Abstract
The inflammatory disease ulcerative colitis (UC) is multifaceted, immune-mediated, chronic, and relapsing, which is considered to be mainly driven by dysregulated mucosal immune response. The remission of the inflammatory response is a marker of mucosal healing, relating to the low risk of hospitalizations, colorectal cancer, and colectomy. In spite of this, it is still unclear what the key immunological mechanism is which contributes to UC. Here, we explored the immune mechanism and related key genes underlying the state of inflammation in UC. Co-expression networks were constructed based on the expression profiles of immune-related genes in GSE179285. Using Weighted Gene Co-expression Network Analysis and Protein-protein interactions analysis, common hub genes were identified in the module of interest. Then, screening of real hub genes, significantly differentially expressing in inflamed UC, was carried out by Differential Expression Genes Analysis of GSE75214, GSE53306, and GSE6731datasets and immunohistochemistry of clinical samples. The diagnosis Capacity of the hub gene was identified by “glm” function in R. The potential key immune-related mechanisms were investigated using functional enrichment analysis and gene set enrichment analysis (GSEA). Bioinformatics tools were used to predict potential upstream transcription factors (TF), including the UCSC genome browser, correlation analyses, and JASPAR browser. The analysis revealed the blue module, consisting of 227 immune-related genes, showed the highest correlation with inflamed UC. And then, forty-three common candidates were distinguished. S100A9 was identified within the key module as a real hub gene with good diagnostic performance. The immune genes in the blue module were markedly enriched in the Cytokine-Cytokine receptor interaction. S100A9 most likely gets involved NOD-like receptor (NLR) signaling pathway. SPI1 showed the strongest likelihood to be the regulator. S100A9 was identified as the real immune-related hub gene for inflamed UC. Both diagnosis and remission may be aided by its high expression in the inflamed UC.
Keywords: biomarker, immune-related mechanism, integrated bioinformatic analyses, remission, ulcerative colitis, weighted gene co-expression network analysis
1. Introduction
Crohn disease and ulcerative colitis (UC) belong to the category of inflammatory bowel disease, which were considered independent risk factors for colorectal cancer.[1,2] Of those, UC is multifaceted, immune-mediated, chronic, and relapsing, which affects the rectum and colorectal mucosa.[3] The patients suffer from painful and serious complicated conditions, such as varying degrees of abdominal pain, recurrent bloody diarrhea, and rectal emergencies.[4] The incidence of UC has been steadily rising worldwide in recent years, especially in Asia.[5]
Accumulating studies have provided evidence that the pathogenesis of UC involves multifactorial, complex, and interrelated processes, mainly characterized by epithelial barrier defects and dysregulated immune responses.[6] Immune cells and inflammatory mediators may consecutively disrupt barrier disruption and culminate in progressive damage and insufficient repair of the intestinal tract, which attracted extensive attention from investigators.[7] Camoglio L et al[8] first proposed that type 2 T helper cell is an important driver of UC. T cells migrate and accumulate in the gut through the homing mechanism, then together constitute a complex immune context of UC with local immune cells, including innate lymphoid cells, neutrophils, macrophages, and dendritic cells.[9] The increased production of pro-inflammatory cytokines by mucosal immune cells in UC has been demonstrated by several studies. The levels of IL-13, TNF, IL-23, IL-9, and IL-36 were reported to specifically increase in the local intestinal mucosa of patients with UC.[10–15] Mantovani A et al[16] revealed that IL-1 family cytokines have pro-inflammatory signaling effects on chronic intestinal inflammation by activating NF-κB pathway. Despite some accomplishments in current research, it is still unclear what the key immunological mechanism is which contributes to UC.
Over the past decade, therapeutic goals for UC have changed from treating symptoms only to histological remission, which was related to the low risk of hospitalizations, colorectal cancer, and colectomy.[17] In patients failing this first-line therapy, immunosuppressive agents and biologics, which suppressed and modulate the aberrant intestinal immune response, achieved some efficacy.[18] Thus, it is of interest to understand the immune mechanism and related key genes underlying the state of inflammation in UC.
In the present study, we collected datasets consisting of inflamed UC, uninflamed UC, and healthy control, then explored the key immune-related genes and biological processes contributing to the inflammation of UC by weighted gene co-expression network analysis (WGCNA). The hub gene was selected and identified by its differential expression in the inflamed UC group. Furthermore, Functional enrichment analysis of the hub module and hub gene revealed the associated crucial immune-related processes. The association of clinical data with molecular mechanisms may lead to the identification of new biomarkers for diagnosis and remission.
2. Materials and Methods
2.1. Data collection, filtering, and preprocessing
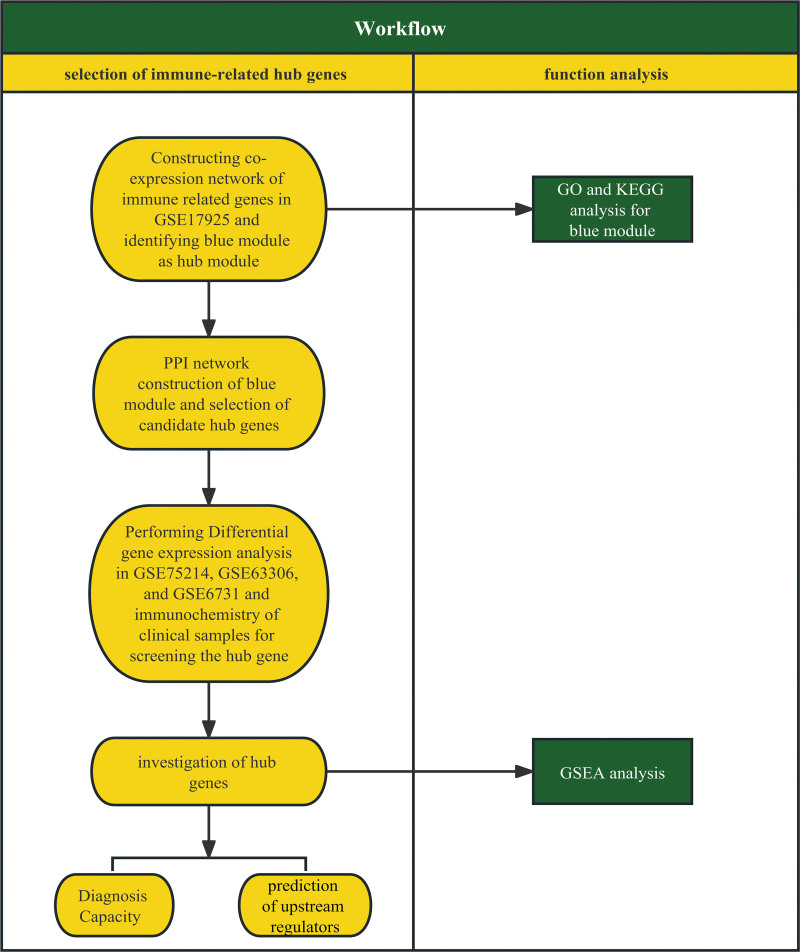
A public repository of microarray data GEO was searched for UC datasets according to the workflow (Fig. S1, http://links.lww.com/MD/K307). Related datasets were rigorously screened to remove non-compliant datasets, and 4 datasets (GSE179285, GSE75214, GSE53306, and GSE6731) remained for further integrated bioinformatics analyses. GEOquery package was used to acquire Normalized datasets from the GEO database.[19] Besides, corresponding annotation files were downloaded directly from the GEO. The clinical information of the included 4 datasets were provided in Table 1. In Figure 1, a swimlane diagram shows the study design.
Table 1.
Characteristics of the 4 studies included in the bioinformatic anal.
Figure 1.
A swim lane diagram of the study design. Methods regarding the selection of the real hub gene were shown in the left lane, and methods regarding function analyses and exploration of mechanisms were shown in the right lane.
2.2. Co-expression network construction of immune-related genes
GSE179285 with a similar and relatively large sample size for each group was used to construct WGCNA.[20] Including chemokine, MHC, cytokine, cytokine receptor, HLA, and many other immune gene signatures, 1577 immune gene signatures were obtained.[21] To explore characteristic immune-related genes or gene modules highly related to inflamed intestinal areas of patients with UC, the immune genes of GSE179285 were extracted to generate the signature matrix, which was applied to construct a gene co-expression network.
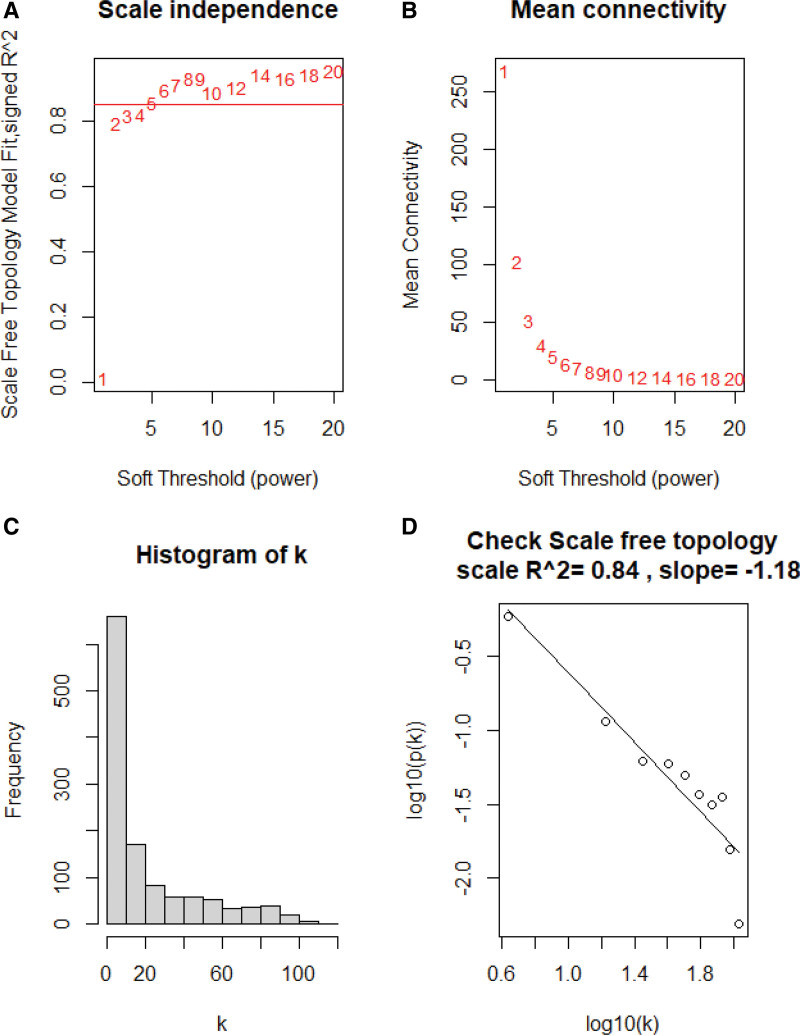
For WGCNA, we first conducted the hierarchical cluster analysis to check the samples’ heterogeneity. Then, we performed the pickSoftThreshold function to calculate the value achieving approximate scale-free topology (R2 > 0.85), which was used to construct a scale-free network. Scale-free network behavior was examined using histograms of connectivity k and scale-free topology plots.
2.3. Functional enrichment analysis
The genes in the hub module were input into the Metascape database (https://metascape.org/) for integrated gene enrichment analysis and visualization, which aggregates multiple databases (Kyoyo Encyclopedia of Genes and Genomes, Gene Ontology, DrugBank, UniProt).[22] After that, interpretable outputs were generated using the “Express Analysis” option. Based on P < .01 taken as significantly different, the analysis results were ranked according to the enrichment score.
2.4. Identification of candidate key immune genes
For the interesting module, genes with module membership >0.8 and gene significance >0.2 were chosen. The immune genes in the interesting module were used in the construction of a protein-protein interaction (PPI) network using STRING (https://string-db.org/).[23] In the following steps, PPI networks were analyzed and visualized by Cytoscape (v3.9.1).[24] The degree of the genes was calculated by plug-in CytoHubba to predict the hub nodes, which with node connectivity > ratio of total edges to total nodes. Based on the intersection of hub genes analyzed by WGCNA and PPI, candidate immune genes were identified.
2.5. Determination of the final hub gene by GEO and clinical samples
In order to further screen hub genes by external data, we compared the expression of candidate key immune genes in 3 other datasets downloaded from GEO (GSE75214, GSE53306, and GSE6731). Differential Expression Genes (DEGs) Analysis was conducted by the R package “Limma” (UC with inflammation vs UC without inflammation and UC with inflammation vs control).
The DEGs in the group of UC with inflammation were verified by immunohistochemistry on clinical samples. In Hangzhou, Zhejiang, China, Sections of paraffin-embedded tissue samples were collected at the Zhejiang University School of Medicine, Department of Pathology. Ethical review board approval was obtained from Ethics Committee of Sir Run Run Shaw Hospital (2023-501-01). Specimens were collected with informed consent and were diagnosed as UC by professional pathologists. To dewax sections, slides of clinical samples were baked at 60°C for 1h, followed by incubation in xylene I and xylene II, for 10 minutes each time. Gradient alcohol solutions, including 95% ethanol, 85% ethanol, and 75% ethanol, were used to hydrate the sections for 3 minutes each. We blocked endogenous peroxidase activity on the slides by incubating them with 3% hydrogen peroxide for 10 minutes after 3 5-minute washes with PBS. To repair antigens, the slides were then boiled using a pressure cooker with EDTA antigen restore solution for 1.5 minutes. For 4 hours, the tissue was incubated with antibodies against S100A9 (1:250, ab92507, Abcam) at room temperature after being washed with PBS for 3 minutes. Incubation was performed at room temperature for 15 minutes with MaxVision Mouse/Rabbit (Kit-5020, Maixin) after washing the slides with PBS. Then DAB and hematoxylin staining were conducted. Next, the slides were incubated with gradient ethanol (75% ethanol, 85% ethanol, 95% ethanol, absolute ethanol) and xylene for dehydration. Finally, the slides were sealed with neutral balsam. We observed and scanned slides using the scanner KF-PRO-020. To evaluate the staining index (values 0–12), we calculated the product of the positive staining score (negative, 0; weak, 1; moderate, 2; strong, 3) and the staining intensity score (<25%, 1; 25–50%, 2; 50–75%, 3; >75%, 4).
2.6. Investigation of diagnosis capacity of the hub gene
As a result of merging datasets and removing batch effect from these using the Sva R package, we obtained new proportions for training and testing of 0.7 and 0.3, respectively. Then the “glm” function in R was used to measure Logistic regression of the hub gene. The InformationValue R package was used to assess model accuracy.
2.7. Gene set enrichment analysis (GSEA)
A median of hub gene expression levels was used to divide GSE179285 samples into 2 groups (high and low). We then performed GSEA (version 4.3.2) to detect related signaling pathways in the high-expression group. It was recommended that the false discovery rate should be equal to or <0.25 and that the P value should be equal to or <.05.
2.8. Prediction of transcription factor
Predicting transcription factors (TF) of the hub gene was done using the UCSC genome browser (http://genome.ucsc.edu/). Then correlation analyses between the potential TFs and targeted hub gene were performed in GSE179285 and GSE75214. The TFs with correlation coefficient values > 0.6, which show a strong positive correlation, were selected. To further confirm the correlation between the selected TFs and the hub gene, Several binding sites for the hub gene were predicted using JASPAR (http://jaspar.genereg.net/).
3. Results
3.1. Weighed co-expression network construction and hub module identification
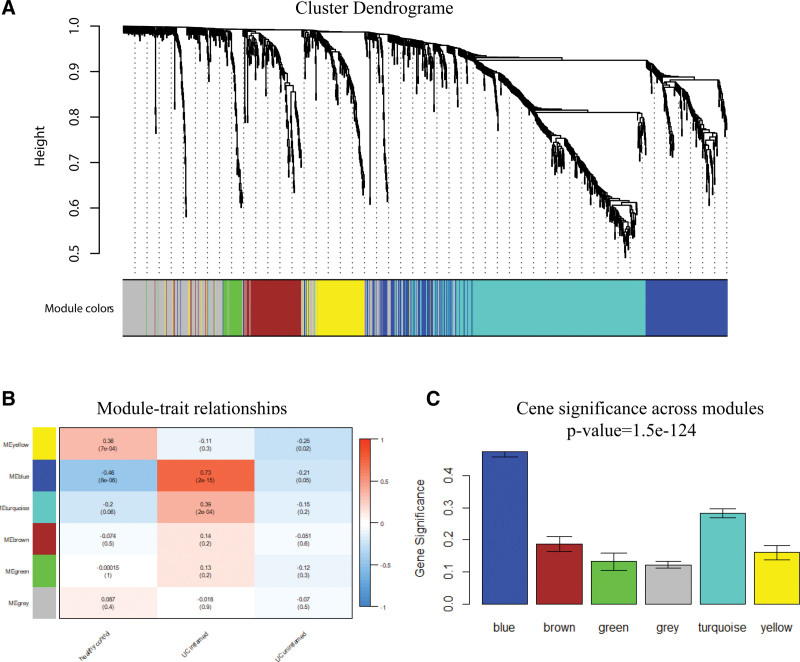
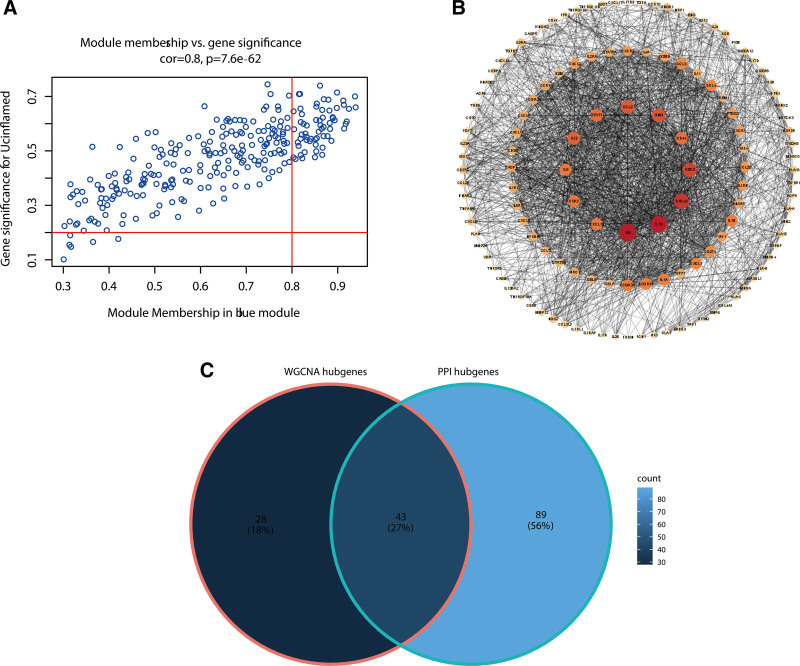
The expression matrix of immune genes in the GSE179285 was obtained after data preprocessing. Figure S2, http://links.lww.com/MD/K308 shows that all samples passed the first quality check (Fig. S2, http://links.lww.com/MD/K308). A scale-free network was constructed using the power of β = 5 (scale-free R2 = 0.84) as the soft threshold (Fig. 2). According to the similarity of expression patterns across samples, the immune genes were clustered into 6 modules. The module names and their corresponding number of expressed genes in that module were listed in Table S1, http://links.lww.com/MD/K309. Among all modules, the blue module showed the highest correlation with inflamed UC based on ME and MS (Fig. 3). The gene list in the blue module was displayed in Table S2, http://links.lww.com/MD/K310.
Figure 2.
Selection soft-thresholding power in the weighted gene co-expression network analysis. (A) Scale-free fit index of various soft-thresholding powers (β). (B) mean connectivity of various soft-thresholding powers(β). (C) Histogram of the connectivity distribution when β = 5. (D) scale-free topology checking when β = 5.
Figure 3.
The correlation between the immune-related modules and the clinical phenotypes of Ulcerative colitis (UC). (A) Dendrogram of all immune-related genes clustered according to a dissimilarity measure. (B) Heatmap of the relevance between module eigengenes and clinical phenotype of UC. (C) Gene significance across the modules.
3.2. Functional enrichment analysis
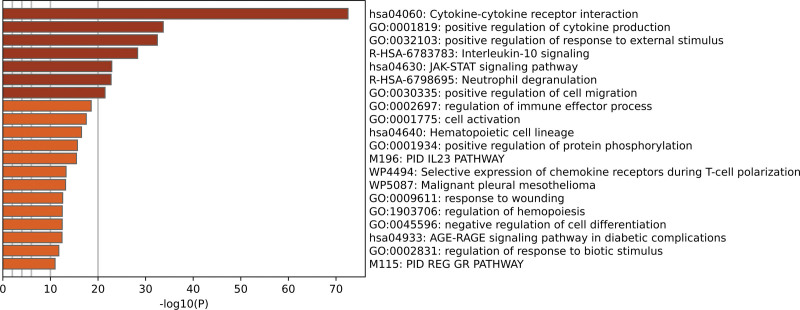
To investigate the role of the blue module in inflamed UC pathogenesis, the Metascape database was utilized to perform functional enrichment analysis. The result of the analysis was presented in the bar graph, which includes the top 20 enriched terms, colored based on P values. Among these, the genes in the blue module were particularly enriched in Cytokine-Cytokine receptor interaction, positive regulation of cytokine production, positive regulation of response to external stimulus, and interleukin-10 signaling (Fig. 4).
Figure 4.
Functional enrichment analysis of immune-related genes in the blue module.
3.3. Identification of candidate key immune genes in the blue module
Our selection of 71 highly connected hub genes in the blue module was based on the module membership of the gene and its significance in the module. In addition, all genes in the blue module were used to construct a network of PPI by Cytoscape, which consisted of 233 nodes and 2139 edges. According to PPI, 132 genes with node connectivity > 9 were identified as potential hub genes. Finally, 43 common candidate hub genes in the co-expression network and PPI network were chosen for further analysis and validation (Fig. 5).
Figure 5.
Candidate hub genes selection by co-expression network construction and protein–protein interaction network construction. (A) Scatter plot of gene significance and module membership in the blue module, the genes within the top left quadrant were identified as candidate hub genes in WGCNA. (B) Candidate hub genes within the blue module in the PPI network. (C) Venn diagram analysis: overlapping candidate hub genes. WGCNA = weighted gene co-expression network analysis.
3.4. Identification of real hub genes
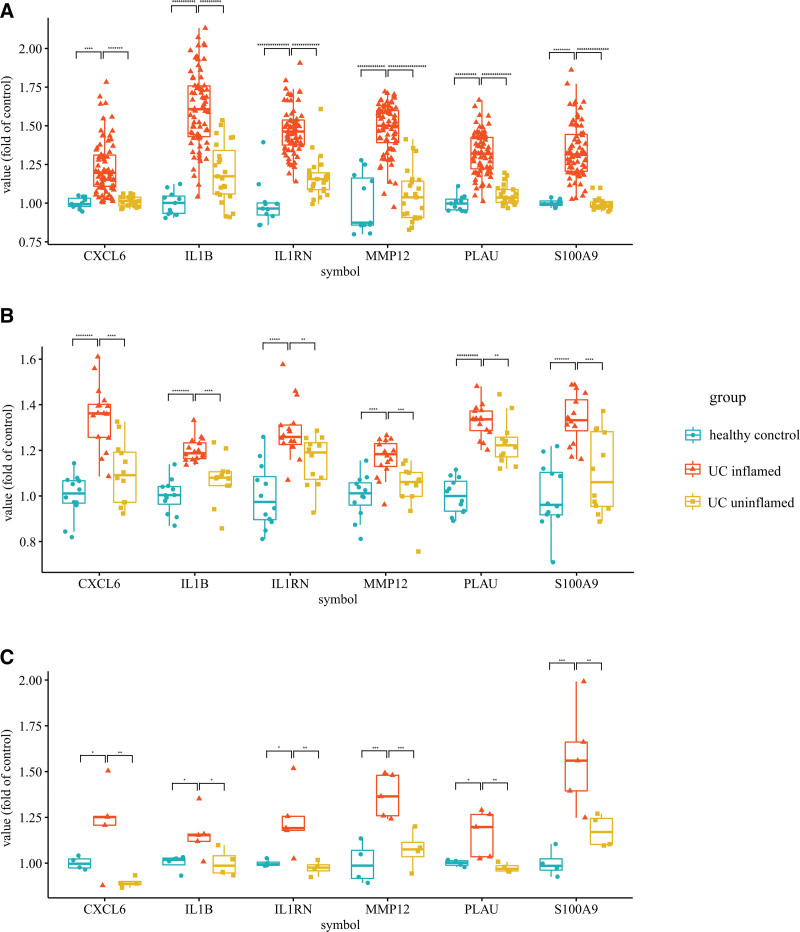
DEG analysis was used to screen DEGs among candidate hub genes. Six genes (CXCL6, IL1B, IL1RN, MMP12, PLAU, and S100A9) were found to be significantly upregulated in the inflamed UC group compared to the healthy control group and the uninflamed UC group (Fig. 6).
Figure 6.
Statistics of genes differentially expressing in the inflamed Ulcerative colitis. Box plots for the expression data in (A) GSE75214, (B) GSE53306, and (C) GSE6731 (*P < .05, **P < .01, ***P < .001, and so on. Student’ t test).
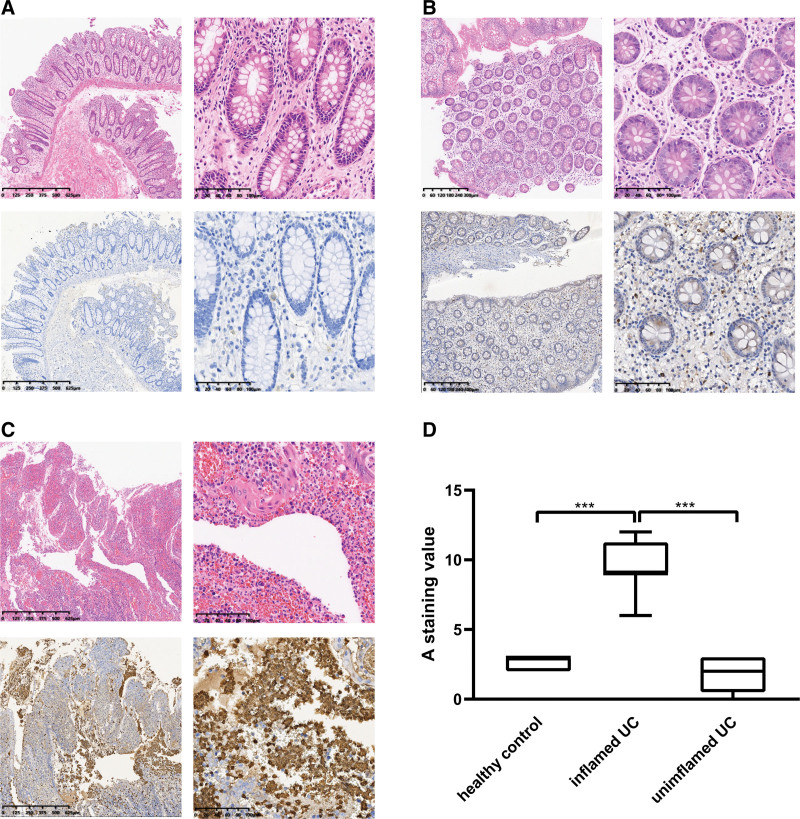
Moreover, twenty-four formalin-fixed paraffin-embedded samples, including 8 healthy control samples, 8 inflamed UC samples, and 8 uninflamed UC samples, were subjected to IHC Immunohistochemical analyses. The immunohistochemistry confirmed the significant elevation of S100A9 at the protein level in inflamed UC group samples. In the optical microscope, strongly-positive expression of S100A9 was observed to especially locate in the nucleus and cytoplasm of inflammatory cells in ulcers. The S100A9 gene was therefore identified as the hub gene in inflamed UC (Fig. 7).
Figure 7.
S100A9 protein expression level assessed by immunohistochemistry (IHC) in Ulcerative colitis samples (UC). (A–C) The representative HE and immunohistochemistry images of the uninflamed UC group, the control healthy group, and the inflamed UC group. (D) Grouped box plots of IHC score. (****P < .001, Student t test).
3.5. Diagnostic capacity of S100A9 for inflamed UC
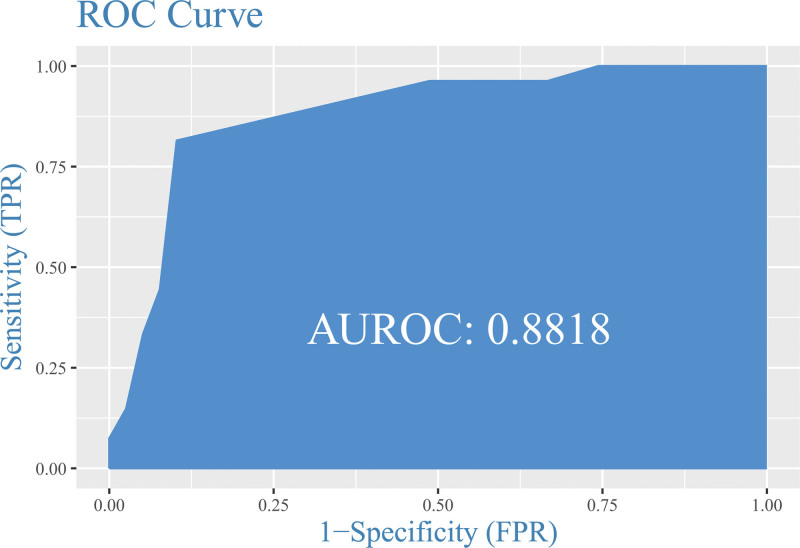
The area under the curve (AUC) of the ROC curve showed an AUC of AUC = 0.88 (95% CI: 0.7977–0.9658, P = .001) which indicated S100A9 has good diagnostic performance for inflamed UC (Fig. 8).
Figure 8.
ROC curve of diagnosis Capacity of the S100A9.
3.6. GSEA
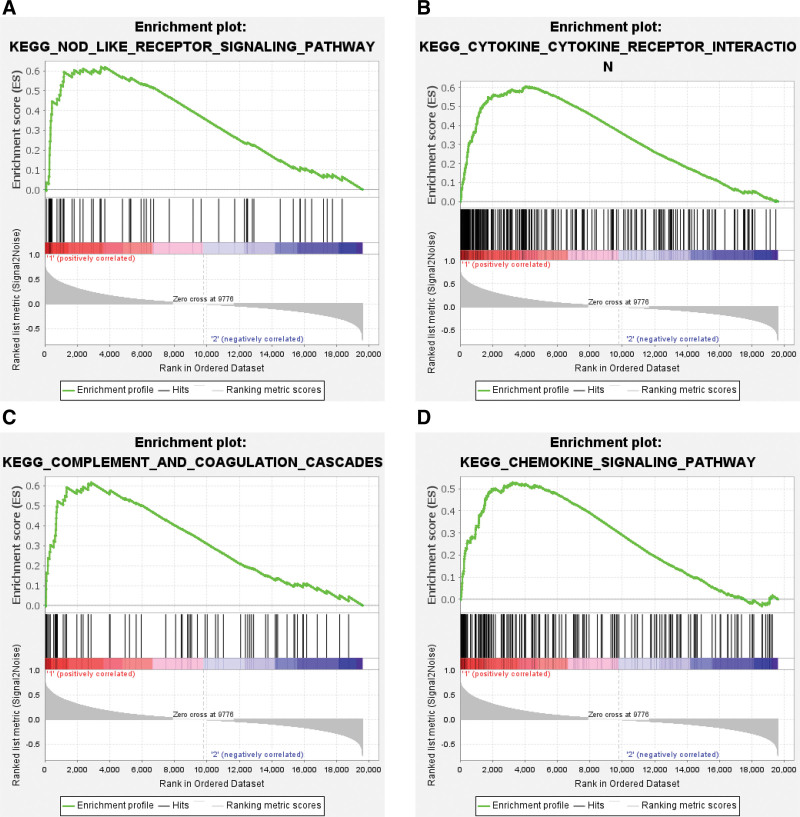
With the significance criteria of false discovery rate ≤ 0.25 and P value ≤ .05, a total of 23 eligible enriched KEGG pathways were identified by GSEA. Pathways were ordered according to normalized enrichment score, the top 4 pathways enriched in the S100A9 high-expression group were the Nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway, Cytokine-Cytokine receptor interaction, Complement and coagulation cascades, and Chemokine signaling pathway (Fig. 9).
Figure 9.
Gene set enrichment analysis of S100A9. (A–D) The top 4 pathways enriched in the S100A9 high-expression group.
3.7. Predicted transcriptional regulators of S100A9
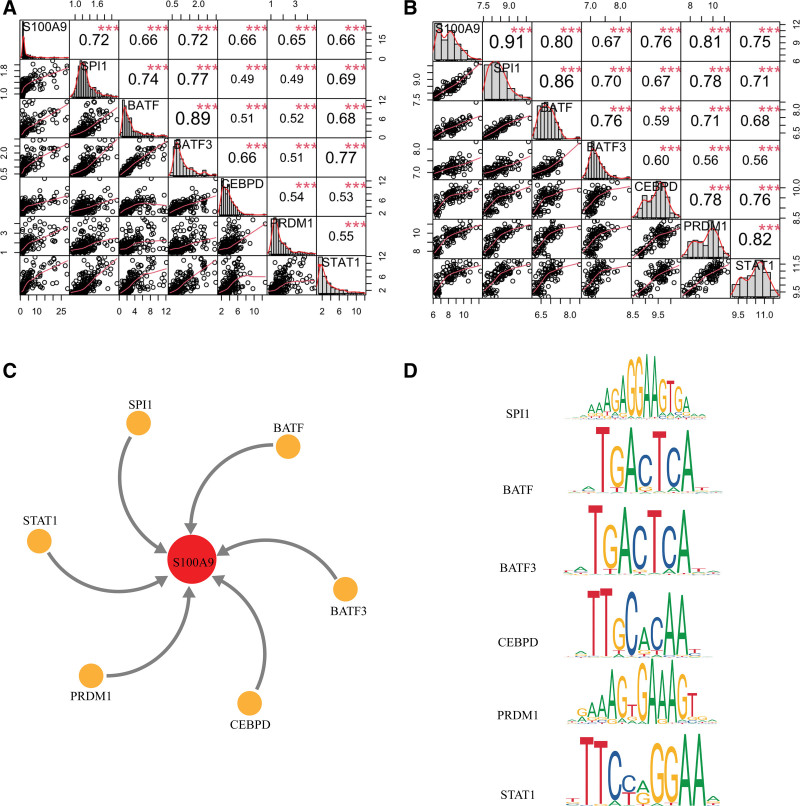
The upstream regulators of S100A9 were predicted as a way to further understand its regulation. A total of 304 TFs were predicted by UCSC. Among these, the expression of 6 TFs (SPI1, BATF, BATF3, CEBPD, PRDM1, and STAT1) strongly positively correlated with the expression of S100A9 (Fig. 10A and B). Next, the binding motifs of the TFs were predicted through JASPAR to confirm their correlation with S100A9. For SPI1, the motif with the highest score was MA0080.5, and the predicted sequence was GGCAGGGAGGTAGTGAAAGG; as for BATF, the motif with the highest score was MA0462.2, and the predicted sequence was TATTACTAATA; as for BATF3, the motif with the highest score was MA0835.2, and the predicted sequence was TATTACTAATA; as for CEBPD, the motif with the highest score was MA0836.2, and the predicted sequence was AATTTCACAAAAA; as for PRDM1, the motif with the highest score was MA0508.1, and the predicted sequence was AGGTAGTGAAAGGGA; as for STAT1, the motif with the highest score was MA0137.3, and the predicted sequence was GTTCCAGGAAC (Fig. 10C and D).
Figure 10.
Prediction of potential regulators of S100A9. (A–B) the results of correlation analyses of 6 transcription factors with R > 0.6 in GSE179285 and GSE75214. (C) the orange nodes in the circle represent 6 transcription factors regulating S100A9. (D) Binding sites of 6 predicted transcription factors using JASPAR database.
4. Discussion
Perpetuation of inflammation and ongoing tissue injury of the intestinal tract of UC patients were supposed to be driven by the dysregulated mucosal immune response, which is characterized by overactive innate immune system, adaptive immune system, and increased production of pro-inflammatory mediators.[18] To investigate the immune-related hub gene and key pathways associated with UC inflammation status, we used multiple datasets and comprehensive bioinformatic analyses in this study.
WGCNA was used as a data exploratory tool to find clusters of highly correlated immune genes with the inflammation of UC by establishing a co-expression network in GSE179285. The analysis revealed the blue module, consisting of 227 immune-related genes, showed the highest correlation with inflamed UC. The PPI and co-expression networks were then analyzed to find 43 common candidates. After a series of analyses and validation of clinical samples, S100A9, significantly differentially expressing in inflamed UC and having good diagnostic performance, was found to be the real hub gene.
S100A9, a bona fide damage-associated molecular pattern molecule, has been linked to both inflammation and cancer for a number of years.[25] Lawrance IC et al[26] proposed the elevated expression of S100A9 in UC for the first time, which was consistent with several studies including ours.[27,28] Calprotectin is a unique heterodimer of S100A8 and S100A9, which are two S100 EF-hand calcium binding proteins. Mortensen JH et al[29] found out that there was an association between neutrophil activity and endoscopic severity from patients with inflammatory bowel disease and a specific calprotectin epitope in serum, CPA9-HNE. Moreover, UC was suggested to be associated with a higher risk of colorectal cancer. Nine gene signatures, including S100A9, were reported to be able to distinguish patients with UC harboring remote neoplastic lesions.[30] In addition, S100A9 has been identified as a potential biomarker to identify patients with UC who will not respond to infliximab.[31] UC is characterized by mucosal superficial inflammation, so the remission of the inflammatory response is a marker of mucosal healing. In our study, we identified that S100A9 could distinguish UC with inflammation from not only healthy control samples but also uninflamed UC. Therefore, we supposed that S100A9 could be a biomarker for both diagnosis and remission based on our results.
To investigate the role of S100A9 in UC, researchers have made efforts. Wu F et al[32] elucidated that S100A9 upregulated in trinitrobenzene sulfonic acid-induced murine colitis and could be regulated by NF-κB inhibition. Leite Dantas R et al[33] demonstrated that S100A8/A9 alarmins in one of the determinants of Spontaneous onset of colonic inflammation which triggered TNFα.
Through functional enrichment analysis and GSEA analysis, the hub module and hub gene were tentatively explored in this study for their biological functions and pathways. An in-depth functional enrichment analysis revealed that the immune genes in the blue module were prominently involved in the Cytokine-Cytokine receptor interaction, positive regulation of cytokine production, positive regulation of response to external stimulus, and other activities. GSEA analysis suggested that S100A9 most likely gets involved NOD-like receptor (NLR) signaling pathway. There are more than twenty members of the NLR family in mammals, which are responsible for detecting pathogens and generating innate immunity against them.[34] The prototypical NLRs NOD1 and NOD2 sense the cytosolic presence of bacterial peptidoglycan fragments escaping from endosomes, which drives the activation of NF-κB and MAPK, cytokine production, and apoptosis.[35] Alternatively, another set of NLRs assembles multiprotein complexes called inflammasomes to activate caspase-1.[36] Combined with Toll-like receptor signaling, the inflammasomes are critical for the generation of mature proinflammatory cytokines.[37] S100A9 can be found in the nucleus, in the cytoplasm, and as secreted proteins depending on the conditions. It involves a number of functional roles by forming homo-dimers, higher order complexes, or hetero-tetramers with S100A8, which have been revealed to interact with toll-like receptor 4 and/or the receptor for advanced glycation end-products.[38,39] Our immunochemical results showed strong positive protein expression of S100A9, located in the nucleus and cytoplasm of inflammatory cells in ulcers, which may indicate S100A9 has both nuclear and cytosolic functions in UC. The specific mechanism should be further explored by experiments.
For the upstream regulatory mechanism of S100A9, Lee MJ et al[40] identified that Interleukin-6 induces S100A9 expression via STAT3 activation in colonic epithelial Cells of experimental UC. Our upstream regulator analysis showed that predicted 6 TFs (SPI1, BATF, BATF3, CEBPD, PRDM1, and STAT1) might regulate the expression of S100A9. in the hub module. Among these, SPI1 showed the strongest likelihood to be the regulator for its strongest correlation with S100A9. SPI1 (PU.1) is regarded as a major regulator of hematopoiesis by controlling the expression of genes, such as adhesion molecules, growth factor receptors, signaling components, and so on.[41]
5. Conclusions
A comprehensive bioinformatics analysis identified S100A9 as the real immune-related hub gene for inflamed UC. Both diagnosis and remission may be aided by its high expression in the inflamed UC. Further analyses revealed that S100A9 might be involved in regulating NLR signaling. SPI1 is most likely to be the TF to control the expression of S100A9 according to our upstream regulator analysis. Our preliminary results provide novel insights into UC immune mechanisms, despite their preliminary nature.
Acknowledgments
A special Acknowledgments goes to Dr Ren Zhou, who kindly gave the antibodies in pre-experiments.
Author contributions
Conceptualization: Lingna Zhou, Xiaotong Hu, Zhinong Jiang.
Data curation: Lingna Zhou, Xiaotong Hu, Zhinong Jiang.
Formal analysis: Lingna Zhou.
Funding acquisition: Lingna Zhou.
Investigation: Lingna Zhou.
Methodology: Lingna Zhou.
Project administration: Lingna Zhou.
Resources: Lingna Zhou, Qianru Gu, Guoxiang Fu.
Software: Lingna Zhou, Qianru Gu, Aihua Huang, Guoxiang Fu.
Supervision: Lingna Zhou, Qianru Gu, Aihua Huang, Guoxiang Fu.
Validation: Lingna Zhou, Qianru Gu, Aihua Huang, Guoxiang Fu.
Visualization: Lingna Zhou, Qianru Gu, Aihua Huang, Guoxiang Fu.
Writing – original draft: Lingna Zhou.
Writing – review & editing: Xiaotong Hu, Zhinong Jiang.
Supplementary Material
Abbreviations:
- AUC
- area under curve
- DEG
- differential expression gene
- GSEA
- gene set enrichment analysis
- NLR
- NOD-like receptor
- PPI
- protein-protein interaction
- TF
- transcription factor
- UC
- ulcerative colitis
- WGCNA
- weighted gene co-expression network analysis
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Zhou L, Gu Q, Huang A, Fu G, Hu X, Jiang Z. Identification of immune-related hub genes contributing to the pathogenesis, diagnosis, and remission of ulcerative colitis by integrated bioinformatic analyses. Medicine 2023;102:43(e35277).
Contributor Information
Lingna Zhou, Email: 21818540@zju.edu.cn.
Qianru Gu, Email: 3318047@zju.edu.cn.
Aihua Huang, Email: hahsrrsh@163.com.
Guoxiang Fu, Email: fgx8228@163.com.
Xiaotong Hu, Email: hxt_hz@zju.edu.cn.
References
- [1].Wang Y, Wang P, Shao L. Correlation of ulcerative colitis and colorectal cancer: a systematic review and meta-analysis. J Gastrointest Oncol. 2021;12:2814–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shokrollah N, Samadi P, Jalali A, et al. A systems biology approach to identify novel biomarkers in progression from crohn’s disease to colorectal cancer. Asian Pac J Cancer Prev. 2023;24:1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ungaro R, Colombel JF, Lissoos T, et al. A treat-to-target update in ulcerative colitis: a systematic review. Am J Gastroenterol. 2019;114:874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sun Y, Zhang Z, Zheng CQ, et al. Mucosal lesions of the upper gastrointestinal tract in patients with ulcerative colitis: a review. World J Gastroenterol. 2021;27:2963–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Park J, Cheon JH. Incidence and prevalence of inflammatory bowel disease across Asia. Yonsei Med J. 2021;62:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nakase H, Sato N, Mizuno N, et al. The influence of cytokines on the complex pathology of ulcerative colitis. Autoimmun Rev. 2022;21:103017. [DOI] [PubMed] [Google Scholar]
- [7].Kobayashi T, Siegmund B, Le Berre C, et al. Ulcerative colitis. Nat Rev Dis Primers. 2020;6:74. [DOI] [PubMed] [Google Scholar]
- [8].Camoglio L, Te Velde AA, Tigges AJ, et al. Altered expression of interferon-gamma and interleukin-4 in inflammatory bowel disease. Inflamm Bowel Dis. 1998;4:285–90. [DOI] [PubMed] [Google Scholar]
- [9].Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 2019;20:970–9. [DOI] [PubMed] [Google Scholar]
- [10].Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–42. [DOI] [PubMed] [Google Scholar]
- [11].Krug SM, Bojarski C, Fromm A, et al. Tricellulin is regulated via interleukin-13-receptor α2, affects macromolecule uptake, and is decreased in ulcerative colitis. Mucosal Immunol. 2018;11:345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rutgeerts P, Sandborn WJ, Feagan BG, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462–76. [DOI] [PubMed] [Google Scholar]
- [13].Kobayashi T, Okamoto S, Hisamatsu T, et al. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn’s disease. Gut. 2008;57:1682–9. [DOI] [PubMed] [Google Scholar]
- [14].Gerlach K, Hwang Y, Nikolaev A, et al. TH9 cells that express the transcription factor PU1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol. 2014;15:676–86. [DOI] [PubMed] [Google Scholar]
- [15].Scheibe K, Kersten C, Schmied A, et al. Inhibiting interleukin 36 receptor signaling reduces fibrosis in mice with chronic intestinal inflammation. Gastroenterology. 2019;156:1082–1097.e11. [DOI] [PubMed] [Google Scholar]
- [16].Mantovani A, Dinarello CA, Molgora M, et al. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 2019;50:778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Peyrin-Biroulet L, Bressenot A, Kampman W. Histologic remission: the ultimate therapeutic goal in ulcerative colitis? Clin Gastroenterol Hepatol. 2014;12:929–34.e2. [DOI] [PubMed] [Google Scholar]
- [18].Ahluwalia B, Moraes L, Magnusson MK, et al. Immunopathogenesis of inflammatory bowel disease and mechanisms of biological therapies. Scand J Gastroenterol. 2018;53:379–89. [DOI] [PubMed] [Google Scholar]
- [19].Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7. [DOI] [PubMed] [Google Scholar]
- [20].Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou L, Ding L, Gong Y, et al. Identification of hub genes associated with the pathogenesis of diffuse large B-cell lymphoma subtype one characterized by host response via integrated bioinformatic analyses. PeerJ. 2020;8:e10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Leanderson T, Liberg D, Ivars F. S100A9 as a pharmacological target molecule in inflammation and cancer. Endocr Metab Immune Disord Drug Targets. 2015;15:97–104. [DOI] [PubMed] [Google Scholar]
- [26].Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445–56. [DOI] [PubMed] [Google Scholar]
- [27].Su S, Kong W, Zhang J, et al. Integrated analysis of DNA methylation and gene expression profiles identified S100A9 as a potential biomarker in ulcerative colitis. Biosci Rep. 2020;40:BSR20202384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Okada K, Okabe M, Kimura Y, et al. Serum S100A8/A9 as a potentially sensitive biomarker for inflammatory bowel disease. Lab Med. 2019;50:370–80. [DOI] [PubMed] [Google Scholar]
- [29].Mortensen JH, Sinkeviciute D, Manon-Jensen T, et al. A specific calprotectin neo-epitope [CPa9-HNE] in serum from inflammatory bowel disease patients is associated with neutrophil activity and endoscopic severity. J Crohns Colitis. 2022;16:1447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pekow J, Dougherty U, Huang Y, et al. Gene signature distinguishes patients with chronic ulcerative colitis harboring remote neoplastic lesions. Inflamm Bowel Dis. 2013;19:461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang J, Wu X, Wei S, et al. Identified potential biomarkers may predict primary nonresponse to infliximab in patients with ulcerative colitis. Autoimmunity. 2022;55:538–48. [DOI] [PubMed] [Google Scholar]
- [32].Wu F, Chakravarti S. Differential expression of inflammatory and fibrogenic genes and their regulation by NF-kappaB inhibition in a mouse model of chronic colitis. J Immunol. 2007;179:6988–7000. [DOI] [PubMed] [Google Scholar]
- [33].Leite Dantas R, Bettenworth D, Varga G, et al. Spontaneous onset of TNFalpha-triggered colonic inflammation depends on functional T lymphocytes, S100A8/A9 alarmins, and MHC H-2 haplotype. J Pathol. 2020;251:388–99. [DOI] [PubMed] [Google Scholar]
- [34].Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. [DOI] [PubMed] [Google Scholar]
- [35].Kienes I, Weidl T, Mirza N, et al. Role of NLRs in the regulation of type I interferon signaling, host defense and tolerance to inflammation. Int J Mol Sci . 2021;22:1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cohen TS, Prince AS. Activation of inflammasome signaling mediates pathology of acute P aeruginosa pneumonia. J Clin Invest. 2013;123:1630–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gautier G, Humbert M, Deauvieau F, et al. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med. 2005;201:1435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vogl T, Tenbrock K, Ludwig S, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13:1042–9. [DOI] [PubMed] [Google Scholar]
- [39].Ghavami S, Rashedi I, Dattilo BM, et al. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol. 2008;83:1484–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lee MJ, Lee JK, Choi JW, et al. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLoS One. 2012;7:e38801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Turkistany SA, DeKoter RP. The transcription factor PU1 is a critical regulator of cellular communication in the immune system. Arch Immunol Ther Exp (Warsz). 2011;59:431–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.