Abstract
The TRIM5α proteins of humans and some Old World monkeys have been shown to block infection of particular retroviruses following virus entry into the host cell. Infection of most New World monkey cells by the simian immunodeficiency virus of macaques (SIVmac) is restricted at a similar point. Here we examine the antiretroviral activity of TRIM5α orthologs from humans, apes, Old World monkeys, and New World monkeys. Chimpanzee and orangutan TRIM5α proteins functionally resembled human TRIM5α, potently restricting infection by N-tropic murine leukemia virus (N-MLV) and moderately restricting human immunodeficiency virus type 1 (HIV-1) infection. Notably, TRIM5α proteins from several New World monkey species restricted infection by SIVmac and the SIV of African green monkeys, SIVagm. Spider monkey TRIM5α, which has an expanded B30.2 domain v3 region due to a tandem triplication, potently blocked infection by a range of retroviruses, including SIVmac, SIVagm, HIV-1, and N-MLV. Tandem duplications in the TRIM5α B30.2 domain v1 region of African green monkeys are also associated with broader antiretroviral activity. Thus, variation in TRIM5α proteins among primate species accounts for the observed patterns of postentry restrictions in cells from these animals. The TRIM5α proteins of some monkey species exhibit dramatic lengthening of particular B30.2 variable regions and an expanded range of susceptible retroviruses.
Following entry into the host cell, retroviruses must negotiate a series of processes to establish a permanent infection of the host cell. These include the uncoating of the viral core, reverse transcription, nuclear access, and integration of the viral DNA into the host genome (1, 45). Early, postentry restrictions to retrovirus infection can determine tropism at the species level (6, 14, 15, 32, 36). Infection by N-tropic murine leukemia virus (N-MLV), for example, is inefficient in most human cells and certain cell lines from African green monkeys (4, 39). Human immunodeficiency virus type 1 (HIV-1) encounters a postentry block in Old World monkeys, whereas simian immunodeficiency virus of macaques (SIVmac) is blocked in most New World monkey cells (14, 15, 32). These species-specific restrictions share several features. First, the block occurs prior to or concurrent with reverse transcription, which occurs in the cytoplasm of the host cell. At most, low levels of early reverse transcripts are made in restricted cells (7, 14, 23, 32). Second, the viral determinant of susceptibility to the block is the capsid protein (7, 11, 19, 25, 26, 39). Other capsid-binding proteins, such as cyclophilin A in the case of HIV-1, can modify the degree of the restriction (25, 27, 41). Third, the restriction is mediated by dominant host factors, the activity of which can be titrated by the introduction of virus-like particles containing proteolytically processed capsid proteins of the restricted viruses (3, 7, 10, 23, 25, 40).
These observations suggested a model in which host restriction factors interact, directly or indirectly, with the viral capsid and prevent its progression along the infection pathway. A tripartite motif (TRIM) protein, TRIM5α, was identified as the major factor in rhesus monkey cells restricting HIV-1 infection (37). The rhesus monkey TRIM5α (TRIM5αrh) was shown to confer potent resistance to HIV-1 infection in otherwise susceptible cells and was necessary for the maintenance of the block to the early phase of HIV-1 infection in Old World monkey cells (37). Cells expressing TRIM5αrh exhibited partial inhibition of SIVmac infection but were as susceptible as control cells to infection by Moloney murine leukemia virus (MoMLV) vectors (37). Humans express a protein, TRIM5αhu, that is 87% identical in amino acid sequence to the rhesus monkey protein, TRIM5αrh. Even when expressed at comparable levels, TRIM5αhu was less potent at suppressing HIV-1 and SIVmac infection than TRIM5αrh (12, 37, 47). Recently, TRIM5αhu was shown to be responsible for the postentry restriction of N-MLV in human cells (12, 17, 27, 47). TRIM5αrh was less effective than TRIM5αhu at blocking these murine leukemia viruses (27). The TRIM5α protein from African green monkeys, TRIM5αagm, inhibited N-MLV, HIV-1, and SIVmac infection (12, 17, 47). These observations underscore the importance of species-specific variation in TRIM5 orthologs to the ability to restrict infection by particular retroviruses.
The tripartite motif that defines the TRIM proteins includes a RING domain, B-box 2 domain, and coiled-coil domain (28). Some TRIM proteins, including TRIM5, contain a C-terminal B30.2 or SPRY domain (13, 28). Differential splicing of the TRIM5 primary transcript gives rise to the expression of several isoforms of the protein product (28). The TRIM5α isoform is the largest product (493 amino acid residues in humans) and contains the B30.2/SPRY domain. The other TRIM5 isoforms (γ and δ are the best substantiated of these) lack an intact B30.2/SPRY domain. The TRIM5γrh isoform does not inhibit HIV-1 or SIVmac infection (37). Thus, the B30.2 domain is critical for the ability of TRIM5α to mediate antiretroviral effects.
Studies of endogenous retroviral sequences indicate that vertebrates, including humans, have been exposed to retroviruses for many millions of years (2, 16, 18, 20-22, 34, 35, 42-44, 46). This long history of host-retrovirus coevolution might favor the selection of particular TRIM5α proteins that effectively suppress lethal retrovirus infections. Conversely, retroviral capsids may have evolved to minimize the impact of the TRIM5α protein produced by the favored host species. Here we clone the TRIM5 cDNAs of several primate species and study the antiretroviral activities of these TRIM5α variants.
MATERIALS AND METHODS
Cells for TRIM5 cDNA synthesis.
SQMK-FP (Pindak) cells from a Bolivian squirrel monkey (Saimiri boliviensis boliviensis), Vero cells from an African green monkey (Cercopithecus aethiops pygerythrus), COS-1 cells from another subspecies (Cercopithecus aethiops tantalus) of African green monkey, murine NIH3T3 cells, and human HeLa cells were obtained from the American Type Culture Collection. Primary rhesus lung fibroblasts were derived from a rhesus macaque (Macaca mulatta) (15). The Coriell Institute for Medical Research (Camden, N.J.) supplied the following cells: AG05352 from a black-handed spider monkey (Ateles geoffroyi), AG05308 from a red-chested mustached tamarin (Saguinus labiatus), AG06209 from a Sumatran orangutan (Pongo pygmaeus abelii), and GM03448 from a chimpanzee (Pan troglodytes verus).
TRIM5 cDNA cloning and sequencing.
First- and second-strand cDNA synthesis employed a cDNA synthesis kit (Clontech), using RNA prepared from the cells described above. Human and primate TRIM5 cDNAs encoding TRIM5α were amplified using TRIMf2, 5′-GCGGAATTCGCCATGGCTTCTGGAATCCTGGTT-3′, and TRIMr2, 5′-GCGATCGATGCCTCAAGAGCTTGGTGAGCACAG-3′.
Amplification was carried out using the Clontech Advantage PCR kit, thermocycling the reactions at 95°C for 30 s followed by 30 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 3 min. Amplified cDNAs were inserted into the pCR-BluntII-TOPO plasmid (Invitrogen) and sequenced using the following primers: M13f (−20), GTAAAACGACGGCCAGT; M13r (−21), AACAGCTATGACCATG; TRIMf3, GGAAGCTGACATCAGAGA; TRIMf4, GATAAGAGACAAGTGAGC; TRIMr3, TCTACCTCCCAGTAATG; and TRIMr4, TCCTTCTCCAGGTTTTGC.
Creation of cells stably expressing TRIM5α variants.
Retroviral vectors encoding the TRIM5α variants were created by using the pLPCX vector (37). The pLPCX-TRIM5α vectors contain only the amino acid-coding sequence of the TRIM5α cDNA. With the exception of human TRIM5α, the TRIM5α proteins possess N-terminal epitope tags derived from influenza virus hemagglutinin (HA). In the human TRIM5α construct, the HA tag is at the C terminus (37). Recombinant viruses were produced in 293T cells by cotransfecting these pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells (5, 15, 48). The resulting virus particles were used to transduce ∼1 × 105 HeLa and NIH3T3 cells in the presence of 5 μg of Polybrene/ml. Cells were selected in 1 μg of puromycin (Sigma)/ml.
Immunoblotting.
HA-tagged TRIM5α proteins expressed stably in HeLa cells were detected by Western blotting of whole-cell lysates. Cells were lysed in 150 mM NaCl, 50 mM Tris-Cl (pH 7.5), and 1% Triton X-100. The primary antibody for detection of the HA tag was HA.11 (Covance, Berkeley, Calif.). As a control, β-actin was detected with the C-2 antibody (Santa Cruz Biotechnology). Following the binding of primary antibody, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) and ECL reagent (Amersham).
Infection with viruses expressing GFP.
Recombinant viruses (HIV-1-GFP, SIVmac-GFP, N-MLV-GFP, B-MLV-GFP, and MoMLV-GFP) expressing the humanized Renilla reniformis green fluorescent protein (GFP) were prepared as previously described (27, 37). The SIVagm-GFP virus was created from the pSIVagm Tan-1 plasmid (33), which was provided by Marcelo Soares and Beatrice Hahn through the National Institutes of Health AIDS Research and Reference Reagent Program. A DNA fragment encoding GFP (Clontech) was inserted into XhoI and NotI sites in place of nef sequences 8900 to 9000 in the SIVagmTan-1 provirus. A deletion from nucleotides 6300 to 7100 was introduced into an env subclone from this provirus, and the Age I-XhoI fragment containing the env deletion was cloned back into the proviral plasmid. Recombinant viruses were produced by transfection of 293T cells with this plasmid, pHCMV-G, and a Rev-expressing plasmid, as described elsewhere (15, 27, 37). HIV-1-GFP, SIVmac-GFP, and SIVagm-GFP viral stocks were quantified by measuring reverse transcriptase activity, as described previously (29). The infectivity of the N-MLV-GFP, B-MLV-GFP, and MoMLV-GFP viruses was determined by titrating the virus stocks on 293T cells. For infection, 3 × 104 HeLa or NIH 3T3 cells expressing the TRIM5α variants were seeded in 24-well plates and incubated with 2 × 104 to 4 × 104 cpm of HIV-1-GFP or SIV-GFP viruses, or MLV-GFP viruses at a multiplicity of infection of 0.5, for 12 to 16 h. Cells were washed and returned to culture for 48 h and then subjected to fluorescence-activated cell sorter (FACS) analysis with a FACScan (Becton Dickinson).
Nucleotide sequence accession numbers.
The primate TRIM5 nucleotide sequences have been deposited in the GenBank database under accession numbers AY740612 to AY740618.
RESULTS
TRIM5α proteins from different primate species.
To investigate the functional importance of variation in the TRIM5α protein, TRIM5 cDNA was cloned from the cells of several primate species. In addition to the previously reported (12, 17, 37, 47) TRIM5 cDNAs from humans and Old World monkeys (rhesus monkeys and African green monkeys), TRIM5 cDNA was cloned from two ape species (chimpanzee and orangutan) and three New World monkeys (squirrel monkey, tamarin, and spider monkey). The predicted amino acid sequences of these primate TRIM5α orthologs are aligned in Fig. 1. We note a few features of the primate TRIM5α sequences potentially relevant to the function of these proteins. Key structural elements comprising the known TRIM domains, such as the cysteines and histidines of the RING and B-box 2 domains, are invariant among the primate TRIM5α proteins studied. The sequences of the RING, B-box 2, and coiled-coil domains align well, with lineage-specific variation in individual amino acid residues apparent (Fig. 1). In the B30.2 domain, interspecies variation in TRIM5α is dramatic. Sequence comparisons of the B30.2 domains of multiple TRIM proteins have revealed four variable regions, designated v1 to v4; in these variable regions, significant length polymorphism as well as individual residue variation occur (33a). The boundaries of v1, v2, and v3 are depicted in Fig. 1; the v4 region of primate TRIM5α proteins exhibits no length polymorphism (33a) and is not shown. Dramatic length polymorphism is observed in the v1 and v3 regions of primate TRIM5α proteins. Old World primate TRIM5α proteins possess relatively long v1 regions, whereas New World primate TRIM5α proteins exhibit relatively long v3 regions (33a). Of particular note, the TRIM5α proteins of African green monkeys and related monkeys possess unusually long v1 regions as a result of tandem sequence duplications (Fig. 1). Similarly, the extremely long v3 region of spider monkeys, a New World species, arises from a tandem triplication. In summary, lineage-specific variation characterizes primate TRIM5α proteins, particularly with respect to the B30.2 domains.
FIG. 1.
Primate TRIM5α sequences. The primary amino acid sequences of the TRIM5α proteins studied herein are aligned. The primate species of origin are indicated to the left of the sequences (agm, African green monkey; pyg, subspecies pygerythrus; tan, subspecies tantalus; m., monkey). Amino acid residues that exhibit variation among the TRIM5α proteins are colored blue in the hominoids, green in the Old World monkeys, and red in the New World monkeys. Gaps in the sequence are indicated by dots. The boundaries of the RING, B-box 2, coiled-coil, and B30.2(SPRY) domains are indicated. The locations of variable regions (v1, v2, and v3) within the B30.2(SPRY) domain are indicated (33a). The tandemly duplicated sequences within the v1 regions of African green monkey TRIM5α proteins are underlined in green and black. The tandemly triplicated sequences in the TRIM5α protein of spider monkeys are underlined in red. The v4 variable region is not marked.
Expression of the primate TRIM5α proteins.
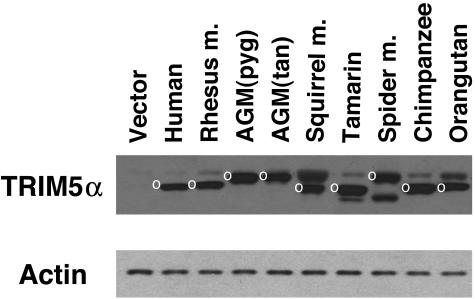
HeLa cells stably expressing the TRIM5α proteins of different primates were established. All of the TRIM5α proteins were engineered to contain an N-terminal epitope tag, except human TRIM5α, which possesses a C-terminal tag (37). As the antiretroviral activity of rhesus monkey TRIM5α has been shown to be slightly affected by the presence of a C-terminal epitope tag (37), we opted to use N-terminal tags for most of the primate TRIM5α constructs. To verify that the TRIM5α proteins were expressed, cell lysates were Western blotted with an antibody that recognizes the epitope tag. In parallel, the lysates were blotted and probed with an anti-β-actin antibody. The results indicated that all of the TRIM5α variants are expressed, and that the levels of expression are within two- to threefold of each other (Fig. 2). In addition to the major TRIM5α band, bands that are presumably the result of posttranslational modification were evident. The nature of these bands is under investigation.
FIG. 2.
Expression of the primate TRIM5α proteins. Lysates from HeLa cells stably expressing the indicated primate TRIM5α proteins, which contained HA epitope tags, were subjected to Western blotting with an antibody against HA. Control lysates from HeLa cells transduced with the empty pLPCX vector were analyzed in parallel (Vector). The major band corresponding to the TRIM5α protein is indicated by a white circle. The lysates were also Western blotted with an antibody directed against β-actin.
Restriction of primate immunodeficiency virus infection by primate TRIM5α proteins.
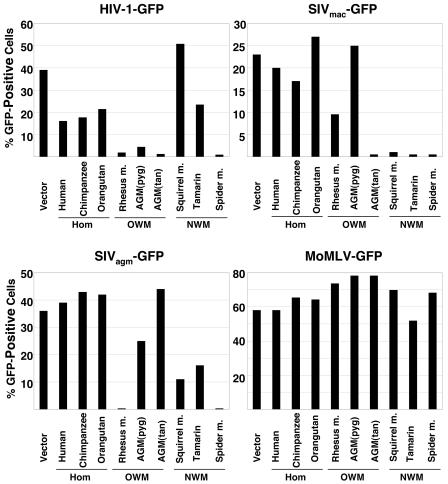
The HeLa cells expressing the TRIM5α proteins of different primate species and control HeLa cells transduced with the empty pLPCX vector were incubated with recombinant viruses containing the GFP gene. The recombinant viruses were derived from HIV-1, SIVmac, SIVagm, and MoMLV proviruses. The efficiency of infection was monitored by measuring the percentage of GFP-positive target cells.
As has been previously observed (37), human TRIM5α moderately inhibited HIV-1-GFP infection but did not significantly affect SIVmac-GFP infection (Fig. 3). Human TRIM5α did not inhibit SIVagm-GFP infection. The patterns of restriction observed for the chimpanzee and orangutan TRIM5α proteins were similar to that of human TRIM5α. Rhesus monkey TRIM5α potently restricted infection by HIV-1-GFP and SIVagm-GFP and partially blocked SIVmac-GFP infection. Both African green monkey TRIM5α proteins restricted HIV-1-GFP infection, but only TRIM5α from the C. aethiops tantalus subspecies blocked SIVmac-GFP infection. Neither African green monkey TRIM5α protein significantly inhibited infection by SIVagm-GFP viruses.
FIG. 3.
Effect of expression of the primate TRIM5α proteins on retrovirus infection. HeLa cells expressing TRIM5α proteins from the various primate species, or control HeLa cells transduced with the empty pLPCX vector (Vector), were incubated with the indicated recombinant viruses expressing GFP. Infected, GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. Hom, hominoid; OWM, Old World monkey; NWM, New World monkey.
The TRIM5α proteins from all three New World monkey species examined in this study potently restricted SIVmac-GFP infection (Fig. 3). The spider monkey TRIM5α protein also efficiently blocked HIV-1-GFP and SIVagm-GFP infections. Modest inhibition of SIVagm-GFP infection was observed for squirrel monkey and tamarin TRIM5α proteins; the latter TRIM5α protein also partially blocked HIV-1-GFP infection.
MoMLV-GFP infection was not significantly affected by expression of any of the primate TRIM5α proteins examined (Fig. 3), indicating the specificity of the observed inhibitory effects on infection by primate immunodeficiency viruses.
Restriction of N-MLV infection by primate TRIM5α proteins.
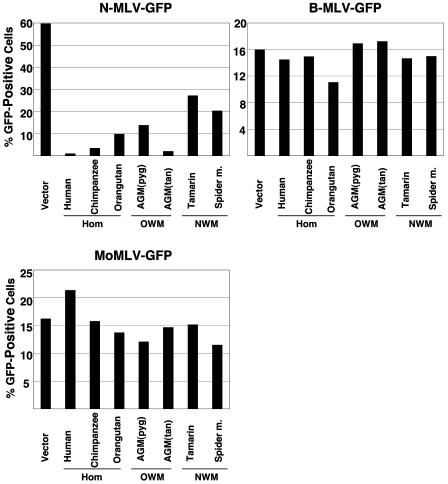
The NIH3T3 cells expressing the different primate TRIM5α proteins were incubated with recombinant murine leukemia viruses N-MLV-GFP, B-MLV-GFP, and MoMLV-GFP. The percentage of GFP-positive target cells was used as an indication of the efficiency of infection.
All of the primate TRIM5α proteins inhibited N-MLV-GFP infection, at least moderately (Fig. 4). The TRIM5α proteins from humans, chimpanzee, orangutan, and African green monkeys significantly reduced the efficiency of N-MLV-GFP infection. The New World monkey TRIM5α proteins less potently inhibited N-MLV-GFP infection. None of the TRIM5α proteins tested significantly affected the efficiency of B-MLV-GFP or MoMLV-GFP infection.
FIG. 4.
Effect of expression of the primate TRIM5α proteins on infection of different MLVs. NIH3T3 cells expressing TRIM5α proteins from the various primate species, or control HeLa cells transduced with the empty pLPCX vector (Vector), were incubated with the indicated recombinant viruses expressing GFP. Infected, GFP-positive cells were counted by FACS. The results of a typical experiment are shown. Similar results were obtained in three independent experiments. Hom, hominoid; OWM, Old World monkey; NWM, New World monkey.
DISCUSSION
Considerable variation among TRIM5α proteins of different primate species exists (33a). This observation is consistent with expectations of variation in host molecules that coevolve with the infectious agents targeted. Despite the observed variation, all of the cloned TRIM5α proteins retain the ability to suppress infection by at least one of the retroviruses tested. Key elements of the TRIM domains, such as the cysteines and histidines of the RING domain, are preserved in all of the primate TRIM5α proteins. RING domain cysteines have been shown to contribute to the anti-HIV-1 activity of rhesus monkey TRIM5α (37).
The patterns of postentry restrictions on the infection of several retroviruses in the cells of various primate species can be explained by TRIM5α variation. HIV-1 infection is blocked soon after entry into Old World monkey cells (14, 15, 32); the TRIM5α proteins from these monkeys potently restrict HIV-1 infection. SIVmac infection encounters postentry restrictions in most New World monkey cells (15). The TRIM5α proteins of three species from distinct branches of the New World monkey phylogenetic tree potently blocked SIVmac infection. Although New World monkeys have not been exhaustively characterized with respect to postentry restriction of immunodeficiency virus infection, owl monkeys represent the only known exception to the block to SIVmac in the New World lineage of monkeys (15). The owl monkey TRIM5 has recently been shown to be disrupted by a retrotransposon-mediated insertion, precluding expression of the complete TRIM5α protein in this species (24, 30). Combined with our data, this observation suggests that TRIM5α is the major postentry restriction factor for SIVmac in New World monkey cells. Given the current geographic isolation of New World monkeys from known SIVs, TRIM5α activity against SIVmac in these species may have been selected by past viral epidemics.
N-MLV infection is restricted at a postentry level in the cells of some but not all primates (4, 40). TRIM5α accounts for this restriction in the cells of humans and African green monkeys (12, 17, 27, 47). Rhesus monkey cells do not restrict N-MLV infection after virus entry (4, 39), consistent with the less potent ability of the TRIM5α protein from this species to block N-MLV infection. We observed that the TRIM5α proteins of chimpanzees and orangutans efficiently inhibited N-MLV infection, suggesting that the cells of some apes will restrict infection by this virus. A gibbon cell line has been reported to be susceptible to N-MLV infection (4, 39), suggesting that quantitative or qualitative differences in TRIM5α expression or antiviral function in some ape cells exist. The TRIM5α proteins of New World monkeys exhibited only moderate inhibition of N-MLV infection. This is consistent with the reported ability of marmoset cells to support the early, postentry steps in N-MLV infection (4, 39). As expected, owl monkey cells, which do not express a complete TRIM5α protein (24, 30), are also susceptible to infection by recombinant N-MLV vectors. Further work will be required to determine if the sporadic restriction of N-MLV infection observed in some mammalian species other than primates (4, 39) is mediated by TRIM5-related proteins.
Some of the observations made in this and related studies (12, 17, 27, 47) support a role for TRIM5α in determining the species tropism of retroviruses in vivo. In no instance does a retrovirus naturally infect a host that encodes a TRIM5α protein with potent restricting activity against the virus. The ubiquitous expression of the TRIM5α protein in many cells of the host (28) probably contributes to the strength of this correlation. Moreover, it appears that naturally infecting immunodeficiency viruses may have evolved to reduce the impact of the TRIM5α protein of the preferred host species to an acceptable minimum. Thus, HIV-1 demonstrates only moderate sensitivity to the TRIM5α proteins of humans and chimpanzees. SIVagm is relatively resistant to inhibition by the TRIM5α proteins from two subspecies of African green monkey. Likewise, SIVmac is only moderately inhibited by the rhesus monkey TRIM5α protein; although rhesus macaques are an Asian species and not naturally infected by SIVs, they are related to mangabeys (Cercocebus species), the natural host of SIVsmm, from which SIVmac was derived (8, 9, 31). The TRIM5α proteins of rhesus macaques and mangabeys are phylogenetically related (33a).
The TRIM5α proteins of the different primate species were all efficiently expressed in the transduced HeLa cells. Therefore, most of the observed specificity for particular retroviruses likely arises from intrinsic differences in the TRIM5α proteins themselves. The most dramatic variation among primate TRIM5α proteins occurs in the B30.2 (SPRY) domain, which has been shown to be essential for the ability of TRIM5αrh to restrict HIV-1 infection (37). Comparative analysis of multiple TRIM proteins has revealed the existence of four variable regions within the B30.2 domain (33a). Considerable length polymorphism in the v1 and v3 variable regions characterizes the TRIM5α proteins of primates. The v1 region of Old World primates is relatively long (33a); the v1 region of TRIM5αrh has been shown to be the major determinant of anti-HIV-1 potency of that protein (38). The v3 region of New World primates is relatively long (33a); this feature might contribute to the potent restriction of SIVmac observed for these TRIM5α variants. An attractive model is that the variable regions of the B30.2 domain represent surface-exposed loops that mediate contacts with the targeted viral capsid. Such a model is supported by the observation that owl monkeys do not express TRIM5α but instead express TRIMCyp, a fusion protein in which the TRIM5α B30.2 domain is replaced by cyclophilin A (24, 30). TRIMCyp acquires the ability to restrict HIV-1 infection by virtue of the binding of the HIV-1 capsid by the cyclophilin A moiety (24, 30). Thus, carboxy-terminal elements that contact retroviral capsids may be functionally complemented for antiviral activity by the TRIM5 amino-terminal elements.
Consistent with a contribution of the B30.2 variable regions to antiretroviral specificity is the observation that the TRIM5α variants with the broadest range of targeted viruses exhibited unusually long variable regions. The African green monkey TRIM5α v1 region is 46 residues long, 18 residues longer than the corresponding region of most Old World monkey TRIM5α proteins (33a). African green monkey TRIM5α variants can restrict HIV-1, SIVmac, N-MLV, and equine infectious anemia virus (12, 17, 47). These restrictions, although broad in range, are not nonspecific, as the SIVagm virus, as well as B-MLV and MoMLV, are not inhibited. Moreover, SIVmac infection is potently blocked by TRIM5αagm(tan) but is not affected by the closely related TRIM5αagm(pyg). Thus, subtle sequence differences between viruses and TRIM5α proteins can exert significant effects on the efficiency of restriction.
The spider monkey TRIM5α v3 region is 96 residues long, much larger than the length of 41 residues typical of New World monkey v3 regions (33a). Like the long v1 region of the African green monkey TRIM5α protein, the spider monkey TRIM5α v3 region apparently arose by the generation of tandem repeat sequences. The spider monkey TRIM5α protein was able to inhibit HIV-1, SIVmac, SIVagm and, partially, N-MLV. Long B30.2 variable loops may provide flexible structures and multiple sequence motifs, allowing interaction with several different viral capsids.
Future studies should clarify the interactions of TRIM5α proteins and retrovirus capsids and might suggest avenues for intervention in retroviral infections.
Acknowledgments
We thank Yvette McLaughlin and Sheri Farnum for manuscript preparation.
This work was supported by grants from the National Institutes of Health (HL54785 and AI063987) and by a Center for AIDS Research grant (PO30 AI28691). We also acknowledge the support of the Bristol-Myers Squibb Foundation, the International AIDS Vaccine Initiative, and the late William F. McCarty-Cooper.
REFERENCES
- 1.Artes, E. J., and M. A. Wainberg. 1996. Human immunodeficiency type 1 reverse transcriptase and early events in reverse transcription. Adv. Virus Res. 46:97-163. [DOI] [PubMed] [Google Scholar]
- 2.Benveniste, R. E., R. Heinemann, G. L. Wilson, R. Callahan, and G. J. Todaro. 1974. Detection of baboon type C viral sequences in various primate tissues by molecular hybridization. J. Virol. 14:56-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besnier, C., L. Ylinen, B. Strang, A. Lister, Y. Takeuchi, S. P. Goff, and G. J. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer, W. R., M. Westphal, W. Ostertag, and D. von Laer. 2002. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range. J. Virol. 76:1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 7.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, M. D., N. L. Letvin, N. W. King, M. Kannagi, P. K. Sehgal, R. D. Hunt, P. J. Kanki, M. Essex, and R. C. Desrosiers. 1985. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228:1201-1204. [DOI] [PubMed] [Google Scholar]
- 9.Desrosiers, R. C. 1990. The simian immunodeficiency viruses. Annu. Rev. Immunol. 8:557-578. [DOI] [PubMed] [Google Scholar]
- 10.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Refl. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatziioannou, T., S. Cowan, U. K. von Schwedler, W. I. Sundquist, and P. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry, J., I. H. Mather, M. F. McDermott, and P. Pontarotti. 1998. B30.2-like domain proteins: update and new insights into a rapidly expanding family of proteins. Mol. Biol. Evol. 15:1696-1705. [DOI] [PubMed] [Google Scholar]
- 14.Himathongkham, S., and P. A. Luciw. 1996. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219:485-488. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, W. E., and J. M. Coffin. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. USA 96:10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keckesova, Z., L. M. J. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. S., O. Takenaka, and T. J. Crow. 1999. Isolation and phylogeny of endogenous retrovirus sequences belonging to the HERV-W family in primates. J. Gen. Virol. 80:2613-2619. [DOI] [PubMed] [Google Scholar]
- 19.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 200:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mager, D. L., and J. D. Freeman. 1995. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology 213:395-404. [DOI] [PubMed] [Google Scholar]
- 21.Mang, R., J. Maas, A. C. van der Kuyl, and J. Goudsmit. 2000. Papio cynocephalus endogenous retrovirus among Old World monkeys: evidence for coevolution and ancient cross-species transmission. J. Virol. 74:1578-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariana-Costantini, R., T. M. Horn, and R. Callahan. 1989. Ancestry of a human endogenous retrovirus family. J. Virol. 63:4982-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munk, C., S. M. Brandt, G. Luccero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to post-entry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perron, M., M. Stremlau, B. Song, W. Ulm, R. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rho, H. M., B. Poiesz, F. W. Ruscetti, and R. C. Gallo. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355-360. [DOI] [PubMed] [Google Scholar]
- 30.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 31.Sharp, P. M., E. Bailes, R. R. Chaudhuri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of acquired immune deficiency syndrome viruses: where and when? Philos. Trans. R. Soc. London B 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata, R., H. Sakai, M. Kawamura, K. Tokunaga, and A. Adachi. 1995. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76:2723-2730. [DOI] [PubMed] [Google Scholar]
- 33.Soares, M., D. L. Robertson, H. Hui, J. S. Allan, G. M. Shaw, and B. H. Hahn. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394-399. [DOI] [PubMed] [Google Scholar]
- 33a.Song, B., B. Gold, C. O’hUigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 34.Steele, P. E., M. A. Martin, A. B. Rabson, T. Bryan, and S. J. O'Brien. 1986. Amplification and chromosomal dispersion of human endogenous retroviral sequences. J. Virol. 59:545-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinhuber, S., M. Brack, G. Hunsmann, H. Schwelberger, M. P. Dierich, and W. Vogetseder. 1995. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum. Genet. 96:188-192. [DOI] [PubMed] [Google Scholar]
- 36.Stoye, J. P. 2002. An intracellular block to primate lentivirus replication. Proc. Natl. Acad. Sci. USA 99:11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts human immunodeficiency virus (HIV-1) infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 38.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 retrovirus restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 42.Tristem, M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 74:3715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1995. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J. Virol. 69:7877-7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Kuyl, A. C., R. Mang, J. T. Dekker, and J. Goudsmit. 1997. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J. Virol. 71:3666-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson, D. A., D. L. Mager, and J.-A. C. Leong. 1994. Endogenous human retroviruses, p. 465-535. In J. A. Levy (ed.), The Retroviridae, vol. III. Plenum Press, New York, N.Y. [Google Scholar]
- 47.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5 protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]