadh2, was transformed with the linear fragments which we previously had used to disrupt PsADH1 in PSU1 and screened by PCR using primers specific for the point at which the PsURA3 DNA fragment was inserted into the Psadh1 gene. Two of the initial Ura+ transformants were found to be Psadh1::PsURA3/Psadh1::PsURA3 and Psadh2::PsLEU2/Psadh2::PsLEU2 homozygous disruptants. The correct integration of each fragment into the corresponding genomic locus was confirmed by Southern blot analysis: 3.2-kbp (Psadh1::PsURA3) and 5.1-kbp (Psadh2::PsLEU2) fragments were detected for both PsADH1 and PsADH2 gene replacement (Fig. 5, lane 6). This Psadh multiple disruptant strain, PLU-1209, was tested along with the previously obtained disruptants for their fermentative and growth properties.
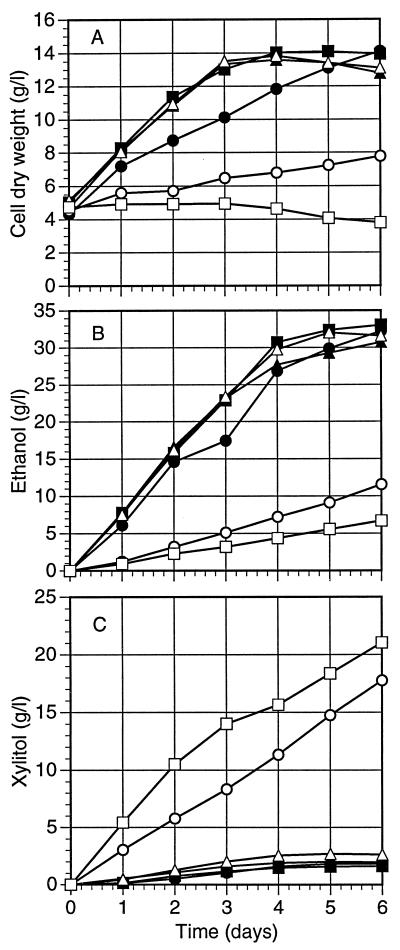
Fermentation of xylose.
The Psadh disruptant strains were compared to appropriate wild-type and parental strains for ethanol production from xylose under oxygen limitation. Compared with those of the control strains, the apparent ethanol production and growth rate of PSU-218 were very low (Fig. 6). At the same time, the level of xylitol production of PSU-218 and double disruptant strain PLU-1209 was much higher than that of the corresponding control strains. On the other hand, disruption of PsADH2 had little effect on growth rate, ethanol production, or xylitol production. Interestingly, Psadh double disruptant strain PLU-1209 failed to grow on xylose but still produced small amounts of ethanol under these conditions.
FIG. 6.
Cell growth (A), ethanol production (B), and xylitol production (C) of wild-type, parental, and disruptant strains of P. stipitis under fermentative conditions. Inocula were grown in flasks for 48 h, and initial cell densities of each strain were adjusted to be similar. Each value represents the mean of two determinations. Symbols: ▪, CBS 6054 (wild type); •, PSU1; ▴, PLU20; ▵, PLU123 (Psadh2 disruptant); ○, PSU218 (Psadh1 disruptant); □, PLU1209 (Psadh double disruptant).
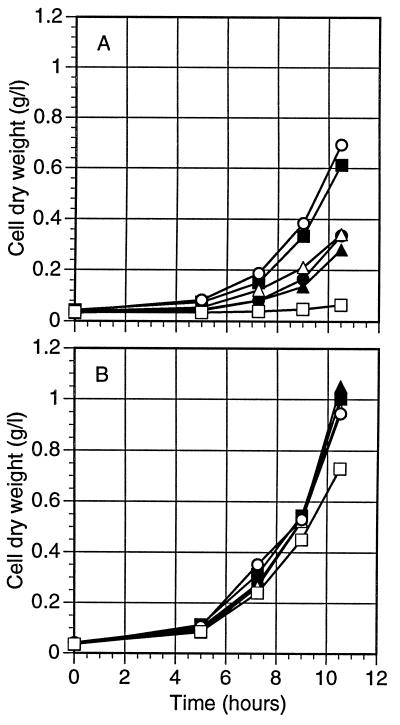
We then analyzed the ability of the Psadh disruptant strains to use ethanol as a sole carbon source under fully aerobic conditions. As can be seen in Fig. 7A, PSU-218 grew as well as CBS 6054 and better than its parent, PSU1, when cultivated in liquid minimal medium (YNB) containing ethanol, while PLU-1209 failed to grow under these conditions. In contrast, PLU-123 grew on ethanol at a rate similar to that of its parental strain, PLU20, indicating that PsADH2 is dispensable for growth of P. stipitis on ethanol unless PsADH1 is disrupted. PLU-123, a Psadh2 disruptant, and PLU-1209, a Psadh1 and Psadh2 double disruptant, were able to grow on glycerol at rates similar to those of their parental and wild-type strains (Fig. 7B). Growth of the double disruptant on glycerol indicated that its lack of growth on ethanol was not attributable to a loss of respiratory capacity.
FIG. 7.
Aerobic growth of P. stipitis wild-type CBS 6054, parental, and Psadh disruptant strains on either ethanol (A) or glycerol (B). Cells were grown in YNB with either 2% ethanol or 3% glycerol plus 100 μg of leucine or uridine/ml, as needed, with shaking in a 125-ml baffled flask at 180 rpm and 30°C. Symbols: ▪, CBS 6054 (wild type); •, PSU1 (ura3-2/ura3-2); ▴, PLU20 (ura3-3/ura303 Psleu2Δ-1/Psleu2Δ-1); ▵, PLU123 (Psadh2 disruptant); ○, PSU218 (Psadh1 disruptant); □, PLU1209 (Psadh double disruptant).
Finally, each of the two ADH activities is able to convert ethanol to acetaldehyde and is sufficient to allow cell growth. Moreover, each Psadh disruptant strain was still able to accumulate ethanol, although in different amounts (PLU-123 > PSU-218 > PLU-1209) when grown on xylose under oxygen-limited conditions.
DISCUSSION
The ADH isozymes in P. stipitis are encoded by multiple genes. The exact number is unknown. Hybridization of ScADH2 to the XbaI genomic fragments indicated that the P. stipitis genome may contain three ADH genes. Two fragments, about 9.0 and 23.0 kb in length, strongly cross-hybridized to ScADH2 (Fig. 1B, lane 8). A third fragment, 4.3 kbp in length, hybridized to a lesser extent. The inability of the Psadh double disruptant to grow on ethanol under full aerobiosis implies that no other ADH activities are present under these conditions. The gene for a third isoenzyme may be present but not expressed on ethanol under aerobic conditions.
We cloned and sequenced two structural genes from P. stipitis, designated PsADH1 and PsADH2. They both have uninterrupted ORFs of 1,047 nucleotides and show high conservation with respect to other yeast group I ADH proteins. The two ADH isozymes of P. stipitis are 80.5% identical at the protein level and 79.5% identical in the coding region at the nucleotide level. PsADH2 more strongly hybridized to ScADH1 and ScADH2 than to PsADH1 (Fig. 1B, lanes 3, 6, and 9). This may be an artifact of DNA conformation on that fragment, since P. stipitis ADH genes are structurally closer to one another than to any of the S. cerevisiae or K. lactis genes. Our designation of the isozymes as PsADH1 and PsADH2 is in keeping with the similarity of their physiological roles to those of ADH1 and ADH2 of S. cerevisiae. However, they appear to have evolved from one another independently of the divergence between ScADH1 and ScADH2 (Fig. 4). Notably, the deduced amino acid sequences of the P. stipitis isozymes showed a conserved amino acid residue (Lys305) that is thought to be involved in the substrate-binding cleft. S. cerevisiae ADH1 has a methionine in this position, which is thought to contribute to the narrow substrate specificity of this enzyme (38).
Analysis of the 5′-flanking region of PsADH1 and PsADH2 revealed consensus sequences for a putative TATA box and an ATG context, which is optimal for highly expressed genes from S. cerevisiae (17a). Several direct repeats, including six direct repeats with a slight variant of the consensus sequence A/TGGAGA, and inverted repeats were found in the 5′-flanking region of PsADH1. The functional significance of these sequences has not been determined, yet it is tempting to speculate that they represent a recognition site(s) for a trans-acting factor(s) involved in the regulation of PsADH1 expression. Dyad symmetry is typical of the sequences recognized by dimeric DNA-binding proteins containing bacterial helix-turn-helix motifs, and the promoter region implicated in transcriptional regulation of S. cerevisiae ADH2 also contains seven repeats with the consensus sequence A/TGGAGA and a 22-bp sequence with perfect dyad symmetry (2, 39, 49).
Bennetzen and Hall (3) suggested that there is a correlation between the expression level of a gene and its codon usage bias. The codon usage rules derived from S. cerevisiae genes seem to apply to P. stipitis as well. The PsADH1 and PsADH2 genes show a highly biased codon usage very similar to that observed in the corresponding genes of S. cerevisiae (data not shown), where only 25 preferred codons are used extensively, whereas the URA3 gene of P. stipitis (47) shows a poorly biased codon usage pattern, as does the same gene of S. cerevisiae and K. lactis, where it is less highly expressed (40).
Disruption of PsADH1 and PsADH2 was more difficult than expected. Melake et al. (24) proposed that wild-type strains of P. stipitis are haploids. Therefore, disruption should have been easy to accomplish. However, disruption studies occasionally showed what appeared to be heterozygotes, indicating that at least some of the parental strains (or resulting disruptants) are diploids. In contrast to that in S. cerevisiae, the frequency of homologous recombination at any given locus in P. stipitis seems to be low and varies somewhat from locus to locus within the genome (6). When these disruption studies were carried out, nonhomologous integration of introduced DNA was more common than homologous integration. The basis for the variability and low frequency is not clear. However, several aspects of DNA metabolism, such as DNA repair and transcription, are closely interrelated with homologous recombination (17, 37).
In the case of the PsADH1 gene disruption, only 1 of 17 Ura+ transformants was a homozygous integrant at the corresponding genomic locus. In addition to this homozygous integration, numerous URA3 gene conversion, heterozygous, and random-integration events were detected (data not shown). Schiestl and Petes (36) found that introducing the restriction enzyme BamHI together with a BamHI-cut DNA fragment increased the number of transformed cells and showed that the DNA fragment was often integrated into BamHI sites of the host genome. This surprising result led us to explore restriction enzyme-mediated integration in P. stipitis for disruption of PsADH2. This approach proved successful.
We created P. stipitis strains disrupted in each or both structural ADH genes. Disruption of PsADH1 clearly affected the growth rate and ethanol production on xylose under oxygen-limiting conditions. The increase in xylitol production by the Psadh1 disruptant indicates that in the absence of PsADH1, NADH accumulates and favors xylitol production. PsADH1 appears to be the principal gene responsible for ethanol production because its loss results in slower growth, lower ethanol production, and much greater xylitol production under oxygen-limited conditions, whereas the loss of PsADH2 has none of these effects. Disruption of PsADH1 in P. stipitis causes this yeast to make even more xylitol than does Candida shehatae or Pachysolen tannophilus, where significant xylitol production has been observed under oxygen-limited conditions (15). Xylitol production has been explained by Debus et al. in terms of an electron sink for NADPH, generated in the phosphogluconate pathway (10). On the basis of our present studies, we propose that in P. stipitis, xylitol production is normally low because PsADH1 suppresses the intracellular level of NADH. When PsADH1 is disrupted, NADH accumulates. Accumulation of NADH would, in turn, shift the equilibrium of the XDH-mediated reaction to favor xylitol over xylulose formation. Fermentative ADH activity is therefore essential not only for ethanol production but also for maintenance of redox balance in the cytoplasm. The accumulation of reducing equivalents in the form of NADH does not seem to be relieved by reoxidation under oxygen-limited conditions or by NADH-linked XR activity. The dependence of xylose metabolism on NADH recycling by PsADH1 could be why relatively slow growth was observed with Psadh1 strains of P. stipitis (PSU-218 and PLU-1209) under oxygen-limited conditions. Psadh1 strains still take up xylose, grow, and accumulate ethanol—albeit to a lesser extent than the parent under oxygen-limited conditions. On the other hand, the Psadh double-disruptant strain failed to grow on xylose and produced even less ethanol and more xylitol under these conditions. These results suggest that the PsADH2 isozyme is also involved in xylose fermentation and growth, although it probably plays a minor role compared to PsADH1.
The analysis of the Psadh disruptant strains, which contained either one or the other of the residual ADH activities, showed that all were able to grow aerobically on media containing ethanol as a sole carbon source. This result suggests that either PsADH1 or PsADH2 can confer the ability to oxidize ethanol. PSU-218 (Psadh1::PsURA3) actually grew better on ethanol than did the parental or wild-type strain, implying that PsADH2 might be overexpressed and more important under these conditions. Because disruption of PsADH2 had no discernible effect on fermentation or growth on xylose, an open question is whether PsADH2 is expressed only when PsADH1 is disrupted or whether it plays some other role.
The simultaneous presence of ethanol-producing and ethanol-oxidizing activities in the cytoplasm could result in a futile cycle. This could be why KlADH4 is compartmentalized within mitochondria in K. lactis (23) and catabolite repression of ADH2 occurs in S. cerevisiae (30). Oxygen-dependent regulation of PsADH expression could avoid futile cycling in the cytoplasm of P. stipitis if respirative enzyme activities were repressed and fermentative enzyme activities were induced under oxygen-limited conditions. In P. stipitis, a Crabtree-negative yeast, fermentation enzymes such as ADH and PDC are induced only under oxygen limitation (25), and no fermentation occurs on medium containing 3% xylose under fully aerobic conditions (7). This is in keeping with a hypothesis of oxygen-dependent regulation of ADH expression in P. stipitis, as discussed above. The different behavior of these yeasts compared to that of S. cerevisiae is most probably explained by differences in the nature of the fundamental mechanisms by which fermentation is regulated.
The Psadh double-disruptant strains are not able to utilize ethanol, indicating that no other important ADH activities are present in P. stipitis when it is growing on ethanol under aerobic conditions. However, the Psadh double disruptants still produce ethanol from xylose, implying that a third enzyme might be expressed under oxygen-limited conditions. ADH null strains of both S. cerevisiae (14) and K. lactis (31) can still produce residual amounts of ethanol when grown aerobically on 2% glucose. The biochemical basis of this phenomenon remains unclear. In any case, the addition of a respiratory inhibitor such as 2,4-dinitrophenol, sodium azide, or potassium cyanide should immediately block the production of ethanol in the Psadh null strain because unimpaired mitochondrial function is necessary for growth on and alcoholic fermentation of xylose in P. stipitis (20). The function of this respiratory activity is not clear, but it could be necessary to generate energy for xylose transport (19). Experiments are under way to unravel the molecular basis for the expression of PsADH genes in this yeast.
ACKNOWLEDGMENTS
This research was supported by National Renewable Energy Laboratory subcontract XAU-4-11193-02 and by USDA NRICGP grant 96-35500-3172.
We thank Mark Davis for technical assistance in fermentation analysis and Brian Davis for useful suggestions.
REFERENCES
- 1.Amore R, Kotter P, Hollenberg C P. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene. 1991;109:89–97. doi: 10.1016/0378-1119(91)90592-y. [DOI] [PubMed] [Google Scholar]
- 2.Beier D R, Sledziewski A, Young E T. Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol Cell Biol. 1985;5:1743–1749. doi: 10.1128/mcb.5.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennetzen J L, Hall B D. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 4.Bennetzen J L, Hall B D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. I. J Biol Chem. 1982;257:3018–3025. [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollag R J, Waldman A S, Liskay R M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–205. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- 7.Bruinenberg P M, de Bot P H M, van Dijken J P, Scheffers W A. NADH-linked aldose reductase: a key to anaerobic alcoholic fermentation of xylose by yeasts. Appl Microbiol Biotechnol. 1984;19:256–260. [Google Scholar]
- 8.Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. I. Isolation and genetic analysis of adh mutants. Mutat Res. 1975;29:315–326. [Google Scholar]
- 9.Dahn K M, Davis B P, Pittman P E, Kenealy W R, Jeffries T W. Increased xylose reductase activity in the xylose-fermenting yeast Pichia stipitis by overexpression of XYL1. Appl Biochem Biotechnol. 1996;57/58:267–276. doi: 10.1007/978-1-4612-0223-3_24. [DOI] [PubMed] [Google Scholar]
- 10.Debus D, Methner H, Schulze D, Dellweg H. Fermentation of xylose with the yeast Pachysolen tannophilus. Eur J Appl Microbiol Biotechnol. 1983;17:287–291. [Google Scholar]
- 11.Denis C L, Ferguson J, Young E T. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983;258:1165–1171. [PubMed] [Google Scholar]
- 12.Denis C L, Ciriacy M, Young E T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981;148:355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drewke C, Thielen J, Ciriacy M. Ethanol formation in adh0 mutants reveals the existence of a novel acetaldehyde-reducing activity in Saccharomyces cerevisiae. J Bacteriol. 1990;172:3909–3917. doi: 10.1128/jb.172.7.3909-3917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du Preez J C, van der Walt J P. Fermentation of d-xylose to ethanol by a strain of Candida shehatae. Biotechnol Lett. 1983;5:357–362. [Google Scholar]
- 16.Du Preez J C, van Driessel B, Prior B A. Effect of aerobiosis on fermentation and key enzyme levels during growth of Pichia stipitis, Candida shehatae and Candida tenuis on d-xylose. Arch Microbiol. 1989;152:143–147. [Google Scholar]
- 17.Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Hamilton R, Wantanable C K, de Boer H A. Compilation and comparison of the sequence context around the AUG start codons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffries T W. Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol. 1983;27:1–32. doi: 10.1007/BFb0009101. [DOI] [PubMed] [Google Scholar]
- 20.Lighthelm M E, Prior B A, du Preez J C. The effect of respiratory inhibitors in the fermentative activity of Pichia stipitis, Pachysolen tannophilus and Saccharomyces cerevisiae under various conditions of aerobiosis. Appl Microbiol Biotechnol. 1988;29:67–71. [Google Scholar]
- 21.Lu P, Davis B P, Jeffries T W. Cloning and characterization of two pyruvate decarboxylase genes from Pichia stipitis CBS 6054. Appl Environ Microbiol. 1998;64:94–97. doi: 10.1128/aem.64.1.94-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, P., J. Hendrick, B. P. Davis, and T. W. Jeffries. Disruption of the β-isopropylmalate dehydrogenase gene (LEU2) of Pichia stipitis with URA3 and recovery of the double auxotroph. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 23.Mazzoni C, Saliola M, Falcone C. Ethanol-induced and glucose-insensitive alcohol dehydrogenase activity in the yeast Kluyveromyces lactis. Mol Microbiol. 1992;6:2279–2286. doi: 10.1111/j.1365-2958.1992.tb01403.x. [DOI] [PubMed] [Google Scholar]
- 24.Melake T, Passoth V, Klinner U. Characterization of the genetic system of the xylose-fermenting yeast Pichia stipitis. Curr Microbiol. 1996;33:237–242. doi: 10.1007/s002849900106. [DOI] [PubMed] [Google Scholar]
- 25.Passoth V, Zimmermann M, Klinner U. Peculiarities of the regulation of fermentation and respiration in the Crabtree-negative, xylose-fermenting yeast Pichia stipitis. Appl Biochem Biotechnol. 1996;57/58:201–212. doi: 10.1007/BF02941701. [DOI] [PubMed] [Google Scholar]
- 26.Pilgrim D, Young E T. Primary structure requirements for correct sorting of the yeast mitochondrial protein ADHIII to the mitochondrial matrix space. Mol Cell Biol. 1987;7:294–304. doi: 10.1128/mcb.7.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 28.Rose M D, Winston F, Hieter P. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 29.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 30.Russel D W, Smith M. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983;258:2674–2682. [PubMed] [Google Scholar]
- 31.Saliola M, Bellardi S, Marta I, Falcone C. Glucose metabolism and ethanol production in adh multiple and null mutants of Kluyveromyces lactis. Yeast. 1994;10:1133–1140. doi: 10.1002/yea.320100902. [DOI] [PubMed] [Google Scholar]
- 32.Saliola M, Gonnella R, Mazzony C, Falcone C. Two genes encoding putative mitochondrial alcohol dehydrogenases are present in the yeast Kluyveromyces lactis. Yeast. 1991;7:391–400. doi: 10.1002/yea.320070409. [DOI] [PubMed] [Google Scholar]
- 33.Saliola M, Shuster J R, Falcone C. The alcohol dehydrogenase system in the yeast, Kluyveromyces lactis. Yeast. 1990;6:193–204. doi: 10.1002/yea.320060304. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz M, Freisem-Rabien U, Jessberger R, Doerfler W. Transcriptional activities of mammalian genomes at sites of recombinations with foreign DNA. J Virol. 1987;61:344–353. doi: 10.1128/jvi.61.2.344-353.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shain D H, Salvadore C, Denis C L. Evolution of the alcohol dehydrogenase (ADH) genes in yeast: characterization of a fourth ADH in Kluyveromyces lactis. Mol Gen Genet. 1992;232:479–488. doi: 10.1007/BF00266253. [DOI] [PubMed] [Google Scholar]
- 39.Shuster J, Yu J, Cox D, Chan R V L, Smith M, Young E T. ADR1-mediated regulation of ADH2 requires an inverted repeat sequence. Mol Cell Biol. 1986;6:1894–1902. doi: 10.1128/mcb.6.6.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuster J, Moyer D, Irvine B. Sequence of the Kluyveromyces lactis URA3 gene. Nucleic Acids Res. 1987;15:8573. doi: 10.1093/nar/15.20.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoog K, Hahn-Hagerdal B. Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl Environ Microbiol. 1990;56:3389–3394. doi: 10.1128/aem.56.11.3389-3394.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 43.Specht C A, DiRusso C C, Novotny C P, Ulrich R C. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem. 1982;119:158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- 44.van Loon A P G M, Young E T. Intracellular sorting of alcohol dehydrogenase isozymes in yeast: a cytosolic location reflects absence of an amino-terminal targeting sequence for the mitochondrion. EMBO J. 1986;5:161–165. doi: 10.1002/j.1460-2075.1986.tb04191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Urk H, Voll W S L, Scheffers W A, van Dijken J P. Transient-state analysis of metabolic fluxes in Crabtree-positive and Crabtree-negative yeasts. Appl Environ Microbiol. 1990;56:281–287. doi: 10.1128/aem.56.1.281-287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson V M, Paquin C E. Homology of Saccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Mol Gen Genet. 1987;209:374–381. doi: 10.1007/BF00329668. [DOI] [PubMed] [Google Scholar]
- 47.Yang V W, Marks J A, Davis B P, Jeffries T W. High-efficiency transformation of Pichia stipitis based on its URA3 gene and a homologous autonomous replication sequence, ARS2. Appl Environ Microbiol. 1994;60:4245–4254. doi: 10.1128/aem.60.12.4245-4254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young E T, Pilgrim D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3024–3034. doi: 10.1128/mcb.5.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J, Donoviel M S, Young E T. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:34–42. doi: 10.1128/mcb.9.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]