Abstract
Human cytomegalovirus (HCMV) glycoprotein B (gB) is an abundant virion envelope protein that has been shown to be essential for the infectivity of HCMV. HCMV gB is also one of the most immunogenic virus-encoded proteins, and a significant fraction of virus neutralizing antibodies are directed at gB. A linear domain of gB designated AD-1 (antigenic domain 1) represents a dominant antibody binding site on this protein. AD-1 from clinical isolates of HCMV exhibits little sequence variation, suggesting that AD-1 plays an essential role in gB structure or function. We investigated this possibility by examining the role of AD-1 in early steps of gB synthesis. Our results from studies using eukaryotic cells indicated that amino acid (aa) 635 of the gB sequence represented the carboxyl-terminal limit of this domain and that deletion of aa 560 to 640 of the gB sequence resulted in loss of AD-1 expression. AD-1 was shown to be required for oligomerization of gB. Mutation of cysteine at either position 573 or 610 in AD-1 resulted in loss of its reactivity with AD-1-specific monoclonal antibodies and gB oligomerization. Infectious virus could not be recovered from HCMV bacterial artificial chromosomes following introduction of these mutations into the HCMV genome, suggesting that AD-1 was an essential structural domain required for gB function in the replicative cycle of HCMV. Sequence alignment of AD-1 with homologous regions of gBs from other herpesviruses demonstrated significant relatedness, raising the possibility that this domain may contribute to multimerization of gBs in other herpesviruses.
Human cytomegalovirus (HCMV) is a well-described human pathogen responsible for a variety of acute disease syndromes, and more recently, several chronic diseases have been associated with HCMV infection (9, 47). The virus is the largest human herpesvirus, and although the exact number of virus-encoded proteins is unknown, recent analysis of clinical strains of HCMV has suggested that the viral genomes could contain over 200 open reading frames (ORFs) (35). In addition, the proteome of the virion may be more complex than previously thought and a large number of host-derived proteins appear to be present in the virion particle (52). Structurally, HCMV exhibits many of the common characteristics of herpesviruses, including a conserved morphology consisting of a nucleocapsid, tegument, and complex envelope (34). Although the composition and the structure of the nucleocapsid have been defined, the structure of the tegument and the envelope of this virus remain poorly characterized (1, 52). A comprehensive description of the protein composition of these latter two structures remains to be accomplished. Accordingly, the function of tegument and envelope proteins is not well understood, with the exception of a limited group of proteins.
Abundant components of the envelope include glycoproteins that are structural and functional homologues of conserved glycoproteins found in other herpesviruses (34). These include glycoprotein B (gB), gH, gL, gM, and gN. Each of these glycoproteins has been shown to be essential for the production of infectious HCMV virions (24). To date, only gB and the gH/gL/gO complex have been demonstrated to have a role in virion attachment and entry (6, 36, 43, 55). Early studies of gB demonstrated that virus neutralizing antibodies could block binding, and other studies have suggested that anti-gB antibodies also blocked gB-mediated fusion (3, 6, 22, 36, 50). More recently, studies have focused on the role of gB in the activation of signal transduction pathways following virus binding to a variety of cells, including permissive human fibroblast (HF) cells (7, 15, 43, 55, 56). Interestingly, HCMV appears to engage Toll-like receptor 2 (TLR-2) and initiate signaling through this cellular receptor (17). Together, these studies have argued for a critical role of gB in the early events of HCMV infection.
The HCMV gB molecule is a type I glycoprotein that is found as a homodimer in the virion (12, 14). The monomer consists of a lumenal or surface (SU) component with an estimated molecular mass of 116 kDa that is linked by disulfide bonds to a transmembrane (TM) component with an estimated molecular mass of 55 kDa (12). The TM component contains a lumenal domain in addition to a membrane-spanning region and a cytoplasmic tail that is over 100 amino acids (aa) in length. The gB molecule is synthesized as a precursor that dimerizes and, following transport into a distal compartment of the secretory pathway, is cleaved by furin into its SU and TM subunits that remain linked by disulfide bonds (12, 14, 16, 19). Much of our understanding of the structural aspects of gB has been derived from studies utilizing monoclonal antibodies (MAbs). The antibody binding sites on gB have been studied by several laboratories, and both conformationally dependent and linear binding sites have been described (25, 33, 38, 41, 51, 53). Linear binding sites have been mapped to two conventional linear epitopes on the SU component of the monomer, and at least one of these epitopes, antigenic domain 2 (AD-2), is a target of virus neutralizing antibodies (33). The TM component also has two linear antibody sites, and one of these, AD-1, is also a target of virus neutralizing antibodies (38, 51). Interestingly, AD-1 is highly immunogenic in humans and animals and readily elicits virus neutralizing antibodies. In fact, antibodies directed at AD-1 represent a disproportionate proportion of virus neutralizing antibodies in human convalescent-phase serum (27).
The AD-1 antibody binding site is unique because of its unconventional structure. A detailed analysis of the AD-1 structure utilizing Escherichia coli-expressed polypeptides revealed that the minimal sequence recognized by AD-1-specific antibodies was represented by aa 552 to 635 of the gB sequence and that internal deletions in this domain eliminated AD-1-specific antibody reactivity (53). These and other findings have argued that AD-1 represented a linear antibody binding domain defined by a continuous stretch of over 75 aa. Additional studies have demonstrated that the disulfide bond between cysteines 573 and 610 within this sequence is required for recognition by AD-1-specific antibodies (46). Sequence analysis of the gene encoding HCMV gB from unrelated clinical isolates revealed a high constraint on sequence variation in AD-1, suggesting that the conservation of the amino acid sequence encoding AD-1 could be secondary to an essential role of this domain in either the structure or the function of gB. In an extension of these and other earlier studies, we introduced a number of mutations in the region of gB containing AD-1, including replacement of Cys with Ser residues at positions 508, 550, 573, and 610, and determined the effect of these mutations on the formation of AD-1 and oligomerization of gB in eukaryotic cells. Our results indicated that a gB molecule containing only the first 635 aa expressed AD-1 and formed oligomers. In contrast, a gB deletion mutant that contained only the first 628 aa failed to react with AD-1-specific antibodies and also did not oligomerize. Expression of the TM component of gB in the absence of the SU component preserved AD-1 formation, but this molecule did not oligomerize. Finally, Cys 573 and 610 were shown to be essential for AD-1 formation, and the mutagenesis of these residues resulted in loss of AD-1, failure of these molecules to oligomerize, and the lack of production of infectious virus from recombinant HCMV bacterial artificial chromosomes (BACs) encoding these mutations. These results provide evidence that AD-1 is essential for oligomerization of gB and that oligomerization is required for intracellular trafficking and the production of infectious virus.
MATERIALS AND METHODS
Cells and viruses.
Primary human dermal fibroblasts (HF) were obtained and maintained as previously described (2, 10). COS-7 cells and 293T human embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% newborn calf serum and penicillin-streptomycin. Primary monkey kidney BSC 40 cells were maintained as described previously (13). Recombinant vaccinia viruses were generated, and virus stocks were produced and titers determined in BSC 40 monkey kidney cells as described previously (13).
Antibodies.
Murine MAbs were produced in our laboratory and have been described in previous reports. MAb 7-17 (immunoglobulin G3 [IgG3]) is an AD-1-specific antibody, MAb 58-15 (IgG2b) has been shown to be reactive with an epitope located between aa 880 and 907 of gB, and MAb 27-39 (IgG1) has been shown to recognize a conformational epitope present on gB oligomers (2, 14). Human MAb 758 is reactive with the AD-2 epitope (aa 68 to 84) and was kindly provided by Scott Koenig, Medimmune Corp., Gaithersburg, Md.
[35S]Meth/Cys labeling, SDS-PAGE, and sedimentation analysis of gB oligomerization.
Recombinant vaccinia virus-infected monkey kidney cells or 293T cells transfected with an expression plasmid encoding gB or gB mutants were radiolabeled with 100 μCi of [35S]methionine/cysteine (Met/Cys) per ml as previously described (2, 14). Following solubilization in standard radioimmunoprecipitation assay buffer (1% NP-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in Tris-buffered saline [TBS; pH 7.5, with 150 mM NaCl]), the labeled proteins were precipitated with MAbs and the precipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) as described in previous publications (2, 14). The assay for multimerization of gB has been described in detail in an earlier publication (14). Briefly, monolayers of recombinant vaccinia virus-infected monkey kidney cells or 293T cells transfected with an expression plasmid were washed and Met/Cys starved for 60 min. The cells were then incubated in Met/Cys-free media containing 100 μCi of [35S]Met/Cys (New England Nuclear, Boston, Mass) per ml for 20 min, and then following extensive washing, the cells were either harvested by placing the dishes at −80°C or chased with media containing Met and Cys and supplemented with 100 μg of cycloheximide per ml for 120 min. These cultures were harvested and placed at −80°C. The labeled monolayers were solubilized in TBS containing 1% NP-40 at 4°C for 30 min. The solubilized proteins were then precleared by incubation with normal goat serum and staphylococcal Cowan I bacteria (Calbiochem, San Diego, Calif.) followed by centrifugation for 15 min at 13,000 × g. The cleared lysate was applied to preformed 5 to 30% linear sucrose gradients and centrifuged for 16 h at 35,000 rpm in a Beckman SW41 rotor. The gradients were fractionated from the bottom into 1-ml fractions, and each fraction was precipitated with the specified antibody. The immune precipitates were collected by first adding rabbit anti-mouse IgG (Cappel Laboratories, Aurora, Ohio) followed by staphylococcal Cowan I bacteria. After extensive washing, the immune precipitates were analyzed by SDS-PAGE in 7.5% gels. Migration of the radiolabeled protein down the gradient, as reflected by recovery of the radiolabeled protein in fractions more proximal to the bottom of the gradient, indicated multimerization of the protein.
Construction of recombinant gB molecules.
The majority of recombinant gB deletion mutants in this report were generated by exonuclease III digestion of a genomic clone of UL55 (gB) by a modification of a previously described method (23). In some cases, cleavage of the UL55 gene at convenient restriction endonuclease sites was utilized to delete the C-terminal residues in the UL55 gene. All constructs were sequenced and designated by the carboxyl-terminal amino acid residue of the gB protein remaining in the deletion mutant. Deleted UL55 genes encoding gB molecules were then cloned into the vaccinia virus shuttle plasmid pSC11, kindly provided by B. Moss (National Institue of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.), as we have described previously (13). For some constructs, the UL55 gene was utilized as a template for PCR construction of site-specific mutations by the Quick Change site-directed mutagenesis procedure (Stratagene, La Jolla, Calif.). In all cases, the resulting mutations in the gB were confirmed by nucleotide sequencing.
Transient expression of gB and gB mutants and image analysis.
Recombinant gB molecules were transiently expressed following subcloning of mutant UL55 genes into the expression plasmid pcDNA3.1 (Invitrogen, Carlsbad, Calif.). Approximately 5 μg of plasmid DNA was transfected into either COS-7 or 293T cells with calcium chloride as described previously (42). Approximately, 48 h after transfection, protein expression was determined by reactivity with MAbs. For image analysis, COS-7 cells were grown on 13-mm-diameter coverslips and transfected as described above. After 48 h, the cells were fixed in 3% paraformaldehyde and permeabilized with DPBS (Dulbecco's modified phosphate-buffered saline, pH 7.4) containing 0.05% NP-40 and 0.002% SDS. Following a blocking step with 10% normal goat serum in DPBS for 1 h, the coverslips were incubated with the specified MAb followed by development with appropriate fluorescein isothiocyanate (FITC) or Texas red-conjugated goat anti-mouse (or human) IgG antibodies (Southern Biotechnology, Birmingham, Ala.). The coverslips were mounted in glycerol and antifade (Molecular Probes, Eugene, Oreg.), and images were captured and processed as previously described (42).
Recombinant HCMV construction and recovery of infectious virus.
The details of the construction of recombinant HCMV utilizing HCMV BACs has been described in an earlier publication (8). The source of the infectious clone of HCMV AD169 maintained as a BAC in E. coli was Martin Messerle and Ulrich Koszinowski (University of Munich, Munich, Germany) (4). The BAC was mutagenized in a two-stop approach in which a LacZ/Ampr cassette was inserted into the UL55 gene, utilizing the lambda RED locus linear recombination system (recombineering) (18). Following isolation and characterization of the HCMV BAC in which the UL55 gene was deleted between nucleotides 1370 and 1982 by insertion of the LacZ/Ampr cassette, we then repaired the UL55 gene by recombining PCR-derived fragments of wild-type gB or fragments containing specific mutations at Cys codons 550 (TGC→TCC), 573 (TGC→TCC), and 610 (TGT→TCT) into the UL55 gene as described previously (8). The BAC DNA was characterized by restriction endonuclease digestion, Southern blotting, and nucleotide sequencing of the UL55 gene. Virus was recovered by electroporation of approximately 5 μg of BAC HCMV DNA into HF cells, and in most cases 12 to 16 days postelectroporation, typical HCMV plaques could be detected. Electroporated cells were maintained for a minimum of 35 days postelectroporation and examined biweekly before designating a mutation in an HCMV gene as lethal based on the lack of recovery of infectious virus.
RESULTS
Domains of gB required for formation of AD-1 and gB oligomerization.
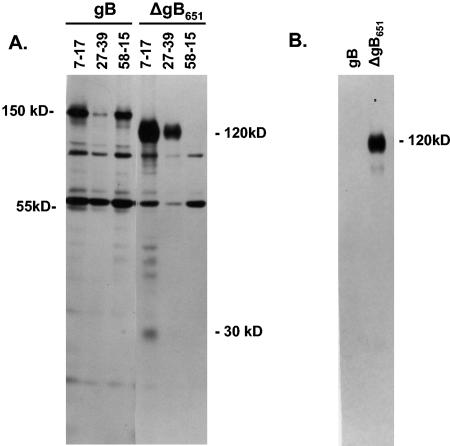
Studies using prokaryotic expression systems have demonstrated that formation of AD-1 was defined by aa 552 to 635 of the gB sequence and that MAb reactivity with AD-1 required disulfide bonding between Cys residues at aa 573 and 610 of gB (46, 53). To extend these findings, we initially determined the domain(s) of gB required for formation of AD-1 in eukaryotic cells, a cellular environment relevant to HCMV infection. A schematic of the gB protein detailing some of the structural landmarks relevant to this study together with the locations of mutations in gB that were generated for this study is shown in Fig. 1. Initially, we generated a series of recombinant gB molecules that contained C-terminal deletions in gB and determined AD-1-specific MAb reactivity for these gB mutants transiently expressed in mammalian cells. In the first series of experiments, we characterized three molecules that had C-terminal deletions in gB at aa 483, 651, and 781. AD-1-specific MAbs recognized only gB mutants truncated at positions 651 and 781, indicating that sequences required for AD-1 formation were located between aa 483 and 651 (data not shown). We selected the gB recombinant with deletion at position 651(ΔgB651) for further characterization. This truncated gB protein lacked both the predicted membrane-spanning domain and the cytoplasmic tail (Fig. 1). When expressed in primary monkey kidney cells, the 120-kDa uncleaved form of the ΔgB651 was precipitated by both the AD-1-specific MAb 7-17 and MAb 27-39, a MAb previously shown to be reactive with a conformational epitope present on oligomeric forms of gB (Fig. 2) (14). MAb 58-15, which is specific for the extreme C-terminal sequence of gB, failed to react with ΔgB651 (Fig. 2). The ΔgB651 molecule was secreted into the media, presumably secondary to the lack of a transmembrane domain (Fig. 2). Wild-type gB was reactive with all three MAbs and was not secreted into the tissue culture medium (Fig. 2). In addition, MAb 7-17 precipitated the gp55 cleavage product of gB and a 30-kDa protein presumably derived from the cleavage of the ΔgB651 protein (Fig. 2).
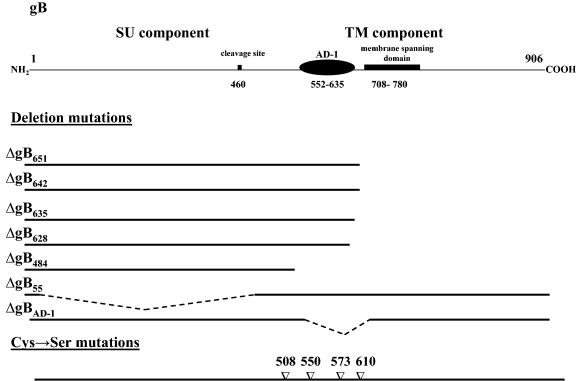
FIG. 1.
Schematic representation of HCMV gB and the mutant forms of the protein utilized in this study. The locations of cleavage site at aa 460 and AD-1 (aa 552 to 635) are both in the lumenal domain of gB. The mutant forms of gB are depicted below. The mutant forms of gB are designated by the last remaining amino acid in the truncated gB reading frame and thus correspond to the location of the C-terminal deletion.
FIG. 2.
Recognition of mutant ΔgB651 by AD-1-specific MAb 7-17. Wild-type (wt) gB and mutant ΔgB651 were expressed in monkey kidney cells infected with recombinant vaccinia viruses as described in Materials and Methods. (A) Following labeling with [35S]Met/Cys, infected cell proteins were immune precipitated with the AD-1-specific MAb 7-17, the gB oligomer-specific antibody 27-39, or antibody 58-15, which is directed at an epitope located at the extreme carboxyl terminus of gB, and the precipitated proteins were analyzed by SDS-PAGE under reducing conditions. The migration of the 150-kDa gB precursor protein of gB and the 120-kDa precursor protein of ΔgB651 is indicated in the margin. The migration of the 55-kDa TM cleavage product of gB and the 30-kDa cleavage product of the ΔgB651 mutant are also indicated. Note that ΔgB651 was not precipitated by MAb 58-15. (B) Supernatant from the infected cells was centrifuged at 13,000 × g for 20 min and then precipitated with MAb 7-17. Precipitated proteins were analyzed by SDS-PAGE as described above.
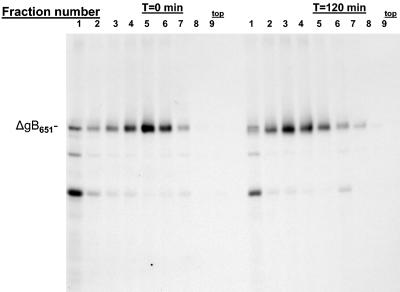
Our earlier findings demonstrated that HCMV gB undergoes oligomerization as part of its folding pathway and that the gB in infectious virions is oligomeric (14). Based on these previous findings and the observation that the MAb 27-39 reacted with ΔgB651, we predicted that this truncated form of gB would also undergo oligomerization as has been described for wild-type gB. Monkey kidney cells infected with a recombinant vaccinia virus expressing ΔgB651 were pulse-labeled with [35S]Met/Cys and then chased by the addition of media without radioactive amino acids. Infected cell proteins were solubilized in nonionic detergent and subjected to centrifugation in sucrose density gradients, and individual fractions were analyzed by immune precipitation with an anti-AD-1-specific antibody, 7-17. After a short pulse-labeling, the truncated ΔgB651 was most abundantly represented in fractions 4, 5, and 6, whereas after a 2-h chase, the ΔgB651 sedimented in fractions 2, 3, and 4 and most abundantly in fraction 3 of the gradient, indicating formation of higher-weight oligomeric forms of the protein (Fig. 3). This result together with the reactivity of ΔgB651 with MAb 7-17 and the conformation-specific antibody 27-39 demonstrated that deletion of the membrane-spanning domain and the cytoplasmic tail of gB did not alter AD-1 formation or oligomerization and folding of gB.
FIG. 3.
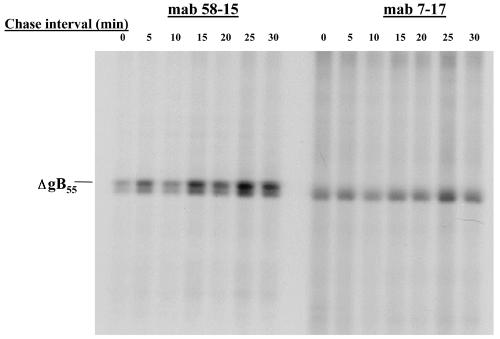
Oligomerization of ΔgB651. Monkey kidney cells were infected with a recombinant vaccinia virus expressing ΔgB651 and pulse-labeled with [35S]Met/Cys for 10 min. The cultures were then washed extensively and either harvested immediately (time [T] = 0 min) or incubated in media for 120 min. Infected cell proteins from both cultures were centrifuged through a 5 to 30% sucrose gradient for 16 h as described in Materials and Methods. One-milliliter fractions were collected from the bottom of the gradient and precipitated with MAb 7-17. Precipitated proteins were analyzed by SDS-PAGE. Note the increased concentration of precipitated ΔgB651 in fraction 3 in the later chase interval, indicating multimerization. The top of the gradient is indicated.
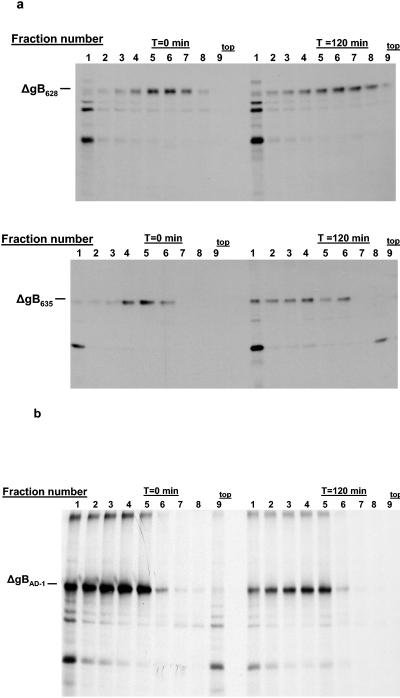
In the next series of experiments, we more precisely investigated sequence requirements for formation of the AD-1 domain and gB oligomerization by generating a panel of gB recombinant molecules with larger C-terminal truncations. As illustrated in Table 1, C-terminal deletions between aa 651 and 635 resulted in gB mutants that were recognized by AD-1-specific MAbs and these gB mutants could be shown to oligomerize (data not shown). In contrast, the loss of reactivity with AD-1-specific MAbs was also associated with a failure of the mutant protein to oligomerize (Table 1). This association between the formation of AD-1 and oligomerization is illustrated by the gB mutants ΔgB628, which was truncated at aa 628, and ΔgB635, which was truncated at aa 635. The ΔgB628 mutant protein was not recognized by MAb 7-17 but was reactive with the AD-2 MAb 758 (Table 1). In contrast, the ΔgB635 mutant protein was recognized by both the AD-1-specific MAb and MAb 758 (Table 1). When these mutant gBs were analyzed by sedimentation through a sucrose gradient followed by precipitation with MAb 758, we found that mutant ΔgB635 migrated into the gradient during the latter chase period, whereas ΔgB628 failed to form a higher-molecular-weight multimer, as illustrated by the lack of sedimentation into the gradient at later time points (Table 1; Fig. 4a). Thus, it appeared that formation of the AD-1 structure was required for oligomerization of gB, a finding consistent with earlier observations that reported that AD-1 forms early in the folding pathway of gB, prior to oligomerization of the molecule (2).
TABLE 1.
HCMV gB mutants utilized for study of the role of AD-1 in oligomerization
| gB mutant | Mutation | Phenotype
|
|||
|---|---|---|---|---|---|
| AD-1a | Mab 27-39b | Oligomerizationc | Cleavage | ||
| Wild type | None | + | + | + | + |
| ΔgB483 | Deletion of aa 484-907 | − | − | − | − |
| ΔgB781 | Deletion of aa 782-907 | + | + | + | + |
| ΔgB651 | Deletion of aa 652-907 | + | + | + | + |
| ΔgB642 | Deletion of aa 643-907 | + | +/− | + | NTe |
| ΔgB635 | Deletion of aa 636-907 | + | − | + | NT |
| ΔgB628 | Deletion of aa 629-907 | − | − | − | NT |
| ΔgBAD-1 | Deletion of aa 560-640 | − | − | − | NT |
| ΔgB55 | Deletion of aa 1-473d | + | − | − | NT |
| ΔgBc508s | Cys→Ser aa 508 | + | − | NT | − |
| ΔgBc550s | Cys→Ser aa 550 | + | − | NT | − |
| ΔgBc573s | Cys→Ser aa 573 | − | − | − | − |
| ΔgBc610s | Cys→Ser aa 610 | − | − | − | − |
Expression of AD-1 defined by reactivity with AD-1-specific Mabs, 7-17, 27-156, 27-180, and 27-78. gB expression was confirmed either by reactivity with the above antibodies, by Mab 58-15, which is directed at epitope in extreme C terminus, or by Mab 758, which is reactive with AD-2 in the amino terminus of gB.
Mab 27-39 has been shown to recognize a conformation-dependent binding site that is expressed on oligomeric forms of gB (14).
Oligomerization determined by sedimentation in sucrose gradients as described in Materials and Methods.
Although the ΔgB55 mutant was shown to be glycosylated, amino acid sequencing has not been carried out to determine if the authentic leader sequence of gB was cleaved during synthesis of this molecule.
NT, not tested.
FIG. 4.
Oligomerization of gB mutants. (a) gB mutants ΔgB628 and ΔgB635 were expressed in monkey kidney cells by infection with recombinant vaccinia viruses and analyzed as described in the legend to Fig. 3, except that the mutant protein was precipitated with MAb 758, which is directed at the NH2 terminus of gB. The mutant ΔgB628 protein failed to sediment further into the gradient at the later chase interval, indicating it did not form higher-molecular-weight multimers. In contrast, the ΔgB635 mutant protein formed higher-molecular-weight oligomers, as evidenced by its migration further into the gradient at the later chase interval. (b) gB mutant ΔgBAD-1 was expressed as a recombinant vaccinia virus and analyzed as described above. In this experiment, ΔgBAD-1 was precipitated with MAb 58-15, which is directed at the C terminus of gB. Note the similar distribution of the ΔgBAD-1 protein near the bottom of the gradient at both 0 and 120 min, indicating that this molecule likely aggregated shortly after its synthesis. The top of the gradient is indicated.
To further investigate the role of the AD-1 of gB in the oligomerization of this glycoprotein, we constructed a recombinant gB molecule with an internal deletion between aa 560 and 640. This deletion in the gB sequence resulted in a mutant gB molecule (gBΔAD-1) that contained the SU component of the gB, the transmembrane domain, and the cytoplasmic tail but lacked AD-1. MAbs reactive with the AD-2 epitope that is located in the extreme amino terminus of gB and MAb 58-15 were reactive with the gBΔAD-1 molecule, indicating that the complete coding sequence was correctly translated (Table 1). The AD-1-specific antibody 7-17 failed to react with gBΔAD-1 (Table 1). Furthermore, the oligomer and conformation-specific MAb 27-39 also failed to recognize the gBΔAD-1 molecule (Table 1). To confirm these findings, we determined if the gBΔAD-1 molecule could oligomerize in the absence of AD-1, using the gradient-based method described in the previous section. We identified species of gBΔAD-1 in both the pulse-labeling and pulse-chase periods that migrated further into the gradient than the oligomeric forms of ΔgB651; however, these species of gBΔAD-1 did not exhibit kinetics of multimerization, suggesting that they represented higher-molecular-weight aggregates that likely formed shortly after synthesis of this mutant form of gB (Fig. 4b). Together, the results presented in these experiments provided direct evidence that AD-1 of gB was essential for the oligomerization of gB in eukaryotic cells.
AD-1 is required for oligomerization of gB but is not sufficient for oligomer formation in the absence of the SU component of gB.
Our results thus far have suggested that AD-1 played an essential role in the gB assembly pathway and that in the absence of AD-1, oligomerization and the subsequent folding of gB failed to occur. Based on the results from studies of other viral glycoproteins, we questioned whether the TM component of gB which contained AD-1 was in itself sufficient to direct oligomerization of the TM component of gB in the absence of the SU component of this type I glycoprotein. To investigate this possibility, we constructed a gB mutant in which the TM components of gB (aa 473 to 903) were fused in frame with the native leader sequence of gB to generate a mutant designated ΔgB55. This recombinant molecule enabled us to study the role of the TM component of gB in oligomerization in the absence of the SU component of the molecule. Image analysis of ΔgB55 following transient expression revealed reactivity with the AD-1-specific MAb 7-17 and with the C-terminal-specific antibody 58-15, but not with antibodies directed at the amino terminus of the SU component of gB (Table 1).
We next investigated the expression of AD-1 in the ΔgB55 protein by a pulse-chase analysis of transiently expressed ΔgB55 followed by immune precipitation with either MAb 58-15 or with an AD-1-specific MAb, 7-17. The precipitated proteins were analyzed under nonreducing conditions to permit detection of disulfide-bonded forms of ΔgB55 as well as the presence of higher-molecular-weight oligomeric or aggregated forms of the protein. As shown in Fig. 5, MAb 58-15 precipitated two forms of Δgp55, whereas the AD-1-specific MAb 7-17 precipitated only the more rapidly migrating form. There were no higher-molecular-weight species detected suggesting that the ΔgB55 did not oligomerize (Fig. 5). Consistent with these results, the ΔgB55 did not oligomerize when analyzed by sedimentation in sucrose gradients, nor was it recognized by MAb 27-39 (Table 1). Interestingly, the signal from the more rapidly migrating form of the ΔgB55 precipitated by the AD-1-specific MAb also appeared to increase in intensity over the course of the chase, with an increase in its signal at chase intervals of 25 and 30 min (Fig. 5). This result suggested that the structure recognized by the AD-1-specific MAb formed during these short chase intervals. Together with the previously reported findings, these results were consistent with the expression of AD-1 by a structural intermediate early in the folding pathway of gB and suggested that formation of AD-1 defined one of the earliest steps in the maturational folding pathway of gB.
FIG. 5.
Pulse-chase analysis of the ΔgB55. The mutant ΔgB55 was expressed as a recombinant vaccinia virus, and infected monkey kidney cells were pulse-labeled for 2 min with [35S]Met/Cys and then chased in media for the indicated times. The infected cell proteins were precipitated with either MAb 58-15 or the AD-1-specific MAb 7-17, and the precipitated proteins were analyzed by SDS-PAGE under nonreducing conditions. The migration of the forms of ΔgB55 is indicated in the left margin.
The formation of AD-1 is dependent on disulfide bonding between Cys 573 and 610.
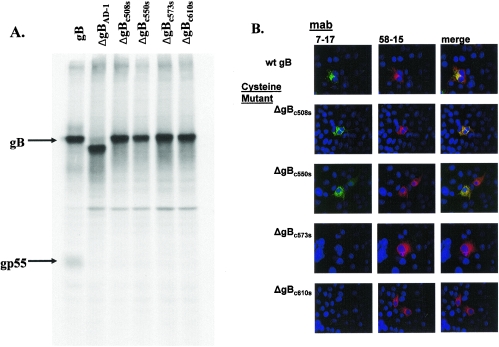
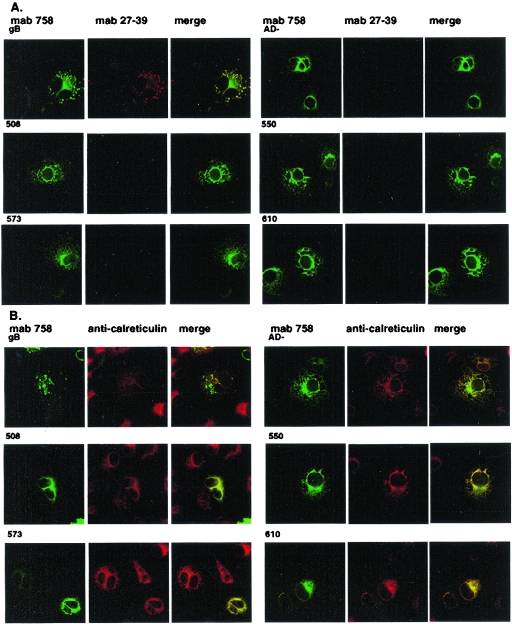
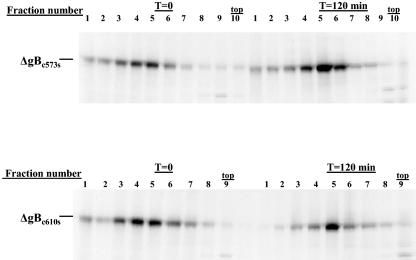
Previous studies utilizing prokaryote-derived fragments of gB have demonstrated that disulfide bonding between Cys 573 and 610 was required for the formation of AD-1 (22). Recent analysis of eukaryote-derived gB has also revealed that Cys 573 and 610 are disulfide bonded (22). We next examined the role of disulfide bonding within AD-1 in the folding of gB by mutating individual cysteines to serines at positions 508, 550, 573, and 610 (designated ΔgBc508s, ΔgBc550s, ΔgBc573s, and ΔgBc610s). When expressed in mammalian cells, all four recombinant proteins reacted with MAb 58-15 (Fig. 6). In contrast to the cleavage of wild-type gB, none of these mutant proteins appeared to be cleaved into the gp55 and gp116 components of gB, suggesting that none of these proteins were transported to the distal secretory compartment (Fig. 6). As predicted from the results of earlier studies using bacterially produced protein, only ΔgBc508s and ΔgBc550s were recognized by the AD-1-specific MAb (Table 1; Fig. 6). In addition, we determined the reactivity of MAb 27-39 for the cysteine mutants (ΔgBc508s, ΔgBc550s, ΔgBc573s, and ΔgBc610s), and the gBΔAD-1 mutant. Although each mutant gB was readily detectable by MAb 758, MAb 27-39 was reactive only with wild-type gB, indicating that the gB mutants failed to form native oligomers (Fig. 7). In agreement with this finding, the mutant forms of gB colocalized with calreticulin, a cellular marker for the endoplasmic reticulum (ER) (Fig. 7). These findings suggested that these gB mutant molecules failed to exit the ER and were not cleaved by host enzymatic activity in the more distal Golgi. We confirmed the effect of the mutation present in ΔgBc573s and ΔgBc610s on oligomerization of gB by transiently expressing these proteins in human embryonic kidney cells, followed by sedimentation of soluble proteins through sucrose gradients as described above. Under these conditions, neither gB Cys mutant formed oligomers (Fig. 8). These data indicated that formation of AD-1 required disulfide bonding between Cys 573 and 610 and in the absence of this disulfide bond, gB failed to oligomerize, acquire conformational epitopes as defined by MAb 27-39, and exit the ER.
FIG. 6.
Analysis of gB cysteine mutants. (A) Wild-type gB and gB mutants were transiently expressed in 293 human embryonic kidney cells as described in Materials and Methods. Following labeling with [35S]Met/Cys, the recombinant proteins were precipitated with MAb 58-15 and analyzed by SDS-PAGE under reducing conditions. Note that only the wild-type gB was cleaved as evidenced by the presence of the gp55 subunit. (B) Image analysis of gB mutants. gB cysteine mutants were generated as described in Materials and Methods and transiently expressed in COS-7 cells grown on glass coverslips. Following fixation in 3% paraformaldehyde, the cells were permeabilized and stained with the AD-1-specific MAb 7-17 or the C-terminal specific MAb 58-15 and developed with FITC-conjugated goat anti-mouse IgG3 or Texas red-conjugated goat anti-mouse IgG2b, respectively. Merged images reveal colocalization of signals resulting in a yellow signal. Wt, wild type.
FIG. 7.
gB cysteine mutants fail to exit the ER and are not recognized by oligomer-specific MAb 27-39. Wild-type gB and gB mutants ΔgBAD-1 (designated AD− in this figure), ΔgBc508s, ΔgBc550s, ΔgBc573s, and ΔgBc610s were transiently expressed in COS-7 cells grown on glass coverslips and fixed at 48 h in 3% paraformaldehyde. (A) The cells were stained with MAb 758, the oligomer-specific MAb 27-39, and developed with an FITC-labeled goat anti-human IgG or Texas red-conjugated goat anti-mouse IgG second antibody. Images were merged to determine colocalization. (B) The cells were stained with MAb 58-15 or with rabbit anti-calreticulin and developed with FITC-conjugated goat anti-mouse IgG or Texas red-labeled anti-rabbit IgG. Images were merged to determine colocalization.
FIG. 8.
Oligomerization of gB cysteine mutants ΔgBc573s and ΔgBc573s. Transiently expressed gB mutants ΔgBc573s and ΔgBc573s were pulse-labeled with [35S]Met/Cys and chased in media as described in Fig. 3. Following lysis, the labeled proteins were sedimented by centrifugation in 5 to 30% sucrose gradients for 16 h as described in Materials and Methods. The gradients were fractionated as described above, and individual fractions were precipitated with MAb 58-15. The precipitated proteins were analyzed by SDS-PAGE under reducing conditions. Note that neither protein migrated further into the gradient in the later time period, indicating that neither the ΔgBc573s nor ΔgBc610s mutant gB formed higher-molecular-weight multimers. The top of the gradient is indicated. T, time.
Construction of recombinant viruses containing mutations in AD-1.
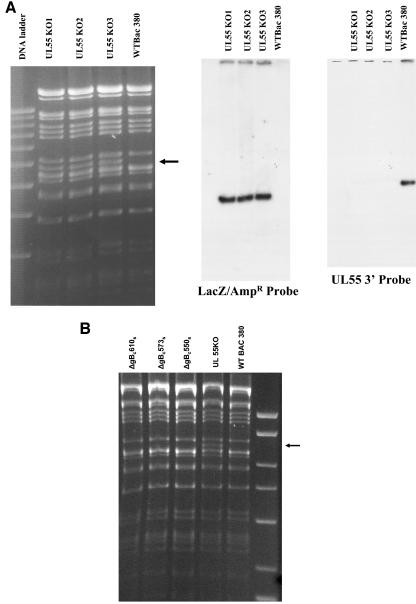
Our findings indicated that formation of AD-1 of gB was required for the oligomerization and early folding pathway of this glycoprotein. In addition, cysteines at aa 573 and 610 were required for AD-1 formation, and mutagenesis of these residues resulted in loss of AD-1-specific MAb reactivity for gB. To determine the importance of this domain in the assembly of infectious virus, we introduced a Cys→Ser mutations at aa 550, 573, and 610 of gB, using lambda phage RED locus-mediated recombination of an infectious clone of HCMV that was maintained as a BAC (4). We chose to insert single-amino-acid changes at these positions to minimize structural changes in gB as well in upstream sequences that could alter expression of the DNA polymerase (UL54) and to directly determine the importance of AD-1 in the generation of infectious virus. The mutation at position 550 was made to confirm the importance of this Cys in the formation of gB oligomers in the assembly of an infectious virion. To generate the mutant HCMV BAC, we initially inserted a LacZ/Ampr gene cassette (β-galactosidase and ampicillin resistance, respectively) into the gB gene between nucleotides (nt) 1370 and 1982 to disrupt the UL55 ORF that encoded gB. Restriction endonuclease digestion of the BAC DNA of three independent colonies of the gB deletion mutant (designated UL55KO) and the wild-type parental BAC revealed the appearance of new fragment corresponding to the contribution of the additional 600 bp from the LacZ/Ampr cassette in the EcoRI M fragment of wild-type AD169 BAC (Fig. 9A). A 32P-labeled probe generated from the LacZ/Ampr cassette hybridized with a single fragment corresponding to the 6.8-kb band in the UL55KO BAC DNA, indicating a single insertion of the LacZ/Ampr cassette into the gB gene (Fig. 9A). A probe specific for nt 1370 to 1982 of UL55 hybridized only with the wild-type BAC, indicating that these sequences of gB were not present in the UL55KO BAC DNAs (Fig. 9A). The sites of insertion of the LacZ/Ampr cassette into the gB sequence were also confirmed by nucleotide sequencing. To generate BACs repaired with mutations in codons 550, 573, and 610 in the gB sequence, we generated 800-bp PCR products that contained specific Cys→Ser mutations at the positions noted above or a gB wild-type sequence and then recombined these into the UL55KO BAC. Successful repair of the UL55K0 HCMV BACs, resulting in replacement of the LacZ/Ampr cassette with wild-type gB sequences or sequences encoding point mutations in gB, was demonstrated by EcoRI restriction endonuclease digestion of the repaired BACS (Fig. 9B). The absence of the approximate 6.8-kb restriction fragment in ΔgBc550s, ΔgBc573s, and ΔgBc610s mutant BACs that was characteristic of the UL55KO BAC resulted in a pattern of restriction fragments that was identical to the wild-type AD169 BAC, wild-type BAC 380 (Fig. 9B). These results were confirmed by probing the Southern transfer with a 32P-labeled LacZ/Ampr cassette and confirmed by sequence analysis of repaired BACs (data not shown). We next electroporated BAC-derived HCMV DNA containing specific mutations in the Cys codon at positions 550, 573, and 610 into human fibroblast cells and observed the monolayers for virus replication. Virus was produced only in cells electroporated with wild-type HCMV BAC DNA, indicating that disruption of AD-1 and loss of gB oligomerization resulted in a lethal mutation in HCMV (Table 2). Infectious virus was not recovered from the supernatant.
FIG. 9.
Insertional deletion of UL55 (gB) gene in HCMV BAC and repair with mutagenic PCR products. (A) The HCMV BAC HV-5 was mutagenized by recombination of a LacZ/Ampr cassette into the UL55 ORF as described previously (8). The LacZ/Ampr cassette was flanked on each side by 40 nt of the UL55 sequences beginning at position 1370 and ending at position 1982. Recombination of the cassette into the UL55 genes at these positions resulted in deletion of the intervening nucleotide. After recombination into the BAC, three independent colonies (UL555KO 1 to 3) and the wild-type parental BAC were analyzed by EcoRI restriction endonuclease digestion followed by Southern transfer and hybridization with either a 32P-labeled LacZ/Ampr probe or a 32P-labeled probe specific for the 3′ end of the UL55 gene (nt 1320 to 1982). Note the appearance of the new restriction fragment in the UL55KO BAC DNA in the ethidium-stained gel and the single insertion of the LacZ/Ampr cassette in the BAC UL55KO DNA. The deletion of nt 1320 to 1982 of the UL55 gene by recombination of the LacZ/Ampr cassette was demonstrated by the failure of the UL55 probe to hybridize with the UL55KO BAC DNA. (B) The UL55KO BAC was repaired with PCR products containing point mutations in codons 550, 573, and 610 to yield the mutant BACs designated ΔgB c550s, ΔgB c573s, and ΔgB c610s. Nucleotide sequence analysis confirmed the predicted mutations. DNA from the mutant BACs, the UL55 KO BAC, and the wild-type AD169 BAC (designated WT BAC 380 on the figure) were digested with EcoRI restriction endonuclease, and the resulting DNA fragments were separated on a 1% agarose gel and stained with ethidium bromide. The migration of the mutant BAC DNA is identical to that of the wild-type BAC 380, and the repair of UL55KO BAC is demonstrated by the loss of the additional restriction fragment at approximately 6.8 kb, designated by the arrow in the right margin. The migration of DNA standards (10, 8, 6, 5, 4, and 3 kb) is shown in the far right lane.
TABLE 2.
Recovery of infectious virus from HCMV BAC
| Mutation | Virus recoverya |
|---|---|
| Wild type | + |
| UL55KO(ΔgB) | − |
| ΔgBc550s(TGC→TCC) | − |
| ΔgBc573s(TGC→TCC) | − |
| ΔgBc610s(TGT→TCT) | − |
Approximately 5 μg of BAC DNA and 1 μg of an expression plasmid encoding HCMV pp71 were electroporated into HF cells as described previously (8). Virus production was monitored by plaque formation and confirmed by Mab staining of cells scraped from the monolayer for late protein (pp28) production.
DISCUSSION
In this report, we have described an essential role of the AD-1 domain for HCMV gB in the oligomerization of this glycoprotein. Several lines of evidence have been presented, including the finding that recombinant gB molecules containing carboxyl-terminal deletions that extended into the AD-1 eliminated both the anti-AD-1-specific MAb recognition of the molecule as well as gB oligomerization. Similarly, when AD-1 was deleted from gB, the resulting molecule was not reactive with AD-1-specific MAbs and also failed to oligomerize. Engineering of a molecule that consisted of only the TM component of gB together with an intact AD-1 retained reactivity with AD-1-specific MAbs, but this molecule failed to oligomerize. Although we have not mapped additional sequences within gB that are required for oligomer formation, presumably these sequences are contained within the SU component of gB. Together these data argue that the AD-1 of gB is an essential structural feature of this glycoprotein that must form prior to oligomerization of the molecule and before gB can complete its folding pathway, as evidenced by the lack of reactivity of the oligomer-specific MAb 27-39 for gB mutants that lack AD-1. Mutations in Cys 573 and 610, two residues which have been shown to be disulfide bonded in gB, resulted in the loss of AD-1 and failure of the resulting gB molecules to oligomerize. When these specific mutations (ΔgBc573s and ΔgBc610s) were introduced into the HCMV genome, virus could not be recovered, indicating that gB oligomerization is required for virus replication. Although we confirmed that the introduction of point mutations at codons 550, 573, and 610 did not alter the nucleotide sequence of gB at positions other than those selected for mutation, we cannot say with absolute certainty other mutations that could affect recovery of infectious virus from these BACs was not introduced by this recombination. Having said this, it is important to note that we have made a large number of mutant HCMVs by this technique, including the repair of mutant sequences with wild-type sequences and have not observed additional deletions or insertions in the BAC. When deletion mutants containing the LacZ/Ampr cassette are repaired with wild-type sequences, recovered viruses exhibit similar growth kinetics in vitro as compared to the wild-type parent HCMV derived from the BAC. Thus, our data would argue that AD-1 represents an essential structural domain of gB and its formation is required for HCMV replication in vitro. We believe that mutations leading to the loss of AD-1 result in the lack of gB in the assembly compartment of virus-infected cells. The loss of gB in the envelope of HCMV almost certainly results in a viral particle that cannot infect HF cells, although there is little data to support this conjecture. In fact, it is unclear if a gB deletion mutant of HCMV can form intracellular particles.
In previous studies, we demonstrated that gB oligomerized as part of its folding program and, based on cosedimentation in sucrose gradients with molecular standards, reported that the oligomer consisted of a homodimer of gB (14). Furthermore, in a study utilizing both AD-1-specific and conformation-dependent anti-gB MAbs, we determined that AD-1 forms prior to acquisition of additional conformational determinants that are defined by a gB-specific MAb, 27-39 (2). Results presented in this report together with these previous findings argue that AD-1 represented a structural domain that defined an early folding intermediate of gB. This interpretation was in agreement with studies in which the synthesis of the ΔgB55 molecule was studied by a pulse-chase analysis. Findings from this experiment indicated that at least two forms of this molecule were synthesized very rapidly, but that only one form expressed AD-1. The identification of a second form of ΔgB55 that was not recognized by AD-1-specific MAbs suggested that both native and nonnative disulfide bonds were generated during the synthesis of the TM component of gB. The form of ΔgB55 that was reactive with anti-AD-1 MAbs exhibited kinetics of synthesis that suggested that it was derived from this pool of ΔgB55 molecules. Whether AD-1 was expressed only by a distinct subpopulation of these molecules and thus represented only a fraction of the newly synthesized gB or, alternatively, was derived by the remodeling of disulfide bonds present in forms of gB55 containing nonnative disulfide bonds is not known.
Studies from two laboratories have independently demonstrated that Cys 573 and 610 of gB form a disulfide loop (31, 46). In earlier studies from our laboratories, disulfide bonding within AD-1 was considered, but the initial studies that mapped AD-1 utilized fragments of gB expressed in E. coli, and these experiments were carried out under reducing conditions. Under these experimental conditions, we assumed that disulfide bonds were reduced and that AD-1-specific MAbs recognized linear sequences in this region of the molecule. Thus, it remained unclear whether the Cys in AD-1 represented antibody binding sites in the AD-1 epitope or whether the predicted Cys bridge was required for formation of the antibody binding site. Subsequent studies have demonstrated that the disulfide bond that forms between Cys 573 and 610 occurs very rapidly and can only be disrupted by the near continuous presence of an efficient reducing agent during its analysis (46). These findings provided an explanation for our previous inability to demonstrate the dependence of AD-1-specific MAb reactivity on the disulfide bonds in the domain of gB. Our current results have demonstrated that mutations of the Cys at either position 573 or 610 prevent recognition by an AD-1-specific MAb and provide definitive evidence that AD-1 is generated by the disulfide bond formation between these two amino acids. This result also demonstrated that MAb recognition of AD-1 cannot be explained by a sequence requirement for either Cys 573 or Cys 610 in a conventional linear antibody binding site.
A function for Cys at positions 573 and 610 in the formation of AD-1 and the subsequent oligomerization of gB has been suggested in previous studies (20). Eickmann et. al. generated recombinant molecules in which Cys→Ser mutations were introduced at position 508, 550, 573, or 610 (20). Although several results from our study confirmed those presented in this earlier report, our results also differed from this study in several key aspects. First, these investigators did not determine the effect of these mutations on the formation of AD-1; however, from our findings it is unlikely that mutations other than those at Cys 573 or 610 would have resulted in the loss of AD-1. Second, in contrast to their findings, we failed to see processing of ΔgBc508s or ΔgBc550s, as evidenced by cleavage into the SU and TM components of gB. Cleavage of gB has been shown to occur in a distal compartment of the secretory system that appears to be closely apposed to the trans-Golgi network (32). In our study, image analysis of these gB mutant molecules following transient expression in COS-7 cells revealed that they were retained within the ER and failed to colocalize with cellular markers of more distal sites in the secretory pathway (data not shown). Thus, our results suggested that these molecules folded into an altered conformation that prevented their export from the ER. Consistent with this observation, none of the cysteine mutants (ΔgBc508s, ΔgBc550s, ΔgBc573s, and ΔgBc610s) generated for this study were reactive with the conformation-specific MAb 27-39, a MAb that is reactive with a conformationally dependent epitope present on oligomeric forms of gB (14). Although there are a number of possible explanations for the discrepancy in the results from these two studies, a likely one is that the oligomerization of gB mutants was based on detection of higher-molecular-weight forms following SDS-PAGE in the absence of reducing agents in the study reported by Eickmann et al. (20). The finding of higher-molecular-weight forms of the gB cysteine mutants could reflect either oligomers of gB or aggregated, misfolded forms of this molecule. We would argue that the gB Cys mutations at positions 508 and 550 resulted in misfolded molecules that failed to exit the ER and did not oligomerize, based on their lack of reactivity with MAb 27-39. Similar findings have been reported by Singh et al., who described linker insertion mutants of gB with mutations outside of AD-1 that lead to loss of oligomerization and cleavage (44). Interestingly, these investigators also noted that a linker insertion mutant within AD-1 (aa 627) leads to loss of reactivity by an AD-1-specific MAb and loss of oligomer formation (44). This finding independently demonstrated that AD-1 formation was required for oligomer formation and transport of gB from the ER. However, our study did confirm some aspects of these earlier studies in that the Cys at positions 573 and 610 appeared essential for oligomerization of gB, presumably through formation of AD-1. We also found that the presence of AD-1 in the TM component of gB was in itself not sufficient for oligomerization and that an interaction between sequences in both the SU and the ectodomain of the TM component of the molecule were also required for oligomerization and folding of gB. This result was consistent with the location of Cys bridges between Cys 550 and Cys 94 and Cys 508 and Cys 111 of gB (31). Finally, and perhaps most importantly, our studies demonstrated that mutations that prevented AD-1 formation could result in the loss of gB function, as evidenced by the failure to recover virus from HCMV BACs containing Cys→Ser mutations at aa 573 and 610.
Studies of other type I viral glycoproteins have demonstrated the domains within the ectodomain of the TM component (lumenal domains) are essential for multimerization of monomeric forms of this glycoprotein through protein-protein interactions. In addition, early studies on glycoprotein transport demonstrated the requirement for oligomerization prior to transport from the ER for both influenza virus hemagglutinin (HA) and vesicular stomatitis virus (VSV) G protein (5, 26, 48). More recent examples of such interactions include gB of herpes simplex virus (HSV), the env glycoproteins of Rous sarcoma virus and murine leukemia virus and gp41 of human immunodeficiency virus (21, 40, 49, 54). Expression of the TM component of Rous sarcoma virus in the absence of the SU component resulted in a protein that was terminally glycosylated and that was transported out of the ER (21). In addition, the TM subunit was shown to oligomerize independently of expression of the SU component of this type I glycoprotein (21). Similar results have been noted in studies of human immunodeficiency virus (40, 54). Studies of HSV gB have indicated that a region in the lumenal domain of the molecule between aa 630 to 720 is essential for multimerization of the molecule (49). In addition, it was demonstrated that deletion of either the transmembrane or the cytoplasmic domain of HSV gB had no effect on its multimerization (49). Utilizing an assay that detected heterodimer formation, Laquerre et al. demonstrated that a 28-aa domain between aa 626 and 653 could direct oligomerization of gB (30). This 28-aa sequence of HSV is in the membrane-proximal region of the lumenal portion of the molecule, a position similar to that of the AD-1 region of HCMV gB. Interestingly, these investigators also demonstrated that this domain in HSV gB could be transplanted to either side of the membrane and still direct multimerization of the molecule (30). Furthermore, this region of HSV gB contains a conserved Cys at position 633 that was shown to contribute to oligomerization, but in contrast to the results from our studies in HCMV gB, was not absolutely required for oligomerization of HSV gB (30). A subsequent report from this same group demonstrated that the cysteine at position 633 was disulfide bonded to cysteine 596 and that this disulfide loop was required for both native folding and transport from the ER, findings that paralleled the results of our studies (29). However, in contrast to our results, these investigators suggested that this disulfide loop was not required for oligomerization but for postoligomerization folding. These authors have argued that residues between aa 626 and 653 of HSV gB direct multimerization of the molecule through hydrophobic interaction and not through any predicted structural motifs that have been shown to lead to multimerization in other proteins (30). It has been suggested that hydrophobic domains in HCMV that include AD-1 may represent the functionally homologous domain in HCMV (20). In contrast to the results from studies of HSV gB, HCMV gB oligomerization is dependent on disulfide bond formation between cysteines located in AD-1 and also on interactions between both the TM and SU component of the molecule. These results suggest that the function of AD-1 in oligomerization of gB may be one of initiating a protein-protein interaction between the two gB polyprotein precursors and that additional disulfide bonding between the remaining 14 Cys residues in gB and additional protein-protein interactions lead to oligomerization and folding.
The AD-1 of gB was first identified by the shared reactivity among a large number of anti-gB MAbs. These antibodies were generated by a variety of immunization methods, including immunization with purified virus, infected cells, and gel-purified gB. In all cases, antibodies reactive with AD-1 were isolated. In addition, HCMV-specific MAbs and immune serum from patients with HCMV infections were also shown to recognize AD-1 (25, 38, 45). A large panel of AD-1-specific MAbs was shown to contain both virus neutralizing and nonneutralizing antibodies, suggesting that substructures of AD-1 exist (45). In a study utilizing a series of recombinant AD-1 molecules in which single (and in some cases two and three) amino acids were mutated within AD-1, it was demonstrated that both murine and human antibodies recognized a complex pattern of antigenic substructures in AD-1 (45). This result suggested that although AD-1 has been described as a linear antibody binding site on gB, it is likely to be structurally complex and presents a unique conformation(s) to the host immune system. This group of conformationally dependent antibody binding sites have not been fully defined at this time but are thought to be dependent on both the amino acid sequences inside and outside of the loop formed by disulfide binding between Cys 573 and 610 (45). Interestingly, antibodies found in human convalescent-phase serum were also shown to be AD-1 specific with assays employing bacterially derived AD-1 and in antibody blocking experiments (45). From these data, it appeared that AD-1 represented a dominant and, perhaps the principal virus neutralizing antibody-binding determinant of gB (45). Morover, analysis of multiple clinical HCMV isolates with a panel of anti-AD-1 MAbs revealed conservation of this antibody binding site in all HCMV isolates thus far studied. Sequence comparison of the predicted amino acid sequence from clinical HCMV isolates has confirmed the sequence conservation in this region in HCMV gB. Thus, AD-1 is conserved even in the face of potent neutralizing antibody antibodies directed at binding determinants expressed on this domain (49). Sequence variations in other HCMV envelope glycoproteins have been reported, and at least in the case of HCMV gN (UL73), this variation has been suggested to reflect selective pressure, presumably secondary to the immune response to this glycoprotein (39). The conservation of AD-1 in the face of persistent antibody responses suggested that antibody responses to this region of gB could have little consequence in vivo in terms of modifying virus replication and spread. Alternatively, it could be argued that AD-1 is conserved among strains of HCMV because it has an essential role in gB folding and assembly of infectious virus. It is possible that variations in the amino acid sequence encoding AD-1 are infrequent because changes in primary amino acid sequence could lead to loss of AD-1 formation and failure of gB to oligomerize, fold, and be transported to sites of virus assembly.
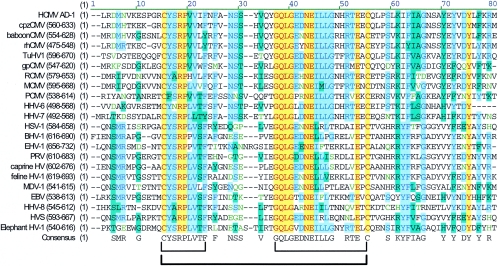
Similar primary sequences with positional homology are found in the gBs of other CMVs, such as rhesus CMV (rhCMV), murine CMV (MCMV), and guinea pig CMV (gpCMV). We have produced a virus neutralizing MAb reactive with gpCMV gB that appears to be reactive with an homologous region of gpCMV gB (11). Perhaps more relevant is the finding that several of the AD-1-specific MAbs derived against HCMV gB also reacted with rhCMV gB (28). In the latter case, the putative rhCMV AD-1 is approximately 67% identical to the amino acid sequence of HCMV AD-1 with primarily conservative amino acid substitutions accounting for the sequence divergence. The Cys at positions 573 and 610 of HCMV can be aligned to Cys 486 and 523 of rhCMV, and there is a remarkable conservation of amino acid sequence surrounding the Cys 610 of HCMV and Cys 523 of rhCMV. Furthermore, the cysteine loop between Cys 573 and 610 in HCMV is separated by 36 aa, the same number of amino acids between the cysteines that form the homologous loop of rhCMV gB, chimpanzee CMV gB, and MCMV gB. The amino acid sequences within the cysteine loops of HCMV and rhCMV are about 80% identical, with only 4 of 36 aa having nonconservative amino acid substitutions. In addition, the amino acid sequence following a conserved proline located at +4 from the Cys 573 of HCMV AD-1 is nearly identical for the gB of rhCMV. Similarly, the homologous region from the chimpanzee gB reveals an overall 87% identity and even greater conservation in this region with HCMV gB than the rhCMV. Thus, it is not surprising that AD-1 MAbs demonstrate significant cross-reactivity for primate AD-1. Together, these findings argue that AD-1 has an essential role in the structure or function of CMV gBs and provide at least one explanation for the conservation of AD-1 in gBs from primate and rodent CMVs. In fact, when we aligned the AD-1 sequence from HCMV gB with those from gBs from CMVs, other beta-herpesviruses, as well as the gBs from both alpha- and gamma-herpesviruses, we were surprised by the relative conservation of this region (Fig. 10). Of particular interest were the conservation of the cysteine and the proline at position +4 from the amino-terminal cytseine and the similar number of amino acids between the conserved cysteines (ranging from 35 to 39 residues, with 16 of 22 of the gBs having 36 aa between the conserved cysteines; Fig. 10). Interestingly, at least one study has shown that a cysteine loop forms between these two cysteines in HSV type 2 gB, and as noted earlier, this cysteine loop was required for native folding of the HSV gB and for production of infectious virus (29, 37). Furthermore, there is a remarkable conservation of the CYSRPL(V)V and GQLGEDNEILL consensus sequences within this intervening stretch of amino acids (Fig. 10). Although our study has only explored the function of this domain in HCMV gB, this degree of homology could suggest a similar function for this region of gB in these herpesviruses.
FIG. 10.
Alignment of HCMV AD-1 with homologous regions of other herpesviruses. The HCMV AD-1 amino acid sequence (aa 560 to 635) was aligned with a desktop alignment software (Vector NTi; Invitrogen, Carlsbad, Calif.) using available gB sequences. The location of the homologous region for each viral gB is shown in parentheses, and the computer-derived consensus sequence is displayed at the bottom. Note the conservation of the cysteines and two motifs in the intervening amino acid sequence indicated by brackets below the alignment table.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (M.M.) and by grants from the HHS, NIH, and NIAID (W.J.B. and M.A.J.).
REFERENCES
- 1.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billstrom, M. A., and W. J. Britt. 1995. Postoligomerization folding of human cytomegalovirus glycoprotein B: identification of folding intermediates and importance of disulfide bonding. J. Virol. 69:7015-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bold, S., M. Ohlin, W. Garten, and K. Radsak. 1996. Structural domains involved in human cytomegalovirus glycoprotein B-mediated cell-cell fusion. J. Gen. Virol. 77:2297-2302. [DOI] [PubMed] [Google Scholar]
- 4.Borst, E.-M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulay, F., R. W. Doms, R. G. Webster, and A. Helenius. 1988. Posttranslational oligomerization and cooperative acid activation of mixed influenza hemagglutinin trimers. J. Cell Biol. 106:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, K. A., and T. Compton. 1998. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J. Virol.. 72:1826-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt, W. J., M. Jarvis, J.-Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, N.Y.
- 10.Britt, W. J., and D. Auger. 1986. Synthesis and processing of the envelope gp55-116 complex of human cytomegalovirus. J. Virol. 58:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britt, W. J., and C. Harrison. 1994. Identification of an abundant disulfide-linked complex of glycoproteins in the envelope of guinea pig cytomegalovirus. Virology 201:294-302. [DOI] [PubMed] [Google Scholar]
- 12.Britt, W. J., and M. Mach. 1996. Human cytomegalovirus glycoproteins. Intervirology 39:401-412. [DOI] [PubMed] [Google Scholar]
- 13.Britt, W. J., L. Vugler, E. J. Butfiloski, and E. B. Stephens. 1990. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J. Virol. 64:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britt, W. J., and L. G. Vugler. 1992. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116). J. Virol. 66:6747-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brucher, K. H., W. Garten, H. D. Klenk, E. Shaw, and K. Radsak. 1990. Inhibition of endoproteolytic cleavage of cytomegalovirus (HCMV) glycoprotein B by palmitoyl-peptidyl-chloromethyl ketone. Virology 178:617-620. [DOI] [PubMed] [Google Scholar]
- 17.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769-779. [DOI] [PubMed] [Google Scholar]
- 19.Crump, C. M., C.-H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eickmann, M., R. Lange, M. Ohlin, M. Reschke, and K. Radsak. 1998. Effect of cysteine substitutions on dimerization and interfragment linkage of human cytomegalovirus glycoprotein B (gp UL55). Arch. Virol. 143:1865-1880. [DOI] [PubMed] [Google Scholar]
- 21.Einfeld, D. A., and E. Hunter. 1994. Expression of the TM protein of Rous sarcoma virus in the absence of SU shows that this domain is capable of oligomerization and intracellular transport. J. Virol. 68:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gicklhorn, D., M. Eickmann, G. Meyer, M. Ohlin, and K. Radsak. 2003. Differential effects of glycoprotein B epitope-specific antibodies on human cytomegalovirus-induced cell-cell fusion. J. Gen. Virol. 84:1859-1862. [DOI] [PubMed] [Google Scholar]
- 23.Henikoff, S. 1984. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351-359. [DOI] [PubMed] [Google Scholar]
- 24.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol.. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneiss, N., M. Mach, J. Fay, and W. J. Britt. 1991. Distribution of linear antigenic sites on glycoprotein gp55 of human cytomegalovirus. J. Virol. 65:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreis, T. E., and H. F. Lodish. 1986. Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell 46:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kropff, B., M. P. Landini, and M. Mach. 1993. An ELISA using recombinant proteins for the detection of neutralizing antibodies against human cytomegalovirus. J. Med. Virol. 39:187-195. [DOI] [PubMed] [Google Scholar]
- 28.Kropff, B., and M. Mach. 1997. Identification of the gene coding for rhesus cytomegalovirus glycoprotein B and immunological analysis of the protein. J. Gen. Virol. 78:1999-2007. [DOI] [PubMed] [Google Scholar]
- 29.Laquerre, S., D. B. Anderson, R. Argnani, and J. C. Glorioso. 1998. Herpes simplex virus type 1 glycoprotein B requires a cysteine residue at position 633 for folding, processing, and incorporation into mature infectious virus particles. J. Virol. 72:4940-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laquerre, S., S. Person, and J. C. Glorioso. 1996. Glycoprotein B of herpes simplex virus type 1 oligomerizes through the intermolecular interaction of a 28-amino-acid domain. J. Virol. 70:1640-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopper, M., and T. Compton. 2002. Disulfide bond configuration of human cytomegalovirus glycoprotein B. J. Virol. 76:6073-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, G. S., D. P. Fenger, G. G. Stout, M. E. Knights, and L. A. Hunt. 1996. Processing of human cytomegalovirus glycoprotein B in recombinant adenovirus-infected cells. J. Gen. Virol. 77:1549-1557. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, H., Y. Masuho, and M. Mach. 1990. The gp116 of the gp58/116 complex of human cytomegalovirus represents the amino-terminal part of the precursor molecule and contains a neutralizing epitope. J. Gen. Virol. 71:2443-2450. [DOI] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., and C. Tan Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 35.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding capacity of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Netterwald, J. R., T. R. Jones, W. J. Britt, S.-J. Yang, I. P. McCrone, and H. Zhu. 2004. Postattachment events associated with viral entry are necessary for induction of interferon-stimulated genes by human cytomegalovirus. J. Virol. 78:6688-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norais, N., D. Tang, S. Kaur, S. H. Chamberlain, F. R. Masiarz, R. L. Burke, and F. Marcus. 1996. Disulfide bonds of herpes simplex virus type 2 glycoprotein gB. J. Virol. 70:7379-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohlin, M., V.-A. Sundqvist, M. Mach, B. Wahren, and C. A. Borrebaeck. 1993. Fine specificity of the human immune response to the major neutralization epitopes expressed on cytomegalovirus gp58/116 (gB), as determined with human monoclonal antibodies. J. Virol. 67:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pignatelli, S., P. Dal Monte, G. Rossini, S. Chou, T. Gojobori, K. Hanada, J. J. Guo, W. Rawlinson, W. Britt, M. Mach, and M. P. Landini. 2003. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 84:647-655. [DOI] [PubMed] [Google Scholar]
- 40.Poumbourios, P., W. El Ahmar, D. A. McPhee, and B. E. Kemp. 1995. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J. Virol. 69:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qadri, I., D. Navarro, P. Paz, and L. Pereira. 1992. Assembly of conformation-dependent neutralizing domains on glycoprotein B of human cytomegalovirus. J. Gen. Virol. 73:2913-2921. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, J., and T. Compton. 2000. Characterization of a panel of insertion mutants in human cytomegalovirus glycoprotein B. J. Virol. 74:1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speckner, A., D. Glykofrydes, M. Ohlin, and M. Mach. 1999. Antigenic domain 1 of human cytomegalovirus glycoprotein B induces a multitude of different antibodies which, when combined, results in incomplete virus neutralization. J. Gen. Virol. 80:2183-2191. [DOI] [PubMed] [Google Scholar]
- 46.Speckner, A., B. Kropff, S. Knor, and M. Mach. 2000. The antigenic domain 1 of human cytomegalovirus glycoprotein B contains an intramolecular disulphide bond. J. Gen. Virol. 81:2659-2663. [DOI] [PubMed] [Google Scholar]
- 47.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S-2804S. [DOI] [PubMed] [Google Scholar]
- 48.Tatu, U., C. Hammond, and A. Helenius. 1995. Folding and oligomerization of influenza hemagglutinin in the ER and the intermediate compartment. EMBO J. 14:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tucker, S. P., R. V. Srinivas, and R. W. Compans. 1991. Molecular domains involved in oligomerization of the Friend murine leukemia virus envelope glycoprotein. Virology 185:710-720. [DOI] [PubMed] [Google Scholar]
- 50.Tugizov, S., D. Navarro, P. Paz, Y. Wang, I. Qadri, and L. Pereira. 1994. Function of human cytomegalovirus glycoprotein B: syncytium formation in cells constitutively expressing gB is blocked by virus-neutralizing antibodies. Virology 201:263-276. [DOI] [PubMed] [Google Scholar]
- 51.Utz, U., W. J. Britt, L. Vugler, and M. Mach. 1989. Identification of a neutralizing epitope on glycoprotein gp58 of human cytomegalovirus. J. Virol. 63:1995-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. (Erratum, 78:13395.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner, B., B. Kropff, H. Kalbacher, W. Britt, V.-A. Sundqvist, L. Ostberg, and M. Mach. 1992. A continuous sequence of more than 70 amino acids is essential for antibody binding to the dominant antigenic site of glycoprotein gp58 of human cytomegalovirus. J. Virol. 66:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weissenhorn, W., S. A. Wharton, L. J. Calder, P. L. Earl, B. Moss, E. Aliprandis, J. J. Skehel, and D. C. Wiley. 1996. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 15:1507-1514. [PMC free article] [PubMed] [Google Scholar]
- 55.Yurochko, A. D., E.-S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E.-S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]