Abstract
Since 1997, outbreaks of highly pathogenic (HP) H5N1 and circulation of H9N2 viruses among domestic poultry in Asia have posed a threat to public health. To better understand the extent of transmission of avian influenza viruses (AIV) to humans in Asia, we conducted a cross-sectional virologic study in live bird markets (LBM) in Hanoi, Vietnam, in October 2001. Specimens from 189 birds and 18 environmental samples were collected at 10 LBM. Four influenza A viruses of the H4N6 (n = 1), H5N2 (n = 1), and H9N3 (n = 2) subtypes were isolated from healthy ducks for an isolation frequency of over 30% from this species. Two H5N1 viruses were isolated from healthy geese. The hemagglutinin (HA) genes of these H5N1 viruses possessed multiple basic amino acid motifs at the cleavage site, were HP for experimentally infected chickens, and were thus characterized as HP AIV. These HA genes shared high amino acid identities with genes of other H5N1 viruses isolated in Asia during this period, but they were genetically distinct from those of H5N1 viruses isolated from poultry and humans in Vietnam during the early 2004 outbreaks. These viruses were not highly virulent for experimentally infected ducks, mice, or ferrets. These results establish that HP H5N1 viruses with properties similar to viruses isolated in Hong Kong and mainland China circulated in Vietnam as early as 2001, suggest a common source for H5N1 viruses circulating in these Asian countries, and provide a framework to better understand the recent widespread emergence of HP H5N1 viruses in Asia.
Influenza A viruses that reside naturally in wild bird species comprise all known subtypes and provide viral genes from which influenza viruses that infect both domestic poultry and mammalian species, including humans, arise (49, 59, 61). In the last 10 years, the incidence of highly pathogenic (HP) avian influenza in domestic poultry has increased substantially, while subtypes of low pathogenicity (LP), such as H9N2, have become endemic in Europe and Asia (1, 12, 13, 41). These events have increased the frequency of human exposure to poultry infected with avian influenza viruses (AIV), which has resulted in poultry-to-human transmission of these viruses. Substantial human illness and death have occurred in the case of HP H5N1 and H7N7 strains, while H9N2 viruses have been isolated from individuals with mild influenza (18, 20, 26, 46, 63, 66). These events highlight the potential of domestic poultry to act as an intermediate host to facilitate transmission of AIV to humans. To date, the AIV that have transmitted to humans have not acquired human influenza virus genes and lacked the ability to spread efficiently from person to person (22, 26, 58). Nevertheless, as the number of human infections with AIV increases, so too does the likelihood of human coinfection with both avian and human viruses and the potential for a novel avian-human reassortant virus to emerge. Such a virus could have the ability to transmit efficiently in human populations with little or no immunity against the novel hemagglutinin (HA) and give rise to a pandemic. Such was the case in 1957 and 1968, when human-avian reassortant viruses, having acquired avian influenza virus HA, neuraminidase (NA), and PB1 or HA and PB1 genes, respectively, emerged to cause pandemics in humans (24, 61).
Most of the pandemic strains in the last century first appeared in southern China, a hypothetical influenza epicenter (51, 52). In 1997 in Hong Kong, during the first known outbreak of human infections with HP H5N1 AIV (7, 8, 55, 56), exposure to live poultry at retail stalls or markets during the preceding week was identified as a significant risk factor for human illness (40). At that time, multiple species of waterfowl and land-based birds were housed together in Hong Kong live bird markets (LBM), creating an ideal environment for genetic reassortment among influenza viruses harbored by different avian species and the generation of avian strains with the potential to infect humans. Serological evidence for H5 infection was found in 10% of LBM poultry workers tested (3). Depopulation of domestic poultry in Hong Kong in late 1997 halted the transmission of the H5N1 AIV to humans, ending the outbreak. Since then, HP H5N1 viruses have reemerged in domestic poultry in Hong Kong on several occasions, most recently in late 2002, when viruses were also isolated from dead wild birds in Hong Kong wildlife parks (14, 15, 16, 53). In February 2003, H5N1 viruses similar to that isolated from a wild water bird were isolated from a father and son with respiratory illness; the father subsequently died, while the son recovered fully (17, 47, 62).
The cultural preference for the consumption of freshly slaughtered poultry is not unique to Hong Kong or mainland China, and LBM are common in many other Asian countries, including Vietnam. In October 2001, we initiated a study to better understand the extent of avian influenza virus circulation and the risk of infection among workers in LBM in Hanoi, Vietnam. The results of the human seroepidemiological study will be reported elsewhere. Here we report the first isolation of multiple subtypes of AIV, including HP H5N1 viruses, from specimens collected from healthy domestic waterfowl in LBM in Hanoi, Vietnam, in 2001. This is the first report of HP H5N1 viruses in LBM outside of Hong Kong, prior to the recent widespread outbreaks of HP H5N1 in Vietnam and other parts of Asia that occurred since December 2003. We describe the antigenic, genetic, and pathogenic properties of the H5N1 viruses isolated in Vietnam in LBM in 2001. The characteristics of these viruses provide important insights for understanding the origins of the HP H5N1 influenza viruses in Asia. Our results highlight the need for further study of the influenza ecology in the region as a component of pandemic preparedness.
MATERIALS AND METHODS
Sample collection and virus isolation.
Over 2 days in October 2001, 396 specimens were collected from five species of domestic poultry at 10 LBM in urban Hanoi, Vietnam. Oropharyngeal and cloacal swabs were collected from each of 189 randomly chosen birds. Additionally, 18 fecal specimens were collected either from the bottom of cages housing at least two bird species or environmental surfaces of the markets. The number of samples collected from each type of poultry was estimated to be proportional to the number and type of poultry in the market. Most specimens were collected from apparently healthy birds, although specimens were also collected from two chickens showing signs of disease (depression, ruffled feathers) and two dead birds. Swabs were placed in transport medium, consisting of phosphate-buffered saline (PBS) containing 50% glycerol, penicillin (2,000 U/ml), gentamicin (250 μg/ml), polymixin B (2,000 U/ml), nystatin (500 U/ml), ofloxacin HCl (60 μg/ml), and sulfamethoxazole (200 μg/ml). Specimens were immediately chilled and then frozen at −70°C on return to the laboratory. Individual specimens from pooled samples that were positive by PCR (see below) were cultured in 10-day-old embryonated chicken eggs for 24 to 48 h at 37°C (and were frozen in aliquots at −70°C). The allantoic fluids were harvested and tested for HA activity as previously described (25). The 50% egg infectious dose (EID50) was determined by serial titration of virus in eggs and was calculated by the method of Reed and Muench (48). All experiments using infectious virus were conducted in a biosafety level 3 (BSL-3) facility with enhancements approved for work with HP AIV by the U.S. Department of Agriculture.
Identification of specimens positive for influenza A viruses by RT-PCR.
RNA was extracted from pools of like specimens (cloacal, oropharyngeal, or fecal) from five birds by using a MagNA Pure LC Total Nucleic Acid Extraction kit and a MagNA Pure LC instrument (Roche, Mannheim, Germany). Samples were amplified using a One-Step reverse transcription-PCR (RT-PCR) kit (QIAGEN, Valencia, Calif.) and either influenza A virus NP-specific primers previously designed by Lee et al. (30) or primers specific for the M and F genes of Newcastle disease virus (NDV) (50), another avian virus frequently isolated from avian species.
Subtyping and antigenic characterization of AIV.
Virus isolates were subtyped by the hemagglutination inhibition (HAI) test, using a panel of reference antisera against 15 HA subtypes obtained either from the National Institutes of Allergy and Infectious Diseases (Bethesda, Md.) reagent repository or kindly provided by R. Webster (St. Jude Children's Research Hospital, Memphis, Tenn.) and using 4 hemagglutinating units (HAU) of virus and 0.5% turkey red blood cells. Additional antigenic characterization of viruses was conducted by HAI using postinfection ferret antisera as previously described (25). To increase titers to some avian viruses, ferrets were boosted 14 days after intranasal infection with approximately 1,000 HAU of virus mixed with TiterMax (CytRx Corporation, Norcross, Ga.) inoculated into the rear footpads in a total volume of 0.5 ml and/or sera were concentrated 1:4 by centrifugation under vacuum. The NA subtypes of viruses were determined genetically. The NA genes of these viruses were amplified using primers and RT-PCR conditions described by Hoffmann et al. (21). The PCR products were directly sequenced as described below, and the NA subtypes were identified by nucleotide BLAST search on viral nucleotide sequences that were available from the National Center for Biotechnology Information, Bethesda, Md. (http://www.ncbi.nlm.nih.gov/BLAST/).
Genetic and phylogenetic analysis of surface glycoproteins.
Viral RNA was extracted from virus-infected allantoic fluids with the One-Step RT-PCR kit (QIAGEN) and was used to amplify HA and NA genomic segments as full-length or overlapping fragments by RT-PCR. The amplicons were sequenced on an automated Applied Biosystems 373 AI 3100 system using cycle sequencing dye terminator chemistry (Perkin-Elmer, Foster City, Calif.). Primer sequences are available upon request. The nucleotide sequences were edited using the Seqman module of the DNAStar package. The nucleotide sequences for the HA genes were aligned by the Clustal W method, using the MegAlign module of DNAStar software. Phylogenetic analysis was performed using the PHYLIP 3.5c software package, implementing the neighbor joining method with Kimura 2-distance parameters (10). The tree topology was evaluated by 1,000 bootstrap analyses. The tree was drawn using TreeView 1.6.6 (45). The nucleotide sequence regions used for phylogenetic analysis for H5 HA, H9 HA, and H4 HA were 179 to 1130, 139 to 1068, and 20 to 1309, respectively.
Pathogenicity tests of H5N1 viruses in avian species.
Four-week-old White Plymouth Rock (WPR) chickens were used in pathogenicity studies using standard procedures (43). Briefly, eight chickens were inoculated by the intravenous (i.v.) route with 0.2 ml of a 1:10 dilution of bacteria-free allantoic fluid containing 107.6 EID50 of A/Goose/Vietnam/113/2001 (Gs/VN/113/01) or A/Goose/Vietnam/324/2001 (Gs/VN/324/01) virus. In addition, eight birds were inoculated by the intranasal (i.n.) route with 106.0 EID50 of each of the above H5N1 viruses. Birds were evaluated for signs of illness for up to 10 days. For studies with ducks, 10 2-week-old Pekin white ducks (Anas platyrhynchos) (Privett hatchery, Portales, N. Mex.) were inoculated i.n. with 0.1 ml of 106.0 EID50 of either Gs/VN/113/01 or A/Egret/Hong Kong/757.2/2002 ([H5N1] Eg/HK/757.2/02). The latter virus was isolated from a wild egret at a Hong Kong waterfowl park in December 2002 and was kindly provided by Trevor Ellis, Agriculture Fisheries and Conservation Department, Tai Lung Veterinary Laboratory, Hong Kong. Two ducks from each group were euthanatized on day 2 postinfection (p.i.), and tissues were collected and frozen at −70°C for subsequent virus isolation in 10-day-old embryonated eggs.
Pathogenicity tests of H5 viruses in mammalian species.
Lightly anesthetized 6- to 10-week-old female BALB/c mice (Charles River Laboratories, Wilmington, Mass.) were inoculated i.n. with 50 μl of virus diluted in PBS to give doses of 100 to 107 EID50. The 50% mouse infectious dose (MID50) and the 50% lethal dose (LD50) were determined as previously described (37). Mice were monitored daily for changes in weight and signs of illness for 14 days p.i. Three mice per group were euthanatized on day 3 or day 6 p.i., and organs were harvested and frozen at −70°C for subsequent virus isolation. Whole tissues were thawed and homogenized in 1 ml of cold PBS and were considered a 100% tissue suspension. Clarified tissue homogenates were titrated in embryonated eggs for infectivity. Virus titers were calculated using the method of Reed and Muench (48).
Six young adult Fitch ferrets (Triple F Farms, Sayre, Pa., or Marshall Farms, North Rose, N.Y.) were inoculated i.n. with 107 EID50 of virus in 1 ml of PBS and were monitored for 14 days as previously described (67). Three animals were euthanatized on day 3 p.i., and portions of organs were harvested and stored at −70°C for subsequent virus titration in embryonated eggs. Tissues were fixed in 10% neutral buffered formalin solution, sectioned, and stained by hematoxylin and eosin.
Nucleotide sequence accession numbers.
The sequence data described in this report were submitted to GenBank with accession numbers ISDN38259 through ISDN38261 (H5) and ISDN68679 through ISDN68681 (H4 and H9) (38).
RESULTS
Multiple subtypes of AIV isolated from LBM.
Cloacal and oropharyngeal specimens from 189 birds and 18 environmental specimens were collected from 10 retail markets in Hanoi, Vietnam. The numbers of specimens collected from each species reflected their approximate overall frequency in the LBM. No AIV were isolated from chickens, from which the most specimens were collected. In contrast, NDV was isolated from 16 of 98 (16%) chickens. Six influenza viruses, representing four subtypes, were isolated from four markets (Table 1). One H4N6, one H5N2, and two H9N3 strains were isolated from 4 different ducks of the 13 sampled, giving a 31% isolation rate in this avian species. H5N1 viruses were isolated from 2 of 33 geese (6.1%) sampled. All viruses were recovered from apparently healthy birds. No AIV were isolated from quail, pigeons, or environmental materials. The H4N6 virus (A/Duck/Vietnam/14/2001 [Dk/VN/14/01]) and one H9N3 virus (A/Duck/Vietnam/68/2001 [Dk/VN/68/01]) were isolated from one market, the largest in Hanoi at the time. Another smaller market yielded the H5N2 virus (A/Duck/Vietnam/342/2001 [Dk/VN/342/01]) and two H9N3 isolates (A/Duck/Vietnam/339/2001 [Dk/VN/339/01] and A/Duck/Vietnam/340/2001 [Dk/VN/340/01]) from oropharyngeal and cloacal swabs, respectively, collected from the same bird. The latter H9N3 isolates are considered a single virus isolation, and for clarity only results for Dk/VN/340/01 will be presented. The H5N1 viruses Gs/VN/113/01 and Gs/VN/324/01 were isolated from geese sampled at two additional markets.
TABLE 1.
Summary of avian influenza viruses isolated from Hanoi LBM by host speciesa
| Species | No. of birds sampled | No. of viruses isolated and virus subtype (market)b
|
Total (%) | |||
|---|---|---|---|---|---|---|
| H4N6 | H5N1 | H5N2 | H9N3 | |||
| Chicken | 98 | |||||
| Pigeon | 39 | |||||
| Goose | 33 | 2d (B, C) | 2 (6.1) | |||
| Duck | 13 | 1c (A) | 1c (D) | 2e (A, D) | 4 (30.8) | |
| Quail | 6 | |||||
| Total (%) | 189 | 1 (0.5) | 2 (1.1) | 1 (0.5) | 2 (1.1) | 6 (3.2) |
Cloacal and oropharyngeal specimens were collected from each bird sampled. In addition, 18 fecal specimens were collected from the bottom of cages housing mixed birds (6) or from environmental surfaces of the market (12). All 18 fecal specimens were negative for influenza A virus by PCR. Newcastle disease virus was isolated from 16 chickens, 4 pigeons, 2 geese, 1 duck, and 1 fecal specimen.
Markets are designated A, B, C, and D.
Isolated from a cloacal swab.
One virus isolated from a cloacal swab and the other from an oropharyngeal swab.
One virus isolated from a cloacal swab; second virus isolated from both cloacal and oropharyngeal swabs.
Genetic and antigenic analysis of H4N6 and H9N3 viruses isolated from LBM.
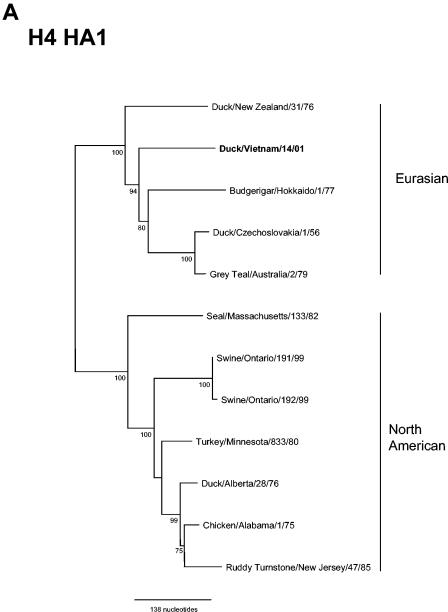
Phylogenetic analysis of the HA1 gene demonstrated that the H4N6 virus, Dk/VN/14/01, grouped with the Eurasian H4 lineage represented by A/Duck/Czechoslovakia/1/56 virus (Fig. 1A). The Dk/VN/14/01 isolate shared 89.9% nucleotide identity with A/Duck/Czechoslovakia/1/56 virus. When compared with additional H4 nucleotide sequences for which only a portion of the HA1 sequence (nucleotides 270 to 418) was available (34), Dk/VN/14/01 was found to be closely related (93.3% identity) to H4 viruses from ducks and chickens isolated in Nanchang, China, LBM in 2000 (Table 2). Ferret antiserum raised against Dk/VN/14/01 with a homologous HAI titer of 640 reacted to a titer of 320 with A/Duck/Czechoslovakia/1/56, and it also cross-reacted with North American H4 viruses A/Duck/New Jersey/5406.27/94 and A/Duck/New Jersey/14190.23/96 but not with an earlier North American H4 virus, A/Chicken/Alabama/1/75 (data not shown).
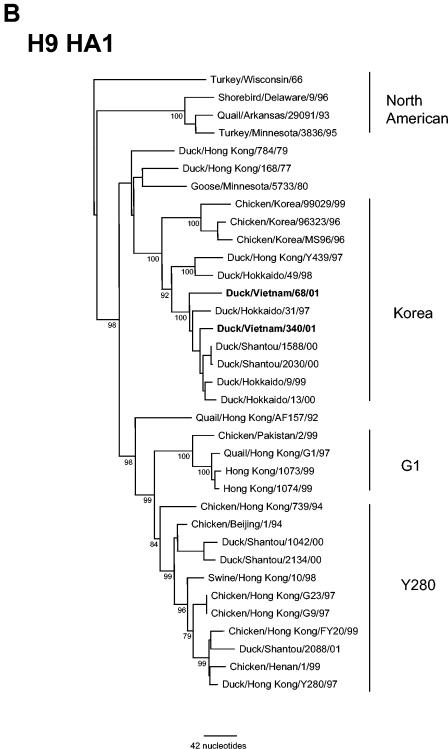
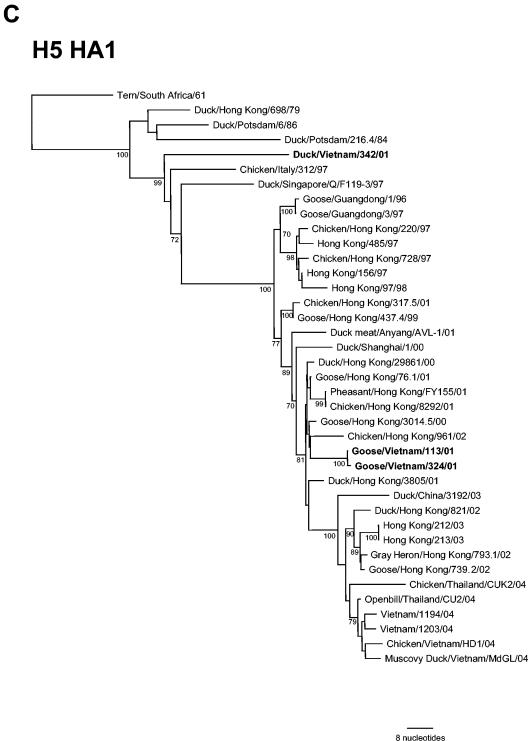
FIG. 1.
Phylogenetic relationships of hemagglutinin genes from (A) H4, (B) H9, and (C) H5 viruses isolated from LBM in Vietnam in 2001 and other representative AIV of these subtypes. The trees were drawn using the TreeView 1.6.6 program (45). Numbers below branches indicate the bootstrap values from 1,000 analyses; values less than 70% are not shown. The nucleotide sequence regions used for phylogenetic analysis were 20 to 1309 (1,290 bp), 179 to 1130 (952 bp), and 139 to 1068 (930 bp) for the H4, H5, and H9 genes, respectively. The H14N5 subtype virus A/Mallard/Gurjev/263/82, H12N5 virus A/Duck/Alberta/60/76, and H5N1 virus A/Chicken/Scotland/59 were used as outgroups for the H4, H9, and H5 genes, respectively. Eurasian and North American lineage and sublineages (Korea, G1, and Y280) are indicated. The viruses characterized in this study are indicated in boldface. The number of nucleotide changes represented by the scale bar is given for each figure.
TABLE 2.
Nucleotide sequence identity of HA1 gene region of AIV isolated in Hanoi LBM
| Virus (subtype) | Region compared | Virus with greatest identity | Identity (%) |
|---|---|---|---|
| Dk/VN/14/01 (H4N6) | 20-1309 | A/Duck/Czechoslovakia/1/56 | 89.9 |
| 270-418 | A/Chicken/Nanchang/20527/00 | 93.3 | |
| 270-418 | A/Duck/Nanchang/4-173/00 | 93.3 | |
| Gs/VN/113/01 (H5N1) | 179-1130 | A/Goose/Hong Kong/76.1/01 | 98.3 |
| A/Goose/Hong Kong/3014.5/00 | 98.3 | ||
| A/Goose/Vietnam/324/01 | 99.7 | ||
| Gs/VN/324/01 (H5N1) | 179-1130 | A/Goose/Hong Kong/76.1/01 | 98.4 |
| A/Goose/Hong Kong/3014.5/00 | 98.4 | ||
| A/Goose/Vietnam/113/01 | 99.7 | ||
| Dk/VN/342/01 (H5N2) | 179-1130 | A/Chicken/Italy/312/97 | 93.5 |
| Dk/VN/68/01 (H9N3) | 139-1068 | A/Duck/Hokkaido/9/99 | 96.7 |
| Dk/VN/340/01 (H9N3) | 139-1068 | A/Duck/Hokkaido/9/99 | 98.4 |
Viruses of the H9 subtype from Eurasia that have circulated since the mid-1990s can be distinguished into three sublineages on the basis of antigenic and genetic properties (13, 19, 31). The HA1 genes of two Vietnam H9N3 viruses were sequenced and compared phylogenetically with other H9 viruses from the Eurasian lineage. Figure 1B illustrates the three distinct genetic groups within the Eurasian lineage. The H9N3 viruses from Vietnam LBM belonged to the Korean group represented by A/Duck/Hong Kong/Y439/97 (13, 19, 31) (Fig. 1B). The HA1 of H9 viruses from Vietnam, Dk/VN/68/01 and Dk/VN/340/01, shared nucleotide sequence identities of 96.7 and 98.4%, respectively, with A/Duck/Hokkaido/9/99 (Table 2). The two H9N3 viruses from Vietnam shared 97.6% amino acid identity in HA1 (data not shown). One amino acid difference between the viruses was located at the HA cleavage site. Dk/VN/68/01 possessed a sequence (PAASSR/GL) similar to that found in other H9 viruses isolated in recent years from southern China and Hong Kong (35, 36), whereas Dk/VN/340/01 possessed a PAASGR/GL cleavage site sequence motif. The H9N3 viruses isolated in the present study possessed a glutamine (Q) at residue 216 in HA1 (H9 numbering; 226 in H3 numbering), a receptor binding site residue typical of AIV and associated with the preferential binding of sialic acid (SA) in α2,3 linkage to galactose (35, 36, 39).
The antigenic characteristics of the H9N3 viruses were determined by the HAI assay using postinfection ferret sera. As shown in Table 3, the two H9N3 viruses isolated in Hanoi LBM were antigenically similar to each other and were most similar to the A/Chicken/Korea/96323/96 reference virus. The H9N3 viruses reacted poorly with antisera to viruses representing the G1- and Y280-like antigenic groups, although sera raised to Dk/VN/340/01 was more broadly cross-reactive with viruses from each of the H9 sublineages.
TABLE 3.
Hemagglutination-inhibition (HAI) reactivity of Vietnam H9N3 viruses compared to that of other H9 subtype viruses
| Virusa | HAI reactivity of reference ferret antiserum against H9 virusesc
|
|||||||
|---|---|---|---|---|---|---|---|---|
| G1-like
|
Y280-like
|
Korea-like
|
||||||
| HK/G1/97 | HK/1073/99 | HK/G9/97 | HK/Y280/97 | Sw/HK/98 | HK/2108/03 | Ck/Kor/96 | VN/340/01 | |
| Qu/HK/G1/97 | 1,280 | 640 | 10 | 80 | <b | 20 | < | 1,280 |
| HK/1073/99 | 640 | 1,280 | 20 | 80 | < | 40 | 10 | 2,560 |
| Ck/HK/G9/97 | 80 | 160 | 2,560 | 5,120 | 2,560 | 320 | 320 | 5,120 |
| Dk/HK/Y280/97 | 40 | 40 | 1,280 | 2,560 | 1,280 | 160 | 160 | 2,560 |
| Sw/HK/10/98 | 20 | 40 | 1,280 | 2,560 | 1,280 | 160 | 160 | 2,560 |
| HK/2108/03 | 40 | 40 | 640 | 640 | 320 | 640 | 320 | 320 |
| Ck/Kor/96323/96 | < | < | < | 20 | < | < | 640 | 640 |
| Dk/VN/340/01 | < | < | < | 80 | < | 20 | 320 | 5,120 |
| Dk/VN/68/01 | < | < | 10 | 80 | < | < | 640 | 5,120 |
Virus abbreviations: Qu, Quail; Ck, Chicken; Dk, Duck; Sw, Swine; HK, Hong Kong; Kor, Korea; VN, Vietnam.
<, No inhibition detected at the lowest serum dilution of 1:10.
HAI titers against homologous virus are shown in boldface type.
Genetic analysis of H5 HA1 and NA genes.
The HA cleavage site (PRIERRRKKR/GL) identified the two H5N1 viruses isolated from apparently healthy geese in the LBM as HP AIV. The two H5N1 viruses differed by only a single amino acid residue in HA1 (E189K, H5 numbering; 194 in H3 numbering), which may contribute to the minor antigenic differences detected in the HAI assay. The H5N2 duck virus possessed a cleavage site (PQRETR/GL) characteristic of an LP AIV. A phylogenetic comparison with HA1 genes from H5 viruses isolated in Asia between 1979 and 2004 is shown in Fig. 1C. The two H5N1 viruses from the Vietnam LBM were genetically related to A/Goose/Guangdong/1/1996 (Gs/Gd/1/96) virus, the progenitor of the H5N1 viruses isolated from poultry and humans in Hong Kong in 1997 and domestic geese in South China between 1999 and 2002 (5). Gs/VN/113/01 and Gs/VN/324/01 were most similar (>98%) in nucleotide sequence to viruses isolated from waterfowl in Hong Kong in 2000 to 2001 (Table 2). In contrast, the HA1 gene of the H5N2 virus Dk/VN/342/01 was most similar in nucleotide (93.5% identity) and amino acid sequence to an earlier Eurasian H5 virus, A/Chicken/Italy/312/97. The NA genes of the two H5N1 isolates were identical to each other and did not possess a deletion in the stalk region found in the H5N1 viruses that infected humans in Hong Kong in 1997 and in Vietnam and Thailand in 2004 (2, 17, 32). Further analysis of the NA genes of the H5N1 viruses and internal genes of all viruses isolated during this study will be described elsewhere (S. Jadhao and J. M. Katz, unpublished data).
Antigenic characterization of H5N1 and H5N2 isolates.
The antigenic relationships among the three H5 viruses isolated from LBM in Vietnam and other H5 viruses isolated in Asia from 1997 to 2003 were evaluated by HAI assay using postinfection ferret antisera (Table 4). The two H5N1 strains (Gs/VN/113/01 and Gs/VN/324/01) were antigenically similar to each other, with the exception of a fourfold lower reactivity of VN/324/01 with antisera raised to Gs/VN/113/01. Both Vietnam H5N1 isolates were antigenically distinct from the H5N2 isolate (Dk/VN/342/01) and other H5N1 viruses isolated from avian species or humans in Hong Kong between 1997 and 2003. Dk/VN/342/01 was antigenically more closely related to H5 Eurasian viruses isolated before 2000. All H5 viruses isolated from Vietnam LBM in 2001 were antigenically distinct from two H5N1 viruses, A/Vietnam/1203/2004 and A/Vietnam/1194/2004, isolated from humans in early 2004.
TABLE 4.
HAI reactivity of Vietnam H5 viruses compared to that of other H5 subtype viruses
| Virusa | HAI reactivity of reference ferret antiserum against H5 virusesc
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HK/97/98 | Dk/Sgl/97 | Gs/HK/437/99 | Dk/An/01 | Gs/VN/113/01 | Gs/VN/324/01 | Dk/VN/342/01 | Gs/HK/02 | RPB/HK/02 | HK/213/03 | VN/1203/04 | VN/1194/04 | |
| HK/156/97 | 320 | 40 | 160 | 80 | 640 | 640 | 1,280 | 40 | 160 | 40 | 640 | 160 |
| HK/97/98 | 640b | 40 | 320 | 80 | 1,280 | 640 | 1,280 | 40 | 320 | 160 | 320 | 160 |
| Dk/Sing/Q/F119-3/97 | 640 | 80 | 160 | 20 | 320 | 160 | 640 | <b | 160 | 80 | 160 | 40 |
| Gs/HK/437,4/99 | 640 | 80 | 640 | 320 | 5,120 | 2,560 | 2,560 | 320 | 640 | 640 | 640 | 320 |
| Dkmt/An/AVL-1/01 | 80 | 10 | 40 | 80 | 1,280 | 160 | 160 | 80 | 160 | 160 | 160 | 160 |
| Gs/VN/113/01 | 80 | 10 | 40 | 80 | 2,560 | 1,280 | 320 | 40 | 80 | 160 | 20 | 10 |
| Gs/VN/324/01 | 80 | 10 | 40 | 40 | 640 | 1,280 | 160 | 20 | 40 | 80 | 20 | 20 |
| Dk/VN/342/01 | 320 | 80 | 320 | 20 | 160 | 320 | 2,560 | 20 | 160 | < | 320 | 80 |
| Gs/HK/739.2/02 | 80 | 10 | 80 | 80 | 320 | 80 | 320 | 160 | 1,280 | 160 | 160 | 160 |
| RBP/HK/821/02 | <b | < | 20 | < | 80 | < | 20 | 40 | 640 | 40 | 80 | 40 |
| HK/213/03 | 320 | 80 | 320 | 1,280 | 10,240 | 5,120 | 1,280 | 640 | 2,560 | 2,560<’b> | 1,280 | 320 |
| VN/1203/04 | < | < | 10 | < | 40 | 20 | < | < | 80 | < | 320 | 160 |
| VN/1194/04 | < | < | 20 | < | 80 | 20 | < | 20 | 80 | < | 320 | 160 |
Virus abbreviations are the same as those in Table 3, plus Sing or Sg, Singapore; Dkmt, duck meat; An, Anyang; Gs, Goose; RBP, Rosy-billed Pochard.
<, No inhibition detected at the lowest serum dilution of 1:10.
HAI titers against the homologous virus are shown in boldface type.
Pathogenicity of H5N1 isolates for avian species.
Based on the sequence of the multibasic amino acid sequence motif at the HA cleavage site, the two H5N1 viruses isolated from the Vietnam LBM were identified as HP strains. To evaluate their virulence for chickens, WPR chickens were infected by the i.v. or i.n. route with the two H5N1 viruses (Table 5). By the i.v. route, both Gs/VN/113/01 and Gs/VN/324/01 caused >75% lethality within 24 h and 100% mortality within 48 h. By the i.n. route, each of the viruses was lethal for seven of eight birds within 72 h. The mean death time (MDT) of chickens was 27 and 24 h following i.v. inoculation and 62 and 57 h following i.n. inoculation with Gs/VN/113/01 and Gs/VN/324/01, respectively. Therefore, the two H5N1 viruses isolated from healthy geese at two separate LBM in Hanoi in 2001 fulfilled the criteria for HP AIV (43). Because the H5N2 virus did not possess multiple basic amino acids at the HA cleavage site, its pathogenicity for chickens was not evaluated.
TABLE 5.
Pathogenicity of H5N1 viruses for avian species
| Species (age) | Virus | Route of inoculation | Infection dose (log10EID50) | No. dead/No. inoculated | MDTa |
|---|---|---|---|---|---|
| WPR chickens (3-4 wk) | Gs/VN/113/01 | i.v. | 7.6 | 8/8 | 27 |
| i.n. | 6.0 | 8/8 | 62 | ||
| Gs/VN/324/01 | i.v. | 7.6 | 8/8 | 24 | |
| i.n. | 6.0 | 8/8 | 57 | ||
| Pekin white ducks (2 wk) | Gs/VN/113/01 | i.n. | 6.0 | 0/8 | NAb |
| Eg/HK/757.2/02 | i.n. | 6.0 | 8/8 | 127 |
MDT, mean death time given in hours after virus inoculation.
NA, not applicable; all birds survived the 14-day observation period.
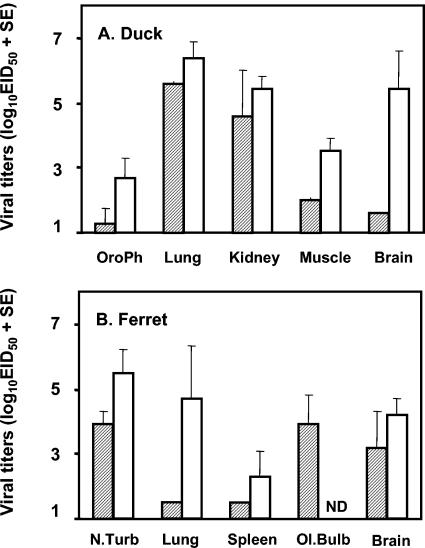
The pathogenicity of GsVN/113/01 (H5N1) virus for Pekin white ducks was also determined. The Eg/HK/757.2/02 (H5N1) virus, known to be lethal for wild aquatic birds, was also included in these studies for comparison (9). Ten 2-week-old ducks were inoculated i.n. with 106.0 EID50 of either virus. Two birds were euthanatized on day 2 p.i. to determine the extent of viral replication in tissues. The remaining eight birds were observed for signs of illness over a 14-day period. None of the birds inoculated with Gs/VN/113/01 showed signs of illness or died (Table 5). Nevertheless, high titers of virus were detected on day 2 p.i. in the lung and kidney of ducks inoculated with this virus (Fig. 2A). In contrast, Eg/HK/757.2/02 was highly lethal for eight of eight ducks, with an MDT of 5.3 days (Table 5). High titers of virus were detected in the lung, kidney, muscle, and brain of birds inoculated with this lethal strain (Fig. 2A). Viral titers in the oropharynx and cloaca were also monitored for the first week of infection. Cloacal swabs from birds infected with either virus yielded minimal (≤101.6 EID50/ml) virus at any day p.i., whereas oropharyngeal titers were highest on day 3 p.i. and were 101.8 and 102.7 EID50/ml for Gs/VN/113/01 and Eg/HK/757.2/02, respectively. Thus, although the H5N1 virus isolated from healthy geese in LBM in Vietnam was highly pathogenic for experimentally infected chickens and could replicate in some tissues of Pekin white ducks, the virus was not lethal for this avian species.
FIG. 2.
Virus titers in tissues from (A) ducks and (B) ferrets infected with H5N1 viruses. (A) Ducks were inoculated i.n. with 106.0 EID50 of Gs/VN/113/01 (shaded bars) or Eg/HK/757.2/02 (open bars) virus. The mean tissue titers plus standard errors (SE) from two birds on day 2 p.i. are shown. Titers are expressed as mean log10 EID50/g except for the oropharyngeal swab (OroPh), for which titers are expressed as mean log10 EID50/ml. (B) Ferrets were inoculated i.n. with 107.0 EID50 of Gs/VN/113/01 (shaded bars) or Hong Kong/486/97 (open bars) virus. The mean tissue titers plus SE for three animals per group on day 3 p.i. are shown. Titers are expressed as mean log10 EID50/g except for the nasal turbinate samples (N.Turb), for which titers are expressed as mean log10 EID50/ml. Ol.Bulb, olfactory bulb; ND, not done.
Pathogenicity of H5N1 viruses in mice and ferrets.
Mice and ferrets have been used previously to evaluate the pathogenicity of HP H5N1 viruses for mammals (37, 67). In these models, HP H5N1 viruses isolated from humans in Hong Kong in 1997 were found to cause severe and/or lethal disease (37, 67) and are used here as comparative controls for virulence. The MID50 and LD50 titers were determined in BALB/c mice infected i.n. with HP Gs/VN/113/01 (H5N1), the LP Dk/VN/342/01 (H5N2), or A/Hong Kong/483/97 (HK/483) (H5N1), previously shown to be highly lethal for mice (37) (Table 6). The MID50 of both the H5N1 and the H5N2 viruses isolated from birds in Vietnam LBM were approximately 10,000-fold higher than the MID50 of HK/483 virus, indicating that the Vietnam isolates exhibited a substantially reduced ability to infect mice compared to the 1997 H5N1 strain. Correspondingly, the LD50s for the two Vietnam isolates were >106.0 EID50, indicating their low pathogenicity for mice. In contrast, the LD50 of HK/483 virus used as a control was 101.8 EID50, indicative of a high-pathogenicity phenotype in mice (23, 37). Weight loss and viral tissue titers were determined in groups of mice inoculated with 106 EID50. Gs/VN/113/01 and Dk/VN/342/01 viruses caused only modest weight loss, reaching a maximum of approximately 4% on days 7 to 9 p.i. In contrast, mice infected with HK/483 virus lost up to 25% of weight and died by day 6 or 7 p.i. Both Gs/VN/113/01 and Dk/VN/342/01 replicated efficiently in the lungs of mice, although with slower kinetics than those of HK/483 virus. Neither Gs/VN/113/01 nor Dk/VN/342/01 was detected in the brain, either at day 3 or day 6 p.i., although virus was isolated from the thymus of mice infected with Gs/VN/113/01 at the latter time point. HK/483 virus was isolated from the brain and lymphoid tissues at both time points. Taken together, these results demonstrated that, compared to HK/483 virus, which was highly pathogenic for BALB/c mice, Gs/VN/113/01 and Dk/VN/342/01 were of low pathogenicity in this mammalian model.
TABLE 6.
Pathogenicity of Vietnam H5 viruses in BALB/c mice
| Virus | MID50 (log10EID50) | LD50 (log10 EID50) | Maximum % weight loss (day) | Mean virus titer ± SEb (log10 EID50/ml)a
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Day 3
|
Day 6
|
||||||||
| Lung | Brain | Spleen | Lung | Brain | Thymus | ||||
| HK/483/97 (H5N1) | 0.5 | 1.8 | 25 (4) | 7.1 ± 1.0 | 1.6 ± 0.5 | 3.5 ± 0.2 | 5.9 ± 0.3 | 3.7 ± 0.4 | 2.9 ± 0.1 |
| Gs/VN/113/01 (H5N1) | 4.3 | 6.5 | 3.7 (9) | 3.9 ± 1.2 | ≤0.8 | ≤0.8 | 5.2 ± 0.5 | ≤0.8 | 3.9 ± 0.6 |
| Dk/VN/342/01 (H5N2) | 4.8 | >7.0 | 3.5 (7) | 4.8 ± 1.2 | ≤0.8 | ≤0.8 | 7.1 ± 1.0 | ≤0.8 | ≤0.8 |
Mice were infected with 106 EID50 of the various viruses. On the indicated days, mice were euthanatized and tissues were collected and subsequently homogenized and titered in eggs for the presence of infectious virus. The limit of detection for lung tissues was 101.5 EID50/ml and for other tissues was 100.8 EID50/ml.
SE, standard error.
We next compared the virulence of Gs/VN/113/01 for ferrets to that of a 1997 H5N1 isolate from humans, A/Hong Kong/486/97 (HK/486), which was previously shown to cause severe and lethal disease in this animal model (67). Groups of four to six ferrets were infected i.n. with 107 EID50 of either virus. Animals were monitored for signs of illness for 14 days, and on day 3 p.i. three animals per group were euthanatized to collect tissues for the determination of virus replication. Gs/VN/113/01 replicated in the upper respiratory tract of ferrets, but nasal turbinate titers were 40-fold lower than those in ferrets infected with HK/486 virus, which also replicated to high titers in the lungs of ferrets (Fig. 2B). In contrast, no virus was isolated from the lungs of any ferret infected with Gs/VN/113/01 virus (Fig. 2B). This virus also caused minimal weight loss in ferrets (mean of 4% on days 5 and 7 p.i.), with some sneezing noted in ferrets on day 7 p.i. In contrast, the positive control animal inoculated with HK/486 virus lost 15% of its body weight by day 7 p.i. and exhibited symptoms of severe lethargy, sneezing, dyspnea, and diarrhea, consistent with our previous findings (67). Thus, the Gs/VN/113/01 virus caused little apparent disease in ferrets. Therefore, it was somewhat surprising that Gs/VN/113/01 was isolated from the olfactory bulb and brain of ferrets on day 3 p.i. In a previous study, a human influenza A H3N2 virus which caused only mild disease in ferrets was also isolated from ferret brains (67). In the absence of neurological symptoms and histopathology, the clinical significance of isolation of this H5N1 virus, and indeed other nonvirulent influenza A viruses from the ferret brain, remains unclear.
DISCUSSION
In 2001 we initiated a seroepidemiological study to investigate the potential for the transmission of AIV to humans working in LBM in Hanoi, Vietnam. To better understand the extent of avian influenza infection of domestic poultry and thus the potential risk of occupational exposure to the LBM workers, we collected virologic specimens from a range of poultry species in the same markets targeted for the seroepidemiologic investigation. The results of the serologic investigation will be published elsewhere. To our knowledge, this was the first virologic investigation for the detection of AIV in domestic poultry in LBM in Vietnam. Our results established that multiple subtypes of AIV were circulating in healthy ducks and geese in Hanoi LBM in late 2001 and identified the domestic duck as the primary host harboring AIV in this setting. A major finding of this study was the existence of HP H5N1 viruses which were present in Vietnam as early as 2001, although no outbreaks of highly pathogenic avian influenza among poultry were reported until early 2004 (64). Prior to this study and the outbreaks in 2003 to 2004, the isolation of H5N1 viruses was reported only from Hong Kong and, most recently, from mainland China (5). Thus, these results provide a context for the recent widespread emergence of HP H5N1 viruses in Asia.
Our study was limited to 2 days in October 2001 and was intended to reflect the status of avian influenza virus circulation at the time of a seroepidemiologic study conducted with poultry workers. The timing of our study may have been fortuitous, because Li et al. (32) recently reported the seasonal peak of H5N1 virus isolation in southern China to be the coolest months (October to March), which is also the coolest season in northern Vietnam. The overall isolation rate of AIV in the Hanoi LBM was approximately 3%. This rate is similar to that observed during a 6-month survey conducted in LBM in Taiwan in 1999 to 2000 (65), but it is higher than the isolation frequency in a 16-month surveillance study during 2000 to 2001 conducted in LBM in Nanchang, China (34). All AIV from the Taiwan LBM were isolated from ducks, although the majority of specimens collected were from chickens (65). Similar to a virologic surveillance study conducted in LBM in Nanchang, China, the Taiwan LBM study established that a given subtype may be present for only a limited period, after which it may be replaced by other subtypes (34, 65). Thus, the subtypes identified in the present study likely do not represent the full diversity of AIV that circulate in the Hanoi LBM.
When the entire HA1 region of the H4 virus Dk/VN/14/01 was compared to available H4 HA1 region sequences, the highest nucleotide identity was observed with A/Duck/Czechoslovakia/1/56 and, over a smaller nucleotide region, with A/Duck/Nanchang/4-173/00 (Fig. 1A). H4N6 viruses represented 14% of AIV isolated from ducks in Nanchang LBM over a 16-month period in 2000 to 2001 and over a 6-month period in Taipei in 1999 to 2000 (34, 65). In fact, the H4 subtype accounted for 50% of all isolates in Taipei during this period (65). A similar rate of H4N6 virus isolation was reported for wild ducks in Siberia between 1996 and 1998 (44), confirming the H4 subtype to be a major subtype prevalent in wild ducks worldwide (27).
Viruses of the H9 subtypes, in particular H9N2 viruses, are endemic in domestic poultry in Asia, Europe, and the Middle East (1, 4, 13, 29, 31, 41). The isolation of H9N2 viruses in 1998 to 1999 and 2003 from humans with respiratory illness identifies this avian influenza virus subtype as one with pandemic potential (4, 31, 46, 63). The H9 viruses isolated from ducks in the Hanoi LBM were of the H9N3 subtype, an HA and NA combination not frequently isolated in Asian LBM or in wild bird surveillance programs (1, 34). Genetically and antigenically, the HAs of these viruses were similar to that of the Korea lineage of H9 viruses (13, 31). Korea lineage H9 viruses were isolated only sporadically in Hong Kong LBM in 1997, but since then the G1 and, most recently, the Y280 lineages have predominated there (6, 13). Korea lineage viruses were also isolated from ducks in LBM in southern China in 2000 (31) and from migratory ducks in Japan in 1997 to 2000 (36). While both the G1 and Y280 lineage of H9N2 viruses have been associated with human infection, the Korea lineage, to date, has not (18, 31, 33, 46).
The most significant finding in the present study was the isolation of HP H5N1 viruses from healthy geese in the LBM, which establishes that H5N1 viruses were circulating outside of Hong Kong and mainland China as early as 2001. The HAs of two H5N1 viruses were genetically related to that of Gs/Gd/1/96 virus, the progenitor of the H5N1 viruses isolated from poultry and humans in Hong Kong in 1997, and were most similar (>98%) in nucleotide sequence to viruses isolated from waterfowl in Hong Kong in 2000 to 2001. However, the genetic, antigenic, and pathological properties of these viruses all differ from those that infected avian species and humans in Vietnam in early 2004, indicating that the H5N1 viruses that circulated in Vietnam LBM in 2001 were not the immediate progenitors of these latter H5N1 viruses. The fact that no AIV, including H5N1, were isolated from chickens during the course of this study is consistent with the lack of any reports of HP H5N1 among domestic poultry in Vietnam in 2001 to 2002. Chen et al. (5) recently reported that HP H5N1 viruses circulated in farmed domestic ducks in mainland China since at least 1999. The HP H5N1 viruses were isolated from apparently healthy ducks and did not cause severe disease in experimentally infected outbred Sheldrake ducks (5). Gs/VN/113/01 virus showed a similar lack of virulence in intranasally infected Pekin white ducks in the present study, despite the fact that the virus replicated efficiently in the lower respiratory tract and was also detected at substantial titers in the kidneys of infected birds. An HP H5N1 virus isolated from frozen duck meat imported into South Korea from China in 2001 was similarly nonvirulent in Pekin ducks, but it was isolated from the brain for up to 4 days p.i. (57). In contrast to these findings, we showed here that another HP H5N1, isolated from a dead wild bird in a Hong Kong wildlife park in late 2002 (Eg/HK/757.2/02), exhibited a high level of virulence and was isolated in substantial titers from the brain. This result is consistent with those recently reported for other viruses isolated from wild waterfowl in late 2002 in Hong Kong (54).
The HP H5N1 virus Gs/VN/113/01 also displayed a low level of virulence for two mammalian species, inbred BALB/c mice and outbred ferrets. In mice, the H5N1 virus, like the LP H5N2 virus Dk/VN/342/01, replicated in the lungs of mice but with slower kinetics than observed for a 1997 H5N1 virus shown to be highly lethal for this species (Table 6) (11, 37). Although neither Vietnam H5 virus was isolated from mouse brains, consistent with a low-pathogenicity phenotype in mice, Gs/VN/113/01 was detected in the thymus of all three mice sampled on day 6 p.i., suggesting some tropism for mammalian lymphoid tissue. The HP H5N1 viruses isolated from ducks in mainland China since 1999 exhibited increasing virulence for BALB/c mice, with more recent isolates from 2002 inducing a systemic and highly lethal infection similar to that observed with the H5N1 viruses isolated from humans in Hong Kong in 1997 (5). Thus, the pathogenicity of the Vietnam H5N1 virus is typical of other H5N1 viruses isolated in Asia before late 2002, when virulence for multiple avian species was first recognized in H5N1 viruses isolated in Hong Kong (54).
The isolation of an LP H5N2 virus from domestic ducks at one Hanoi LBM was of interest, because LP H5 viruses have been rarely reported in Asia since 1997, when low-pathogenicity strain Duck/Singapore/Q/F119-3/97 (H5N3) was used as a surrogate vaccine strain for the 1997 H5N1 viruses isolated from humans (42). The LP H5N2 virus was antigenically and genetically more closely related to H5 viruses that circulated around 1997 in Italy, and it shared only 91% amino acid sequence identity in the HA1 region with the H5N1 viruses isolated in this study. These results also established that the H5N2 virus was not a low-pathogenicity progenitor for the H5N1 viruses isolated in Vietnam in 2001, and neither was the human H5N1 isolates from Hong Kong in 2003 or Vietnam and Thailand in 2004. In fact, an LP H5 strain that is antigenically and genetically similar to these human 2003 and 2004 H5N1 strains has not been identified to date, and the use of reverse genetics has been required to generate a vaccine strain bearing an antigenically matched H5 HA modified by the deletion of the multibasic amino acid motif which contributes to virulence in both avian and mammalian species (60).
The isolation of multiple subtypes of AIV from ducks in Hanoi LBM is consistent with the domestic duck being the major reservoir of the avian influenza virus gene pool in nature. Recently, it has become clear that the domestic duck also plays a major role in the generation and maintenance of HP H5N1 viruses in southern China (5, 32), while a role for migratory wild birds in the recent dissemination of H5N1 viruses throughout much of Asia has been proposed. Our study demonstrates that LBM in Asian countries, such as Vietnam, as well as those in mainland China and Hong Kong, are a suitable environment for potential avian virus reassortment and transmission of H5N1 virus from waterfowl to chickens and humans. Moreover, LBM in Hanoi have not adopted the stringent precautions enforced in Hong Kong after 1997 to limit the spread of HP H5N1 viruses from aquatic to terrestrial poultry species and from avian species to humans (28, 53). The serological investigation that initiated this virologic study demonstrated that in 2001 the seroprevalence for antibody to either HP H5N1 (Gs/VN/113/01) or LP H5N2 (Dk/VN/342/01) in poultry workers in the Hanoi LBM was low (≤1%) and similar to that detected in an urban control population not occupationally exposed to domestic poultry (T. Uyeki, unpublished data). Further human serological studies in Asian countries that recently experienced HP H5N1 outbreaks in poultry would greatly improve our understanding of the true capacity for these viruses to infect humans. Since 1997, it has been recognized that humans themselves may become the mixing vessel for a reassortment event between human and avian influenza viruses that could generate a pandemic strain. Because HP H5N1 viruses are now endemic in Asia, heightened surveillance in wild and domestic avian species as well as humans will be an essential component of pandemic preparedness.
Acknowledgments
We acknowledge support for Doan C. Nguyen and Samadhan Jadhao from the Emerging Infectious Diseases Laboratory Fellowship Program, administered by the United States Association of Public Health Laboratories.
We thank Robert Webster and Trevor Ellis for avian viruses, Joan Beck for technical assistance, Kanta Subbarao for technical advice, and Ruben Donis for review of the manuscript.
REFERENCES
- 1.Banks, J., E. C. Speidel, P. A. Harris, and D. J. Alexander. 2000. Phylogenetic analysis of influenza A viruses of H9 hemagglutinin subtype. Avian Pathol. 29:353-360. [DOI] [PubMed] [Google Scholar]
- 2.Bender, C., H. Hall, J. Huang, A. Klimov, N. Cox, A. Hay, V. Gregory, K. Cameron, W. Lim, and K. Subbarao. 1999. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997-1998. Virology 254:115-123. [DOI] [PubMed] [Google Scholar]
- 3.Bridges, C. B., W. Lim, J. Hu-Primmer, L. Sims, K. Fukuda, K. H. Mak, T. Rowe, W. W. Thompson, L. Conn, X. Lu, N. J. Cox, and J. M. Katz. 2002. Risk of influenza A (H5N1) infection among poultry workers, Hong Kong, 1997-1998. J. Infect. Dis. 185:1005-1010. [DOI] [PubMed] [Google Scholar]
- 4.Cameron, K. R., V. Gregory, J. Banks, I. H. Brown, D. J. Alexander, A. J. Hay, and Y. P. Lin. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:37-41. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., G. Deng, Z. Li, G. Tian, Y. Li, P. Jiao, L. Zhang, Z. Liu, R. G. Webster, and K. Yu. 2004. The evolution of H5N1 influenza viruses in ducks in southern China. Proc. Natl. Acad. Sci. USA 101:10452-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, Y. K., H. Ozaki, R. J. Webby, R. G. Webster, J. S. Peiris, L. Poon, C. Butt, Y. H. C. Leung, and Y. Guan. 2004. Continuing evolution of H9N2 influenza viruses in Southeastern China. J. Virol. 78:8609-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claas, E. C. J., A. D. M. E. Osterhaus, R. Van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472-477. [DOI] [PubMed] [Google Scholar]
- 8.De Jong, J. C., E. C. J. Claas, A. D. M. E. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, T. M., R. B. Bousfield, L. Bissett, K. C. Dyrting, G. S. M. Luk, Y. Guan, S. T. Tsim, K. Sturm-Ramirez, R. G. Webster, and J. S. M. Peiris. 2004. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wide birds in Hong Kong in late 2002. Avian Pathol. 33:492-505. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1993. PHYLIP (Phylogeny Interference Package), version 3.5c. University of Washington, Seattle, Wash.
- 11.Gao, P., S. Watanabe, T. Ito, H. Goto, K. Wells, M. McGregor, A. J. Cooley, and Y. Kawaoka. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J. Virol. 73:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses, were they the donors of the “internal” genes of H5N1 in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, Y., J. S. M. Peiris, K. F. Kong, K. C. Dyrting, T. M. Ellis, T. Sit, L. J. Zhang, and K. F. Shortridge. 2002. H5N1 influenza viruses isolated from geese in southeastern China, evidence for genetic reassortment and interspecies transmission to ducks. Virology 292:16-23. [DOI] [PubMed] [Google Scholar]
- 15.Guan, Y., J. S. M. Peiris, A. S. Lipatov, T. M. Ellis, K. C. Dyrtings, S. Krauss, L. J. Zhang, R. G. Webster, and K. F. Shortridge. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 99:8950-8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, Y., J. S. Peiris, L. L. Poon, K. C. Dyrting, T. M. Ellis, L. Sims, R. G. Webster, and K. F. Shortridge. 2003. Reassortant of H5N1 influenza viruses recently isolated from aquatic poultry in Hong Kong SAR. Avian Dis. 47:911-913. [DOI] [PubMed] [Google Scholar]
- 17.Guan, Y., L. L. Poon, C. Y. Cheung, T. M. Ellis, W. Lim, A. S. Lipatov, K. H. Chan, K. M. Sturm-Ramirez, C. L. Cheung, Y. H. Leung, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. H5N1 influenza: a protean pandemic threat. Proc. Natl. Acad. Sci. USA 101:8156-8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, Y. J., J. W. Li, and I. Cheng. 1999. Discovery of humans infected by avian influenza A (H9N2) virus. Chin. J. Exp. Clin. Virol. 15:105-108. [PubMed] [Google Scholar]
- 19.Guo, Y. J., S. Krauss, D. A. Senne, I. P. Mo, K. S. Lo, X. P. Xiong, M. Norwood, K. F. Shortridge, R. G. Webster, and Y. Guan. 2000. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267:279-288. [DOI] [PubMed] [Google Scholar]
- 20.Hien, T. T., N. T. Liem, N. T. Dung, L. T. San, P. P. Mai, N. V. V. Chau, P. T. Suu, V. C. Dong, L. T. Q. Mai, N. T. Thi, D. B. Khoa, L. P. Phat, N. T. Truong, H. T. Long, C. V. Tung, L. T. Giang, N. D. Tho, L. H. Nga, N. T. K. Tien, L. H. San, L. V. Tuan, C. Dolecek, T. T. Thanh, M. de Jong, C. Schultsz, P. Cheng, W. Lim, P. Horby, the World Health Organization International Avian Influenza Investigative Team, and J. Farrar. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179-1188. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 22.Katz, J. M. 2003. The impact of avian influenza viruses on public health. Avian Dis. 47:914-920. [DOI] [PubMed] [Google Scholar]
- 23.Katz, J. M., X. Lu, T. M. Tumpey, C. B. Smith, M. W. Shaw, and K. Subbarao. 2000. Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 74:10807-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kendal, A. P., M. S. Pereira, and J. J. Skehel. 1982. Concepts and procedures for laboratory-based influenza surveillance. U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, Atlanta, Ga.
- 26.Koopmans, M., B. Wilbrink, M. Conyn, G. Natrop, H. van der Nat, H. Vennema, A. Meijer, J. van Steenbergen, R. Fouchier, A. Osterhaus, and A. Bosman. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363:587-593. [DOI] [PubMed] [Google Scholar]
- 27.Krauss, S., D. Walker, S. P. Pryor, L. Niles, L. Chenghong, V. S. Hinshaw, and R. G. Webster. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector-Borne Zoonot. Dis. 4:177-189. [DOI] [PubMed] [Google Scholar]
- 28.Kung, N. Y., Y. Guan, N. R. Perkins, L. Bissett, T. Ellis, L. Sims, R. S. Morris, K. F. Shortridge, and J. S. M. Peiris. 2003. The impact of a monthly rest day on avian influenza virus isolation rates in retails live poultry markets in Hong Kong. Avian Dis. 47:1037-1041. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. W., C. S. Song, Y. J. Lee, I. P. Mo, M. Garcia, D. L. Suarez, and S. J. Kim. 2000. Sequence analysis of the hemagglutinin gene of H9N2 Korean avian influenza viruses and assessment of the pathogenic potential of isolate MS96. Avian Dis. 44:527-535. [PubMed] [Google Scholar]
- 30.Lee, M. H., P. C. Chang, J. H. Shien, M. C. Cheng, and H. K. Shieh. 2001. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J. Virol. Methods 97:13-22. [DOI] [PubMed] [Google Scholar]
- 31.Li, K. S., K. M. Xu, J. S. M. Peiris, L. M. L. Poon, K. Z. Yu, K. Y. Yen, K. F. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China, a candidate for the next influenza pandemic in humans? J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, K. S., Y. Guan, J. Wang, G. J. D. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. S. Estoepangestie, A. Chaisingh, P. Auewarakul, H. T. Long, N. T. H. Hanh, R. W. Webby, L. L. M. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209-213. [DOI] [PubMed] [Google Scholar]
- 33.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses, relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, M., S. He, D. Walker, N. Zhou, D. R. Perez, B. Mo, F. Li, X. Huang, R. G. Webster, and R. J. Webby. 2003. The influenza virus gene pool in a poultry market in south central China. Virology 305:267-275. [DOI] [PubMed] [Google Scholar]
- 35.Liu, J., K. Okazaki, H. Ozaki, Y. Sakoda, Q. Wu, F. Chen, and H. Kida. 2003. H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathol. 32:551-560. [DOI] [PubMed] [Google Scholar]
- 36.Liu, J. H., K. Okazaki, W. M. Shi, and H. Kida. 2003. Phylogenetic analysis of hemagglutinin and neuraminidase genes of H9N2 viruses isolated from migratory ducks. Virus Genes 27:291-296. [DOI] [PubMed] [Google Scholar]
- 37.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. Osterhaus, N. Cox, and A. Hampson (ed.), International Congress Series 1219, Proceedings of the World Congress on Options for the Control of Influenza IV. Excerpta Medica, Amsterdam, The Netherlands.
- 39.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 40.Mounts, A. W., H. Kwong, H. S. Izurieta, Y. Ho, T. Au, M. Lee, C. Bridges, S. W. Williams, K. H. Mak, J. M. Katz, W. W. Thompson, N. J. Cox, and K. Fukuda. 1999. Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong, 1997. J. Infect. Dis. 180:505-508. [DOI] [PubMed] [Google Scholar]
- 41.Naeem, K., A. Ullah, R. J. Manvell, and D. J. Alexander. 1999. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet. Rec. 145:560. [DOI] [PubMed] [Google Scholar]
- 42.Nicholson, K. G., A. E. Colegate, A. Podda, I. Stephenson, J. Wood, E. Ypma, and M. C. Zambon. 2001. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomized trial of two potential vaccines against H5N1 influenza. Lancet 357:1937-1943. [DOI] [PubMed] [Google Scholar]
- 43.Office International des Epizooties. 2000. (updated in 2002). Highly pathogenic avian influenza (fowl plaque), p. 155-160. In G. A. Cullen and S. Linnance (ed.), Manual of standards for diagnostic tests and vaccines, 3rd ed. Office International des Epizooties, Paris, France.
- 44.Okazaki, K., A. Takada, T. Ito, M. Imai, H. Takakuwa, M. Hatta, H. Ozaki, T. Tanizaki, T. Nagano, A. Ninomiya, V. A. Demenev, M. M. Tyaptirganov, T. D. Karatayeva, S. S. Yamnikova, D. K. Lvov, and H. Kida. 2000. Precursor genes of future pandemic influenza viruses are perpetuated in ducks nesting in Siberia. Arch. Virol. 145:885-893. [DOI] [PubMed] [Google Scholar]
- 45.Page, R. D. M. 1996. Treeview: An application to display phylogenetic tree on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 46.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. S. Ip, R. W. M. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 47.Peiris, J. S., W. C. Yu, C. W. Leung, C. Y. Cheung, W. F. Ng, J. M. Nicholls, T. K. Ng, K. H. Chan, S. T. Lai, W. L. Lim, K. Y. Yuen, and Y. Guan. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 49.Rohm, C., N. Zhou, J. Suss, J. Mackenzie, and R. G. Webster. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508-516. [DOI] [PubMed] [Google Scholar]
- 50.Seal, B. S., D. J. King, and J. D. Bennett. 1995. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J. Clin. Microbiol. 33:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shortridge, K. F., and C. H. Stuart-Harris. 1982. An influenza epicentre? Lancet 320:812-813. [DOI] [PubMed] [Google Scholar]
- 52.Shortridge, K. F. 1992. Pandemic influenza: a zoonosis? Semin. Respir. Infect. 7:11-25. [PubMed] [Google Scholar]
- 53.Sims, L. D., T. M. Ellis, K. K. Liu, K. Dyrting, H. Wong, M. Peiris, Y. Guan, and K. F. Shortridge. 2003. Avian influenza in Hong Kong 1997-2002. Avian Dis. 47:832-838. [DOI] [PubMed] [Google Scholar]
- 54.Sturm-Ramirez, K. M., T. Ellis, B. Bousfield, L. Bissett, K. Dyrting, J. E. Rehg, L. Poon, Y. Guan, M. Peiris, and R. G. Webster. 2004. Reemerging H5N1 influenza virus in Hong Kong in 2002 are highly pathogenic to ducks. J. Virol. 78:4892-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suarez, D. L., M. L. Perdue, N. J. Cox, T. Rowe, C. Bender, J. Huang, and D. E. Swayne. 1998. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J. Virol. 72:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 57.Tumpey, T. M., D. L. Suarez, L. E. Perkins, D. A. Senne, J. G. Lee, Y. J. Lee, I. P. Mo, H. W. Sung, and D. E. Swayne. 2002. Characterization of a highly pathogenic H5N1 avian influenza A virus isolated from duck meat. J. Virol. 76:6344-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uyeki, T. M., Y. H. Chong, J. M. Katz, W. Lim, Y. Y. Ho, S. S. Wang, T. H. F. Tsang, W. W. Y. Au, S. C. Chan, T. Rowe, J. Hu-Primmer, J. C. Bell, W. W. Thompson, C. B. Bridges, N. J. Cox, K. H. Mak, and K. Fukuda. 2002. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China, 1999. Emerg. Infect. Dis. 8:154-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webby, R. J., and R. G. Webster. 2001. Emergence of influenza A viruses. Philos. Trans. R. Soc. Lond. B 356:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webby, R. J., D. R. Perez, J. S. Coleman, Y. Guan, J. H. Knight, E. A. Govorkova, L. R. McClain-Moss, J. S. Peiris, J. E. Rehg, E. I. Tuomanen, and R. G. Webster. 2004. Responsiveness to a pandemic alert: use of reverse genetics for rapid development of influenza vaccines. Lancet 363:1099-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.World Health Organization. 2003. Influenza A(H5N1), Hong Kong SAR of China—update 2. Wkly. Epidemiol. Rec. 9:57-58. [Google Scholar]
- 63.World Health Organization. 2003. Influenza A(H9N2), Hong Kong SAR of China. Wkly. Epidemiol. Rec. 51/52:439. [Google Scholar]
- 64.World Health Organization. 2004. Preliminary epidemiological summary of influenza A(H5N1), Vietnam and Thailand. Wkly. Epidemiol. Rec. 7:68. [Google Scholar]
- 65.Yen, H. L., M. C. Cheng, J. L. Liu, C. L. Kao, S. R. Shih, N. J. Cox, R. G. Webster, and C. C. King. 2001. Influenza surveillance in poultry market and its interspecies transmission in Taiwan, p. 201-211. In A. Osterhaus, N. Cox, and A. Hampson (ed.), International Congress Series 1219, Proceedings of the World Congress on Options for the Control of Influenza IV. Excerpta Medica, Amsterdam, The Netherlands.
- 66.Yuen, K. Y., P. K. S. Chan, M. Peiris, D. N. C. Tsang, T. L. Que, K. F. Shortridge, P. T. Cheung, W. K. To, E. T. F. Ho, R. Sung, A. F. B. Cheng, and members of the H5N1 study group. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467-471. [DOI] [PubMed] [Google Scholar]
- 67.Zitzow, L. A., T. Rowe, T. Morken, W. J. Shieh, S. Zaki, and J. M. Katz. 2002. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J. Virol. 76:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]