Abstract
Objectives:
People with HIV-1 (PWH) on effective antiretroviral therapy (ART) continue to exhibit chronic systemic inflammation, immune activation, and persistent elevations in markers of HIV-1 infection [including HIV-DNA, cell-associated HIV-RNA (CA HIV-RNA), and antibodies to HIV-1 proteins] despite prolonged suppression of plasma HIV-RNA levels less than 50 copies/ml. Here, we investigated the hypothesis that nonreplicating but transcriptionally and translationally competent ‘defective’ HIV-1 proviruses may be one of drivers of these phenomena.
Design:
A combined cohort of 23 viremic and virologically suppressed individuals on ART were studied.
Methods:
HIV-DNA, CA HIV-RNA, western blot score (measure of anti-HIV-1 antibodies as a surrogate for viral protein expression in vivo), and key biomarkers of inflammation and coagulation (IL-6, hsCRP, TNF-alpha, tissue factor, and D-dimer) were measured in peripheral blood and analyzed using a combined cross-sectional and longitudinal approaches. Sequences of HIV-DNA and CA HIV-RNA obtained via 5′-LTR-to-3′-LTR PCR and single-genome sequencing were also analyzed.
Results:
We observed similar long-term persistence of multiple, unique, transcriptionally active ‘defective’ HIV-1 provirus clones (average: 11 years., range: 4–20 years) and antibody responses against HIV-1 viral proteins among all ART-treated participants evaluated. A direct correlation was observed between the magnitude of HIV-1 western blot score and the levels of transcription of ‘defective’ HIV-1 proviruses (r = 0.73, P < 0.01). Additional correlations were noted between total CD8+ T-cell counts and HIV-DNA (r = 0.52, P = 0.01) or CA HIV-RNA (r = 0.65, P < 0.01).
Conclusion:
These findings suggest a novel interplay between transcription and translation of ‘defective’ HIV-1 proviruses and the persistent immune activation seen in the setting of treated chronic HIV-1 infection.
Keywords: antibodies to viral proteins, HIV-1 defective proviruses, HIV-RNA transcripts, immune activation, inflammation, persistence
Introduction
The advent, refinement, and widespread implementation of antiretroviral therapy (ART) has dramatically reduced the morbidity and mortality for people with HIV (PWH) [1,2]. Though unable to fully eradicate or ‘cure’ chronic HIV-1 infection, the use of ART has successfully decreased the progression to AIDS and associated illnesses in the vast majority of treated individuals by blocking HIV-1 viral replication. However, despite these critical advances, PWH remain at a disproportionately higher risk of incident disease and death from serious non-AIDS complications when compared with age-matched uninfected individuals [1,3,4]. Among the conditions seen with increased frequency are a number of non-AIDS associated cancers and chronic cardiovascular, liver, and kidney diseases. This elevated risk is notably observed irrespective of sustained suppression of plasma viremia on appropriate ART and is associated with the now well-described phenomenon of elevated markers of chronic inflammation and immune activation as reflected in persistently elevated levels of IL-6 and D-dimer [5–10]. The reasons behind this remain unclear, and it is likely that mechanisms other than ongoing HIV-1 viral replication are responsible [11–13].
To date, the cause of HIV-associated chronic immune activation in virologically suppressed PWH remains elusive. Multiple processes are theorized to play a role; these include microbial translocation, chronic co-infection with other pathogens such as cytomegalovirus (CMV), host genetics, lifestyle factors (e.g. smoking), side effects of drugs, and persistence of low-level HIV-1 replication that is not ‘detectable’ via standard clinical assays [14–17]. We have hypothesized that another potential source of this chronic inflammation may be the persistence, expansion, and biologic activity of replication-incompetent, yet transcriptionally competent ‘defective’ HIV-1 proviruses [18,19]. These ‘defective’ proviruses comprise the majority of the HIV-1 provirus (HIV-DNA) pool in peripheral blood and persist despite years or even decades of continued use of suppressive ART [18–28]. Although previously suspected to be irrelevant ‘silent’ species based on their inability to reproduce infectious virions, it has been shown that these proviruses do in fact result in both transcriptional and translational activities, capable of producing cell-associated HIV-RNA (CA HIV-RNA) transcripts and expressing novel HIV-1-related proteins in vivo or in vitro[18,29–32]. This latter finding provides a potential mechanism(s) by which these proviruses may chronically perturb both the innate and adaptive elements of the host's immune system and contribute to persistent immune activation.
A better understanding of chronic immune activation and inflammation during ART is key to identifying ways to minimize it and by extension improve the health and quality of life for PWH. In the present study, we have extended our prior observations that defective HIV-1 proviruses are transcriptionally active [18] and encode viral proteins [29] to provide evidence that ongoing production of HIV-1 proteins from non-full-length, defective HIV-1 proviruses is a potential source of a persistent immune response to HIV-1; and that the biologic activity of these ‘defective’ HIV-1 proviruses is likely to play an important role in the persistent low-level immune activation and inflammation seen in PWH treated successfully with ART.
Materials and methods
Study design
Twenty-three participants enrolled in the National Institute of Allergy and Infectious Diseases Institutional Review Board-approved HIV-1 clinical research protocols formed the basis for this study (Supplementary Table 1). All provided written informed consent before study participation. Among these participants, 20 participants were sampled at a single timepoint during the course of their HIV-1 infection: five participants (Pts 1–5) were sampled prior to ART at a time when plasma HIV-RNA levels (pVL) were at least 50 copies/ml, and 15 participants (Pts 6–20) were sampled on ART with pVL less than 50 copies/ml (average duration of pVL less than 50 copies/ml = 5.7 years, range: 1.1–10.8 years) (Supplementary Figure S1). Three additional participants (Pts 21–23) were assessed longitudinally and sampled sequentially over 17 years (average: 17.7, range: 14.5–20.2), including prior to initiation of ART, at or within approximate 6 months of first documented virologic suppression after initiating of ART, and at approximate 3-year intervals thereafter during chronic infection with pVL less than 50 copies/ml.
Quantification and amplification of HIV-DNA and cell-associated HIV-RNA
Total HIV-DNA and CA HIV-RNA were isolated from peripheral blood mononuclear cells (PBMCs) using Gentra Puregene Cell Kit (Qiagen, Germantown, Maryland, USA) and the RNeasy Mini kit (Qiagen) with an on-column DNase I digestion step per manufacturer's instructions, respectively. Levels of HIV-DNA and CA HIV-RNA copy numbers were quantified with 7900HT Fast-Real time PCR system (ThermoFisher Scientific, Waltham, Massachusetts, USA) using primers and probes as previously reported [33,34]. HIV-1 copy numbers are expressed as copies per milliliter of blood. The limit of detection of the quantitative PCR (qPCR) is 2 copies/μg of genomic DNA for HIV-DNA and 10 copies/μg of cellular RNA for CA HIV-RNA. Genomic DNA and cDNA were subjected to limiting dilution before amplification with a near full-length PCR as previously described [18]. For participants 21–23, the following alternate reverse primers were used for RNA-PCR: -9591(HXB2 coordinate: 9591–9612) 5′-TGAGGCTTAAGCAGTGGGTTCC-3′ and -9567 (HXB2 coordinate: 9567–9588) 5′-AGTTAGCCAGAGAGCTCCCAGG-3′. On average, 24 (range 10–40) amplicons were obtained for each sampling time point.
Sequence analyses
Single-molecule direct sequencing was performed using the 3500xL Genetic Analyzer (Applied Biosystems by ThermoFisher Scientific) with a BigDye Terminator v3.1 Cycle Sequencing Kit. Intactness of the HIV-1 genome was assessed with the HIVAlign program (www.hiv.lanl.gov) [35]. AliView (v1.26) [36] was used to generate sequence alignments. ‘Full-length intact’ HIV-1s are defined as species that are intact in length (PCR amplicons of 9.0 kb in size for HIV-DNA; and those of 8.8 kb in size for CA HIV-RNA) and possess all nine intact protein-coding genes (gag, pol, env, nef, tat, rev, vpr, vif, vpu). ‘Full-length defective’ HIV-1s are intact in length but contain out-of-frame indels, premature stop codons, hypermutations, or inversions. ‘Defective’ HIV-1s are less than intact in length and may contain large internal deletions with/without out-of-frame indels, premature stop codons, hypermutations, or inversions.
Soluble biomarker and western blot analyses
D-dimer levels were measured by an enzyme-linked fluorescent assay on a VIDAS instrument (BioMerieux, Durham, North Carolina, USA) sCD14 by a traditional ELISA (R&D Systems, Minneapolis, Minnesota, USA), and IL-6, hsCRP, TNFα by an electrochemiluminescence assay (Meso Scale Discovery, Gaithersburg, Maryland, USA). Western blots were performed using the HIV-1 western blot kit (Cambridge Biotech, Worcester, Massachusetts, USA) for participants 1–20 (Pts 1–20); and the GS HIV-1 Western Blot kit (Bio-Rad, Hercules, California, USA) for participants 21–23 (Pts 21–23) following the manufacturer's specifications. Frozen serum samples were used for Pts 1–20 in the cross-sectional group; plasma samples were used for Pts 21–23 in the longitudinal group. Specific antibody responses were measured in 20 μl of serum samples against 9 (the Cambridge Biotech kit) or 10 (the Bio-Rad kit) different HIV-1 viral proteins, as previously described [37]. Densitometric analysis using ImageJ software [38] was performed to individually quantify the intensity of the bands relative to the intensity calculated in the corresponding band of the positive control. The intensities of the p51 and p55 bands were combined to a single value. Thus, the total western blot score was calculated as the sum of 8 or 9 bands, with a total score of 8 or 9 for the control.
Statistical analyses
Nonparametric rank-based correlations (Spearman) using the Prism software (version 9.3.1; and generalized linear regression using R software were performed for analyses. Tables and figures were constructed using the Prism and Adobe Illustrator software. All HIV-1 copy numbers and biomarker measurements fell within the range of quantitation as described above or per assay specification with the exception of three (out of 23 specimens) measurements of IL-6: These were below limit of detection and assigned the low cutoff value of 0.179 pg/ml for statistical calculations.
Results
Long-term persistence of cell-associated HIV-RNA and antibody responses to HIV-1 viral proteins in the absence of ongoing HIV-1 replication during prolonged suppression of plasma HIV-RNA on antiretroviral therapy
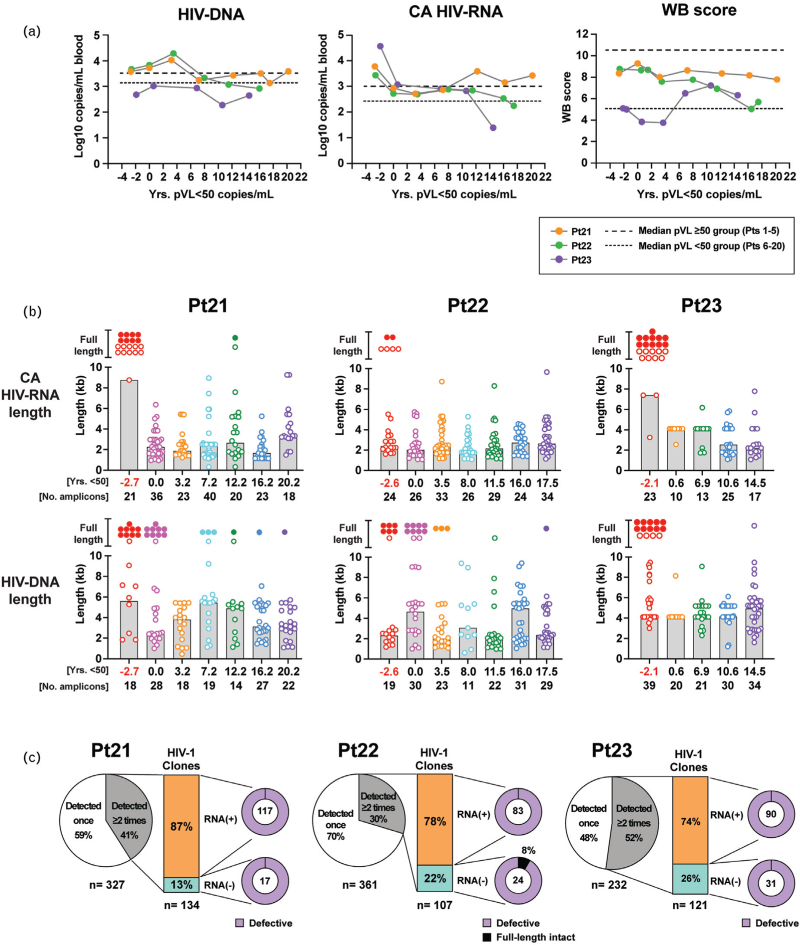
Levels of HIV-DNA, CA HIV-RNA, and western blot score remained virtually constant over a period of 15–22 years despite prolonged suppression of plasma HIV-RNA levels to less than 50 copies/ml on ART among the three longitudinally followed participants (Pts 21–23) (Fig. 1a). Similar findings were observed among the 20 individuals examined cross-sectionally (Supplementary Fig. S2). Levels of HIV-DNA indicate the precursor frequencies of cells that had been infected with HIV-1, whereas levels of CA HIV-RNA indicate the degree of transcription from these HIV-1 proviruses. The western blot score measure antibody responses to specific HIV-1 proteins and thus serves as a surrogate maker for the presence of those viral proteins in vivo. Longitudinal assessments of HIV-1 western blot scores demonstrated a long-term persistence of antibodies to HIV-1 viral proteins despite up to 20 years of plasma HIV-RNA levels less than 50 copies/ml (Fig. 1a, Supplementary Figures S3 and S4). Among the three participants in the longitudinal group, only one full-length intact HIV-RNA sequence was detected during virologic suppression (at 12.2 years in Pt 21) out of a total of 397 CA HIV-RNA sequences identified during HIV-RNA levels less than 50 copies/ml (Fig. 1b). These data suggest persistent exposure to HIV-1 viral proteins expressed by sources other than intact replicating viruses. Extensive sampling of HIV- DNA and CA HIV-RNA sequences from the entire cohort revealed an abundance of cells harboring defective proviruses and corresponding RNA transcripts both in the presence and absence of detectable plasma viremia (Fig. 1b, Supplementary Fig. S3b and c). For the three participants followed longitudinally (Pts 21–23), 30–52% of these unique proviruses were identified in more than one cell and thus were reflective of clonal expansion (Fig. 1c). Given the anticipated precursor frequencies of different CD4+ T-cell clones, it is reasonable to assume that the majority of cells detected in peripheral blood are the product of in-vivo clonal expansion.
Fig. 1.
Long-term persistence of antibody responses to HIV-1 antigens in the absence of active viral replication during plasma HIV-RNA less than 50 copies/ml on antiretroviral therapy.
(a) HIV-DNA levels, CA HIV-RNA levels, and western blot score were longitudinally analyzed for three participants (Pts 21–23). A color-coding scheme to indicate each participant is provided in the key. (b) Distributions of HIV-DNA and CA HIV-RNA sequence lengths for these three participants are shown. Filled circles represent full-length intact species; and open circles represent non-full-length HIV-1 species or full-length containing out-of-frame indels, premature stop codons, or hypermutations. The height of the shaded box represents the median length (kb) calculated for non-full-length HIV-1 species at a given timepoint. Duration of years with plasma HIV-RNA less than 50 copies/ml at time of sampling and numbers of amplicons obtained for HIV-DNA or CA HIV-RNA for each time point are indicated below the plots. Time points when plasma HIV-RNA level was at least 50 copies/ml are indicated in red. (c) Pie charts illustrate the relative abundance of expanded HIV-1 clones (i.e. those detected ≥2 times). Relative abundance of the expanded HIV-1 clonal populations either associated with HIV-RNA transcripts: RNA(+) or without HIV-RNA transcripts: RNA(−) are shown in bar-of-pie charts. Proportions of full-length intact and defective HIV-1 species associated with or without HIV-RNA transcripts are shown in doughnut pie charts.
This is further supported by the observation that as the number of cells/clones examined in a given individual increases, the fraction representing clonal expansions increases (Supplementary Fig. S5). Interestingly, among the clonally expanded proviral species, 74–87% of them were detected with their corresponding RNA transcripts, suggesting that transcriptionally-active ‘defective’ proviruses may be more common than previously thought [39,40].
Long-term persistence of multiple transcriptionally-active ‘defective’ HIV-1 provirus clones during suppressive antiretroviral therapy
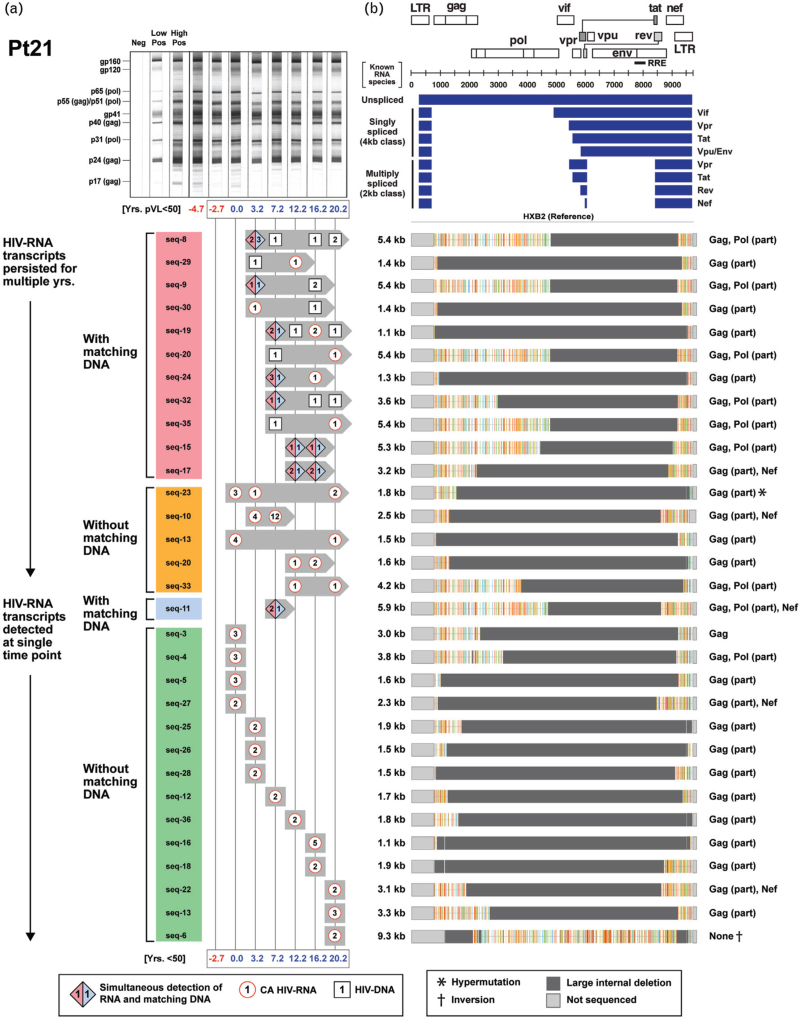
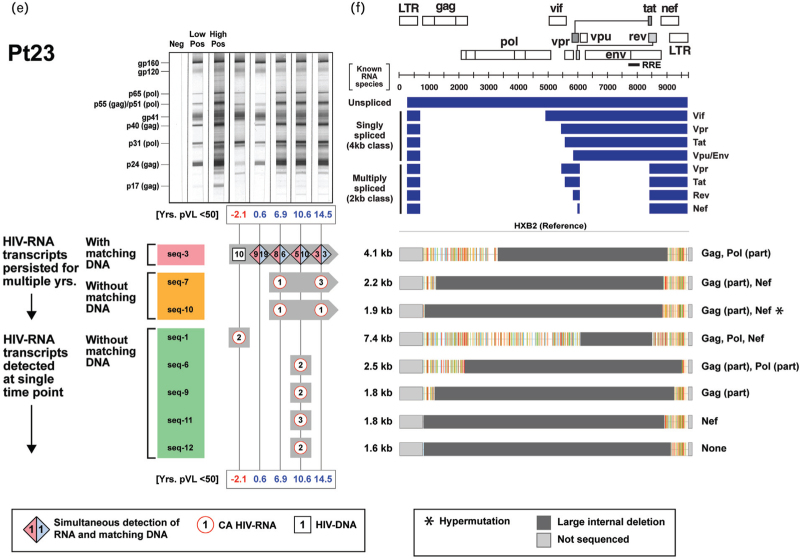
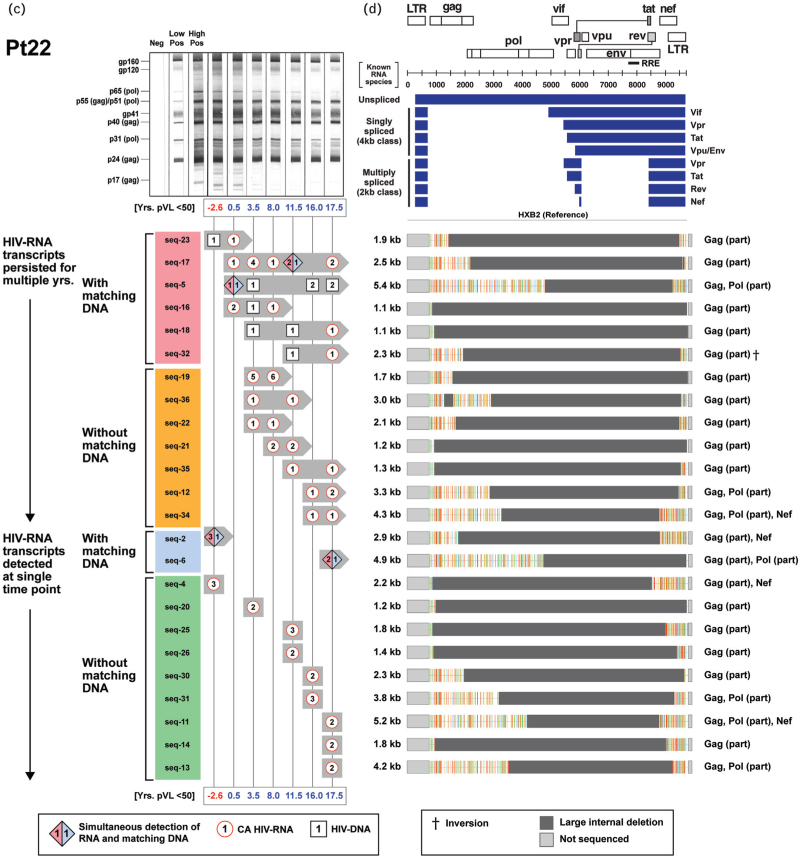
To determine whether any of the transcriptionally active ‘defective’ HIV-1 provirus clones possessed novel HIV-RNA structures capable of producing viral proteins in the setting of ART-induced prolonged viral suppression, we examined their genomes and monitored their persistence within each of the three participants in the longitudinal group (Fig. 2 ). We previously reported a case of 17-year-persistence of a transcriptionally-active ‘defective’ HIV-1 provirus clone in a PWH on suppressive ART [19]. In the present study, we were able to identify a total of 31 unique and transcriptionally active HIV-1 provirus clones in a single individual (Pt 21, Fig. 2 a). We further demonstrated long-term persistence of transcriptionally active expanded clones in all three individuals for an average of 11 years (range: 4–20 years) (Fig. 2 a, c, and e). Sequence analyses of the HIV-RNA transcripts from the expanded HIV-1 clones revealed that they were different from those of known spliced HIV-RNA species (Fig. 2 b, d, f). These novel HIV-RNA transcripts with open reading frames ranged in size from 1.1 to 7.4 kb. A part of the gag and nef genes were retained in 81% of these novel HIV-RNA transcripts (Fig. 2 b, d, f). The long-term persistence of these transcriptionally active ‘defective’ HIV-1 provirus clones was mirrored with a similar persistence of antibody responses directed against multiple HIV-1 viral proteins (Fig. 2 a, c, and e).
Fig. 2.
Long-term persistence of multiple transcriptionally active ‘defective’ HIV-1 provirus clones during suppression of plasma HIV-RNA less than 50 copies/ml on antiretroviral therapy.
(a, c, and e) Longitudinal changes in the number of sequences from expanded HIV-1 clones obtained from Pts 21–23 (detected as either HIV-RNA transcripts with or without corresponding HIV-DNA) and corresponding western blot patterns were assessed over the period of observation. Timepoints when plasma HIV-RNA level was at least 50 copies/ml are indicated in red. Distinct expanded provirus clones are assigned different sequence IDs. Symbols denote type of sequences detected at a given time point; numbers inside each symbol indicate the number of sequences detected at a given time point. (b, d, and f) Genome structures and protein-coding regions of the expanded HIV-1 clones are shown; the Highlighter analysis indicates nucleotide substitutions compared with HIV-1 HXB2. Schematic diagrams of known unspliced and spliced HIV-RNA species (represented by blue bars) are shown at the upper right panel for comparison.
Fig. 2 (Continued).
Long-term persistence of multiple transcriptionally active ‘defective’ HIV-1 provirus clones during suppression of plasma HIV-RNA less than 50 copies/ml on antiretroviral therapy.
(a, c, and e) Longitudinal changes in the number of sequences from expanded HIV-1 clones obtained from Pts 21–23 (detected as either HIV-RNA transcripts with or without corresponding HIV-DNA) and corresponding western blot patterns were assessed over the period of observation. Timepoints when plasma HIV-RNA level was at least 50 copies/ml are indicated in red. Distinct expanded provirus clones are assigned different sequence IDs. Symbols denote type of sequences detected at a given time point; numbers inside each symbol indicate the number of sequences detected at a given time point. (b, d, and f) Genome structures and protein-coding regions of the expanded HIV-1 clones are shown; the Highlighter analysis indicates nucleotide substitutions compared with HIV-1 HXB2. Schematic diagrams of known unspliced and spliced HIV-RNA species (represented by blue bars) are shown at the upper right panel for comparison.
Persistence and correlation between markers of HIV-1 transcription, protein production, and inflammation during prolonged suppression of plasma HIV-RNA levels less than 50 copies/ml on antiretroviral therapy
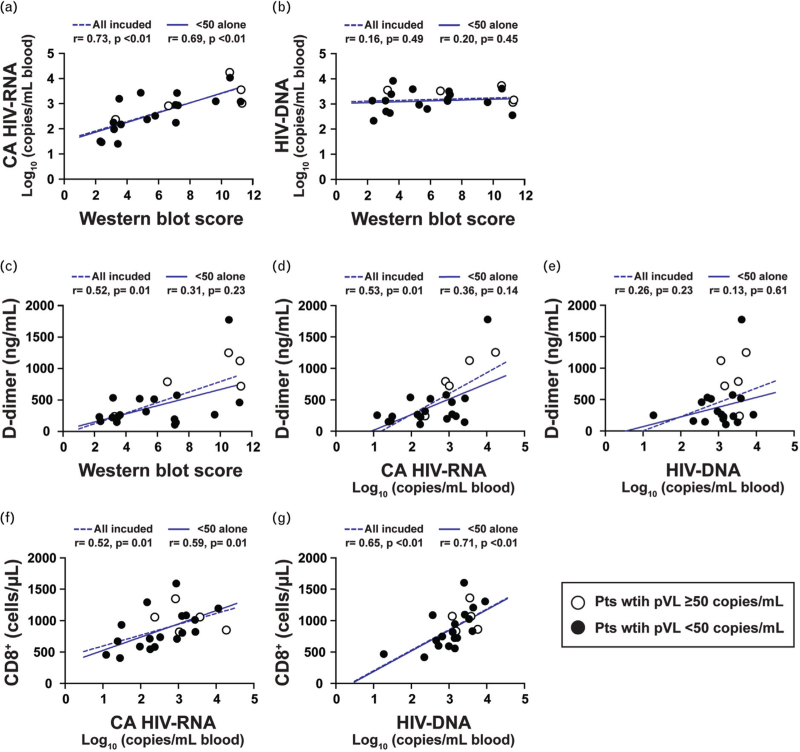
We next looked for relationships between HIV-DNA, CA HIV-RNA, western blot score, and a panel of biomarkers. With the exception of TNF-alpha, there was no decline in the biomarkers examined (IL-6, hsCRP, tissue factor, and D-dimer) when analyzed as a function of duration of viral suppression (Supplementary Figure S6). However, with the exception of D-dimer, they all remained elevated, compared with the levels observed in the HIV-negative controls (P < 0.05, Supplementary Fig. S6a–e). Consistent with the trends observed in the cross-sectional cohort, no significant changes in biomarker measurements were seen in up to 20 years of follow-up within the longitudinal cohort (Supplementary Fig. S6f–i). Taken together, these results suggest that levels of these biomarkers (except TNF-alpha) were only modestly impacted by the long-term use of ART once suppression of viral replication was achieved. Supplementary Table 2 shows Spearman correlation coefficients (r) and P values; and Fig. 3 shows Spearman correlograms. Correlations were assessed for the entire cohort of 23 participants (Pts 1–23) and separately among the subgroup of virologically suppressed individuals with HIV-RNA less than 50 copies/ml (Pts 6–23). Strong positive correlations were noted between western blot score and CA HIV-RNA (r = 0.73, P < 0.01), suggesting that the CA HIV-RNA derived from “defective” proviruses was associated with persistent production of HIV-1-related proteins and an ongoing antibody response to those proteins (Fig. 3a). No relationship was found between western blot score and HIV-DNA levels (Fig. 3b). Positive correlations were noted between D-dimer levels and western blot score or CA HIV-RNA (Fig. 3c and d). These reached statistical significance for western blot score and D-dimer (r = 0.52, P = 0.01) and CA HIV-RNA and D-dimer (r = 0.53, P < 0.01) for the cohort that included all participants. No significant associations were found between HIV-DNA and D-dimer levels (Fig. 3e). Additional correlations were noted between total CD8+ T-cell counts and HIV-DNA (r = 0.52, P = 0.01) or CA HIV-RNA (r = 0.65, P < 0.01), implying the persistent production of intracellular proteins were recognized by the host as foreign (Fig. 3f and g, Supplementary Table 2).
Fig. 2 (Continued).
Long-term persistence of multiple transcriptionally active ‘defective’ HIV-1 provirus clones during suppression of plasma HIV-RNA less than 50 copies/ml on antiretroviral therapy.
(a, c, and e) Longitudinal changes in the number of sequences from expanded HIV-1 clones obtained from Pts 21–23 (detected as either HIV-RNA transcripts with or without corresponding HIV-DNA) and corresponding western blot patterns were assessed over the period of observation. Timepoints when plasma HIV-RNA level was at least 50 copies/ml are indicated in red. Distinct expanded provirus clones are assigned different sequence IDs. Symbols denote type of sequences detected at a given time point; numbers inside each symbol indicate the number of sequences detected at a given time point. (b, d, and f) Genome structures and protein-coding regions of the expanded HIV-1 clones are shown; the Highlighter analysis indicates nucleotide substitutions compared with HIV-1 HXB2. Schematic diagrams of known unspliced and spliced HIV-RNA species (represented by blue bars) are shown at the upper right panel for comparison.
Fig. 3.
Associations between levels of HIV-1, western blot score, D-dimer, and T cells.
Associations between levels of: (a) WB (western blot) score and cell-associated (CA) HIV-RNA; (b) WB score and HIV-DNA; (c) WB score and D-dimer; (d) CA HIV-RNA and D-dimer; (e) HIV-DNA and D-dimer; (f) CA HIV-RNA and CD8+ T cells; and (g) HIV-DNA and CD8+ T cells for Pts 1–23. For Pts 21–23 (Longitudinal group), the time points from the last available sample during plasma HIV-RNA less than 50 copies/ml were used (Pt 21: 20.2 years.; Pt 22: 17.5 years.; Pt 23: 14.5 years). WB score from Pt 19 could not be obtained because of high background in the western blot assay and thus not included in (a–c). Open circles represent data derived from sampling time points with plasma HIV-RNA at least 50 copies/ml; filled circles represent data derived from time points during suppressive ART with plasma HIV-RNA less than 50 copies/ml. Spearman correlations were estimated separately for a group including all participants (dotted blue lines) and a group excluding the five individuals (Pts 1–5) with HIV-RNA at least 50 copies/ml (solid blue lines). The Spearman correlation coefficient (r) and P value are shown.
Discussion
In the present study, we demonstrated the long-term persistence of both translationally competent non-full-length CA HIV-RNA and antibodies to specific HIV-1 proteins among a cohort of virologically suppressed PWH on ART. Furthermore, we showed that the presence and persistence of these non-full-length CA HIV-RNA transcripts in the peripheral circulation correlated with ongoing antibody-responses to HIV-1 viral proteins, CD8+ T-cell counts, and the levels of D-dimer. Taken together, our findings demonstrate a novel interplay between transcription and translation from ‘defective’ HIV-1 proviruses and the persistent immune activation even in the setting of long-standing, treated HIV-1 infection.
The frequency of cells with HIV-1 proviruses expressing CA HIV-RNA has been estimated to be approximately 7% (range: 2–18%) [39]. Whether or not CA HIV-RNAs detected during suppressive ART are full-length intact species has been unclear in many of the previous studies [39–44]. This is because of the fact that the detection of CA HIV-RNA by widely available quantitative PCR-based method alone does not distinguish full-length intact replication-competent forms from non-full-length or full-length defective forms. In the present study, using a combination of quantitative PCR and a 5′-LTR-to-3′-LTR PCR, we were able to demonstrate that an overwhelming proportion of CA HIV-RNA (99.8%, 481 out of 482 amplicons analyzed in a total of 12 participants: Pts 6, 7, 10–13, 15, 16, 20, 21–23) detected during suppressive ART were transcripts from ‘defective’ HIV-1 proviruses. Having identified multiple clones harboring transcriptionally active ‘defective’ proviruses in the longitudinal cohort, we have used the proviruses as molecular markers to monitor the fate of these clones and were able to demonstrate that 51% of the clones (32 out of 63 different clone types) persisted an average 10 years (range: 2–20 years). The other 49% of the clones were detected only at a single time point. All HIV-RNA transcripts detected in the participants followed longitudinally were in novel unspliced forms, faithfully reflecting the genome structures of their template of defective proviral DNA. The long-term persistence of transcriptionally active, translationally competent ‘defective’ HIV-1 provirus clones was mirrored in the long-term persistence of antibody responses directed against HIV-1 viral proteins as reflected in the western blot banding profiles and western blot scores.
In the present study, we used the western blot score as a surrogate marker of the expression of up to 10 different HIV-1 viral proteins in vivo. While the western blot scoring system is an indirect semi-quantitative measure of in-vivo production of HIV-1 viral proteins, it allows for a dynamic characterization of specific antibody responses generated against various HIV-1 viral proteins. A similar application has shown that changes in western blot scores positively correlate with HIV-1 reservoir size in perinatally infected infants [37]. Consistent with our previous findings [18,19], we found that asynchronous fading in the western blot banding pattern was characteristic of suppressed individuals on long-term ART. A similar stochastic element to the fading of western blot bands was also noted and interpreted as a lack of recent exposure to viral antigens in the Berlin, London, and Dusseldorf patients [45–47] who underwent curative hematopoietic stem cell transportation (HSCT). While it is possible that the elimination of all the HIV-specific B cells upon ablation was the reason for the observed decreasing levels of specific antibody responses after the HSCT, we feel this unlikely, as the total IgG levels did not change substantially before and after HSCT in the Dusseldorf patient (personal communication).
It is well established that ART is markedly effective at suppressing viral replication, increasing CD4+ T-cell counts, and reducing all-cause mortality among PWH [1,2]. However, despite the use of effective ART, this population remains at a disproportionately higher risk of comorbid disease and mortality associated with chronic inflammation and coagulation dysregulation, as noted by persistent elevations in biomarkers such as IL-6 and D- dimer [1,3–10]. Although multiple studies have attempted to discover associations between the levels of persistent HIV-1 and biomarkers associated with inflammation and immune activation, results have remained mixed [48–52]. The present study shows a clear linkage between levels of CA HIV-RNA, antibodies to HIV-1 proteins, and total CD8+ T-cell counts in virologically suppressed individuals, providing evidence to suggest the pool of transcriptionally active ‘defective’ proviruses may be contributing to the state of persistent antigenic stimulation in the absence of active viral replication.
Our study has some limitations. Our sample size for HIV-1 sequencing was relatively small with the exception of the three participants sampled longitudinally (Pts 21–23). We also acknowledge that the panel of biomarkers may be limited and could be expanded in future studies to include other markers of increasing interests in the field. In addition, although the HIV-1 western blot scoring system remains effective, its indirect and semi-quantitative nature might have limited our ability to assess the relationships between the levels of transcriptional/translational activity and the levels of markers of immune activation and inflammation. Utilization of a system that allows a direct measurement of the levels of in-vivo HIV-1 viral proteins in PWH with suppressed viral replication would be of value in future studies.
Collectively, our results suggest that HIV-RNA transcripts from persistent ‘defective’ HIV-1 proviruses are a persistent feature of ‘successfully’ treated HIV infection and may act as one of the drivers for chronic inflammation and immune activation during suppressive ART. Better clarification of the translational competence of ‘defective’ proviruses is a key direction for future work. In addition, assessing the distribution of these proviruses in other tissue compartments may help delineate any additional roles they may play in the pathogenesis of chronic HIV-1 infection. Although ART is capable of suppressing HIV-1 replication to levels ‘below detectable’ by clinical diagnostic assays, it does not suppress expression of HIV-RNA transcripts from all HIV-1 proviruses. As such, any contribution to inflammation and immune activation via CA HIV- RNA would not necessarily be abated by current ART regimens or strategies targeting latently infected cells. Therefore, therapeutic options directed toward a cure may need to include interventions directed at suppressing CA HIV-RNA transcription or eliminating cells harboring the transcriptionally competent HIV-1 proviruses, given their role in persistent immune activation and the theoretical possibility that, following discontinuation of ART, they could undergo recombination to generate competent virus.
Acknowledgements
We thank J. Kovacs, F. Maldarelli, C. Morse, and M. Wright for providing clinical specimens and A.S. Fauci for his guidance and support.
Funding: this work was funded in part through the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH, and in part with federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The views expressed in this article are those of the authors and do not necessarily represent the views of the National Institute of Allergy and Infectious Diseases (NIAID) or Frederick National Laboratory for Cancer Research.
Authors’ contributions: K.S. performed experiments, analyzed data, and wrote the manuscript. V.N., R.D., A.R., I.D., Y.B., T.Z., N.W., F.S., M.S., I.D., P.L., A.G., J.H., T.B., and B.G. performed experiments. C.R. coordinated collection of clinical specimens. Z.H. performed statistical analysis. H.C.L. designed the study and wrote the manuscript. H.I. supervised the project, designed experiments, analyzed data, and wrote the manuscript.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
References
- 1.Antiretroviral Therapy Cohort Colloboration. Causes of death in HIV-1-infected patients treated with antiretroviral therapy, 1996-2006: collaborative analysis of 13 HIV cohort studies. Clin Infect Dis 2010; 50:1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, et al. Insight Start Study Group. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med 2015; 373:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingle SM, May MT, Gill MJ, Mugavero MJ, Lewden C, Abgrall S, et al. Antiretroviral Therapy Cohort Collaboration. Impact of risk factors for specific causes of death in the first and subsequent years of antiretroviral therapy among HIV-infected patients. Clin Infect Dis 2014; 59:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trickey A, May MT, Vehreschild J, Obel N, Gill MJ, Crane H, et al. Antiretroviral Therapy Cohort Collaboration (ART-CC). Cause-specific mortality in HIV-positive patients who survived ten years after starting antiretroviral therapy. PLoS One 2016; 11:e0160460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulware DR, Hullsiek KH, Puronen CE, Rupert A, Baker JV, French MA, et al. INSIGHT Study Group. Higher levels of CRP, D-dimer, IL-6, and hyaluronic acid before initiation of antiretroviral therapy (ART) are associated with increased risk of AIDS or death. J Infect Dis 2011; 203:1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grund B, Baker JV, Deeks SG, Wolfson J, Wentworth D, Cozzi-Lepri A, et al. Relevance of interleukin-6 and D-dimer for serious non-AIDS morbidity and death among HIV-positive adults on suppressive antiretroviral therapy. PLoS One 2016; 11:e0155100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 2008; 5:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. INSIGHT SMART Study Group. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 2011; 203:780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverberg MJ, Neuhaus J, Bower M, Gey D, Hatzakis A, Henry K, et al. Risk of cancers during interrupted antiretroviral therapy in the SMART study. AIDS 2007; 21:1957–1963. [DOI] [PubMed] [Google Scholar]
- 10.Sereti I, Krebs SJ, Phanuphak N, Fletcher JL, Slike B, Pinyakorn S, et al. RV254/SEARCH 010, RV304/SEARCH 013 and SEARCH 011 protocol teams. Persistent, albeit reduced, chronic inflammation in persons starting antiretroviral therapy in acute HIV infection. Clin Infect Dis 2017; 64:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Coninck Z, Hussain-Alkhateeb L, Bratt G, Ekstrom AM, Gisslen M, Petzold M, et al. Non-AIDS mortality is higher among successfully treated people living with HIV compared with matched HIV-negative control persons: a 15-year follow-up cohort study in Sweden. AIDS Patient Care STDS 2018; 32:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity 2013; 39:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis 2003; 187:1534–1543. [DOI] [PubMed] [Google Scholar]
- 14.Freeman ML, Lederman MM, Gianella S. Partners in crime: the role of CMV in immune dysregulation and clinical outcome during HIV infection. Curr HIV/AIDS Rep 2016; 13:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser P, Joos B, Niederost B, Weber R, Gunthard HF, Fischer M. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol 2007; 81:9693–9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman BT, Hu X, Singh K, Haine L, Rupert AW, Neaton JD, et al. ESPRIT, SMART and START Study Groups. Genome-wide association study of high-sensitivity C-reactive protein, D-dimer, and interleukin-6 levels in multiethnic HIV+ cohorts. AIDS 2021; 35:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirley DK, Kaner RJ, Glesby MJ. Effects of smoking on non-AIDS-related morbidity in HIV-infected patients. Clin Infect Dis 2013; 57:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A 2016; 113:8783–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamichi H, Natarajan V, Adelsberger JW, Rehm CA, Lempicki RA, Das B, et al. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 2014; 28:1091–1099. [DOI] [PubMed] [Google Scholar]
- 20.Cho A, Gaebler C, Olveira T, Ramos V, Saad M, Lorenzi JCC, et al. Longitudinal clonal dynamics of HIV-1 latent reservoirs measured by combination quadruplex polymerase chain reaction and sequencing. Proc Natl Acad Sci U S A 2022; 119:e2117630119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falcinelli SD, Kilpatrick KW, Read J, Murtagh R, Allard B, Ghofrani S, et al. Longitudinal dynamics of intact HIV proviral DNA and outgrowth virus frequencies in a cohort of individuals receiving antiretroviral therapy. J Infect Dis 2021; 224:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandhi RT, Cyktor JC, Bosch RJ, Mar H, Laird GM, Martin A, et al. AIDS Clinical Trials Group A5321 Team. Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis 2021; 223:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication- competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs JL, Halvas EK, Tosiano MA, Mellors JW. Persistent HIV-1 viremia on antiretroviral therapy: measurement and mechanisms. Front Microbiol 2019; 10:2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol 1991; 65:3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peluso MJ, Bacchetti P, Ritter KD, Beg S, Lai J, Martin JN, et al. Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight 2020; 5:e132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez G, Xu X, Chermann JC, Hirsch I. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol 1997; 71:2233–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin HA, Kadiyala GN, Telwatte S, Wedrychowski A, Chen TH, Moron-Lopez S, et al. New assay reveals vast excess of defective over intact HIV-1 transcripts in antiretroviral therapy-suppressed individuals. J Virol 2022; 96:e0160522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imamichi H, Smith M, Adelsberger JW, Izumi T, Scrimieri F, Sherman BT, et al. Defective HIV-1 proviruses produce viral proteins. Proc Natl Acad Sci U S A 2020; 117:3704–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duette G, Hiener B, Morgan H, Mazur FG, Mathivanan V, Horsburgh BA, et al. The HIV- 1 proviral landscape reveals that Nef contributes to HIV-1 persistence in effector memory CD4+ T cells. J Clin Invest 2022; 132: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe 2017; 21:494.e4–506.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White JA, Wu F, Yasin S, Moskovljevic M, Varriale J, Dragoni F, et al. Clonally expanded HIV-1 proviruses with 5’-leader defects can give rise to nonsuppressible residual viremia. J Clin Invest 2023; 133:e165245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 2003; 41:4531–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somsouk M, Dunham RM, Cohen M, Albright R, Abdel-Mohsen M, Liegler T, et al. The immunologic effects of mesalamine in treated HIV-infected individuals with incomplete CD4+ T cell recovery: a randomized crossover trial. PLoS One 2014; 9:e116306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaschen B, Kuiken C, Korber B, Foley B. Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics 2001; 17:415–418. [DOI] [PubMed] [Google Scholar]
- 36.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014; 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocca S, Zangari P, Cotugno N, De Rossi A, Ferns B, Petricone D, et al. Human immunodeficiency virus (HIV)-antibody repertoire estimates reservoir size and time of antiretroviral therapy initiation in virally suppressed perinatally HIV-infected children. J Pediatric Infect Dis Soc 2019; 8:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiegand A, Spindler J, Hong FF, Shao W, Cyktor JC, Cillo AR, et al. Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc Natl Acad Sci U S A 2017; 114:E3659–E3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musick A, Spindler J, Boritz E, Perez L, Crespo-Velez D, Patro SC, et al. HIV infected t cells can proliferate in vivo without inducing expression of the integrated provirus. Front Microbiol 2019; 10:2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong F, Aga E, Cillo AR, Yates AL, Besson G, Fyne E, et al. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 2016; 54:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massanella M, Ignacio RAB, Lama JR, Pagliuzza A, Dasgupta S, Alfaro R, et al. Long- term effects of early antiretroviral initiation on HIV reservoir markers: a longitudinal analysis of the MERLIN clinical study. Lancet Microbe 2021; 2:e198–e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasternak AO, Grijsen ML, Wit FW, Bakker M, Jurriaans S, Prins JM, et al. Cell- associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART. JCI Insight 2020; 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, et al. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog 2018; 14:e1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 2009; 360:692–698. [DOI] [PubMed] [Google Scholar]
- 46.Gupta RK, Peppa D, Hill AL, Galvez C, Salgado M, Pace M, et al. Evidence for HIV-1 cure after CCR5Delta32/Delta32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet HIV 2020; 7:e340–e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen BO, Knops E, Cords L, Lubke N, Salgado M, Busman-Sahay K, et al. In-depth virological and immunological characterization of HIV-1 cure after CCR5Delta32/Delta32 allogeneic hematopoietic stem cell transplantation. Nat Med 2023; 29:583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cockerham LR, Siliciano JD, Sinclair E, O’Doherty U, Palmer S, Yukl SA, et al. CD4+ and CD8+ T cell activation are associated with HIV DNA in resting CD4+ T cells. PLoS One 2014; 9:e110731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, et al. ACTG A5321 Team. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 2017; 13:e1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med 2010; 7:e1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatano H, Jain V, Hunt PW, Lee TH, Sinclair E, Do TD, et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J Infect Dis 2013; 208:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poizot-Martin I, Faucher O, Obry-Roguet V, Nicolino-Brunet C, Ronot-Bregigeon S, Dignat-George F, et al. Lack of correlation between the size of HIV proviral DNA reservoir and the level of immune activation in HIV-infected patients with a sustained undetectable HIV viral load for 10 years. J Clin Virol 2013; 57:351–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.