Abstract
Poliovirus selectively replicates in neurons in the spinal cord and brainstem, although poliovirus receptor (PVR) expression is observed in both the target and nontarget tissues in humans and transgenic mice expressing human PVR (PVR-transgenic mice). We assessed the role of alpha/beta interferon (IFN) in determining tissue tropism by comparing the pathogenesis of the virulent Mahoney strain in PVR-transgenic mice and PVR-transgenic mice deficient in the alpha/beta IFN receptor gene (PVR-transgenic/Ifnar knockout mice). PVR-transgenic/Ifnar knockout mice showed increased susceptibility to poliovirus. After intravenous inoculation, severe lesions positive for the poliovirus antigen were detected in the liver, spleen, and pancreas in addition to the central nervous system. These results suggest that the alpha/beta IFN system plays an important role in determining tissue tropism by protecting nontarget tissues that are potentially susceptible to infection. We subsequently examined the expression of IFN and IFN-stimulated genes (ISGs) in the PVR-transgenic mice. In the nontarget tissues, ISGs were expressed even in the noninfected state, and the expression level increased soon after poliovirus infection. On the contrary, in the target tissues, ISG expression was low in the noninfected state and sufficient response after poliovirus infection was not observed. The results suggest that the unequal IFN response is one of the important determinants for the differential susceptibility of tissues to poliovirus. We consider that poliovirus replication was observed in the nontarget tissues of PVR-transgenic/Ifnar knockout mice because the IFN response was null in all tissues.
The replication of many viruses is restricted to certain cells and tissues in the host. This tissue tropism results in a distinct disease pattern unique for each virus. Since virus infection initiates after binding of the virion to a receptor on the cell surface, cellular receptors for viruses have been considered the primary determinants of tissue tropism. However, following the identification of receptors for a number of viruses, it became apparent that receptor distribution in the host is wider than the virus replication sites (38). This indicates that virus tropism may be determined by another factor(s) in addition to the virus receptor.
Poliovirus, belonging to the genus Picornaviridae, is the causative agent of an acute human central nervous system disease, poliomyelitis (33). After poliovirus infection, the virus first multiplies in the oropharyngeal and intestinal mucosa and then in the lymphatic tissues, such as the tonsils and Peyer's patches. The virus drains into the blood and circulates within the body. Because visceral tissues, except for adipose tissues, seem to be nonpermissive for poliovirus infection, apparent pathological lesions are not observed in the nonneural tissues. Therefore, the site of virus multiplication during the viremic phase has not been identified. Finally, poliovirus reaches the central nervous system, which leads to the development of a paralytic disease in less than 1% of persons naturally infected with wild-type poliovirus (4, 23, 35). Even in the central nervous system, the poliovirus antigen, nerve cell changes, and inflammatory reactions are localized mainly in motor neurons in the anterior horn of the spinal cord and neurons in the brainstem. The brainstem as far as the hypothalamus and thalamus bears most of the cerebral pathological changes in poliomyelitis. The cerebral cortex (except for the motor cortex), basal ganglia (except occasionally for the globus pallidus), and the cerebellar cortex (except for the vermis) are rarely affected (2, 6). In terms of modern molecular biology, tissue tropism may be determined by interactions between host and viral factors. It is therefore important to elucidate the molecular mechanism responsible for tropism.
The poliovirus receptor (PVR) has been considered a major determinant of poliovirus tissue tropism (13). The molecular cloning of the human PVR gene was reported more than a decade ago (18, 24). With cultured cells, susceptibility of poliovirus infection completely correlates with the presence of functional PVR. However, in vivo, this rule is not always true. Analyses of PVR expression in humans revealed that there are many tissues, such as the liver and kidneys, that express PVR but are not involved in the infection (18, 24). Transgenic mice expressing the human PVR gene with its natural promoter (PVR-transgenic mice) were produced as a new animal model for the study of poliovirus pathogenicity (21, 32). PVR-transgenic mice exhibited a paralytic disease that resembled human poliomyelitis after poliovirus infection. PVR mRNA was detected in all tissues of PVR-transgenic mice by Northern blot hybridization, although expression was restricted to certain cells, such as neurons in the central nervous system (20, 32) and Bowman's capsule and tubules in the kidney (32). PVR was also detected in the glomerulus in the kidney by immunofluorescent staining (15). These data suggest that PVR is necessary for poliovirus infection but may not be the sole determinant of tissue tropism (reviewed in reference 29). The results also suggest the existence of other factors that determine tissue tropism concomitantly with PVR expression. It is possible that host factors required for poliovirus replication are abundant only in target tissues or that host factors inhibiting virus replication are present in nontarget tissues.
To identify possible host factors, we produced another transgenic mouse strain in which there is ubiquitous PVR expression under the control of the CAG promoter (14). If we hypothesize that host factors required for poliovirus replication are present only in susceptible tissues, the distribution of poliovirus replication sites in the new transgenic mice would be the same as or wider than that in the transgenic mice previously produced, since the distribution of the PVR has broadened. However, after intracerebral infection, poliovirus propagated to a slight degree in neurons and glial and ependymal cells near the inoculation sites on day 1 postinfection (p.i.) but the virus titer decreased from day 2 p.i. without development of fatal encephalitis and poliomyelitis. After intraperitoneal and intravenous infection, no apparent signs of illness or virus replication were observed. It seemed to us that an unknown factor(s) that prevented virus replication and spreading was induced. This led us to hypothesize that an innate immune response, such as the production of interferon (IFN), may influence the pathogenesis of poliovirus.
Picornaviruses are sensitive to IFNs (7, 8, 25, 27, 49). IFN plays a central role in the innate immune antiviral response. Infected cells produce alpha/beta IFNs, which induce a number of genes, called IFN-stimulated genes (ISGs), that confer an antiviral state (36, 39, 41). In some viruses, including coxsackievirus and Theiler's virus, alpha/beta IFN plays an important role in the pathogenicity and tissue tropism of the virus (9, 10, 28, 34, 44). However, little is known about the role of alpha/beta IFNs in poliovirus pathogenesis.
Here we report the results of experiments that we conducted to assess the contribution of alpha/beta IFN in the pathogenesis of poliovirus infection with a PVR-transgenic mouse model (20, 21). PVR-transgenic mice were crossed with IFN-α/β receptor (IFNAR) knockout mice in which alpha/beta IFN signaling is disrupted (26). Poliovirus infection of the resulting PVR-transgenic/Ifnar knockout mice revealed that the IFN system is an important determinant of poliovirus tissue tropism and poliovirus pathogenesis.
MATERIALS AND METHODS
Cells, viruses, and mice.
African green monkey kidney (AGMK) cell line JVK-03 (19) was maintained in Eagle's minimal essential medium supplemented with 5% fetal bovine serum. The poliovirus type 1 Mahoney strain was obtained by transfection of in vitro-synthesized RNA from infectious cDNA clone pOM (40) into JVK-03 cells. Virus titer was determined by a plaque assay on JVK-03 cells. A transgenic mouse strain, ICR-PVRtg21 (20, 21), was backcrossed for 10 generations with C57BL/6 mice, and then N10 mice were mated to obtain homozygotes (PVR-transgenic mice). The mouse strain deficient in the Ifnar gene, A129 (26), was purchased from B&K Universal Limited (United Kingdom) with the permission of M. Aguet. A129 was backcrossed for five generations with C57BL/6 mice and then with PVR-transgenic mice for two generations. PVR+/+ Ifnar −/− mice were obtained by intercrossing these PVR-transgenic/Ifnar knockout mice.
Until the infection experiments, mice were maintained in an animal facility free of the following pathogens: Citrobacter rodentium, Corynebacterium kutscheri, Mycoplasma pulmonis, Pasteurella pneumotropica, Salmonella spp., cilia-associated respiratory bacillus, Helicobacter hepaticus, Pseudomonas aeruginosa, Clostridium piliforme, Mycoplasma pulmonis, ectromelia virus, lymphocytic choriomeningitis virus, mouse hepatitis virus, Sendai virus (HVJ), EDIM virus (rotavirus), minute virus of mice, mouse encephalomyelitis virus, pneumonia virus of mice, mouse adenovirus, reovirus type 3, ectoparasites, intestinal protozoa, and pinworm. Six-week-old mice were used for the experiments. All experiments with mice were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of the Tokyo Metropolitan Institute for Neuroscience.
Poliovirus infection in mice.
Poliovirus at the indicated doses was inoculated intracerebrally, intravenously, or intraperitoneally as described previously (21). The mice were observed daily for 3 weeks or sacrificed at the indicated time p.i. The 50% lethal dose (LD50) was calculated by the method of Kärber (45). In poly(I):poly(C) [poly(I:C)] protection experiments, 200 μg of poly(I:C) (Calbiochem) in phosphate-buffered saline was administered intracerebrally, and poliovirus challenge (104 PFU) was performed with the same inoculation routes on the next day.
Determination of virus titer in mouse tissues.
Six mice inoculated with poliovirus were sacrificed on day 3 p.i. They were anesthetized and blood was collected from the heart. After perfusion with 20 ml of phosphate-buffered saline, the tissues were removed and frozen at −80°C. The tissues were thawed and homogenized in 5 to 10 ml of minimal essential medium. After centrifugation for 20 min at 3,000 × g, the virus titer of the supernatant was determined.
Histological and immunological examinations.
Three to six mice were sacrificed on day 1, 2, or 3 p.i. or when the mice showed paralysis and were then used for histological examination. The mice were anesthetized and perfused with 10 ml of phosphate-buffered saline followed by 10 ml of 4% paraformaldehyde in phosphate-buffered saline. The fixed tissues were embedded in paraffin, from which 3-μm-thick sections were prepared. The poliovirus antigen was detected with an immunoperoxidase method as described previously (21).
Alanine aminotransferase and amylase activity.
To evaluate the extent of liver injury in the mice, the serum alanine aminotransferase (ALT) level was measured with the Transnase Nissui kit (Nissui Pharmaceutical Co. Ltd.). The serum amylase level in the mice was measured by the method of Henkel et al. (12) with the Cica Auto amylase kit (Kanto Chemical Co. Inc.).
Quantitative real-time PCR.
Mice tissues were separated immediately after sacrifice and stored at −80°C. Total RNA was isolated from the tissue with the RNeasy mini RNA isolation kit (Qiagen, Valencia, Calif.) according to the supplier's instructions. After DNase I treatment, cDNA primed with a random hexamer was synthesized with the Taqman reverse transcription reagent (Applied Biosystems, Foster City, Calif.). The quantification of RNAs was performed with ABI Prism 7900HT. 18S rRNA was quantified by the SYBR Green method with 18S-rRNA-F (5′-GTA ACC CGT TGA ACC CCA TT-3′) and 18S-rRNA-R (5′-CCA TCC AAT CGG TAG TAG CG-3′) as primers. Poliovirus RNA was quantified by the Taqman method with PV c-2493F (5′-TGG TTG GTG ACA GTT CTT ACA CAT T-3′) and PV c-2629R (5′-CCA CTG TGG CAC ACA GTG ATG-3′) as primers and PV c-2579T (5′-FAM-CCA TGT CGA ACG CAA AGC GCC-TAMRA-3′) as the probe. The detection of IFN-β, 2′-5′ oligoadenylate synthetase (OAS)1a, OAS1g, OAS2, OAS3, OASL2, protein kinase R, RIG-I, and helicard mRNAs was performed with Assay-on-Demand PCR probes (Applied Biosystems). The amount of mRNA was determined by comparison with standard templates of cloned cDNAs of known copy number. The expression levels were then normalized to the level of 18S rRNA.
RESULTS
Increased susceptibility of PVR-transgenic/Ifnar knockout mice to poliovirus.
We previously produced transgenic mice expressing the human PVR gene (20, 21). PVR mRNA was detected in the brain, spinal cord, thymus, lungs, heart, stomach, and muscle at high levels, in the spleen and kidneys at intermediate levels, and in the liver at low levels (data not shown). The PVR-transgenic mice were susceptible to poliovirus infection via the intracerebral, intraperitoneal, and intravenous routes. The infected mice developed a paralytic disease resembling human poliomyelitis. Unlike humans, however, PVR-transgenic mice were not highly susceptible to oral infection with poliovirus. We performed subsequent experiments with this transgenic mouse model.
Mice lacking the alpha/beta IFN response become highly susceptible to several virus infections (26). We compared the susceptibility of PVR-transgenic mice with that of PVR-transgenic/Ifnar knockout mice to poliovirus infection via intracerebral, intravenous, and intraperitoneal inoculations. The mice showed paralysis and died within 2 weeks p.i. in a dose-dependent manner following inoculation via all routes. This suggested that the mice died mainly due to poliovirus infection in the central nervous system irrespective of the inoculation route. The results and the LD50 values are shown in Table 1. The PVR-transgenic/Ifnar knockout mice were more sensitive to fatal poliovirus infection than the PVR-transgenic mice by any infection route. The LD50 value for PVR-transgenic/Ifnar knockout mice inoculated intracerebrally was 50-fold lower than that for PVR-transgenic mice. Notably, PVR-transgenic/Ifnar knockout mice became 3,000- and >20,000-fold more sensitive to poliovirus via intravenous and intraperitoneal inoculation, respectively. Furthermore, 106 PFU of poliovirus inoculated orally caused paralysis in 50% of the PVR-transgenic/Ifnar knockout mice (data not shown). On the contrary, PVR-transgenic mice with the disrupted IFN-γ gene (42) did not exhibit a significant increase in susceptibility to poliovirus infection (data not shown). The results indicate that alpha/beta IFN strongly influences poliovirus pathogenesis.
TABLE 1.
Susceptibility to poliovirus infection
| Inoculation route | Mouse straina | No. of mice dead/no. inoculated at inoculum dose (log10 PFU/mouse):
|
LD50 (log10) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −1 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| Intracerebral | PVR-tg | 0/6 | 1/6 | 1/6 | 1/6 | 4/6 | 5/6 | 6/6 | 2.5 | |||
| PVR-tg/lfnar KO | 0/6 | 1/6 | 3/6 | 6/6 | 6/6 | 0.8 | ||||||
| Intravenous | PVR-tg | 0/6 | 1/6 | 1/6 | 4/6 | 3/6 | 6/6 | 5.0 | ||||
| PVR-tg/lfnar KO | 0/6 | 0/6 | 5/6 | 6/6 | 1.7 | |||||||
| Intraperitoneal | PVR-tg | 0/6 | 1/6 | 3/6 | 5/6 | 5/6 | >6.2 | |||||
| PVR-tg/lfnar KO | 0/6 | 3/6 | 5/6 | 6/6 | 1.2 | |||||||
tg, transgenic; KO, knockout.
Distribution of poliovirus infection in PVR-transgenic/Ifnar knockout mice.
Increased susceptibility to poliovirus by peripheral infection routes suggested that the poliovirus replicated efficiently in nonneural tissues. We compared poliovirus titers in the various tissues of nontransgenic C57BL/6 mice, PVR-transgenic mice, and PVR-transgenic/Ifnar knockout mice intravenously inoculated with 2 × 107 PFU of poliovirus. After intravenous inoculation, poliovirus was immediately delivered to all tissues, including the central nervous system, independently of the presence of PVR (47). Most of the PVR-transgenic mice and PVR-transgenic/Ifnar knockout mice developed paralysis by day 3 or 4 p.i., while the nontransgenic mice did not.
Figure 1 shows the virus load of various tissues on day 3 p.i. The virus titers in the brain and spinal cord of PVR-transgenic mice were much higher than those recovered from the same tissues of nontransgenic mice. The virus titer in the pancreas of PVR-transgenic mice was also higher than that of nontransgenic mice but not as high as the titers in the neural tissues of transgenic mice. The viral load of most of the other tissues of PVR-transgenic mice was slightly higher than that recovered from nontransgenic mice. These results suggest that poliovirus replicated efficiently in the central nervous system and less efficiently in the pancreas and did not replicate or replicated to only a slight degree in other nonneural tissues of PVR-transgenic mice. On the contrary, in PVR-transgenic/Ifnar knockout mice, poliovirus titers in all of the tissues examined were very high (Fig. 1), suggesting that the virus can replicate in nonneural tissues if alpha/beta IFN signaling is disrupted.
FIG. 1.
Comparison of poliovirus titers in tissues of nontransgenic, PVR-transgenic, and PVR-transgenic/Ifnar knockout mice. The mice were inoculated intravenously with 2 × 107 PFU of poliovirus type 1 Mahoney strain. The tissues of nontransgenic mice (hatched bars), PVR-transgenic mice (open bars), and PVR-transgenic/Ifnar knockout mice (solid bars) were separated on day 3 p.i., and virus titers were determined by a plaque assay. The values represent the mean virus titer + standard deviation of six mice. The asterisk indicates that the values were below the limit of detection (2.0 log10 PFU/g).
Poliovirus replication site in the visceral tissues of PVR-transgenic/Ifnar knockout mice.
We investigated the localization of the poliovirus antigen and corresponding pathological changes to determine the virus replication sites. In the liver of PVR-transgenic mice, cells positive for the poliovirus antigen were detected occasionally, after careful observation. The antigens were found sporadically as a single cell or as a group of a few poliovirus antigen-positive cells with cellular damage in the liver on day 1 p.i., but were rarely detected after day 2 p.i. Inflammatory cell infiltration was also observed around the infected cells (Fig. 2A). Strong poliovirus antigen staining was not detected in the spleen, but a few very weakly stained cells were observed in the marginal zone (Fig. 2C). In the pancreas, groups of a few poliovirus antigen-positive cells were sporadically observed on day 1 p.i., with the number increasing slightly on day 3 p.i. (Fig. 2E). Although the infected area in the pancreas was not large, the antigens were always detected in all the mice. This observation is consistent with a higher poliovirus titer in the pancreas than in the other visceral tissues (Fig. 1).
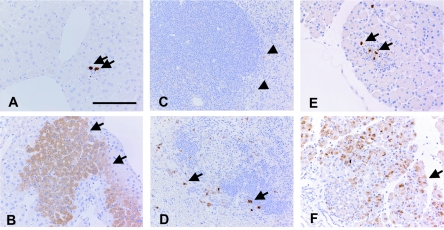
FIG. 2.
Immunohistochemical detection of poliovirus antigen in infected mice. Poliovirus antigens were detected in PVR-transgenic (A, C, and E) and PVR-transgenic/Ifnar knockout (B, D, and E) mice with a rabbit polyclonal antibody recognizing the poliovirus capsid antigen. The mice were intravenously inoculated with 2 × 107 PFU of poliovirus. (A) Liver of the PVR-transgenic mice on day 1 p.i. Poliovirus antigen-positive cells, indicated by arrows, were focally observed with slight cellular infiltration around the infected cell. (B) Liver of PVR-transgenic/Ifnar knockout mice on day 1 p.i. Hepatic cells positive for poliovirus were observed in a zonal pattern. (C) Spleen of PVR-transgenic mice on day 1 p.i. A few very weakly stained cells are observed in the marginal zone, indicated by arrowheads. (D) Spleen of PVR-transgenic/Ifnar knockout mice on day 1 p.i. Many poliovirus antigen-positive large cells are localized in the marginal zone. The cells were identified as macrophages on the basis of the detection of CD11. (E) Pancreas of PVR-transgenic mice on day 3 p.i. A small cluster of cells positive for poliovirus antigen was observed in the lobulus in association with a slight inflammatory reaction. The poliovirus antigen was observed constantly in all mice. (F) Pancreas of PVR-transgenic/Ifnar knockout mice on day 3 p.i. Numerous acinar cells positive for the poliovirus antigen were distributed in many lobuli of the pancreas. Only a few poliovirus antigen-positive cells were observed in Langerhans' islets in the bottom left. (A) Bar, 125 μm.
In contrast, in PVR-transgenic/Ifnar knockout mice, zonal areas or clusters of poliovirus antigen-positive cells were observed in the liver on day 1 p.i. (Fig. 2B). These infected cells were identified morphologically as hepatocytes. The number of poliovirus antigen-positive cells decreased on day 3 p.i., but the inflammatory infiltrate became more evident. In the spleen, poliovirus antigen was detected mainly in large mononuclear cells in the marginal zone (Fig. 2D). These large mononuclear cells were positive for CD11 in serial sections and were shown to be macrophages (data not shown). In the pancreas, massive infection was observed in acinar cells (Fig. 2F).
The destruction of hepatocytes and pancreatic acinar cells was also confirmed by biochemical examination. Serum ALT and amylase levels were measured in intravenously inoculated mice on day 3 p.i. ALT values increased markedly in the PVR-transgenic/Ifnar knockout mice compared to the nontransgenic mice (P < 0.05, t test), whereas there was only a slight increase in the PVR-transgenic mice (Fig. 3A). This result indicates destruction of hepatocytes in the PVR-transgenic/Ifnar knockout mice. The amylase activity values in the serum of PVR-transgenic mice were 1.3- to 1.5-fold (mean, 1.4-fold) higher than that of the nontransgenic mice (P < 0.05, t test), while those of PVR-transgenic/Ifnar knockout mice were 1.2- to 2.3-fold higher (mean, 1.7-fold) than that of nontransgenic mice (P < 0.05, t test) (Fig. 3B). The results indicated acinar cell destruction in the pancreas and/or salivary glands in PVR-transgenic and PVR-transgenic/Ifnar knockout mice. These observations further indicate that the loss of alpha/beta IFN signaling apparently alters the tissue tropism. The hepatocytes, acinar cells, and macrophages in the spleen became potentially permissive for poliovirus infection, indicating that they express host factors required to support poliovirus replication.
FIG. 3.
Serum ALT and amylase activities in infected mice. The mice were inoculated intravenously with 2 × 107 PFU of poliovirus. The sera of nontransgenic (hatched bars), PVR-transgenic (open bars), and PVR-transgenic/Ifnar knockout (solid bars) mice were collected on day 3 p.i., and their ALT activity (A) and amylase activity (B) were determined. The mean values plus standard deviation of four mice are shown. The asterisks indicate that the values are significantly higher than those observed in the nontransgenic C57BL/6 mice (P < 0.05, t test).
With PVR-transgenic/Ifnar knockout mice, the virus antigens were not clearly detected in the kidneys, heart, and lungs of poliovirus-inoculated mice. Similar experiments were performed with another PVR-transgenic mouse strain, PVRtg25 (46). In this mouse strain, PVR mRNA expression levels are higher than those of PVRtg21. We further crossed this strain with Ifnar knockout mice and examined the susceptibility of PVRtg25/Ifnar knockout mice to poliovirus. All of the inoculated mice became moribund with jaundice on day 1 p.i. Immunohistological examination revealed that viral antigen-positive cells were detected in the liver, spleen, pancreas, kidneys, and heart (data not shown). Poliovirus infection in the liver was associated with massive necrosis of the parenchymal cells, which was correlated with liver failure. This confirmed that poliovirus can also replicate in the kidneys and heart when IFN signaling is disrupted.
Role of IFN in poliovirus spreading in the body.
The above data indicate that the IFN response is particularly effective in restricting virus replication in the visceral tissues. Viremia occurred as a consequence of virus multiplication in extraneural sites. It is possible that the IFN response influences virus titer and, accordingly, that IFN also contributes in decreasing the incidence of paralytic and fatal poliovirus infection by lowering the chance of poliovirus entry into the central nervous system. To demonstrate this, we compared the virus titers in PVR-transgenic and PVR-transgenic/Ifnar knockout mice. We determined virus titers in the plasma of PVR-transgenic and PVR-transgenic/Ifnar knockout mice infected intraperitoneally with poliovirus (103 PFU). This dose was employed because it is below the LD50 of the PVR-transgenic and above that of the PVR-transgenic/Ifnar knockout mice (see Table 1).
Expectedly, a very high titer (108 to 109 PFU/ml) of poliovirus was detected on day 3 p.i. in the PVR-transgenic/Ifnar knockout mice, resulting in the death of the mice at day 4 p.i. In contrast, less than 103 PFU of poliovirus/ml was detected in the PVR-transgenic mice between days 2 and 5 p.i., and it was no longer detected at day 7 p.i. (Fig. 4). These results suggest that some cells, if not all, that express PVR can act as reservoirs of poliovirus during the progression of the disease in the viremic phase. However, only a small proportion of these cells produce poliovirus with a low efficiency because of the inhibitory effect of IFN, and high-titer viremia is prevented in animals with a normal IFN system. We consider that virus replication sites during the viremic phase have not been histologically identified because poliovirus replication levels are normally low in these cells in PVR-transgenic mice, monkeys, and humans.
FIG. 4.
Viremia in PVR-transgenic mice and PVR-transgenic/Ifnar knockout mice. Poliovirus (103 PFU) was inoculated intraperitoneally. Plasma from three or four infected mice was collected at the indicated day p.i., after which the virus titer in the plasma was determined. Open circles, PVR-transgenic mice; solid circles, PVR-transgenic/Ifnar knockout mice. Note that the virus titer in the PVR-transgenic/Ifnar knockout mice is very high. The data for PVR-transgenic/Ifnar knockout mice on days 5 and 7 p.i. were not available because all mice died on the fourth day after inoculation.
Expression of IFN-β and ISGs in the host.
The susceptibility to poliovirus varied among the tissues. We therefore determined if the expression profiles of genes that confer an antiviral state in the IFN response were different between target and nontarget tissues. We determined the expression of mRNAs for IFN-β and ISGs with a quantitative real-time reverse transcription-PCR technique (Fig. 5). Since IFN-β is the first alpha/beta IFN induced after virus infection (36, 39, 41) and picornaviruses are known to be sensitive to OAS (7, 8) and protein kinase R (25), we focused on expression of IFN-β, OAS, and protein kinase R mRNAs. Of the ten genes that are similar to human OAS in the mouse genome, OAS1a, OAS1g, OAS2, OAS3, and OASL2 were shown to synthesize 2′-5′oligoadenylate (16).
FIG. 5.
Expression of IFN-β and ISGs in PVR-transgenic mice. The expression levels of IFN-β and ISG mRNAs in noninfected PVR-transgenic mice and PVR-transgenic mice infected intravenously with poliovirus (2 × 107 PFU) were determined by real-time quantitative PCR. The amounts of IFN-β (A), OAS1a (B), OAS1g (C), OAS2 (D), OAS3 (E), OASL2 (F), and protein kinase R (G) mRNAs normalized to 107 copies of 18S rRNA are shown. Open bars, gray solid bars, and black solid bars indicate the results for noninfected mice, infected mice at 1 day p.i., and infected mice at 3 days p.i., respectively. The mean values for three to six mice are indicated. The numbers above each figure indicate values that could not be represented within the figures. Note that the open bars in A are not visible because IFN-β mRNA expression in the noninfected mice was very low.
We first determined the expression levels of IFN-β, OAS, and protein kinase R mRNAs in the noninfected PVR-transgenic mice. Very little expression of IFN-β mRNA was observed in all tissues of the noninfected mice (Fig. 5A). The expression levels of the mRNAs for OAS1a, OAS1g, OAS2, OAS3, OASL2, and protein kinase R are shown in Fig. 5B to G (open bars). The expression level of each ISG mRNA and the tissue distribution profiles were different. However, in general, they were expressed in the nontarget tissues more abundantly than in the target tissues of noninfected PVR-transgenic mice. No ISG was expressed at high levels in the central nervous system, an observation consistent with previous reports (1, 22, 30).
We then determined the changes in the expression levels of IFN-β and ISG mRNAs in the infected mice (Fig. 5A to G). On day 1 p.i., IFN-β mRNA expression in the spleen was observed at very high levels. Low-level IFN-β expression was observed in the heart, lungs, liver, kidneys, and muscle. No significant increase of IFN-β mRNA was observed in the brain and spinal cord on day 1 p.i. (Fig. 5A, gray solid bars). On day 3 p.i., IFN-β mRNA levels in the heart, lungs, liver, kidneys, and muscle decreased to nearly basal levels. In contrast, they increased to very high levels in the brain and spinal cord (Fig. 5A, solid black bars). Thus, the IFN-β mRNA expression profiles are different between target tissues and nontarget tissues in poliovirus-infected mice.
The expression levels of ISG mRNAs also changed after poliovirus infection, consistent with the change in IFN-β mRNA levels (Fig. 5B to G, gray and solid black bars). The ISG expression pattern was clearly different between the target and nontarget tissues. ISG mRNAs increased in most of the visceral tissues on day 1 p.i. The expression level of ISG mRNAs increased most efficiently in the spleen. The levels of ISG mRNAs were relatively high in the heart, lungs, liver, kidneys, and skeletal muscle. In these tissues, the viral load was low (Fig. 1), and the poliovirus antigen was not detected. This indicates that ISG induction in these tissues was sufficient to inhibit poliovirus replication. The expression levels of ISG mRNAs in these tissues decreased on day 3 p.i., with a corresponding decrease in IFN-β mRNA levels. In the brain and spinal cord, however, significant induction of ISG mRNAs was not observed on day 1 p.i. The induction became evident only on day 3 p.i., when poliovirus destroyed a large number of neurons. We also noticed that the ratio of ISG and IFN-β mRNAs (ISG/IFN-β mRNA) in the brain and spinal cord on day 3 p.i. was much lower than that in nontarget tissues on day 1 p.i. (Fig. 5A to G). This indicates that the IFN response did occur in neurons in the brain and spinal cord but was not sufficient and failed to inhibit viral growth in the early phase of infection.
Expression of RIG-I, MDA-5/helicard, and IRF-7 in target and nontarget tissues.
The data suggests that neurons in the target tissues failed to respond sufficiently to poliovirus infection. It is possible that there is a difference in the expression mechanism of the IFN response. We proceeded to examine a regulatory factor required for IFN response. Yoneyama et al. recently found that RIG-I functioned as a detector of intracellular double-stranded RNA. The cells that express this gene at high levels in vitro can induce IFN in an accelerated fashion and can survive against encephalomyocarditis virus and vesicular stomatitis virus infection (48). MDA-5/helicard is another caspase recruitment domain (CARD)-containing helicase, which is implicated as having a function similar to that of RIG-I (48). Both RIG-I and MDA-5/helicard are inducible by IFNs (17, 48).
Figures 6A and B show the changes in the RIG-I and MDA-5/helicard levels, respectively. Like that of other ISGs, the expression of these genes is low in the brain and spinal cord but high in nontarget tissues in the noninfected mice. The response of these genes after poliovirus infection is similar to that of other ISGs. They were induced at high levels in the nontarget tissues on day 1 p.i. but not in the target tissues. Thus, the nontarget tissues that expressed RIG-I and MDA-5/helicard at high levels may have an advantage in inducing IFN-β soon after poliovirus infection. We also examined the expression of IRF-7, another regulatory factor involved in the activation of IFN-α genes. IRF-7 thus is important to amplify the IFN response (37). The expression profile of IRF-7 was also similar to those of other ISGs (Fig. 6C).
FIG. 6.
Expression of RIG-I, helicard, and IRF-7 mRNAs in PVR-transgenic mice. The expression levels of RIG-I, helicard, and IRF-7 mRNAs of noninfected PVR-transgenic mice and PVR-transgenic mice infected intravenously with poliovirus (2 × 107 PFU) were determined by real-time quantitative PCR. The mean values for three mice are indicated. The amounts of RIG-I (A), helicard (B), and IRF-7 (C) mRNAs were determined. Open bars, gray solid bars, and black solid bars indicate the results of noninfected mice, infected mice at 1 day p.i., and infected mice at 3 days p.i., respectively. The amounts of mRNA per 107 copies of 18S rRNA are shown.
Protection of mice from poliovirus infection by poly(I:C) treatment.
The preceding data suggest that neurons in the brain and spinal cord were highly susceptible to poliovirus because expression levels of ISGs, including OASs and RIG-I, were low in the noninfected state. If this is the case, pretreatment to induce the antiviral state in the central nervous system would increase the survival rate of poliovirus-infected mice. Hence, treatment with poly(I:C) is expected to induce IFNs and ISGs and establish an antiviral state.
Poly(I:C) (200 μg) was administered intracerebrally to PVR-transgenic mice, and on the next day, RNA was prepared from the brain and spinal cord. The levels of OAS1a and RIG-I were determined by real-time quantitative PCR. As expected, expression of the mRNAs for OAS1a and RIG-I was elevated to high levels by poly(I:C) (Fig. 7A and B). PVR-transgenic mice treated with poly(I:C) or mock treated were challenged with poliovirus (104 PFU) by the same route. The survival of the mice is shown in Fig. 7C. Mock-treated PVR-transgenic mice died at 2 to 6 days p.i., with a survival rate of 7.7%. After poly(I:C) treatment, mice died at 4 to 16 days p.i., increasing the survival rate to 30.8%. The clinical symptoms observed in the poly(I:C)-treated PVR-transgenic mice were almost the same as those observed in mock-treated mice, suggesting the same pathology. The results indicate that the poly(I:C)-treated mice survived longer than mock-treated mice, with the survival rate of poly(I:C)-treated mice being higher than that of mock-treated mice (P < 0.05, log rank test). These data suggest that the antiviral state induced by treatment with poly(I:C) was effective in preventing poliovirus replication in the central nervous system.
FIG. 7.
Induction of mRNAs for OAS1a (A) and RIG-I (B) after poly(I:C) treatment. PVR-transgenic mice was administered poly(I:C) (solid bars) or mock treated (open bars). RNA was prepared from the mice 1 day after administration. The amounts of RNA were determined by real-time quantitative PCR. (C) Survival of infected mice. PVR-transgenic mice administered poly(I:C) (solid circles) or mock treated (open circles) (13 mice each) were challenged intracerebrally with 104 PFU of poliovirus. Mice were observed for 3 weeks. The survival rate of poly(I:C)-treated mice was significantly higher than that of mock-treated mice (P < 0.05, log-rank test).
DISCUSSION
IFN system is a host factor that inhibits poliovirus replication.
Picornaviruses are sensitive to IFNs. However, little is known about the role of alpha/beta IFN in the pathogenesis of poliovirus in vivo. We have shown the importance of the IFN response in poliovirus infection in vivo with a transgenic mouse expressing human PVR. In the PVR-transgenic mice, poliovirus replicates and produces severe lesions in the brain and spinal cord, while other tissues did not show severe pathological changes.
It would be reasonable to assume that some host factors required for poliovirus replication are lacking in the nontarget cells and tissues. PVR was thought to be such a determinant (13). However, previous studies revealed that many nontarget tissues expressed PVR (18, 21, 24, 31, 32). It is therefore impossible to explain poliovirus tissue tropism solely by the presence of PVR. Gromeier et al. (11) and Yanagiya et al. (46) proposed a hypothetical mechanism called the internal ribosome entry site (IRES)-dependent mechanism to explain the tissue tropism of viruses. The IRES controls the efficiency of protein synthesis in some viruses. Chimeric viruses containing the IRES of human rhinovirus and hepatitis C virus instead of the poliovirus IRES do not replicate in the central nervous system. Therefore, the poliovirus IRES confers the ability to replicate in neurons on the chimeric virus, while the IRESs of human rhinovirus and hepatitis C virus do not. It is possible that the poliovirus IRES, particularly in virulent strains, is designed to exhibit full activity in neurons. In this hypothesis, host factors related to poliovirus IRES function should be restricted to neurons. However, poliovirus can replicate in cultured cells of nonneural origin. This hypothesis does not completely explain why poliovirus does not replicate efficiently in nontarget tissues in vivo.
It is also possible to assume that some host factors that inhibit poliovirus replication are present in the nontarget tissues. The liver, spleen, and pancreas were spared severe poliovirus infection only when the IFN system was functional. These data demonstrate that the IFN system is one of the major factors that confers resistance against poliovirus infection. Limitation of poliovirus tissue tropism is achieved by inhibition of poliovirus replication by the IFN system in nontarget tissues. An altered tissue distribution of viral replication was observed in mice deficient in the Ifnar gene or in mice deficient in the signal transducer and activator of transcription 1 (Stat-1) gene infected with viruses other than poliovirus (9, 10, 28, 34, 44). In these animals, virus replication was observed in tissues that were normally considered nontarget tissues. Therefore, it is a general principle that the tissue tropism of viruses is determined, at least in part, by an IFN-dependent mechanism.
Unequal IFN response selectively inhibits poliovirus replication in nontarget tissues.
In cultured cells, encephalomyocarditis virus replication occurs rapidly within 6 h, and the infected cells are usually destroyed by lytic replication of virus before they produce IFNs. However, constitutive expression of OAS or protein kinase R (7, 8, 25), which are effectors of antiviral activity, and expression of RIG-I (48), a regulator of IFN induction, inhibited viral replication. It is very likely that the same mechanism operates during poliovirus infection.
We determined the expression of ISGs in the tissues. The distribution of ISG mRNAs in the noninfected mice was not equal among tissues. They were expressed in the nontarget tissues more abundantly than in the target tissues (Fig. 5). These existing ISG products may help restrict virus replication and spread in the nontarget tissues during the initiation of infection in vivo. In PVR-transgenic/Ifnar knockout mice, the expression levels of mRNAs for ISGs were greatly reduced. The mRNAs for OASs were detected only in the intestine and thymus (data not shown). This is also consistent with the result of Ueda et al. (43), which showed that OAS expression in most tissues was reduced in p48 (IRF-9)-deficient mice. This result suggests that ISG expression in most of the tissues, even in the noninfected state, is mainly dependent on the IFNAR-dependent pathway. It also suggests that these tissues were continuously exposed to IFN stimulation at low levels (43). Visceral tissues such as the intestine and lungs are continuously at risk of exposure to pathogens. These tissues may be programmed to respond readily to viral infection. Alternatively, they are constantly stimulated by nonpathogenic microorganisms present in the body and thus are already primed.
Furthermore, the IFN response after poliovirus infection was also different among tissues. High-level response was observed in the spleen but was not observed in the spinal cord. This suggests that the IFN response profile may differ depending on the cells and tissues. Since RIG-I and MDA-5/helicard are also IFN inducible (17, 48), they also existed more abundantly in the nontarget tissues (Fig. 6), like other ISGs (Fig. 5). Some of the important regulators of the IFN response, such as IRF-7 and IRF-9, are also IFN inducible. The cells that are primed even at low levels of IFNs may be equipped with all the machinery necessary for the IFN response. Thus, the nontarget tissues may be ready to respond to viral infection. Unequal distribution of the regulators of the IFN response is again consistent with the idea that poliovirus replication is selectively inhibited in nontarget tissues.
On the contrary, neurons in the brainstem and spinal cord could not induce a sufficient antiviral state after poliovirus infection. However, pretreatment with poly(I:C) increased the survival of PVR-transgenic mice against poliovirus challenge (Fig. 7). This suggests that neurons also became resistant to poliovirus infection as long as they were treated. It is therefore possible that the status of ISG expression in the early phase of infection is critical in determining the fate of infected cells and an unequal IFN response may be one of the reasons for the differential susceptibility of cells and tissues to poliovirus. Although the IFN response was not equal in PVR-transgenic mice, both the basal expression of ISGs and induction of ISGs after poliovirus infection are equally null in PVR-transgenic/Ifnar knockout mice. Without these unequal protective responses, replication of poliovirus was observed in PVR-transgenic/Ifnar knockout mice in the nontarget tissues as well.
Incidence of paralytic disease is influenced by the IFN response.
In a natural poliovirus infection, less than 1% of infected individuals develop paralytic disease, and virus clearance occurs in most persons with asymptomatic or mild infections (4, 23, 35). Viremia is observed only transiently in experimentally infected chimpanzees and monkeys, with titers of less than 105 tissue culture infective doses per ml with virulent strains (3, 5). Viremia was not observed in a chimpanzee and a human volunteer administered attenuated vaccine strains (35). Thus, viremia titers seem to correlate with central nervous system invasion in primates.
Our data showed that viral replication in visceral tissues is inhibited by the IFN response in PVR-transgenic mice. Poliovirus can enter the central nervous system, penetrating the blood-brain barrier. This pathway is considered the main pathway of poliovirus entry into the central nervous system in the PVR-transgenic mice after poliovirus infection via peripheral routes (47). Inhibition thus results in reduction of the virus titer in the blood and reduction of the chance of virus entry into the central nervous system. In contrast, PVR-transgenic/Ifnar knockout mice showed viremia with a very high titer and a high incidence of paralytic disease. It is therefore possible to speculate that the low incidence of paralytic poliomyelitis in humans is also a result of inhibition of poliovirus replication in nonneural tissues by the host IFN response, although we have no experimental evidence on humans. Paralytic poliomyelitis may occur when the alpha/beta IFN response does not work sufficiently in patients with certain conditions. Individuals who have a defect(s) in a gene(s) that contributes to the IFN response would be more susceptible to paralytic poliomyelitis.
Conclusion.
The tissue tropism and pathogenesis of viruses are determined by a combination of several factors. In the case of poliovirus infection, poliovirus replication sites are primarily determined by the presence of the receptor, with the capture and entry of the virus into the cells supported by the PVR. Cells expressing PVR at high levels may be favored for poliovirus infection (20). Thus, the tropism of poliovirus may be dependent on the amount of PVR. After virus entry into cells, efficient replication of poliovirus may be dependent on the milieu of infected cells. If the environment is optimal for RNA and viral protein synthesis, a large number of viral particles will be produced per cell. If antiviral activities, such as the IFN response, are sufficiently high, virus replication will be inhibited. Thus, the fate of infected cells is determined by the balance of the replicating capacity of poliovirus and the antiviral activity of the host. Visceral tissues will then fail to serve as a massive factory of poliovirus, and the chance of viral entry into the central nervous system is greatly reduced.
If the virus enters the central nervous system, virulent poliovirus strains can replicate in neurons, where the antiviral defense is not sufficiently ready, and the patient develops paralytic disease. Therefore, the innate antiviral defense is an important determinant of tissue tropism and pathogenicity of poliovirus. It is of interest to investigate if the alpha/beta IFN response also contributes to selective poliovirus infection in the motor neurons in the central nervous system or to infection in the gastrointestinal tract. These questions will be elucidated in future studies. In the case of other viruses, situations such as distribution of the receptor molecule, replication capacity in each tissue, and resistance to the IFN system may differ from those of poliovirus. It therefore seems likely that each virus displays a distinct disease pattern unique to that particular virus.
Acknowledgments
We thank Masayoshi Kohase, Seii Ohka, Akio Nomoto, Hitoshi Horie, Shinobu Abe, Bunshichi Shimizu, Sou Hashizume, Michiaki Masuda, Masahiko Takada, Eckard Wimmer, and Yoshiyuki Nagai for providing materials, technical assistance, and helpful discussion.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology and by a Grant-in-Aid for Research on Emerging and Re-emerging Infectious Diseases from the Japanese Ministry of Health, Labour and Welfare.
REFERENCES
- 1.Asada-Kubota, M., T. Ueda, M. Shimada, K. Takeda, and T. Sokawa. 1995. Distribution of immunoreactive 2′, 5′-oligoadenylate synthetase in mouse digestive tract. J. Interferon Cytokine Res. 15:863-867. [DOI] [PubMed] [Google Scholar]
- 2.Bodian, D. 1949. Histopathologic basis of clinical findings in poliomyelitis. Am. J. Med. 6:563-578. [DOI] [PubMed] [Google Scholar]
- 3.Bodian, D. 1954. Viremia in experimental poliomyelitis. I. General aspects of infection after intravascular inoculation with strains of high and of low invasiveness. Am. J. Hyg. 60:339-357. [PubMed] [Google Scholar]
- 4.Bodian, D. 1955. Emerging concept of poliomyelitis infection. Science 12:105-108. [DOI] [PubMed] [Google Scholar]
- 5.Bodian, D. 1956. Poliovirus in chimpanzee tissues after virus feeding. Am. J. Hyg. 64:181-197. [DOI] [PubMed] [Google Scholar]
- 6.Bodian, D., and H. A. Howe. 1940. An experimental study of the role of neurones in the dissemination of poliomyelitis virus in the nervous system. Brain 63:135-167. [Google Scholar]
- 7.Chebath, J., P. Benech, M. Revel, and M. Vigneron. 1987. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature 330:587-588. [DOI] [PubMed] [Google Scholar]
- 8.Coccia, E. M., G. Romeo, A. Nissim, G. Marzaili, R. Albertini, E. Affabris, A. Battistini, G. Fiorucci, R. Orsatti, G. B. Rossi, and J. Chebath. 1990. A full-length murine 2-5A synthetase cDNA transfected in NIH-3T3 cells impairs EMCV but not VSV replication. Virology 179:228-233. [DOI] [PubMed] [Google Scholar]
- 9.Fiette, L., C. Aubert, U. Mueller, S. Huang, M. Aguet, and M. Brahic, and J.-F. Bureau. 1995. Theiler's virus infection of 129Sv mice that lack the interferon α/β or interferon γ receptors. J. Exp. Med. 181:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Sastre, A., R. K. Durbin, H. Zheng, P. Palese, R. Gertner, D. E. Levy, and J. E. Durbin. 1998. The role of interferon in influenza virus tissue tropism. J. Virol. 72:8550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkel, E., S. Morich, and R. Henkel. 1984. 2-chloro-4-nitrophenyl-β-d-maltoheptaoxide: A new substrate for the determination of α-amylase in serum and urine. J. Clin. Chem. Clin. Biochem. 22:489-495. [DOI] [PubMed] [Google Scholar]
- 13.Holland, J. J. 1961. Receptor affinities as major determinants of enterovirus tissue tropisms in humans. Virology 15:312-326. [DOI] [PubMed] [Google Scholar]
- 14.Ida-Hosonuma, M., T. Iwasaki, C. Taya, Y. Sato, J. Li, N. Nagata, H. Yonekawa, and S. Koike. 2002. Comparison of neuropathogenicity of poliovirus in two transgenic mouse strains expressing human poliovirus receptor with different distribution patterns. J. Gen. Virol. 83:1095-1105. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki, A., R. Welker, S. Mueller, M. Linehan, A. Nomoto, and E. Wimmer. 2002. Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J. Infect. Dis. 186:585-592. [DOI] [PubMed] [Google Scholar]
- 16.Kakuta, S., S. Shibata, and Y. Iwakura. 2002. Genomic structure of the mouse 2′, 5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 22:981-993. [DOI] [PubMed] [Google Scholar]
- 17.Kang, D.-C., R. V. Gopalkrishnan, Q. Wu, E. Jankowsky, A. M. Pyle, and P. B. Fisher. 2002. mda-5: An intereferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 99:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koike, S., H. Horie, I. Ise, A. Okitsu, M. Yoshida, N. Iizuka, K. Takeuchi, T. Takegami, and A. Nomoto. 1990. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9:3217-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koike, S., I. Ise, Y. Sato, H. Yonekawa, O. Gotoh, and A. Nomoto. 1992. A second gene for the African green monkey poliovirus receptor that has no putative N-glycosylation site in the functional N-terminal immunoglobulin-like domain. J. Virol. 66:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koike, S., C. Taya, J. Aoki, Y. Matsuda, I. Ise, H. Takeda, T. Matsuzaki, H. Amanuma, H. Yonekawa, and A. Nomoto. 1994. Characterization of three different transgenic mouse lines that carry human poliovirus receptor gene -influence of the transgene expression on pathogenesis. Arch. Virol. 139:351-363. [DOI] [PubMed] [Google Scholar]
- 21.Koike, S., C. Taya, T. Kurata, S. Abe, I. Ise, H. Yonekawa, and A. Nomoto. 1991. Transgenic mice susceptible to poliovirus. Proc. Natl. Acad. Sci. USA 88:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mashimo, T., P. Glasser, M. Lucas, D. Simon-Chazottes, P. E. Ceccaldi, X. Montagutelli, P. Despres, and J.-L. Guénet. 2003. Structural and functional genomics and evolutionary relationship in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics 82:537-552. [DOI] [PubMed] [Google Scholar]
- 23.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.) Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 24.Mendelson, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 25.Meurs, E. F., Y. Watanabe, S. Kadereit, G. N. Barber, M. G. Katze, K. Chong, B. R. Williams, and A. G. Hovanessian. 1992. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J. Virol. 66:5805-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller, U., U. Stainhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 27.Munoz, A., and L. Carrasco. 1984. Action of human lymphoblastoid interferon on HeLa cells infected with RNA-containing animal viruses. J. Gen. Virol. 65:377-390. [DOI] [PubMed] [Google Scholar]
- 28.Mrkic, B., J. Pavlovic, J., T. Ruelicke, P. Volpe, C. J. Buchholz, D. Hourcade, J. P. Atkinson, A. Aguzzi, and R. Cattaneo. 1998. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 72:7420-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomoto, A., S. Koike, and J. Aoki. 1994. Tissue tropism and species specificity of poliovirus infection. Trends Microbiol. 2:47-51. [DOI] [PubMed] [Google Scholar]
- 30.Perelygan, A. A., S. V. Scherbik, I. B. Zhulin, B. M. Stockman, Y. Li, and M. A. Brinton. 2002. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. USA 99:9322-9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren, R., F. Constantini, E. J. Gorgacz, J. J. Lee, and V. R. Racaniello. 1990. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63:353-362. [DOI] [PubMed] [Google Scholar]
- 32.Ren, R., and V. R. Racaniello. 1992. Human poliovirus receptor gene expression and poliovirus tissue tropism in transgenic mice. J. Virol. 66:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rueckert, R. 1996. Picornaviridae: the viruses and their replication. p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.) Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 34.Ryman, K., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabin, A. 1956. Pathogenesis of poliomyelitis: reappraisal in light of the new data. Science 123:1151-1157. [DOI] [PubMed] [Google Scholar]
- 36.Samuel, C. E. 1991. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology 183:1-11. [DOI] [PubMed] [Google Scholar]
- 37.Sato, M., H. Suemori, N. Hata, M. Asargiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 38.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: link to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 39.Sen, G. C., and P. Lengyel. 1992. The interferon system. A bird's eye view of its biochemistry. J. Biol. Chem. 267:5017-5020. [PubMed] [Google Scholar]
- 40.Shiroki, K., H. Kato, S. Koike, K. Odaka, and A. Nomoto. 1993. Temperature-sensitive mouse cell factors for strand-specific initiation of poliovirus RNA synthesis. J. Virol. 67:3989-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schriber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 42.Tagawa, Y., K. Sekikawa, and Y. Iwakura. 1997. Suppression of concanavalin A-induced hepatitis in IFN-gamma−/− mice, but not in TNF-alpha−/− mice: role for IFN-gamma in activating apoptosis of hepatocytes. J. Immunol. 159:1418-1428. [PubMed] [Google Scholar]
- 43.Ueda, T., R. Tatsumi, N. Tanaka, M. Asada-Kubota, K. Hamada, S. Noguchi, T. Taniguchi, and Y. Sokawa. 1998. Production of immunoreactive 2′,5′-oligoadenylate synthetase in p48-deficient mice. J. Interferon Cytokine Res. 18:181-185. [DOI] [PubMed] [Google Scholar]
- 44.Wessely, R., K. Klingel, K. U. Knowlton, and R. Kandolf. 2001. Cardioselective infection with coxsackievirus B3 requires intact type 1 interferon signaling: implications for mortality and early viral replication. Circulation 103:765-771. [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization. 1990. Manual for the virological investigation of poliomyelitis. World Health Organization, Expanded Programme on Immunization and Division of Communicable Diseases. W.H.O. Publication no. W.H.O./EPI/CDS/POLIO/ 90.1. World Health Organization, Geneva, Switzerland.
- 46.Yanagiya, A., S. Ohka, N. Hashida, M. Okamura, C. Taya, N. Kamoshita, K. Iwasaki, Y. Sasaki, H. Yonekawa, and A. Nomoto. 2003. Tissue-specific replicating capacity of a chimeric poliovirus that carries the internal ribosome entry site of hepatitis C virus in a new mouse model transgenic for the human poliovirus receptor. J. Virol. 77:10479-10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, W. X., T. Terasaki, K. Shiroki, S. Ohka, J. Aoki, S. Tanabe, T. Nomura, E. Terada, Y. Sugiyama, and A. Nomoto. 1997. Efficient delivery of circulating poliovirus to the central nervous system independently of poliovirus receptor. Virology 17:421-428. [DOI] [PubMed] [Google Scholar]
- 48.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oliogoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]