Abstract
The catalase gene katA of Lactobacillus sakei LTH677 was cloned and expressed in Escherichia coli UM2, Lactobacillus casei LK1, and Lactobacillus curvatus LTH1432. The last host is a catalase-deficient plasmid-cured derivative of a starter organism used in meat fermentation. The regulation of katA expression was found to be the same in L. sakei LTH677 and the recombinant strains. The addition of H2O2 to anaerobic cultures, as well as a switch to aerobic conditions, resulted in a strong increase in KatA activity. The expression was investigated in more detail with L. sakei LTH677 and L. curvatus LTH4002. The recombinant strain LTH4002 did not accumulate H2O2 under glucose-limited aerobic conditions and remained viable in the stationary phase. Under inductive conditions, the katA-specific mRNA and the apoenzyme were synthesized de novo. Deletion derivatives of the katA promoter were produced, and the regulatory response was investigated by fusion to the β-glucuronidase reporter gene gusA and expression in L. sakei LTH677. The fact that gene expression was subject to induction was confirmed at the level of transcription and protein synthesis. A small putative regulatory sequence of at least 25 bp was identified located upstream of the −35 site. Competition experiments performed with L. sakei LTH677 harboring the fusion constructs consisting of the katA promoter and gusA revealed that an activator protein is involved in the transcriptional induction of katA.
Lactic acid bacteria (LAB) play an important role in food fermentations. In these processes the effects of LAB are beneficial, but malfermentation may occur when, for example, the ecological factors and technological conditions are unfavorable. The presence of oxygen is a factor that greatly affects the outcome of a fermentation process. In general, LAB tolerate oxygen but grow best under nearly anaerobic conditions. In the presence of oxygen hydrogen peroxide is formed. This strongly oxidizing compound may accumulate and have undesired effects in foods (for example, it may cause color defects and rancidity [28]), and, in addition, it kills the H2O2-producing organisms (7). Numerous species of LAB contain peroxidase and/or catalase to prevent these deleterious effects (10). One group of enzymes, the true catalases, are active when hematin is added. A second group of enzymes is the so-called nonheme catalases, pseudocatalases, or manganese catalases, which are found in only a few species. Recently, the gene encoding the manganese catalase of Lactobacillus plantarum was characterized (16). The gene katA, encoding the true catalase of Lactobacillus sakei LTH677, was cloned and characterized by Knauf et al. (20).
Lactobacillus curvatus and L. sakei are the most prevalent organisms in meat fermentations. In contrast to L. curvatus, L. sakei contains a heme-dependent catalase, which can function in meat products as these substrates contain abundant heme sources. The katA gene of L. sakei is a potential candidate for improving meat starter organisms, such as L. curvatus, because it could give the organisms the ability to produce catalase. Such a manipulation has the potential to simplify starter preparations as there would no longer be a need to combine L. curvatus with catalase-positive species, such as Staphylococcus carnosus or Kocuria varians (12). In order to use catalase-containing LAB, knowledge of the regulation of catalase is very important. Data on the effects of ecological factors on the activity of this enzyme have been presented by Engesser and Hammes (10). For example, the pseudocatalase activity of Pediococcus pentosaceus is affected by glucose and oxygen. In Escherichia coli (31) and Bacillus subtilis (9) the expression of different catalases is growth phase dependent or regulated by oxidative stress. The aim of this study was to gain insight into the regulation of katA expression in the natural host L. sakei LTH677, as well as in recombinant strains of L. curvatus, L. casei, and E. coli. To do this, the effects of oxygen and hydrogen peroxide on catalase expression were investigated at the transcription and protein synthesis levels and by performing a detailed analysis of the subcloned katA promoter.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are shown in Table 1. Lactobacilli were grown in MRS medium (8) at 30°C, and E. coli and B. subtilis were cultivated in Luria-Bertani medium (29) at 37°C. Selective media contained ampicillin (100 μg/ml) or chloramphenicol (10 μg/ml). For detection of catalase activity in lactobacilli an aqueous solution (sterilized by filtration) of hematin was added to the medium to obtain a final hematin concentration of 31.5 μM. Lactobacilli were incubated either aerobically by using shaking cultures in Erlenmeyer flasks or anaerobically in 98% N2–2% H2 atmosphere.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| B. subtilis UM1013 | tryC2 katA1 spo kat-6 | 21 |
| E. coli NM554 | recA13 araD139 Δ(ara-leu)7696 Δ(lac)17A galU galK hsdR rpsL (Strr) mcrA mcrB | Stratagene, La Jolla, Calif. |
| E. coli TG1 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/supE Δ(hsdM-mcrB)5 (rK−mK−McrB−) thi Δ(lac-proAB) | Amersham, Buckinghamshire, United Kingdom |
| E. coli UM2 | katE2 katG15 | 22 |
| L. casei LK1 | Formerly referred to as strain WS97 | 37 |
| L. curvatus LTH1432 | Plasmid-cured derivative of strain LTH683, a component of a starter preparation used for fermented sausages | 14 |
| L. sakei LTH677 | Wild-type strain, a component of a starter preparation used for fermented sausages, formerly referred to as strain Ls8 | 13 |
| Plasmids | ||
| pBluescript II KS(+) | E. coli cloning vector, Apr | Stratagene, La Jolla, Calif. |
| pHK1150 | pUC19 containing katA of L. sakei LTH677, Apr | 20 |
| pJK356 | Lactobacillus cloning vector on the basis of pLC2, Cmr | 19 |
| pNZ272 | Transcriptional fusion vector containing the promoterless gusA of E. coli, Cmr | 25 |
| pLSC300 | pJK356 containing katA on the 2.7-kb PstI fragment of pHK1150 | This study |
| pLSC400 | pBluescript II KS(+) containing katA on the 2.7-kb PstI fragment of pHK1150 | This study |
| pGS100 | pNZ272 containing the 128-bp PCR product generated with primers kat32 and kat31 | This study |
| pGS101 | pNZ272 containing the 97-bp PCR product generated with primers kat32 and kat33 | This study |
| pGS102 | pNZ272 containing the 72-bp PCR product generated with primers kat34 and kat33 | This study |
DNA techniques.
Plasmid DNAs were isolated from E. coli and B. subtilis by the methods of Birnboim and Doly (3) and Ish-Horowicz and Burke (18), respectively. Each DNA was further purified by treatment with phenol-chloroform as described by Sambrook et al. (29). Plasmid DNA was isolated from lactobacilli by a method described previously (5). DNA manipulations were performed as described by Sambrook et al. (29). DNA sequences were determined by the dideoxy chain termination method with sequencing kits (Sequenase, version 2.0 [U.S. Biochemicals, Cleveland, Ohio] or AutoRead [Pharmacia, Freiburg, Germany]). The following primers were used: universal primers T3 and T7, kat16 (5′CAGTATCTCTACATCGG3′), and gus1 (5′GGGTTTCTACAGGACGTA3′).
PCR amplification.
Amplification reactions were performed in a total volume of 100 μl containing 200 μM (each) dATP, dCTP, dGTP, and dTTP, 50 pmol of each primer, 2 ng of pLSC400 DNA, 2.5 U of Pwo DNA polymerase (Boehringer, Mannheim, Germany), and the corresponding 1× Pwo buffer. Reactions were carried out with a Perkin-Elmer thermocycler by using initial denaturation at 92°C for 2 min, followed by 30 cycles consisting of 92°C for 1 min, 45°C for 1 min, and 72°C for 1 min and a final extension step consisting of 72°C for 7 min.
Transformation.
E. coli was transformed by the method of Ausubel et al. (1), and the method of Chang and Cohen (6) was used for B. subtilis. L. curvatus and L. casei were transformed by electroporation by the method of Bringel and Hubert (4), and L. sakei was transformed by the method of Berthier et al. (2).
Analyses of mRNA.
For dot blot hybridization total RNA was isolated with an RNeasy minikit (Qiagen, Hilden, Germany), with the following modifications. The cells were resuspended in TE buffer containing lysozyme (25 mg/ml) and mutanolysin (10,000 U/ml) and incubated for 30 min at 37°C. The RNA (5 μg) was denatured at 65°C for 15 min in 20 μl of a buffer containing 50% deionized formamide, 7.4% formaldehyde, and 1× SSC (standard saline citrate; 1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate, pH 7.0). After 130 μl of 15× SSC was added, the denatured RNA was transferred to uncharged nylon membranes (Qiabrane; Qiagen) with a dot blot apparatus (Bio-Rad, Munich, Germany). The membranes were prehybridized at the hybridization temperature (see below) for 1 h in a solution containing 5× SSC, 20 mM disodium phosphate dihydrate, 7% sodium dodecyl sulfate (SDS), and 10× Denhardt’s solution (1× Denhardt’s solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% bovine serum albumin). Hybridization was performed for 4 h in the same solution containing 10 pmol of probe labeled as described previously (15). The incubation temperatures used were 40°C for the katA-specific probe kat13 (5′CCTCGTTAGTCGTTAGTT3′) and the gusA-specific probe gus1 and 45°C for the universal probe 1028R (5′CCTTCTCCCGAAGTTACGG3′) (23). The membranes were washed twice for 5 min in 2× SSC containing 0.1% SDS at the hybridization temperatures and once at the probe-dependent temperatures, which were 44°C for kat13, 47°C for gus1, and 51°C for 1028R. After autoradiography, the hybrids were denatured twice in 0.1× SSC containing 0.5% SDS for 15 min at 90°C.
The site of transcription initiation was determined by primer extension analysis as described by Obst et al. (24), with the following modification: synthesis was performed in the presence of 40 U of RNase inhibitor (Boehringer). The oligonucleotide kat13, which is complementary to the 5′ region of the katA mRNA, was used as the primer. Northern hybridization analysis was performed as described previously (24), with the following modification: a 696-bp DNA fragment of katA was labeled with digoxigenin (DIG)-dUTP by using a PCR DIG probe synthesis kit (Boehringer), the katA-specific primers kat1 (5′GACAATCAACATTCG3′) and kat2 (5′TGCGTCGAATAAATC3′), and plasmid pLSC300 DNA as the template. The DIG-labeled probe was used and hybridization was performed as recommended by the supplier.
Preparation of cell extracts.
To determine catalase activities, aliquots (20 ml) of the cultures were harvested by centrifugation (3,000 × g) at 4°C and washed with 10 ml of ice-cold phosphate buffer (50 mM, pH 7). The cells were resuspended in 1 to 2 ml of the same buffer (final optical density at 578 nm, 20) and disrupted with a precooled miniature French pressure cell (SLM Instruments, Urbana, Ill.) at 1,500 lb/in2 and 4°C. The procedure was repeated three times, and the cell fragments were removed by centrifugation at 13,000 × g and 4°C. To determine β-glucuronidase activity, the cell extracts were prepared as described by Platteeuw et al. (25), with the following modifications. The harvested cells were washed in GUS buffer, resuspended, and disrupted with glass beads (diameter, 0.5 mm) in a cell mill (Bühler, Tübingen, Germany) for 10 min at 4°C. After centrifugation, the cell extracts were assayed immediately. The protein content of the crude extracts was determined with a Bio-Rad protein assay.
Enzyme assays and determination of hydrogen peroxide content.
Catalase-positive transformants were detected after the colonies on agar replica plates were flooded with 0.87 M hydrogen peroxide and the effervescence was recorded. A quantitative analysis of the catalase activities in intact cells or crude cell extracts of lactobacilli was performed as described by Engesser and Hammes (10). Formation of hydrogen peroxide was determined as described previously (36), with the following modification: aliquots (250 μl) of the culture broth were added to 1.25 ml of ABTS peroxidase reagent. For the β-glucuronidase assay cell extracts (50 to 100 μl) were added to GUS buffer (see above) supplemented with 1.25 mM para-nitrophenyl-β-d-glucuronide. The activity was determined at 420 nm with a spectrophotometer at 37°C.
RESULTS
Cloning and expression of katA.
The gene katA was isolated as a 2.7-kb PstI fragment from plasmid pHK1150 (20). To further characterize this fragment, the DNA sequences upstream and downstream of the coding region of katA were determined. To do this, the fragment was introduced into plasmid pBluescript II KS(+), and the resulting plasmid, pLSC400, was transferred to E. coli NM554. The sequence analysis revealed that upstream of katA were a terminator sequence and an incomplete 680-bp open reading frame. No open reading frame longer than 123 bp was found downstream of the terminator of katA. To clone katA in L. curvatus LTH1432, the PstI fragment was purified and introduced into the vector pJK356 (19). The resulting plasmid, pLSC300, was transferred to the catalase-deficient strain B. subtilis UM1013 by protoplast transformation. Transformants were screened for formation of an active catalase. Subsequently, plasmid pLSC300 was reisolated and transferred to L. curvatus LTH1432 by electroporation. Transformants were screened for catalase-positive colonies on hematin-containing agar plates. The resulting recombinant strain, containing plasmid pLSC300, was designated L. curvatus LTH4002. The plasmid DNA was reisolated and subjected to a restriction analysis. The resulting restriction pattern was identical to the pattern obtained with the plasmid of B. subtilis.
Recombinant strain LTH4002 was grown under aerobic and anaerobic conditions, and the catalase activities of intact cells and crude extracts were determined quantitatively. For purposes of comparison, L. sakei LTH677 was investigated in parallel. As shown in Table 2, the catalase activity of strain LTH4002 was ca. fourfold higher than that of L. sakei LTH677. Finally, the stability of plasmid pLSC300 in L. curvatus LTH1432 was investigated under conditions simulating essential elements of sausage fermentation (i.e., batch fermentation without shaking and addition of antibiotic). The plasmid was retained in more than 95% of the cells after 20 generations, and no structural instability was observed.
TABLE 2.
Comparison of catalase activities of L. curvatus LTH4002 and L. sakei LTH677 containing the gene katAa
| Strain | Growth conditions | Catalase activities
|
|
|---|---|---|---|
| Intact cells (mg of O2 · liter−1 · min−1 · OD578−1)b | Crude extracts (mg of O2 · min−1 · mg of protein−1) | ||
| L. sakei LTH677 | Aerobic | 134.3 | 1.03 |
| Anaerobic | 9.5 | 0.06 | |
| L. curvatus LTH4002 | Aerobic | 596.9 | 4.11 |
| Anaerobic | 68.3 | 1.12 | |
Cultures were incubated for 3.5 h under aerobic conditions in MRS medium, and catalase activities were measured by using intact cells or crude extracts.
OD578, optical density at 578 nm.
Characteristics of aerobically grown cultures.
The effect of catalase expression on the growth of L. curvatus LTH4002 was investigated, and the growth characteristics were compared with the growth characteristics of recipient strain LTH1432. As glucose represses the formation of hydrogen peroxide (7, 10), an experiment to determine the effect of glucose was included in the study. With L. curvatus LTH1432 increasing concentrations of glucose (1.25, 2.5, 5.0, 7.5, and 10 g/liter) not only resulted in an increase in the growth yields of aerobic cultures, without affecting the growth rate, but also decreased remarkably the formation of hydrogen peroxide. No production of H2O2 was observed in the presence of 7.5 and 10 g of glucose per liter, whereas in the presence of 1.25 g of glucose per liter about 9 mmol of H2O2 per liter was formed by recipient strain LTH1432. On the other hand, no hydrogen peroxide was detected with the recombinant strain L. curvatus LTH4002.
The effect of H2O2 production on the viability of L. curvatus LTH1432 was investigated by growing cultures aerobically in media containing 1.25 and 10 g of glucose per liter. In the presence of 10 g of glucose per liter the culture grew to a density of 1.6 × 109 CFU/ml, and the cells remained viable during the stationary phase for up to 25 h of incubation. In the presence of 1.25 g of glucose per liter a viable cell count of 7 × 108 CFU/ml was obtained at the early stationary phase. Subsequently, the number of cells decreased within 2 h to 1 × 106 CFU/ml. With the recombinant strain L. curvatus LTH4002 the culture grew to a density of 1.7 × 108 CFU/ml in the presence of 1.25 g of glucose per liter, and the viable cell count did not decrease in the stationary phase.
Induction of catalase activity by oxygen.
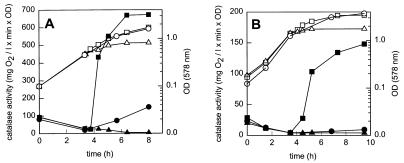
To investigate the regulation of catalase by oxygen, L. curvatus LTH4002 and L. sakei LTH677 were each grown in three batches under strictly anaerobic conditions for approximately 3.5 h. Then one batch of each organism was incubated without any changes, and two batches were aerated by shaking. To one of the aerated batches erythromycin was added 30 min before the culture was exposed to aerobic conditions. As shown in Fig. 1, the change from anaerobic to aerobic conditions resulted in significant increases in the catalase activity from 26 to 670 mg of O2/liter per min per unit of optical density for L. curvatus LTH4002 and from 10 to 150 mg of O2/liter per min per unit of optical density for L. sakei LTH677. On the other hand, the catalase activity of the anaerobically grown culture remained low; a slight increase was observed for L. curvatus LTH4002 in the stationary phase. In the culture treated with antibiotic the catalase activity did not increase when the culture was shifted to aerobic conditions. To confirm the changes in physiological activities at the enzyme level, the catalase activities of L. curvatus LTH4002 and L. sakei LTH677 crude cell extracts were determined after growth for 3.5 h under aerobic and anaerobic conditions, respectively. As shown in Table 2, the activities were high in aerobically grown cultures, and the catalase activity detected in L. curvatus LTH4002 was higher than the catalase activity detected in L. sakei LTH677.
FIG. 1.
Effect of oxygen on growth (open symbols) and catalase activity (solid symbols) of L. curvatus LTH4002 (A) and L. sakei LTH677 (B). ○ and •, anaerobic incubation; □ and ▪, culture shifted to aerobic conditions at 3.75 h; ▵ and ▴, erythromycin (100 μg/ml) added at 3.25 h and culture shifted to aerobic conditions at 3.75 h. l, liter; OD, optical density; OD (578 nm), optical density at 578 nm.
Effect of hydrogen peroxide on catalase activity.
To study the effect of hydrogen peroxide on catalase activity, L. curvatus LTH4002 and L. sakei LTH677 were grown under anaerobic conditions. After 5 h the cultures were divided, and hydrogen peroxide was added to one part of each culture to a final concentration of 0.2 mmol/liter. An increase in catalase activity was observed in both strains after hydrogen peroxide was added. The activity of L. curvatus LTH4002 increased from 95 to 670 mg of O2/liter per min per unit of optical density, and the activity of L. sakei LTH677 increased from 10 to 150 mg of O2/liter per min per unit of optical density. Corresponding experiments were performed with aerobically grown cultures of the strains. After hydrogen peroxide was added, the catalase activity of these cells did not increase above the control value.
Analysis of transcription of katA.
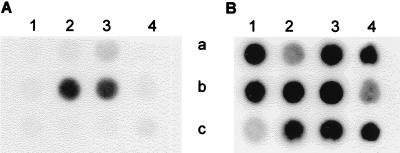
To study the regulation of catalase on the transcriptional level, the experiments described above were repeated with L. curvatus LTH4002, L. sakei LTH677, and (as a control) L. curvatus LTH1432. After 3 h each of the anaerobically grown cultures was divided into three parts. The control part remained under anaerobic conditions, one part was aerated, and the third part was treated with hydrogen peroxide. After 2 h samples were taken, and the total RNA of the cells was isolated and subjected to dot blot hybridization with katA-specific probe kat13 and universal probe 1028R (Fig. 2). Using the latter probe provided a control for the amount of total RNA transferred and the accessibility of RNA to oligonucleotide probes (Fig. 2B). In L. curvatus LTH4002, an increased concentration of katA-specific mRNA was detected in cells grown under aerobic conditions, as well as in cells treated with hydrogen peroxide. A similar difference in the concentrations of katA-specific mRNA was obtained with the wild-type strain L. sakei LTH677 (data not shown). No hybridization of the RNA from recipient L. curvatus LTH1432 was detected, which confirmed the stringency of the hybridization conditions.
FIG. 2.
Dot blot hybridization of RNAs from L. curvatus LTH4002 (lanes 1 and 2) and L. curvatus LTH1432 (lanes 3) with the katA-specific probe kat13 (A) and the universal probe 1028R (B). In row a, one part of an anaerobic culture was shifted for 2 h to aerobic conditions (dot 1a), and the control remained under anaerobic conditions (dot 2a). In row b, 2 h before samples were taken hydrogen peroxide was added to one part of a culture (dot 1b), and the control was not treated (dot 2b). Control strain LTH1432 was treated identically (i.e., aerated [dot 3a] and exposed to hydrogen peroxide [dot 3b]).
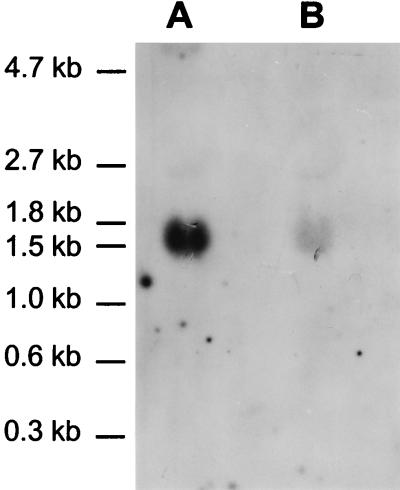
To study the in vivo transcription of katA, RNAs were isolated from aerobically grown cultures of L. curvatus LTH4002 and L. sakei LTH677 and subjected to a Northern hybridization analysis. A single transcript of ca. 1,550 nucleotides was detected for both strains under inducing conditions (Fig. 3). The length of the transcript is consistent with the sequence data for katA determined previously (20). The transcript includes the reading frame of katA, the putative terminator, and the promoter region located between the putative terminator of orfx and the start codon (Fig. 4).
FIG. 3.
Northern hybridization analysis of RNAs isolated from aerobically grown cultures of L. curvatus LTH4002 (lane A) and L. sakei LTH677 (lane B). The sizes of the marker fragments are indicated on the left.
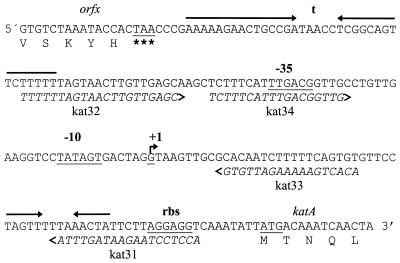
FIG. 4.
DNA sequence of the promoter region of the catalase gene katA of L. sakei LTH677. Important features in the sequence are underlined and marked. The arrows indicate putative stem-loop structures. The locations and sequences of the primers used for amplification of various parts of the promoter are indicated.
To characterize the promoter region of katA in more detail, the start point of transcription of katA was determined by primer extension analysis. To do this, RNAs were isolated from L. curvatus LTH4002 and L. sakei LTH677. The cultures were either grown anaerobically and subsequently subjected to aeration or grown anaerobically or aerobically and subsequently treated with H2O2, as described above. As shown in Fig. 5, analysis of the RNAs of the various cultures revealed only one primer extension product. For the anaerobically grown culture of L. sakei LTH677 (Fig. 5, lane 1), the katA transcript could not be detected under the noninducing conditions. Moreover, the primer extension analysis revealed that the site of transcription initiation was the guanidine residue 67 bp upstream of the katA translational start codon (Fig. 4). This site was identical in cells grown anaerobically and in cells grown under inducing conditions (cells induced either by aeration or by addition of hydrogen peroxide). Based on this result, the −10 and −35 regions upstream of the transcription start point were identified.
FIG. 5.
Primer extension analysis of katA transcripts. The transcripts were generated from total RNAs isolated from cultures of L. curvatus LTH4002 (lanes 2, 4, 5, and 7) and L. sakei LTH677 (lanes 1, 3, 6, and 8) grown under the following conditions: lanes 1 and 2, anaerobic conditions; lanes 3 and 4, aerobic conditions; lanes 5 and 6, anaerobic conditions with added H2O2; lanes 7 and 8, aerobic conditions with added H2O2. The arrow indicates the position of the products obtained; the asterisk indicates the transcription start site.
Cloning and characterization of the katA promoter.
To identify putative regulatory sequences within the promoter of katA, the promoter region and deletion derivatives were characterized by using vector pNZ272 containing the promoterless β-glucuronidase gene gusA of E. coli. To do this, the complete promoter region was amplified with primers kat31 and kat32 (Fig. 4). The fragment was introduced into pNZ272, resulting in plasmid pGS100. A 3′-truncated promoter and a 5′-truncated promoter were constructed by PCR with the primer pairs kat32–kat33 and kat33–kat34, respectively (Fig. 4). Introduction of the amplified fragments resulted in plasmids pGS101 and pGS102, respectively. The correct location and sequence of the promoter fragments were verified by sequencing with the gusA-specific primer gus1. Finally, the plasmids were transferred to the katA donor L. sakei LTH677 by using E. coli TG1 as an intermediate host.
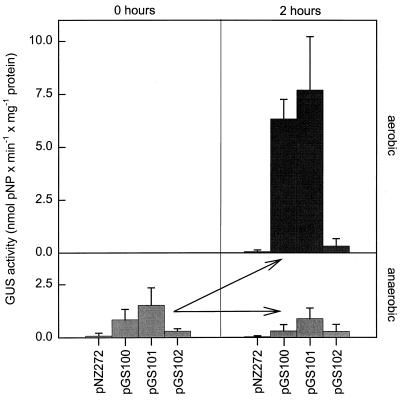
The resulting strains, L. sakei(pGS100), L. sakei(pGS101), and L. sakei(pGS102), were used to investigate the transcriptional induction of gusA. L. sakei(pNZ272) containing promoterless gusA served as a control. The strains were grown anaerobically in MRS broth to an optical density of approximately 0.5. Each culture was divided, and one part was shifted to aerobic conditions. Samples were taken at zero time and at 2 h after the shift to aerobic conditions and were immediately supplemented with erythromycin (100 μg/ml). GusA expression was determined in crude cell extracts. As Fig. 6 shows, in L. sakei(pGS100) and L. sakei(pGS101) a strong increase in β-glucuronidase activity was observed after the shift to aerobic conditions. On the other hand, the activity in L. sakei(pGS102) containing the 5′-truncated katA promoter and the activity in the control organism L. sakei(pNZ272) did not increase.
FIG. 6.
Effect of oxygen on the β-glucuronidase activity of strains of L. sakei. The strains harbored plasmids pGS100, pGS101, and pGS102 containing parts of the katA promoter, as indicated in Table 1. L. sakei(pNZ272) served as a control. Each anaerobic culture was divided, and each part was incubated for 2 h under aerobic or anaerobic conditions. The values are the averages from three independent experiments. The standard deviations are indicated by error bars. pNP, p-nitrophenol.
To determine induction at the mRNA level, RNAs were isolated from aliquots of the cultures and subjected to dot blot hybridization by using the probe gus1 and the universal probe 1028R as a control. As shown in Fig. 7, induction of transcription of gusA occurred under aerobic conditions only with L. sakei(pGS100) and L. sakei(pGS101). The formation of the specific mRNA under inducing conditions was consistent with the GUS activities.
FIG. 7.
Dot blot hybridization of RNAs from L. sakei(pGS100) (lanes 2), L. sakei(pGS101) (lanes 3), L. sakei(pGS102) (lanes 4), and L. sakei(pNZ272) (lanes 1) with the gusA-specific probe gus1 (A) and the universal probe 1028R (B). Each anaerobic culture (row a) was divided, and each resulting portion was incubated for 2 h under aerobic (row b) or anaerobic (row c) conditions.
To elucidate the mechanism of katA regulation in more detail, regulation of the catalase activity of the wild-type strain L. sakei LTH677 was investigated with cells containing the various promoters on plasmids pGS100, pGS101, and pGS102. To do this, the experimental design used to determine the expression of GusA was used, but catalase activity was measured instead of GusA activity. Samples were taken 1 and 2 h after induction by aeration. Table 3 shows the relative catalase activities compared to the wild-type strain L. sakei LTH677 activity. In the strains harboring plasmid pGS100 or pGS101, the level of L. sakei catalase activity induced was ca. 70% of the level of activity determined in strain LTH677. On the other hand, no effect on induction was observed with L. sakei(pGS102) containing the 5′-truncated promoter region of katA.
TABLE 3.
Relative catalase activities after induction by aeration of the wild-type strain L. sakei LTH677 and derivatives harboring plasmids pGS100, pGS101, and pGS102a
| Strain | Relative catalase activity (%) after:
|
|
|---|---|---|
| 1 h | 2 h | |
| L. sakei LTH677 | 100 | 100 |
| L. sakei(pGS100) | 69 ± 8 | 81 ± 8 |
| L. sakei(pGS101) | 66 ± 17 | 72 ± 10 |
| L. sakei(pGS102) | 106 ± 16 | 91 ± 11 |
The plasmids contained parts of the katA promoter fused to the promoterless gene gusA as indicated in Table 1. The values are the averages from three independent experiments.
Regulation of katA in L. casei and E. coli.
The function of the regulatory sequence of katA was studied in the catalase-deficient strains L. casei LK1 and E. coli UM2 containing plasmids pLSC300 and pLSC400, respectively. Transformants were screened for formation of an active catalase. The plasmid DNA was reisolated and subjected to a restriction analysis. The restriction patterns were identical to the corresponding patterns obtained previously. The resulting recombinant strains, L. casei(pLSC300) and E. coli(pLSC400), were grown anaerobically in MRS and Luria-Bertani broth, respectively. After 4.5 h the cultures were divided, and one part of each culture was shifted to aerobic conditions (the preparation was aerated), whereas the control remained under anaerobic conditions. Induction of the catalase activity was observed in both strains (data not shown). During 3 h of aeration the activity of L. casei(pLSC300) increased from 60 to 228 mg of O2/liter per min per unit of optical density and the activity of E. coli(pLCS400) increased from 82 to 308 mg of O2/liter per min per unit of optical density. No increase in catalase activity was observed in the control.
DISCUSSION
This study of regulation of the catalase of L. sakei revealed unique combinations of properties. The catalase activity responds to exposure of the cells to oxygen. Regulation takes place at the transcriptional level, and a unique sequence involved in regulation of the promoter has been identified. The molecular evidence for this response to oxidative stress is consistent with the physiological properties of the cells, as determined with crude extracts. Catalase synthesis remains regulated after the katA gene is transferred to catalase-deficient bacteria, such as L. curvatus, L. casei, and E. coli UM2. We demonstrated that induction of the activity occurred in these organisms after the organisms were transferred from anaerobic conditions to aerobic conditions. Thus, the regulatory mechanism is not restricted to the native enzyme producer species.
Regulation of catalases has been studied previously in detail in E. coli and B. subtilis. In E. coli two distinct catalase species, hydroperoxidase I (HPI) and HPII, are synthesized (31). HPI is encoded by katG, is inducible by H2O2, and is a part of the oxidative stress response regulon which is regulated by OxyR. HPII is induced when the cells enter the stationary phase. Similarly, two well-characterized catalases have been identified for B. subtilis, catalase 1 (KatA) and catalase 2 (KatE). Whereas KatA is a member of the oxidative stress-specific protein group, KatE is a general ςB-dependent stress protein (9). There is a question concerning the relatedness of KatA from L. sakei LTH677 to these enzymes. A comparison revealed that the enzymes are similar with respect to the size of the subunit and the hexameric structure of catalase 1 from B. subtilis (20). This finding is supported by the high level of sequence similarity which was observed in homology studies of amino acid sequences of various catalases (26).
It was pointed out by Condon (7) that LAB respond to exposure to oxygen with a change in sugar metabolism and formation of reactive oxygen species, including H2O2. It has been observed that in the catalase-deficient strain L. curvatus LTH1432 the time until H2O2 accumulation by a culture commenced depended on the glucose concentration in the growth medium. This finding is consistent with the assumption (7) that after glucose is used up, a metabolic change which includes H2O2 production by the cells takes place. Cells with catalase activity do not accumulate this toxic compound. It has been shown that for KatA of B. subtilis (9) and for HPI of E. coli (31) H2O2 induces synthesis of catalase. The catalase of L. sakei LTH677 was also induced by H2O2 under anaerobic conditions. However, this reaction does not necessarily indicate that H2O2 per se is the inducer, as this reactive compound generates oxygen that may be the effective signal. As the level of induction did not increase after aerobically grown cells were exposed to H2O2, we suggest that oxygen, not H2O2, is the inducing agent. On the other hand, E. coli and B. subtilis respond to the addition of H2O2 to aerobically growing cultures by increasing HPI activity and KatA activity, respectively.
With regard to the regulation of KatA of L. sakei by oxygen, it was observed that high catalase activity is found only in aerated cultures. This finding is consistent with aerobic induction of oxidative stress enzymes, such as oxidases, peroxidases, and superoxide dismutase (7). After a switch from anaerobic to aerobic conditions the activity of KatA is induced by de novo synthesis of the apoenzyme. Activation of the enzyme itself does not take place, as shown by addition of erythromycin to a culture. The studies of katA expression at the mRNA level confirmed the results of the physiological studies. It was observed that the synthesis of katA-specific mRNA is induced by aeration or by adding H2O2 to anaerobic cultures. This direct proof that there is regulation at the transcriptional level provides the first example of oxidative stress protein induction in lactobacilli. Direct proof that oxidative stress induction occurs was also obtained with the superoxide dismutase gene sodA of the related organism Lactococcus lactis (30). By fusing the katA promoter to the β-glucuronidase reporter gene, we showed that the regulatory sequence for katA expression is part of the promoter. The synthesis of gusA-specific mRNA, as well as GusA protein, correlated with the expression of katA.
To identify the regulatory sequence, a detailed analysis of the promoter sequence was performed. A stem-loop structure was found 32 bp downstream from the transcription start point (Fig. 4). This sequence showed some similarity to the consensus sequence of potential binding sites for FNR homologs (32, 33). The hypothesis that this type of regulatory sequence might be involved in regulation of katA is supported by the fact that a FNR-like protein is present in L. casei (17) and by the fact that two genes coding for FNR-like proteins were characterized from Lactococcus lactis MG1363 (11). Our promoter fusion experiments revealed that an FNR homolog does not play a role in regulation. On the other hand, no induction of katA was seen when the 5′ region upstream of the −35 region was deleted, and this sequence, therefore, should be responsible for transcriptional induction of katA. To investigate whether an activator or a repressor protein is involved in this regulation, the effect of the presence of the putative regulatory sequence located on a plasmid was studied. In this experiment possible competition for a regulatory protein between the putative regulatory sequence on the plasmids and the putative regulatory sequence on the chromosome was investigated by determining the induction of the catalase of L. sakei LTH677 by aeration. As a decrease in catalase activity was observed only in those strains which harbored plasmids containing the putative regulatory sequence, the results suggested that the putative regulatory protein was titrated by the sequence present on the plasmid. Thus, this sequence should be the binding site for a transcriptional activator.
Attribution of the putative regulatory sequence of katA to a described binding site for oxidative stress regulator proteins was not possible. There was some similarity to the sequences of the OxyR binding sites (34, 35). Remarkably, these binding sites for various OxyR-regulated promoters were found to exhibit very low levels of similarity (three conserved bases) in the enterobacteria. An OxyR binding site was also discovered upstream of the promoter of the NADH peroxidase gene npr of Enterococcus faecalis (27). Ross and Claiborne demonstrated that OxyR from E. coli binds to the site of this LAB, and, furthermore, a protein which cross-reacted with antisera to E. coli OxyR was found in Enterococcus faecalis. On the other hand, the physiological properties argue against the involvement of OxyR regulation, as katA was not inducible by H2O2 in aerobic cultures of L. sakei LTH677 and L. curvatus LTH4002, whereas OxyR controls the expression of H2O2-inducible genes in E. coli.
The specific properties of the katA promoter activity may also be useful in food fermentation. It has been shown that catalase-deficient LAB, such as L. curvatus, can be endowed with this activity, which improves their technical properties. For example, in starter preparations access to oxygen may be less harmful to the viability of the cells, and during food fermentation accumulation of H2O2 can be prevented. Finally, by fusing the promoter with useful genes, gene expression can be controlled, and, when the genes are located on a multicopy plasmid, they can be expressed at high levels. The feasibility of such constructions was demonstrated by our fusion experiments performed with gusA.
ACKNOWLEDGMENTS
We thank E. Herrmann and M. Schramm for excellent technical assistance. We are indebted to W. M. de Vos (NIZO, BA Ede, The Netherlands) for providing plasmid pNZ272 and to P. Loewen (University of Manitoba, Winnipeg, Canada) for providing E. coli UM2 and B. subtilis UM1013.
This work was supported by grant 0319280B from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie and by grant 11-104035 from Fraunhofer-Gesellschaft.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Sedman J A, Struhl K. Current protocols in molecular microbiology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 2.Berthier F, Zagorec M, Champomier-Vergès M, Ehrlich S D, Morel-Deville F. Efficient transformation of Lactobacillus sake by electroporation. Microbiology. 1996;142:1273–1279. doi: 10.1099/13500872-142-5-1273. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bringel F, Hubert J C. Optimized transformation by electroporation of Lactobacillus plantarum strains with plasmid vectors. Appl Microbiol Biotechnol. 1990;33:664–670. [Google Scholar]
- 5.Cavadini C, Hertel C, Hammes W P. Stable expression of the lysostaphin gene in meat lactobacilli by introducing deletions within the prosequence. Syst Appl Microbiol. 1996;19:21–27. [Google Scholar]
- 6.Chang S, Cohen S N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 7.Condon S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev. 1987;46:269–280. [Google Scholar]
- 8.De Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 9.Engelmann S, Hecker M. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol Lett. 1996;145:63–69. doi: 10.1111/j.1574-6968.1996.tb08557.x. [DOI] [PubMed] [Google Scholar]
- 10.Engesser D M, Hammes W P. Non-heme catalase activity of lactic acid bacteria. Syst Appl Microbiol. 1994;17:11–19. [Google Scholar]
- 11.Gasson M J, Benson K, Swindell S, Griffin H. Metabolic engineering of Lactobacillus lactis diacetyl pathway. Lait. 1996;76:33–40. [Google Scholar]
- 12.Hammes W P, Knauf H J. Starters in the processing of meat products. Meat Sci. 1994;36:155–168. doi: 10.1016/0309-1740(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 13.Hammes W P, Bantleon A, Min S. Lactic acid bacteria in meat fermentation. FEMS Microbiol Rev. 1990;87:165–174. [Google Scholar]
- 14.Heidel M, Gaier W, Hammes W P. Plasmids in lactobacilli used as starter organisms. FEMS Microbiol Rev. 1987;46:P25. [Google Scholar]
- 15.Hertel C, Ludwig W, Obst M, Vogel R F, Hammes W P, Schleifer K H. 23S rRNA-targeted oligonucleotide probes for the rapid identification of meat lactobacilli. Syst Appl Microbiol. 1991;14:173–177. [Google Scholar]
- 16.Igarashi T, Kono Y, Tanaka K. Molecular cloning of manganese catalase from Lactobacillus plantarum. J Biol Chem. 1996;271:29521–29524. doi: 10.1074/jbc.271.47.29521. [DOI] [PubMed] [Google Scholar]
- 17.Irvine A S, Guest J R. Lactobacillus casei contains a member of the CRP-FNR family. Nucleic Acids Res. 1993;21:753. doi: 10.1093/nar/21.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ish-Horowicz D, Burke F J. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2999. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein J R, Ulrich C, Plapp R. Characterization and sequence analysis of a small cryptic plasmid from Lactobacillus curvatus LTH683 and its use for construction of new Lactobacillus cloning vectors. Plasmid. 1993;30:14–29. doi: 10.1006/plas.1993.1030. [DOI] [PubMed] [Google Scholar]
- 20.Knauf H J, Vogel R F, Hammes W P. Cloning, sequence, and phenotypic expression of katA, which encodes the catalase of Lactobacillus sake LTH677. Appl Environ Microbiol. 1992;58:832–839. doi: 10.1128/aem.58.3.832-839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loewen P C, Switala J. Genetic mapping of katA, a locus that affects catalase 1 levels in Bacillus subtilis. J Bacteriol. 1987;169:5848–5851. doi: 10.1128/jb.169.12.5848-5851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewen P C, Triggs B L, George C S, Hrabarchuk B E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J Bacteriol. 1985;162:661–667. doi: 10.1128/jb.162.2.661-667.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig W, Kirchhof G, Klugbauer N, Weizenegger M, Betzl D, Ehrmann M, Hertel C, Jilg S, Tatzel R, Zitzelsberger H, Liebl S, Hochberger M, Shah J, Lane D, Wallnöfer P R, Schleifer K H. Complete 23S ribosomal RNA sequences of gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1992;15:487–501. [Google Scholar]
- 24.Obst M, Meding E R, Vogel R F, Hammes W P. Two genes encoding the β-galactosidase of Lactobacillus sake. Microbiology. 1995;141:3059–3066. doi: 10.1099/13500872-141-12-3059. [DOI] [PubMed] [Google Scholar]
- 25.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha E R, Smith C J. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1995;177:3111–3119. doi: 10.1128/jb.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross R P, Claiborne A. Evidence for regulation of the NADH peroxidase gene (npr) from Enterococcus faecalis by OxyR. FEMS Microbiol Lett. 1997;151:177–183. doi: 10.1111/j.1574-6968.1997.tb12567.x. [DOI] [PubMed] [Google Scholar]
- 28.Rozier J. Die Rolle der Katalase-Aktivität des Fleisches bei der Rohwurst-Fabrikation. Fleischwirtschaft. 1971;7:1063–1066. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sanders J W, Leenhouts K J, Haandrikman A J, Venema G, Kok J. Stress response in Lactobacillus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schellhorn H E. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol Lett. 1994;131:113–119. doi: 10.1111/j.1574-6968.1995.tb07764.x. [DOI] [PubMed] [Google Scholar]
- 32.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 33.Spiro S, Guest J R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;75:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 34.Tartaglia L A, Gimeno C J, Storz G, Ames B N. Multidegenerate DNA recognition by the OxyR transcriptional regulator. J Biol Chem. 1992;267:2038–2045. [PubMed] [Google Scholar]
- 35.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 36.Wolf G, Strahl A, Meisel J, Hammes W P. Heme-dependent catalase activity of lactobacilli. Int J Food Microbiol. 1991;12:133–140. doi: 10.1016/0168-1605(91)90062-t. [DOI] [PubMed] [Google Scholar]
- 37.Zink A, Klein J R, Plapp R. Transformation of Lactobacillus delbrueckii ssp. lactis by electroporation and cloning of origins of replication by use of a positive selection vector. FEMS Microbiol Lett. 1991;78:207–212. [Google Scholar]