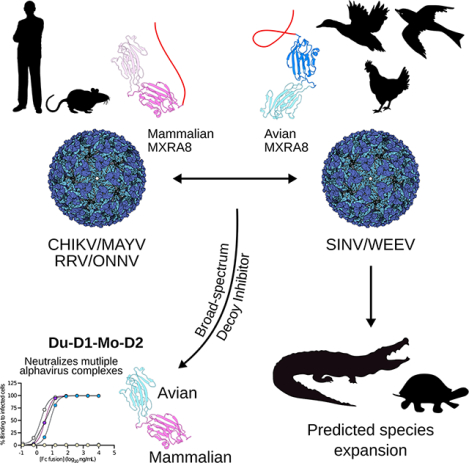
SUMMARY
MXRA8 is a receptor for chikungunya (CHIKV) and other arthritogenic alphaviruses with mammalian hosts. However, mammalian MXRA8 does not bind to alphaviruses that infect humans and have avian reservoirs. Here, we show that avian, but not mammalian, MXRA8 can act as a receptor for Sindbis, western equine encephalitis (WEEV), and related alphaviruses with avian reservoirs. Structural analysis of duck MXRA8 complexed with WEEV reveals an inverted binding mode compared to mammalian MXRA8 bound to CHIKV. Whereas both domains of mammalian MXRA8 bind CHIKV E1 and E2, only domain 1 of avian MXRA8 engages WEEV E1, and no appreciable contacts are made with WEEV E2. Using these results, we generated a chimeric avian-mammalian MXRA8 decoy-receptor that neutralizes infection of multiple alphaviruses from distinct antigenic groups in vitro and in vivo. Thus, different alphaviruses can bind MXRA8 encoded by different vertebrate classes with distinct engagement modes, which enables development of broad-spectrum inhibitors.
Graphical Abstract
eTOC
Avian MXRA8 is an entry receptor for alphaviruses belonging to Western equine encephalitis complex that have bird reservoirs but not for those in the Semliki Forest complex, which instead utilize mammalian MXRA8. Avian MXRA8 and mammalian MXRA8 engage different alphaviruses using inverted binding modes and distinct regions. A duck domain 1- mouse domain 2 chimeric MXRA8 serves as a decoy-receptor and inhibits infection of multiple alphaviruses from distinct antigenic complexes in vitro and in vivo.
INTRODUCTION
Alphaviruses are a group of globally important, mosquito-transmitted, enveloped, positive-sense RNA viruses in the Togaviridae family1, and those infecting humans have several reservoirs including non-human primates, macropods, reptiles, rodents, and birds1. Two clinical syndromes occur in humans after alphavirus infection: acute encephalitis and neurological disease are caused by Venezuelan equine encephalitis (VEEV), eastern equine encephalitis (EEEV), and western equine encephalitis (WEEV) viruses; and acute and chronic musculoskeletal disease and arthritis are caused by chikungunya (CHIKV), Ross River (RRV), O’nyong’nyong (ONNV), Mayaro (MAYV) and Sindbis (SINV) viruses.
Over the last several thousand years, New World alphaviruses (e.g., VEEV and EEEV) evolved and separated from Old World alphaviruses (e.g., SINV and Semliki Forest (SFV) virus)2 (Fig 1A). The western equine encephalomyelitis (WEE) complex is an antigenically related group3 that includes four New World (Aura (AURAV), Fort Morgan (FMV), Highlands J (HJV), and WEEV) and several Old World (e.g., SINV and Whataroa (WHAV)) viruses1. WEEV, HJV, and FMV are considered descendants of a recombination between a SINV-like virus and EEEV that is believed to have occurred in South America4,5. The sequences of non-structural proteins, the capsid protein, and the (untranslated) 3’-terminal 80 nucleotides of WEEV, HJV, and FMV are related most closely to EEEV, whereas those of the E2 and E1 structural glycoproteins are related more to SINV4,5. WEEV has the encephalitic properties of New World alphaviruses but the structural proteins and presumably receptor specificity of Old World alphaviruses. SINV, which causes periodic outbreaks in Africa, Europe, Asia, and Australia, has three antigenic subtypes: Babanki (BBKV; Africa), Ockelbo (OCKV; Sweden), and Kyzylagach (KYZV: Azerbaijan and China)1, all of which share clinical syndromes of fever, rash and arthralgia in humans.
Figure 1. Avian MXRA8 is required for optimal infection of SINV and WEEV alphaviruses.
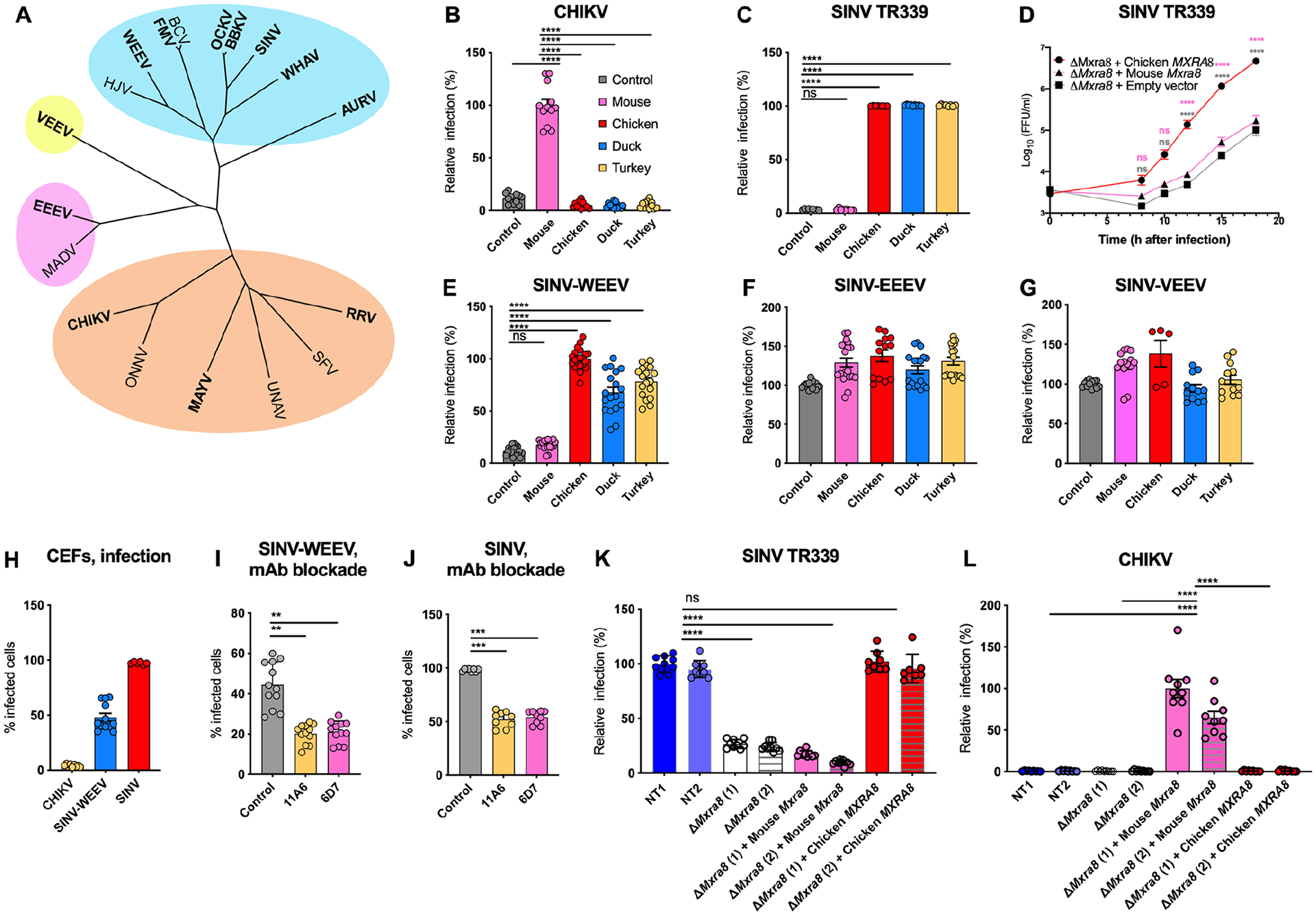
A. A phylogenetic tree based on alphavirus E1 and E2 protein sequences with complexes clustered in blue (WEEV complex), orange (SFV complex), purple (EEEV complex), and yellow (VEEV complex). Viruses in this study are bolded. B-C. ΔMxra8 3T3 cells complemented with mouse (pink), chicken (red), duck (blue) or turkey (yellow) MXRA8 or empty vector control (gray) were inoculated with CHIKV 181/25 (B) or SINV TR339 (C) and stained for CHIKV or SINV antigen (4 and 3 experiments in triplicate, respectively). Infection is normalized to levels detected with cells expressing mouse (B) or chicken (C) MXRA8. D. Multi-step growth curves with SINV TR339 in ΔMxra8 3T3 cells complemented with chicken or mouse MXRA8 or empty vector control. Virus was titrated by focus-forming unit (FFU) assay (4 experiments, duplicate). E-G. ΔMxra8 3T3 cells complemented with mouse, chicken, duck or turkey MXRA8 or empty vector control were inoculated with SINV-WEEV-GFP (E), SINV-EEEV-GFP (F) or SINV-VEEV-GFP (G). Infection was assessed by GFP expression (4 to 7 experiments, triplicate). H. CEFs were inoculated with CHIKV 181/25, SINV-WEEV-GFP, or SINV TR339 and stained for viral antigen. I-J. CEFs were pre-incubated with anti-chicken Mxra8 or an isotype control mAb and inoculated with SINV-WEEV-GFP (I) or SINV TR339 (J). Infection was assessed by GFP expression or E1 staining and compared between infected (red) and non-infected cells (blue) (3 experiments in triplicate). K-L. ΔMxra8 chicken fibroblasts complemented with chicken or mouse MXRA8 (2 independent clones of each shown) or empty vector control and non-targeted (NT1 and NT2) CEFs were inoculated with SINV TR339 (K), CHIKV 181/25 (L). Infection was assessed by flow cytometry, derived from the percent and mean fluorescence intensity of the infected cells, and normalized to the non-targeting control NT1 (for SINV) or ΔMxra8 (1) + Mouse Mxra8 (for CHIKV) (3 experiments, in triplicate). B, C, E, I, J, K and L: one-way ANOVA with Dunnett’s post-test; mean ± standard deviation (SD). D: Two-way ANOVA with Tukey post-test; mean ± SD. ns, non-significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001). See also Fig S1 S2, S3, S4, and Table S1.
Matrix Remodeling Associated 8 (MXRA8) is a conserved cell-surface molecule in mammals, birds, reptiles, and fish, and comprised of two Ig-like domains arranged in a unique head-to-head orientation6–8. MXRA8 serves as an entry receptor for CHIKV and several other arthritogenic alphaviruses that are members of the Semliki Forest (SF) antigenic group (Fig 1A), including MAYV, RRV, and ONNV, but not for encephalitic alphaviruses or WEE complex members6,9. Whereas ectopic expression of many mammalian species of MXRA8 proteins (e.g., mouse, rat, chimpanzee, dog, horse, goat, and sheep) facilitates CHIKV infection in 3T3 cells, MXRA8 from several avian species tested (e.g., chicken, turkey, and duck) do not promote CHIKV infection6.
Given the evolutionary relationships between alphaviruses, their differential utilization of mammalian MXRA8, and distinct dependence on bird reservoirs, we explored whether avian MXRA8 could act as a receptor for alphaviruses that use birds in their enzootic cycle. Here, we demonstrate that avian MXRA8 is an entry receptor for WEE complex alphaviruses. Cryo-electron microscopy (cryo-EM) structural analysis unexpectedly shows that WEEV binds to duck MXRA8 in a flipped orientation relative to mammalian MXRA8 binding to CHIKV. We applied this discovery to generate a chimeric duck-mouse MXRA8 receptor that supports infection of both WEE and SF complex viruses and a soluble decoy receptor inhibitor that blocks infection of viruses from both families in cell culture and in vivo. Cryo-EM analysis of a duck-mouse chimeric MXRA8 bound to CHIKV or WEEV confirmed the flipped binding orientation and revealed that D1 of duck MXRA8 is critical for binding to both WEE and SF complex alphaviruses despite engaging unique locations through different approaches. Overall, our findings demonstrate how zoonotic viruses within the same alphavirus family use distinct binding modes to allow for receptor usage and species tropism, and how this knowledge can be harnessed for the development of possible broad-spectrum therapies against a range of alphaviruses.
RESULTS
WEEV complex alphaviruses with an avian reservoir use avian MXRA8 for infection of mammalian cells.
We complemented mouse 3T3 fibroblasts lacking MXRA8 expression (ΔMxra8) with MXRA8 from chicken (Gallus gallus), duck (Anas platyrhynchos) or turkey (Meleagris gallopavo) (Fig S1A–B), which share approximately 59% amino acid identity with murine MXRA8 (Fig S1C). As expected, murine MXRA8 facilitated infection of the SF complex viruses CHIKV, RRV, and MAYV, whereas expression of chicken, duck, or turkey MXRA8 did not (Fig 1B, S1D–E, and Ref6). We next evaluated the impact of avian MXRA8 expression on other alphaviruses with bird reservoirs. Whereas SINV (strain TR339) infection was low in ΔMxra8 3T3 cells or ΔMxra8 3T3 cells complemented with mouse Mxra8, high levels of SINV infection were detected in ΔMxra8 cells complemented with chicken, duck, or turkey MXRA8 (Fig 1C). Other SINV strains (SINV-Girdwood and SINV-Toto) had similar phenotypes (Fig S1F). Multi-step growth curve analysis corroborated a dependence of SINV infection on avian but not mammalian MXRA8 for optimal virus production (Fig 1D). Infection levels of SINV-WEEV, a chimeric alphavirus encoding the non-structural genes of SINV and structural genes of WEEV10, also were higher in ΔMxra8 3T3 cells complemented with avian MXRA8 than ΔMxra8 cells or ΔMxra8 cells complemented with mouse Mxra8 (Fig 1E). In contrast, expression of avian MXRA8 proteins did not enhance infection of SINV-VEEV or SINV-EEEV chimeric viruses in ΔMxra8 3T3 cells (Fig 1F–G). We confirmed and extended these results with other cell types. SINV-VEEV infection was not enhanced in Jurkat cells expressing mouse or turkey MXRA8, whereas greater infection was observed in cells expressing LDLRAD3, a receptor for VEEV10 (Fig S1G). In comparison, expression of mouse and turkey MXRA8 respectively promoted CHIKV and SINV infection in Jurkat cells (Fig S1G). In K562 cells, mouse and turkey MXRA8 also did not promote infection of SINV-EEEV, whereas expression of VLDLR, a recently described EEEV receptor11, did (Fig S1G). These results are consistent with alphavirus evolutionary relationships, as the structural genes of WEEV are more closely related to SINV than to EEEV or VEEV1,3,12.
Avian MXRA8 enhances infection of WEEV complex alphaviruses with an avian reservoir in avian cells.
We confirmed our findings in a species-relevant cell system using chicken embryonic fibroblasts (CEFs), which express chicken MXRA8 on their surface (Fig S1I). We first tested whether CEFs could support infection of different mammalian and avian alphaviruses including CHIKV, SINV-WEEV, or SINV. Whereas CHIKV antigen was absent in CEFs at 24 h, WEEV and SINV antigens were detected at high levels at 9 h post-infection (Fig 1H and S1J). To determine whether CEFs infection by SINV-WEEV and SINV depended on chicken MXRA8, we pre-incubated CEFs with blocking anti-chicken MXRA8 mAbs (see Methods); this treatment reduced infection of SINV-WEEV (Fig 1I) or SINV (Fig 1J) compared to an isotype control mAb.
To corroborate these findings, we generated ΔMXRA8 CEFs using gene editing and then complemented them with chicken MXRA8 or mouse Mxra8 (Fig S2A). This panel of cells, along with control CEFs edited with non-targeted single-guide RNA, were inoculated with SINV, CHIKV, or SINV-EEEV. ΔMXRA8 CEFs or ΔMXRA8 CEFs complemented with mouse Mxra8 showed less SINV antigen expression than non-targeted CEFs or ΔMXRA8 CEFs complemented with chicken MXRA8 (Fig 1K). At 10 h after CHIKV inoculation, ~50% of ΔMXRA8 CEFs complemented with mouse Mxra8 stained positively for CHIKV antigen, whereas virtually none of the non-targeted CEFs or ΔMXRA8 CEFs complemented with chicken MXRA8 supported CHIKV infection (Fig 1L). SINV-EEEV infection levels were similar in all tested CEFs (Fig S2B). These experiments suggest that avian MXRA8 promotes infection of SINV and WEEV, CHIKV is non-permissive for CEFs unless mouse MXRA8 is expressed ectopically, and EEEV infects CEFs in a MXRA8-independent manner.
Avian MXRA8 facilitates SINV attachment and entry into cells and binds directly to WEEV virus-like particles (VLPs).
We evaluated whether avian MXRA8 has a role in viral attachment and entry as demonstrated for mouse MXRA8 and CHIKV9. Indeed, SINV showed increased binding at 4°C to ΔMxra8 mouse 3T3 cells complemented with chicken MXRA8 compared to ΔMxra8 cells or ΔMxra8 cells complemented with mouse Mxra8 (Fig 2A). Consistent with an effect of avian MXRA8 on virus attachment, pre-incubation of cells with anti-chicken MXRA8 mAbs inhibited binding of SINV (Fig 2B). When virus internalization was allowed to occur after incubation at 37°C, higher levels of SINV RNA were detected within ΔMxra8 cells complemented with chicken MXRA8 than within ΔMxra8 cells or ΔMxra8 cells complemented with mouse Mxra8 (Fig 2C).
Figure 2. Avian MXRA8 modulates SINV attachment and internalization and binds to WEEV VLPs.
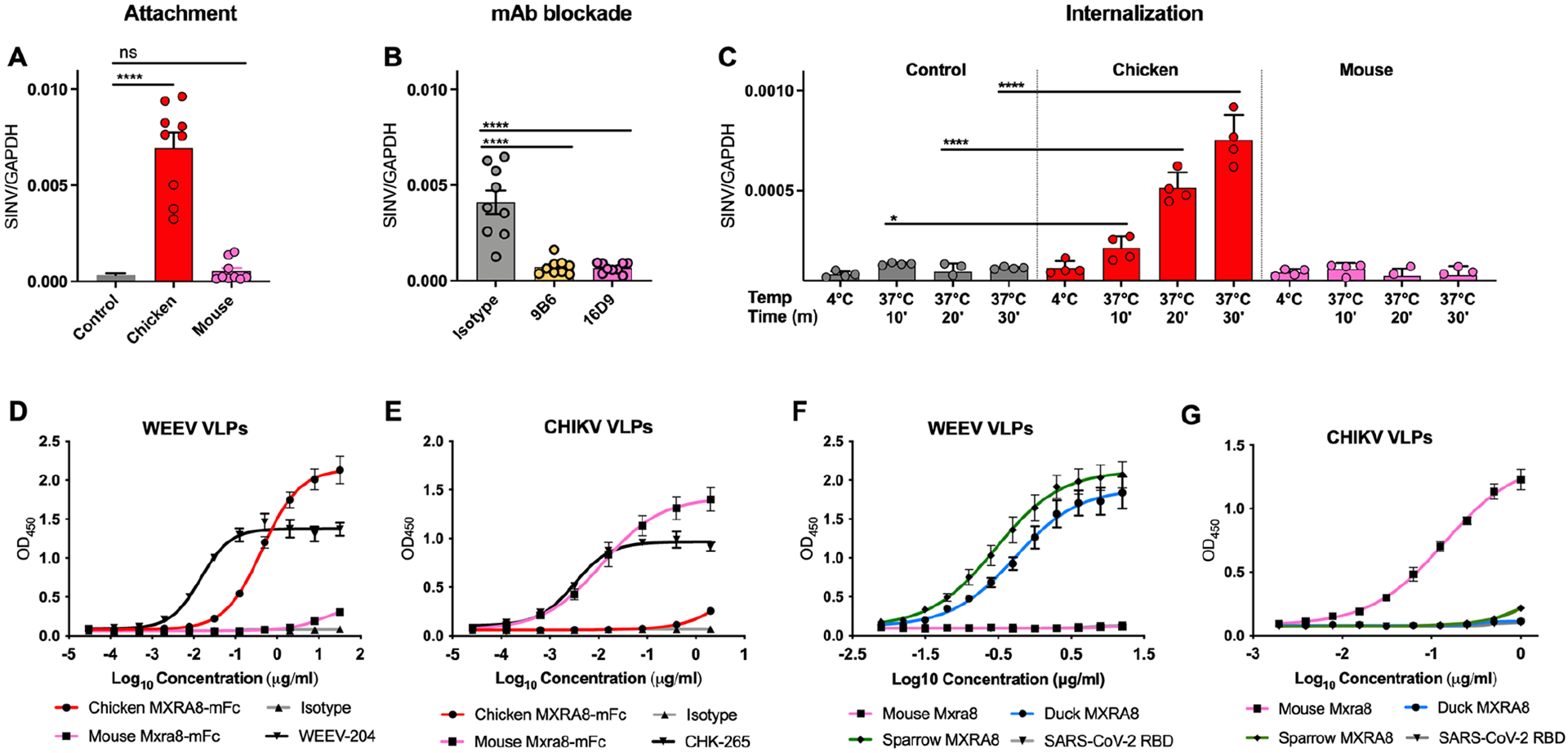
A-C. SINV was incubated with ΔMxra8 3T3 cells (control), chicken Mxra8-complemented ΔMxra8 3T3 cells, or mouse Mxra8-complemented ΔMxra8 3T3 cells at 4 °C without (A) or with (B) pre-incubation with mouse-anti-chicken MXRA8 mAbs. Bound virions were quantified by measuring viral RNA levels and Gapdh levels via qRT–PCR. (C) After removal of unbound virus, the temperature was shifted to 37 °C to allow internalization. Intracellular RNA (SINV and Gapdh) levels were measured by qRT–PCR. (3 experiments, triplicate). D-E. Binding of chicken MXRA8–Fc and murine MXRA8-Fc to CHIKV (D) or WEEV (E) VLPs by ELISA (WEEV positive control, WEEV-204; CHIKV positive control, CHK-265; negative control, anti-HCV H77.39; CHIKV VLP ELISA: 2 experiments, triplicate; WEEV VLP ELISA: 2 experiments, duplicate). F-G. Binding of WEEV VLPs (F) and CHIKV VLPs (G) to duck, sparrow, or mouse MXRA8 by ELISA. SARS-CoV-2 receptor-binding domain served as a negative control (3 experiments, duplicate). A-C: one-way ANOVA with Dunnett’s post-test; mean ± SD. *, P < 0.05; ****, P < 0.0001).
We tested whether avian MXRA8 could bind directly to virus-like particles (VLPs) containing WEEV or CHIKV structural proteins6,13–15. We generated a chicken MXRA8-Fc fusion protein, which bound directly to WEEV but not to CHIKV VLPs, whereas mouse MXRA8-Fc fusion protein bound to CHIKV but not to WEEV VLPs (Fig 2D–E). Since sparrows are reservoirs for some alphaviruses16, and sparrow MXRA8 sequence varies from chicken MXRA8 by 4% at the amino acid level, we also generated this protein. WEEV VLPs bound to both duck and sparrow MXRA8 but did not bind to mouse MXRA8 (Fig 2F). Reciprocally, CHIKV VLPs bound to mouse MXRA8 but not to duck or sparrow MXRA8 (Fig 2G).
Multiple WEE complex members use avian but not mammalian MXRA8.
We assessed whether other WEE complex members could use avian MXRA8 as an entry receptor. Based on phylogenetic relationships (Fig 1A), we performed experiments in control and MXRA8-expressing chicken (CEFs) and human (293T and HeLa) cells with three additional WEE antigenic complex members (WHAV, BBKV, and OCKV) with avian reservoirs that periodically cause infections in humans. We also tested AURAV, a WEE complex member restricted to South America that lacks an established animal reservoir17. Avian MXRA8 promoted infection of SINV, BBKV, OCKV, and WHAV, whereas mouse MXRA8 did not (Table S1 and Fig S2–S3). For AURAV, neither mouse nor chicken MXRA8 enabled infection in mammalian 3T3, 293T or HeLa cells or in CEFs (Table S1 and Fig S2–S3).
Chicken MXRA8 expression in mice enhances SINV infection in vivo.
Since genetic studies are not easily performed in avian hosts, we evaluated the impact of chicken MXRA8 on SINV infection in vivo by transiently expressing it in mice using an adenoviral vector. Five-week-old BALB/c mice were inoculated via an intranasal route with 1010 viral particles of a replication-defective human adenovirus encoding chicken MXRA8 or an empty vector (Fig S4A). Five days later, mice were inoculated via an intranasal route with 104 focus-forming units (FFU) of SINV that was preceded by administration of anti-IFNAR1 mAb18 one day earlier to facilitate virus dissemination. At 40 and 72 h post-inoculation, SINV RNA and infectious virus levels were higher in the spleen and lungs of mice transduced with chicken MXRA8 than animals receiving the empty adenoviral vector (Fig S4B–D). Thus, avian MXRA8 can functionally enhance SINV infection in vivo.
Duck MXRA8 binds WEEV in an inverted orientation compared to mammalian MXRA8 and CHIKV.
To determine how WEE and SF complex members respectively bind avian and mammalian MXRA8, we used cryo-EM to obtain a reconstruction of duck MXRA8 bound to WEEV-VLPs (Table S2) and compared these data to structures of mouse and human MXRA8 bound to CHIKV-VLPs7,8. As expected, the WEEV-VLP structural proteins exhibited T = 4 icosahedral symmetry with 60 asymmetric units (Fig 3A). Each virion has 240 E1-E2 heterodimers arranged into 80 trimeric spikes, located around icosahedral threefold (“i3”, n = 20) and quasi-threefold (“q3”, n = 60) axes. Each asymmetric unit contains a q3 spike and a single i3 E1-E2 heterodimer, with MXRA8 molecules occupying four unique binding sites in each unit, conventionally labeled as sites 1 through 47 (Fig 3B). To account for deviations from ideal icosahedral symmetry, we extracted these asymmetric units for focused refinement, achieving a final resolution of 4.74 Å (Fig S5–6). Local resolution ranged from ~5.0 to 6.0 Å for the transmembrane helices and the MXRA8 domain most distal from the virus to ~4.0 to 4.5 Å for the remainder of the structure. Similar to the binding interface of mouse MXRA8 and CHIKV7, each duck MXRA8 molecule has a complex quaternary binding mode, with three different heterodimer interactions: (1) wrapped; interacting laterally against one E1-E2 heterodimer, near the E1 fusion loop, (2) intraspike; engaging the neighboring E1-E2 heterodimer within a trimeric spike, and (3) interspike, binding across different E1-E2 trimeric spikes (Fig 3C–D).
Figure 3. Cryo-EM reconstruction of duck MXRA8 bound to WEEV.
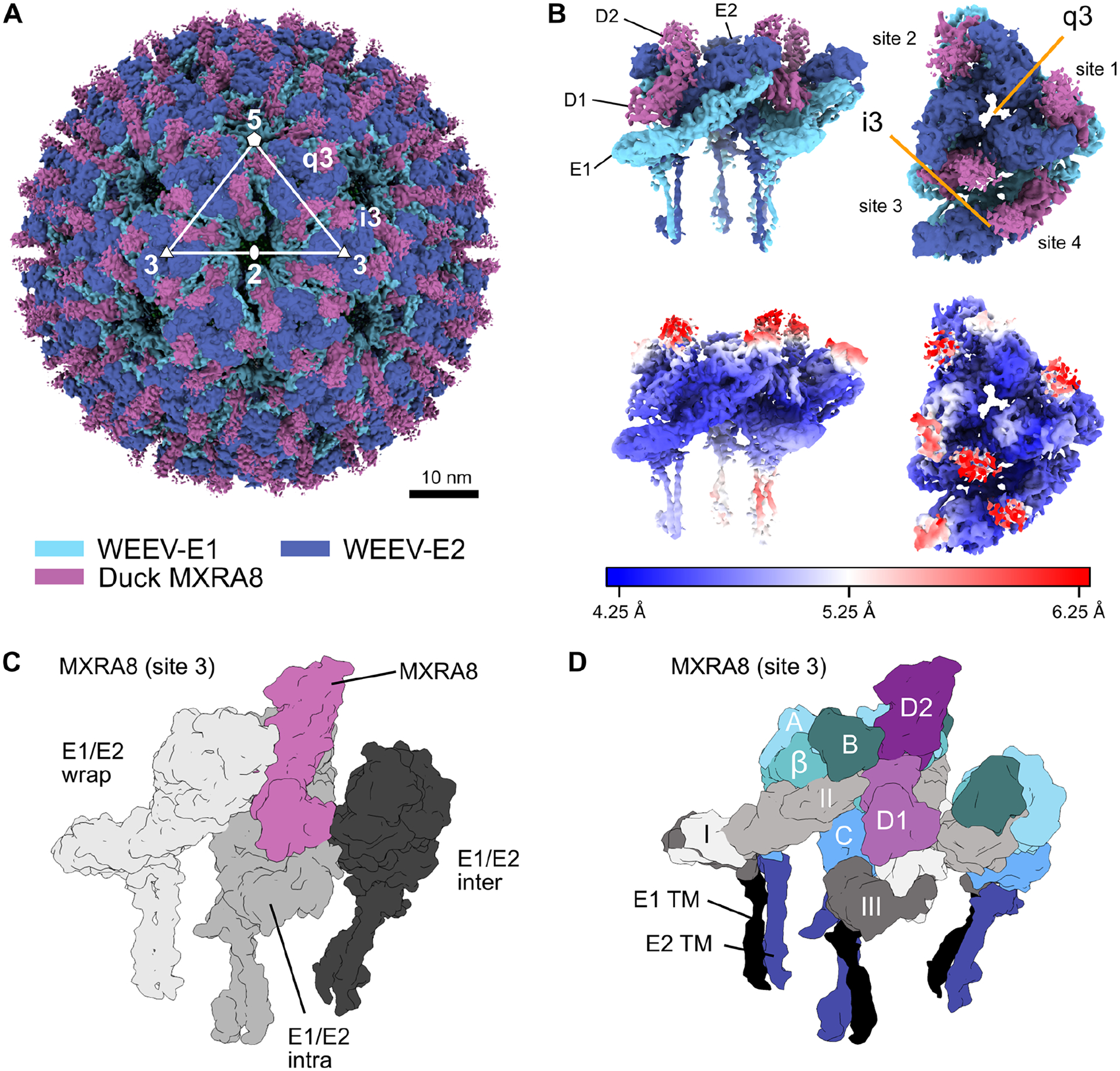
A. Cryo-EM density map of WEEV-VLP bound to duck MXRA8 at 4.74 Å resolution. WEEV-E1, WEEV-E2, and MXRA8 are colored as light-blue, dark-blue, and violet, respectively. Rotational symmetries along the 2-fold, 3-fold, and 5-fold axes are displayed with white numbers. B. Map of WEEV-VLP bound to duck MXRA8. Asymmetric unit contains the entire quasi 3-fold spike (q3), and a single icosahedral spike (i3) E1-E2 heterodimer. Shown are two views of the asymmetric unit, a side view (left, parallel with the viral membrane) and a top-down view (right, perpendicular with the viral membrane). Local resolution of map is colored from blue (4.25 Å) to white (5.25 Å) to red (6.25 Å). Density maps are viewed at contour level = 0.26 (0.96σ). C. Surface diagram of MXRA8 at site 3, detailing the three unique viral E1-E2 heterodimer contacts, wrapped (light gray), intraspike (gray), and interspike (dark gray). D. Surface diagram of MXRA8 at site 3 and interacting E1-E2 heterodimers, termed: E1-E2-wrapped, E1-E2-intraspike, or E1-E2-interspike. MXRA8 D1: light magenta; MXRA8 D2: dark magenta; E1 domain I: light gray; E1 domain II: medium-gray; E1 domain III: dark-gray; E1-TM (black); E2 A domain: light-cyan; E2 β-linker: medium-cyan; E2 B domain: dark-cyan; E2 C domain: medium-blue; E2-TM: dark-blue. See also Fig S5, S6, and Tables S2–S4.
We hypothesized that duck MXRA8 would bind WEEV in an orientation similar to that of mammalian MXRA8 and CHIKV. Unexpectedly, the binding mode of duck MXRA8 to WEEV is inverted with respect to how mammalian MXRA8 binds to CHIKV (Fig 4A–D; Video S1, Table S3)7,8. Whereas domain 1 (D1) of mammalian MXRA8 predominantly contacts CHIKV E2, D1 of duck MXRA8 contacts E1 of WEEV, forming interactions with the wrapped (E1 domain-II), intraspike (E1 domain-I), and interspike (E1 domain-II) heterodimers (Fig S7–9 and Table S4). Additionally, although D2 of mammalian MXRA8 makes substantial contacts with E1 of CHIKV, D2 of duck MXRA8 contributes minimally to WEEV binding. Thus, duck and mouse MXRA8 appear to engage with a domain-inverted binding mode using both distinct MXRA8 domains and different alphavirus structural proteins.
Figure 4. Duck and mouse MXRA8 use a domain-inverted binding paradigm to engage WEEV and CHIKV.
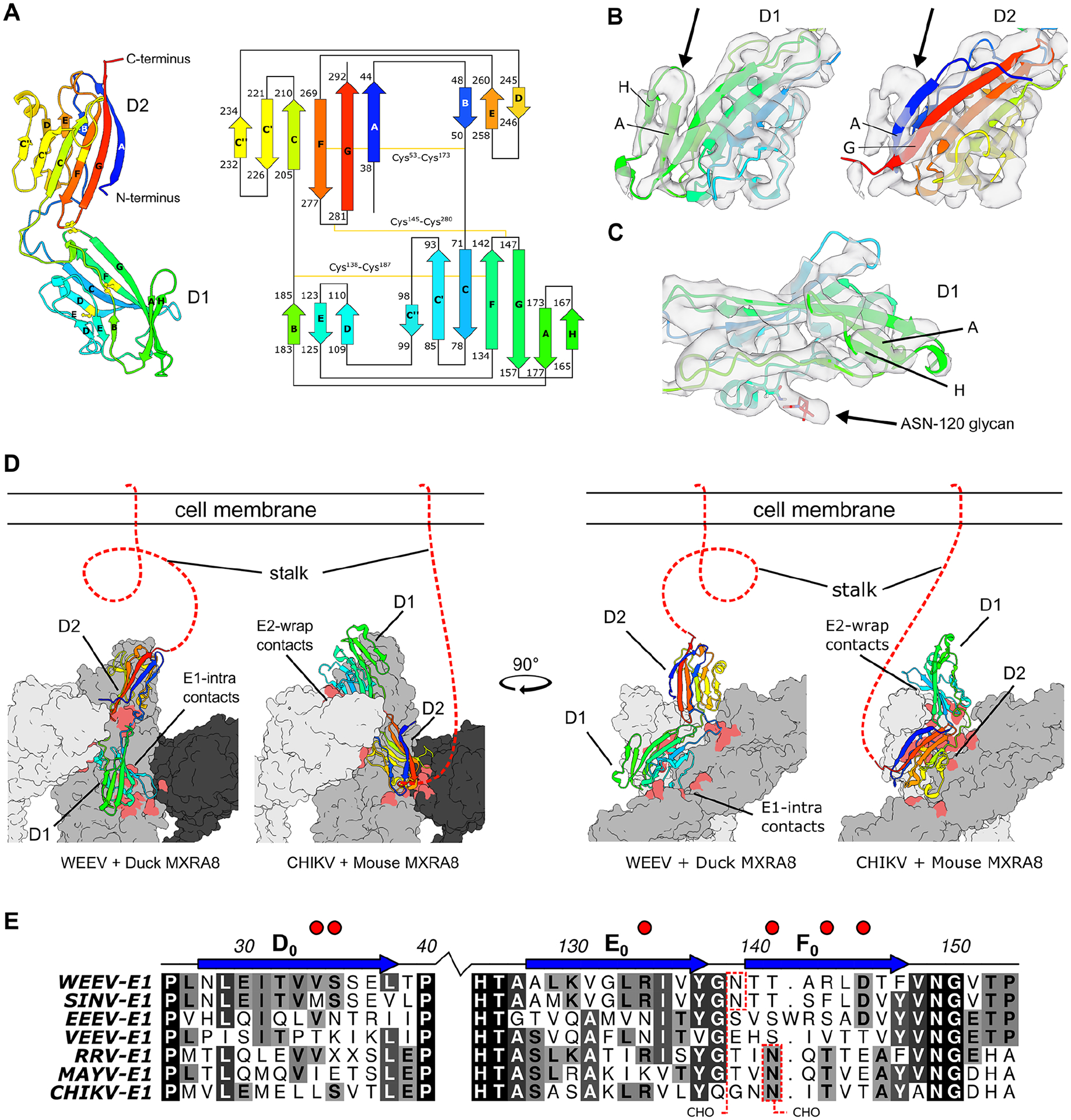
A. Ribbon and topology diagram of duck MXRA8 with β-strands labeled with standard Ig-like fold conventions. The β-strands are colored by a rainbow and are depicted as a cartoon with residue number start/stop locations. Disulfide bonds between cysteines are depicted with a yellow line. B. Cryo-EM density of the duck MXRA8 domain in the binding-groove, modeled as either D1 or D2. D1 has an extra β-strand (strand H) that better fits the density, whereas D2 lacks this β-strand and leaves this density unfilled. Density maps are viewed at contour level = 0.42 (1.56σ). C. Cryo-EM map of duck D1 with clear density in the region expected for N-linked glycan on Asn-120. Density map is viewed at contour level = 0.42 (1.56σ). D. Differential binding modes for avian and mammalian MXRA8 binding to WEEV and CHIKV, respectively. Avian MXRA8 D1 is distal to the cell membrane and binds E1-intraspike heterodimer, whereas mammalian MXRA8 D1 is proximal to the cell and binds E2-wrapped heterodimer. E. Multi-sequence alignment of WEE- and SF-complex E1 proteins with E1-intraspike duck MXRA8 D1 contacts (depicted with red circles). N-linked glycosylation sites are shown with dotted red boxes. Sequence similarities are colored from white to black, with black being most similar. E1 β-strands are labeled as described in Voss et al36. See also Fig S7–S9.
Given this unanticipated finding, we assessed the accuracy of our atomic model and the inverted binding modes of avian and mammalian MXRA8 using multiple methods. Although the substantial pseudo-symmetry between D1 and D2 of duck MXRA8 makes determination of domain orientation challenging, particular domain-specific features support our binding model. First, there is an extra β-strand (strand-H) in D1 of duck MXRA8, antiparallel to strand-A. Second, duck MXRA8 domains have different N-linked glycosylation patterns: D2 has two N-linked glycans (Asn-40 and Asn-245) and D1 has one (Asn-120), each located within different topological regions. Because of these differences, the duck MXRA8 density in the binding-groove of WEEV is fit better by D1, featuring clear density for strand-H and a single N-linked glycan near Asn-120 (Fig 4B–C).
We performed sequence alignments of the alphavirus contact residues to elucidate why WEE and SF complex members show unique MXRA8 species-specific binding. At regions predicted to be sites of contact with duck MXRA8, E1 amino acid sequences of WEE complex members (WEEV and SINV) vary from SF complex members (CHIKV, MAYV, and RRV), and VEEV/EEEV (Fig 4E and Fig S7) with an average sequence identity of only 27.9% for segments within 3 residues of the contact sites, compared to 52.5% for the entire E1 sequence. Analogously, at predicted sites of contact with mouse MXRA8, there is substantial sequence variation in regions of E2 of WEE and SF complex members (Fig S8) with an average sequence identity of 30.9% for segments within 3 residues of the contact sites. These sequence features might explain why mammalian MXRA8 does not bind to the analogous region on WEEV E1-intraspike heterodimer.
Genetic assessment of the atomic model.
Based on our cryo-EM data, duck MXRA8 D1 makes primary contact with WEEV E1 at four distinct sites: C-C’ loop (interspike), C”-D loop (intraspike), and D-E loop and B-C connector (wrapped) (Fig 5A–C). To evaluate our structural model, we designed MXRA8 mutations and insertions in these regions that should disrupt binding to WEEV and attenuate the ability of duck MXRA8 to support WEE complex virus infection. We complemented ΔMxra8 3T3 cells with duck MXRA8 encoding a set of double arginine mutations or 15 amino-acid insertions at the interface of predicted contact sites at the D-E loop, C”-D loop, and C-C’ loop6 in either D1 or D2 (Fig 5A–B and S10). ΔMxra8 3T3 cells complemented with the different duck MXRA8 mutants were inoculated with SINV and analyzed for infection by flow cytometry. As predicted by our model, mutations or insertions in duck D1 (C”-D loop insertion, C”-D loop mutations [S103R/T104R], and D-E loop mutation [T117R/D188R]) but not duck D2 resulted in reduced SINV infection (Fig 5B–D). However, the C-C’ loop insertion in duck MXRA8 D1 or D2, which corresponds to a loop insertion in bovine MXRA8 D1 that blocks CHIKV infection6, had no effect on SINV infection, indicating that the E1 interspike contacts, which are critical for MXRA8-dependent infections by SF complex members6, are less consequential for MXRA8-dependent infection by WEE complex members (Fig 5D).
Figure 5. Assessment of the MXRA8 binding model.
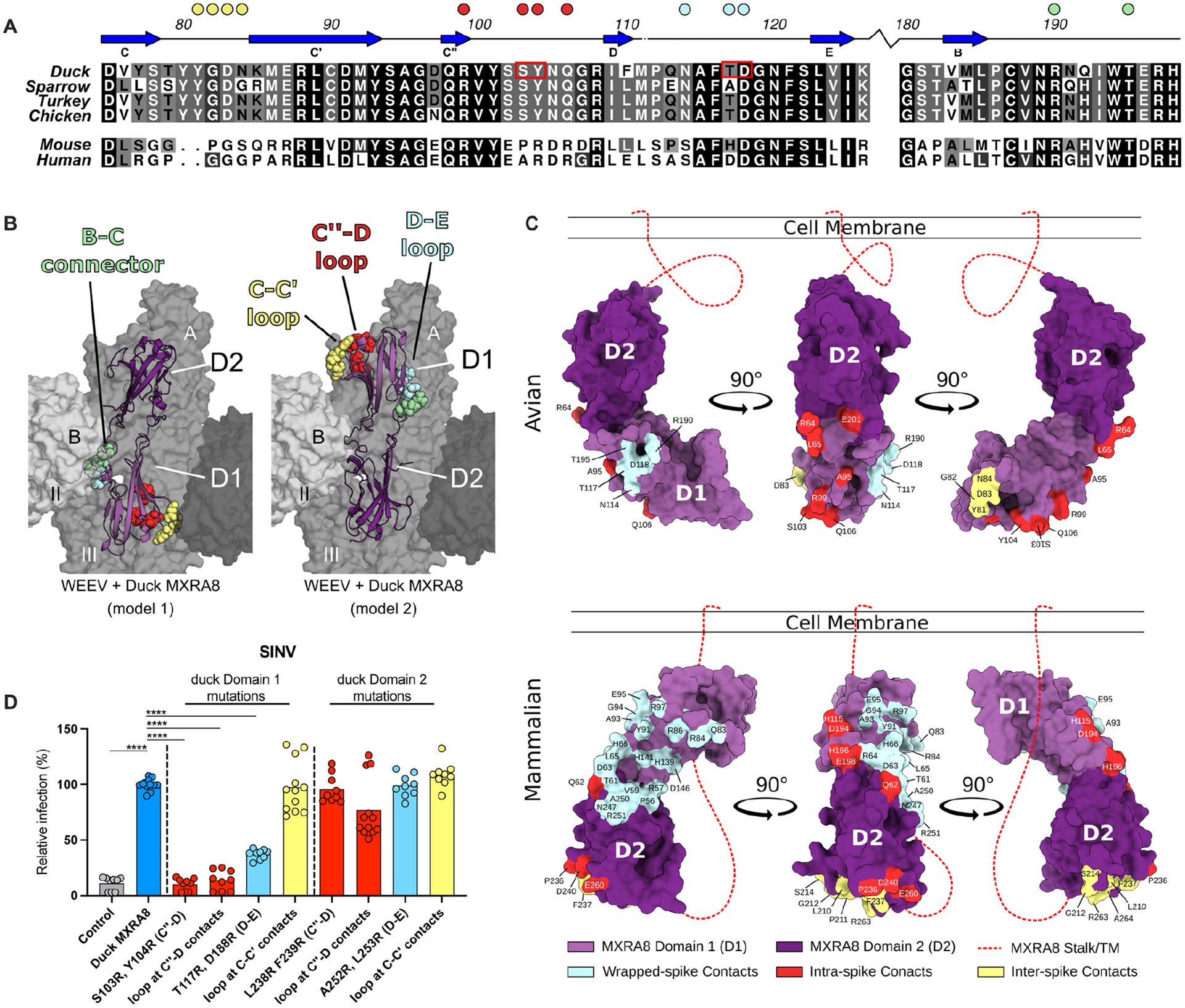
A. Sequence alignment of D1 of MXRA8 at regions that contact WEEV E1. Similar sequence identities are colored by most variable (white) to conserved (black). β-strands are labeled based on topology in Fig 4A. Filled circles above the alignment denote avian MXRA8 contact residues in the C-C’ loop (yellow), C”-D loop (red), D-E loop (cyan), and B-C connector (pale green) are denoted by circles. B. Structure of avian MXRA8 in two possible binding modes, non-flipped (model 1, left) and flipped (model 2, mouse/human-like, right), with contact residues in either D1 or D2 shown as space filling spheres in the C-C’ loop (yellow), C”-D loop (red), D-E loop (cyan), and B-C connector (pale green). C. Binding sites for WEEV and CHIKV on D1 (light purple) and D2 (dark purple) of MXRA8. Structurally defined binding sites and amino acid contact residues are colored according to the viral E2-E1 heterodimer engaged: wrapped (cyan), intraspike (red), or interspike (yellow). Stalk attaching MXRA8 to the cell membrane is represented as a red dashed line and is used to denote mode of binding, with mammalian MXRA8 adopting a flipped orientation relative to avian MXRA8. Avian and mammalian MXRA8 engage E2-E1 heterodimers most proximal to viral membrane with D1 and D2, respectively. D. SINV-GFP infection in ΔMxra8 3T3 cells complemented with wild-type (blue) and indicated mutants (light blue, yellow, or red) duck MXRA8 (3 experiments, triplicate; normalized to wild-type duck MXRA8 infection. Infection and GFP fluorescence were analyzed by flow cytometry. D: one-way ANOVA with Dunnett’s post-test; mean ± standard deviation (SD). ****, P < 0.0001). See also Fig S10.
Chimeric duck-mouse MXRA8 supports infection of alphaviruses from both SFV and WEE complexes.
As a complementary experiment, we engineered chimeric domain-swapped MXRA8 proteins (Fig 6A and Table S5). We used this approach because we were unsuccessful at expressing isolated MXRA8 single domains (D1 or D2), possibly because of the unique strand-swapped domain topology7. Because mouse MXRA8 does not bind WEEV, our model predicts that when D1 of duck is replaced with D1 of mouse MXRA8, binding to WEEV should be abrogated. ΔMxra8 3T3 cells complemented with the duck-mouse (Du-D1-Mo-D2) and mouse-duck (Mo-D1-Du-D2) MXRA8 chimera, both with duck MXRA8 stalk sequences, were inoculated with SINV, SINV-WEEV, SINV-CHIKV, and MAYV. While similar levels of SINV and SINV-WEEV infection were observed in cells complemented with Du-D1-Mo-D2 and wild-type duck MXRA8, substantially less infection was detected in cells expressing Mo-D1-Du-D2 MXRA8 (Fig 6B–C). Unexpectedly, when cells were inoculated with SINV-CHIKV and MAYV, two viruses that use mammalian but not avian MXRA8, high levels of infection were detected in cells expressing Du-D1-Mo-D2 but not Mo-D1-Du-D2 MXRA8 (Fig 6D–E). These experiments with Du-D1-Mo-D2 establish that D1 of duck MXRA8 is compatible with SF complex virus binding and infection when expressed with D2 of mouse MXRA8. Indeed, 8 of the 14 residues in D1 of mouse MXRA8 that contact CHIKV7,8 are conserved in D1 of duck MXRA8. The lack of infection with Mo-D1-Du-D2 MXRA8 and CHIKV suggest either a requisite role of D2 of mouse MXRA8 in facilitating binding to CHIKV or an inhibitory role of D2 of duck MXRA8 in preventing engagement of CHIKV.
Figure 6. Chimeric avian-mammalian MXRA8 interacts with both WEE and SF complex alphaviruses.
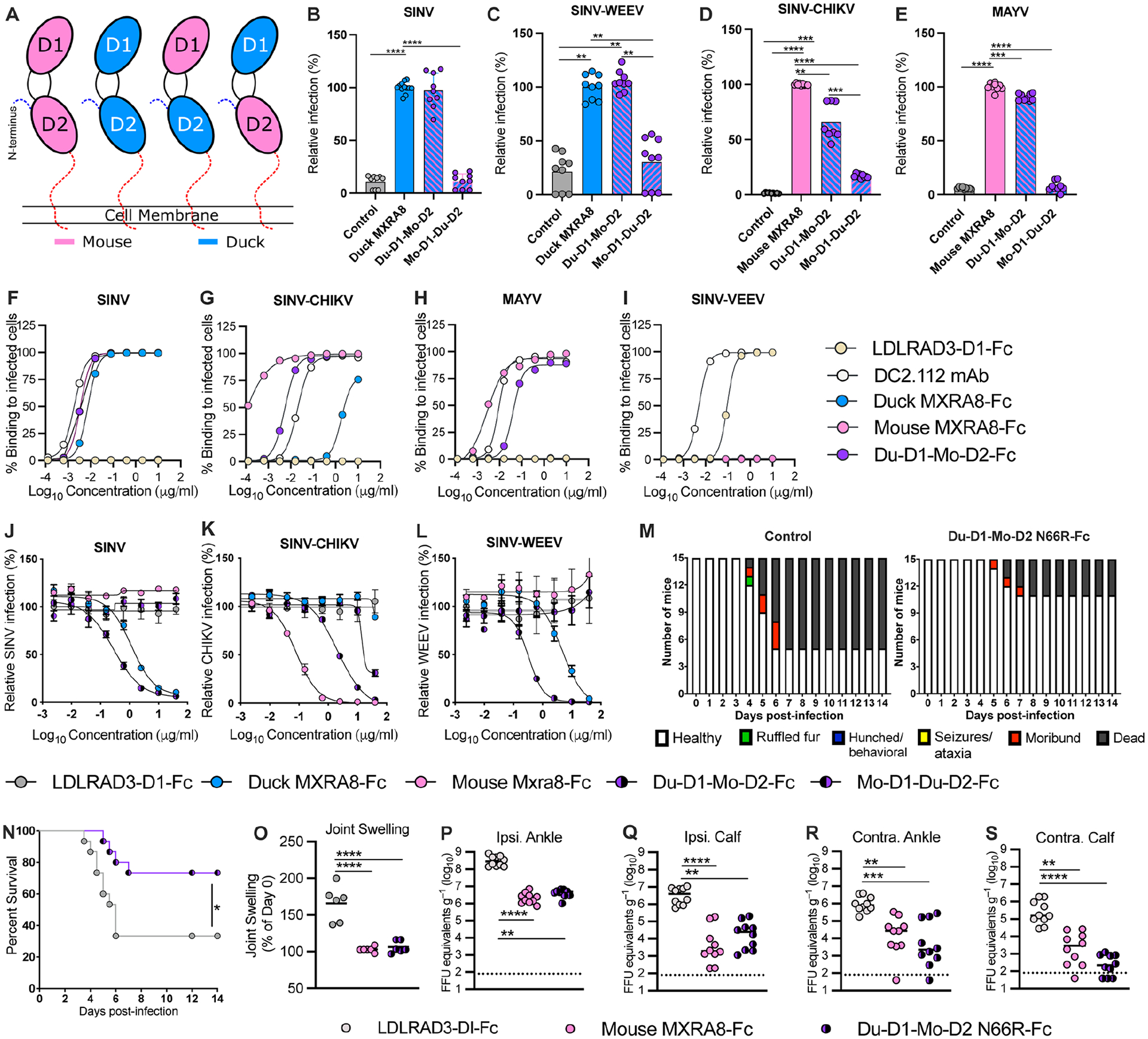
A. Schematic of chimeric MXRA8 proteins used to assess binding modes. In addition to mouse or duck MXRA8, chimeras shown are Du-D1-Mo-D2 and Mo-D1-Du-D2. B-C. SINV-GFP (B) and SINV-WEEV-GFP (C) infection in ΔMxra8 3T3 cells complemented with wild-type duck (blue) and chimeric (blue/pink) duck-mouse MXRA8 (3 experiments in triplicate; normalized to wild-type duck MXRA8 infection. Infection and GFP fluorescence were analyzed by flow cytometry. D-E. SINV-CHIKV (LR 2006 strain) (D) and MAYV (E) infection of ΔMxra8 3T3 cells complemented with an empty vector, or wild-type mouse and chimeric duck-mouse MXRA8 (3 experiments in triplicate; normalized to wild-type mouse MXRA8 infection). Infection was analyzed by flow cytometry by evaluating viral antigen staining with mAbs (CHIKV or MAYV). F-I. Staining of the surface of Vero cells infected with SINV-TR339 (F), SINV-CHIKV (LR 2006 strain), (G) MAYV (H), or SINV-VEEV (I) after incubation with serially diluted mouse MXRA8-Fc, duck MXRA8-Fc, Du-D1-Mo-D2 MXRA8-Fc, LDLRAD3-D1-Fc proteins or cross-reactive anti-E1 mAb (DC2.112, positive control)37) control. Data are expressed as the percentage of infected cells that bound positively to the indicated proteins by flow cytometry (representative of 2 experiments). J-L. Neutralization of SINV TRR399 (J) SINV-WEEV (K) or SINV-CHIKV (L) (all three viruses expressing eGFP) infection by duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo D2-MXRA8-Fc, Mo-D1-Du-D2-MXRA8-Fc, or LDLRAD3-D1-Fc control (3 experiments, duplicates). Infection was analyzed by flow cytometry (GFP expression) and normalized to levels after incubation with LDLRAD3-D1-Fc protein. M-N. Clinical disease (M) and survival (N) of 4-week-old female CD-1 mice inoculated with 103 PFU of WEEV (McMillan strain) mixed with Du-D1-Mo D2 N66R-MXRA8-Fc or LDLRAD3-D1-Fc. O-S. Foot swelling (O) and viral RNA levels in indicated tissues (P-S) of 4-week-old male C57BL/6J mice at 72 h after inoculation with 103 FFU of CHIKV (La Reunion 2006) mixed with 50 μg of mouse Mxra8-Fc, Du-D1-Mo-D2-N66R-MXRA8-Fc, or LDLRAD3-D1-Fc (O: 2 experiments n = 6; P-S: 2 experiments n = 10). B-E and O: one-way ANOVA with Dunnett’s post-test; mean ± standard deviation (SD). N: Log rank test. P-S: Kruskal-Wallis ANOVA with Dunn’s post-test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. See also Fig S11, S12, and Table S5.
To corroborate the infection patterns of chimeric MXRA8 using an orthogonal assay, we inoculated Vero cells with SINV, CHIKV, MAYV, or SINV-VEEV and stained the surface of infected cells, which display viral E1-E2 proteins prior to virion morphogenesis and budding19,20, with mouse MXRA8-Fc, duck MXRA8-Fc, Du-D1-Mo-D2 MXRA8-Fc, or LDLRAD3 D1-Fc10 fusion proteins (Fig 6F–I and Table S5). Whereas mouse MXRA8-Fc showed dose-dependent binding to cells infected with CHIKV and MAYV (Fig 6G–H), duck MXRA8-Fc bound avidly only to SINV-infected cells (Fig 6F), and neither bound to cells expressing VEEV structural proteins (Fig 6I). In contrast to data showing that duck MXRA8 does not support CHIKV infection (Fig 1B), higher (> 1 μg/ml) concentrations of duck-MXRA8-Fc bound to the surface of CHIKV-infected cells (Fig 6G); this binding could reflect differential interaction with cell surface-associated (unencapsidated) and virion-associated CHIKV E1-E2 heterodimers, which have different structural features21,22. In comparison, and consistent with infection experiments, Du-D1-Mo-D2 MXRA8-Fc bound to the surface of cells infected with SF and WEE complex viruses but not to those expressing VEEV structural proteins (Fig 6F–I); SINV-VEEV infected cells bound LDLRAD3-D1-Fc (Fig 6I), as expected10.
Chimeric duck-mouse MXRA8 inhibits infection of alphaviruses from both SFV and WEE complexes in vitro and in vivo.
Given the broader cell-surface reactivity of Du-D1-Mo-D2 for alphavirus E1-E2 proteins, we assessed the inhibitory activity of Du-D1-Mo-D2 MXRA8-Fc and Mo-D1-Du-D2-Fc against viruses displaying SF and WEE complex structural proteins. We pre-incubated SINV TR399, SINV-WEEV, and SINV-CHIKV with LDLRAD3-D1-Fc, duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo-D2 MXRA8-Fc, or Mo-D1-Du-D2 MXRA8-Fc and then inoculated ΔMxra8 3T3 cells expressing either duck or mouse MXRA8 (Fig 6J–L and Table S5). Duck and Du-D1-Mo-D2 MXRA8-Fc dose-dependently inhibited infection of SINV TR399 or SINV-WEEV in ΔMxra8 3T3 cells expressing duck MXRA8, whereas mouse MXRA8-Fc, Mo-D1-Du-D2 MXRA8-Fc, and LDLRAD3 D1-Fc did not (Fig 6J–K). However, Du-D1-Mo-D2 MXRA8-Fc showed more potent inhibition (EC50 values) of SINV TR399 (3-fold, P = 0.05) and SINV-WEEV (20-fold, P < 0.05) infection than duck MXRA8-Fc. Both mouse and Du-D1-Mo-D2 MXRA8-Fc proteins inhibited SINV-CHIKV infection in ΔMxra8 3T3 cells expressing mouse MXRA8, although the potency of mouse-Mxra8-Fc was greater (25-fold, P < 0.0001) than Du-D1-Mo-D2 MXRA8-Fc. In comparison, duck MXRA8-Fc and LDLRAD3-D1-Fc did not inhibit SINV-CHIKV infection. Together, these experiments support the cryo-EM-derived flipped binding orientation model (Fig 5B) and show this information can be used to develop decoy receptors that more broadly inhibit infection of alphaviruses.
To determine the basis for the different neutralization potencies of the MXRA8 decoy molecules we measured the affinity (KD) of monomeric MXRA8 for VLPs. Consistent with the neutralization data, chimeric Du-D1-Mo-D2 MXRA8 showed higher affinity binding than duck MXRA8 to WEEV VLPs (KD of 493 ± 6 nM and 5.2 ± 1.1 μM, respectively; Fig S11A–B), and mouse MXRA8 bound more strongly than Du-D1-Mo-D2 MXRA8 chimera to CHIKV VLPs (KD of 68 ± 17 nM and 545 ± 71 nM, respectively; Fig S11C–D). We also evaluated the influence of the stalk region of chimeric MXRA8 on binding since its length modulated binding of human MXRA8 to CHIKV in a prior study23. We generated Du-D1-Mo-D2 MXRA8-Fc molecules with a mouse (moStalk, 43 amino acids) or duck (duStalk, 38 amino acids) stalk, which are 35% identical at the amino acid level. Notably, Du-D1-Mo-D2 duStalk MXRA8-Fc and Du-D1-Mo-D2 moStalk MXRA8-Fc showed similar inhibitory activity against SINV-CHIKV and SINV-WEEV infection (Fig S11E–F).
To test the ability of chimeric Du-D1-Mo-D2 MXRA8-Fc to protect against alphaviruses in vivo, we generated Du-D1-Mo-D2 MXRA8-Fc with an N66R point mutation that facilitated greater expression in cell culture. Du-D1-Mo-D2 N66R MXRA8-Fc had similar neutralizing activity against CHIKV and WEEV (Fig S11G–H). We evaluated whether co-injection of Du-D1-Mo-D2 N66R MXRA8-Fc with WEEV (McMillan strain) would protect CD-1 mice from infection. Less clinical disease and higher survival rates after WEEV infection were observed in mice that received Du-D1-Mo-D2 N66R MXRA8-Fc than the LDLRAD3-D1-Fc control (Fig 6M–N; and Table S5). In an analogous experimental set-up, we tested whether co-injection of Du-D1-Mo-D2 N66R MXRA8-Fc with CHIKV (La Reunion 2006 strain) would diminish infection in C57BL/6 mice, as reported previously for mouse MXRA8-Fc9. Seventy-two hours after inoculation, joint swelling in the ipsilateral foot and CHIKV RNA levels in the ipsilateral and contralateral ankles and calf muscles were lower in mice treated with Du-D1-Mo-D2 N66R MXRA8-Fc or mouse MXRA8-Fc compared to the LDLRAD3-D1-Fc control (Fig 6O–S).
Reptile MXRA8 supports SINV infection.
To gain further insight into vertebrate class-specific MXRA8 binding, we assessed whether other non-avian MXRA8 species could support WEE complex virus infection. Based on our infection results with Du-D1-Mo-D2 MXRA8, we searched for species with D1 of MXRA8 that were similar to D1 of avian MXRA8, focusing in particular on the C”-D loop, which is a dominant contact area of duck MXRA8 and WEEV E1 (Fig S12). The top ‘hits’ included alligator, crocodile, and sea turtle (96%, 96%, and 93% identical to duck C”-D loop, respectively) MXRA8; for these proteins, no more than a single residue varied from duck MXRA8 in the C”-D loop binding pocket of D1 (Fig S12A). Of note, several species of reptiles can be infected by encephalitic alphaviruses, including WEE complex members24–27. Indeed, ΔMxra8 3T3 cells complemented with alligator or sea turtle MXRA8 (Fig S12B) supported SINV but not CHIKV infection (Fig S12C). These findings support our domain binding model for WEEV and D1 of duck MXRA8 and suggest that alphaviruses that infect reptiles might use MXRA8 to enter cells.
Du-D1-Mo-D2 binds to CHIKV and WEEV in opposing orientations.
To understand better how chimeric MXRA8 inhibits infection, we generated cryo-EM structures of Du-D1-Mo-D2 in complex with CHIKV or WEEV at 3.9 Å for both maps (Fig 7A–E and Table S3–S4). Du-D1-Mo-D2 preferentially binds to CHIKV at site 1, similar to mouse MXRA87, and WEEV at sites 4 and 3, similar to duck MXRA8 (Fig 7F). Du-D1-Mo-D2 binds to CHIKV and WEEV in flipped orientations with respect to each other (Fig 7E), consistent with how mouse and duck MXRA8 bind to CHIKV and WEEV, respectively. Du-D1-Mo-D2 can use duck D1 to bind either CHIKV E2-wrapped or WEEV E1-intraspike heterodimers. This binding topology model is supported by the location of the A and H β-strands in D1 and glycan density at Asn-120 in each model (Fig 7B and 7D).
Figure 7. Cryo-EM structure of Du-D1-Mo-D2 MXRA8 chimera bound to CHIKV VLP.
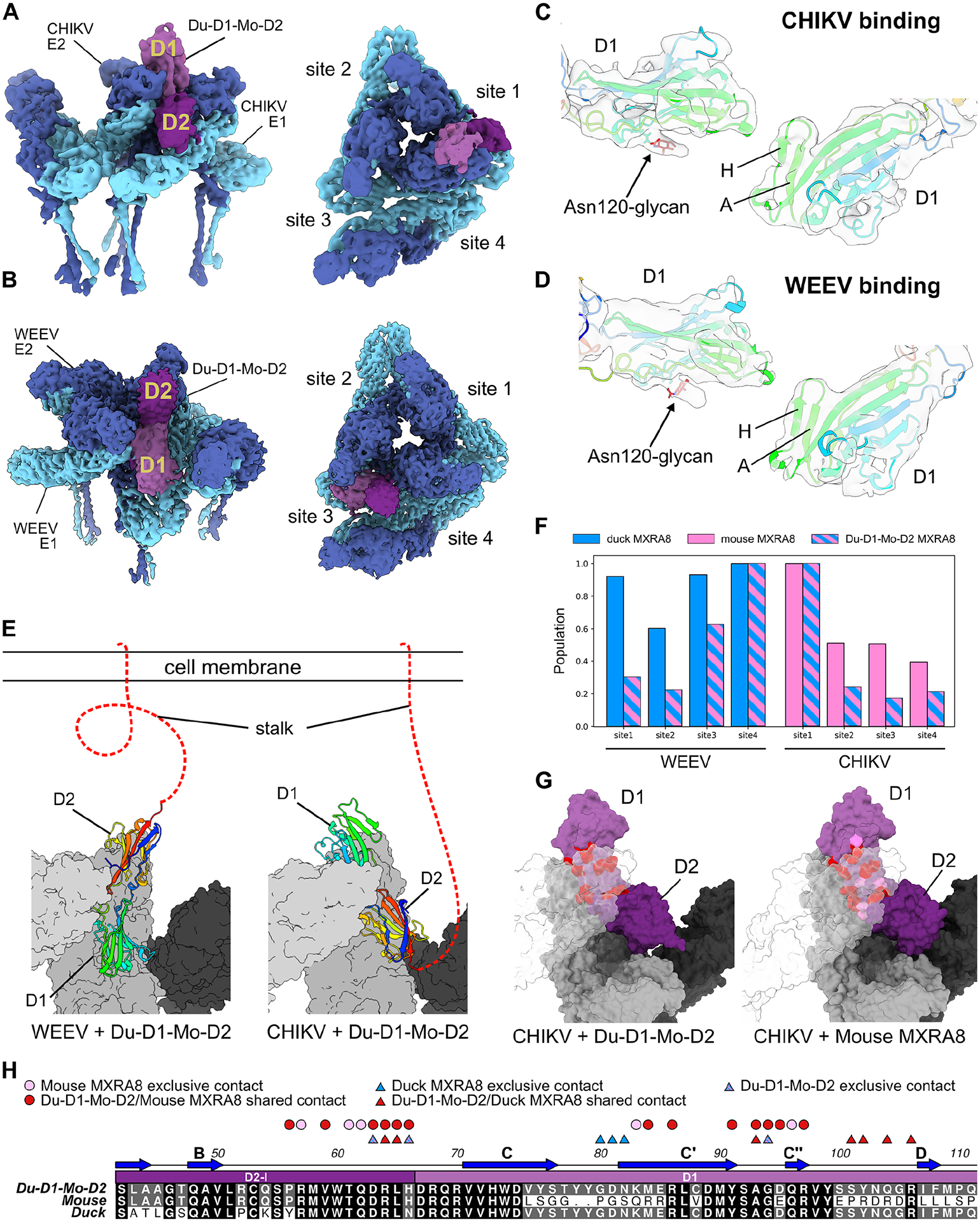
A-B. Cryo-EM density of Du-D1-Mo-D2 bound to CHIKV (A) or WEEV (B) VLPs. Shown are two views of a single asymmetric unit, with E1 (light blue), E2 (dark blue), D1 of Du-D1-Mo-D2 (light purple), and D2 of Du-D1-Mo-D2 (dark purple) at site 1. C-D. Atomic models of Du-D1-Mo-D2 MXRA8 with experimental density highlighting the symmetry breaking N-linked glycan at Asn120 and β-strands A and H in D1 when binding to CHIKV (C) or WEEV (D). E. Atomic models of Du-D1-Mo-D2 MXRA8 binding to WEEV (left) or CHIKV (right) highlighting the flipped orientations. Surfaces are shown for the wrapped E1-E2 heterodimer (light gray), intra E1-E2 heterodimer (gray), and inter E1-E2 heterodimer (dark gray). Du-D1-Mo-D2 is depicted as a ribbon and is rainbow-colored from the N- to C-terminus. Stalk attaching MXRA8 to the cell membrane is represented as a red dashed line. F. MXRA8 site occupancies for duck MXRA8 (light blue), mouse MXRA8 (pink), and Du-D1-Mo-D2 (blue and pink stripes) bound to WEEV (left) or CHIKV (right). G. Molecular models of Du-D1-Mo-D2 and mouse MXRA8 bound to CHIKV. Shown are wrapped E1-E2 heterodimer (light gray, transparent), intraspike E1-E2 heterodimer (gray), interspike E1-E2 heterodimer, MXRA8 D1 (light purple), and MXRA8 D2 (dark purple). Observed contact residues in both Du-D1-Mo-D2 and mouse MXRA8 are colored dark red, and contacts observed only for mouse MXRA8 are colored pink. H. Sequences of Du-D1-Mo-D2, mouse, and duck MXRA8, highlighting conserved contacts between Du-D1-Mo-D2 and mouse MXRA8 to CHIKV (red circles), contacts only in mouse MXRA8 to CHIKV (pink circles), conserved contacts between Du-D1-Mo-D2 and duck MXRA8 to WEEV (red triangles), contacts only in duck MXRA8 to WEEV (blue triangles), and contacts only in Du-D1-Mo-D2 to WEEV (blue and pink stripped triangles). See also Fig S5, S11, and Tables S2–S4.
D1 of duck MXRA8 is a suitable replacement for D1 of mouse MXRA8 when contacting CHIKV. Contacts on D1 of duck MXRA8 with CHIKV E2, when expressed as the Du-D1-Mo-D2 chimera, are congruent with D1 of mouse MXRA8 and CHIKV E2; for every residue of Du-D1 that contacts CHIKV E2, there is an equivalently positioned contact residue in D1 of mouse MXRA8 (Fig 7G–H). There are only 5 contact residues in D1 of mouse MXRA8 for which equivalently positioned residues in Du-D1-Mo-D2 do not contact CHIKV. In comparison, when mouse MXRA8 D2 is expressed in the Du-D1-Mo-D2 chimeric protein, it has no or few observed interspike and intraspike contacts with CHIKV E1, both of which are present in the structure of mouse MXRA8 bound to CHIKV7. In the Du-D1-Mo-D2 chimera, mouse MXRA8 D2 does not alter the binding of duck D1 to WEEV E1 when compared to duck MXRA8. Primary contact residues between β-strands C” and D are identical between D1 of duck MXRA8 and the Du-D1-Mo-D2 chimera. Overall, these studies show that chimeric Du-D1-Mo-D2 MXRA8 can bind to both CHIKV and WEEV in a similar topology as mouse MXRA8 and duck MXRA8, respectively, and that D1 of duck MXRA8 can functionally replace D1 of mouse MXRA8 to bind CHIKV when it is paired with D2 of mouse MXRA8.
DISCUSSION
In this study, we discovered that avian MXRA8 can function as a receptor for members of the WEE complex even though it does not act as a receptor for SF complex members that have mammalian reservoirs and can bind mammalian MXRA8. Members of the SF complex use mammalian but not avian MXRA8, and members of the WEE complex use avian but not mammalian MXRA8. VEEV, EEEV, and at least one virus (AURAV) in the WEE complex do not appear to utilize either avian or mammalian MXRA8.
Although we initially hypothesized that members of the SF and WEE complexes would engage mammalian or avian MXRA8 similarly, our cryo-EM analysis of avian MXRA8 bound to WEEV VLPs revealed a domain-inverted binding mode compared to mammalian MXRA8 and CHIKV7,8. Whereas both D1 and D2 of mammalian MXRA8 contact CHIKV, the binding of avian MXRA8 to WEEV is principally through D1 with minimal contact of D2 with WEEV. CHIKV E2 binds D1 of mammalian MXRA8 distal to the viral membrane, whereas CHIKV E1 binds D2 proximal to the viral membrane. In contrast, WEEV E1 engages a different set of residues in D1 of avian MXRA8 in a reverse orientation proximal to the viral membrane, with WEEV E2 making minimal or no contacts with D1 or D2 of avian MXRA8. A unique feature of MXRA8 is the head-to-head orientation of its two Ig-like domains7,8. This architecture generates a “pseudo-symmetric” molecule, such that a flip results in topological similarity and a broader range of virus engagement opportunities. This could offer a potential explanation as to how different MXRA8-alphavirus binding modes evolved across species.
Our results establish a critical interaction role of D1 of MXRA8, irrespective of the species, binding mode, or alphavirus. D1 of duck MXRA8 forms intraspike contacts with WEEV E1, and D1 of mouse MXRA8 forms wrapped contacts with CHIKV E2, with distinct residues at each interface. This result was unexpected because MXRA8 domains D1 and D2 have pseudo-symmetric folds and sequence relatedness across species: duck and mouse D1 are 43.7% identical, and duck and mouse D2 are 62.2% identical at the amino acid level. Despite the pseudo-symmetry relationships of MXRA8, we show that each virus utilizes independent binding schemes for initiation of infection. Our chimera and mutagenesis experiments, and the cryo-EM co-structures of Du-D1-Mo-D2, supported the domain-inverted binding model, as expression of Du-D1-Mo-D2 MXRA8 promoted infection of both WEE and SF complex members, whereas the Mo-D1-Du-D2 MXRA8 did not. The ability of Du-D1-Mo-D2 MXRA8-Fc protein to neutralize SINV and SINV-WEEV infection suggests a dominant role of D1 of avian MXRA8 in binding WEE complex viruses. The structure of Du-D1-Mo-D2 in complex with CHIKV also revealed that corresponding residues on D1 of duck MXRA8 can serve as replacements for interaction with D1 of mouse MXRA8, which likely contributes to the ability of membrane-associated and soluble Du-D1-Mo-D2 to support or inhibit infection, respectively, of SF-complex members. Overall, the structural data with Du-D1-Mo-D2 MXRA8 and CHIKV and the functional results with Du-D1-Mo-D2 MXRA8 and Mo-D1-Du-D2 MXRA8 and CHIKV are consistent with a required role of D2 of mammalian MXRA8 in facilitating binding and/or an inhibitory role of D2 of duck MXRA8 in preventing binding.
In support of the flipped-domain orientation model, introduction of mutations or loop insertions into D1 but not D2 of duck MXRA8 abrogated or reduced infection of WEE complex members. Associated with the swap in MXRA8 domains was a differential reliance on the viral structural proteins; WEEV principally uses E1 of the intraspike heterodimer to bind duck MXRA8, and based on the number of residue contacts at the binding interface7,8, CHIKV and MAYV predominantly use E2 of the wrapped spike heterodimer to engage murine or human MXRA8. Structural features of the complexes also offer potential explanations for the distinct binding specificities including shifted N-linked glycosylation of E1 at the intraspike contact site, which in the case of CHIKV might clash with avian MXRA8 and prevent engagement. Of note, SFV binds VLDLR through interactions in domain III of E128, which differ from the binding site on E1 that CHIKV and WEEV use to engage mammalian and avian MXRA8, respectively.
Based on deletion experiments, a prior study established that the 48-amino acid stalk length of human MXRA8 was critical for supporting CHIKV entry into cells8. Our swap experiments showed that in the context of the chimeric Du-D1-Mo-D2-Fc decoy receptor, duck (38 amino acids) and mouse (43 amino acids) stalk regions did not alter neutralization potency against SINV-CHIKV or SINV-WEEV. Thus, the stalk sequence does not appear to dictate avian or mammalian MXRA8 binding preferences to different alphaviruses, and the flip of duck-mouse chimeric MXRA8 orientation between its binding modes with WEEV and CHIKV is not sensitive to the small 5-amio acid difference in duck and mouse MXRA8 stalk lengths. However, because our cryo-EM images did not directly visualize the stalk region, we can only speculate on the conformations required for binding the different alphaviruses.
During the evolution of alphaviruses between birds, other vertebrates, and mammals, adaptation to MXRA8 as a receptor likely occurred resulting in changes in binding modes of the viral structural proteins in a vertebrate-class specific-manner. Although speculative, one of three evolutionary scenarios might explain the MXRA8 receptor usage patterns we observed: (a) there was a common viral ancestor of SF and WEE complexes that could use mammalian, avian, and reptile MXRA8. During evolution, the SF branch specialized to infect mammals and lost its capacity to engage avian or reptile MXRA8, whereas the WEE complex lost its ability to use mammalian MXRA8; (b) the ancestor of WEE complex viruses bound only avian or reptile MXRA8. During evolution, the virus adapted to mammalian MXRA8, which enabled the SF complex to branch and infect mammalian hosts. However, during the process of adaptation to mammalian MXRA8, SF complex viruses lost their ability to bind avian MXRA8 and use avian hosts; or (c) WEE and SF complexes alphaviruses adapted independently to use reptile, avian, and mammalian MXRA8.
Avian MXRA8 is not the only receptor reported for SINV in chicken cells, as a 67 kDa protein was identified previously29. Chicken MXRA8 is 50 kDa, which makes it unlikely that it is the previously recognized SINV receptor. Several other attachment or entry factors have been described for SINV in mammalian cells including a high-affinity laminin receptor30, VLDLR11, C-type lectins (DC-SIGN and L-SIGN)31, NRAMP232, and heparan sulfate33–35, although their significance in vivo remains uncertain and role in avian cell infection is unknown. Our findings of residual yet diminished infection by WEE complex alphaviruses in chicken cells after antibody blockade or gene editing of chicken MXRA8 suggests the existence of additional avian receptors.
The finding of an inverted binding mode enabled the engineering of a Du-D1-Mo-D2 chimera that can support infection of both WEE complex and SF complex viruses. Produced as a bivalent soluble decoy receptor, this chimera has neutralizing activity in cell culture and in vivo, and has potential as a broad-spectrum countermeasure against alphaviruses. We have shown using cryo-EM that the Du-D1-Mo-D2 chimeric MXRA8 can bind to CHIKV and WEEV using the respective flipped binding modes observed with mouse and duck MXRA8. We also used our structural and evolutionary analyses to identify other vertebrate MXRA8 proteins from reptiles that support WEE complex virus infection. These results could help to identify potential reservoirs of WEE complex viruses and the sources of future zoonotic outbreaks.
Limitations of the study.
We acknowledge several limitations of our study. (1) Although we establish the usage of avian MXRA8 by WEE complex alphaviruses in mammalian and chicken cells and in a mouse ectopically expressing chicken MXRA8, we did not establish the role of avian MXRA8 in the pathogenesis of these viruses in their enzootic hosts, birds. (2) We did not identify at what point in alphavirus evolution the flip in MXRA8 binding orientation and class specificity occurred. (3) While we generated an avian-mammalian chimeric MXRA8 decoy with broad-spectrum inhibitory activity against alphaviruses, we did not test its post-exposure activity. (4) The moderate resolutions of our cryo-EM reconstructions limit our ability to make mechanistic claims based on specific atomic interactions.
In summary, our study reveals how different domains of the E1 and E2 proteins of zoonotic alphaviruses engage divergent vertebrate receptors in distinct orientations to allow for unique tropisms. These findings enhance our understanding of how viruses evolve to infect new hosts and reveal insight into viral structure, receptor utilization, and class adaptation. Finally, the ability of chimeric Du-D1-Mo-D2 MXRA8-Fc to inhibit infection of both SF and WEE complex members in vitro and in vivo suggests the possible development of decoy proteins that broadly neutralize infection of alphaviruses from different antigenic groups.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact.
Further information and requests for resources and reagents should be directed to the Lead Contact, Michael S. Diamond (mdiamond@wustl.edu).
Materials availability.
All requests for resources and reagents should be directed to the Lead Contact author. This includes viruses, proteins, and cells. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data and code availability.
All data supporting the findings of this study are available within the paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not include original code. Cryo-EM data has been deposited in public databases (PDB: 8DAQ [WEEV VLP], 8DAN [WEEV VLP + Duck MXRA8], 8SQN [WEEV VLP + Du-D1-Mo-D2], and 8EWF [CHIKV VLP + Du-D1-Mo-D2]).
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cells.
NIH-3T3 (CRL-1658), HEK-293 (CRL-1573), BHK-21 (CCL-10), HeLa (ATCC CCL-1), K562 (CRL-3344), Jurkat (ATCC TIB-152), and Vero (CCL-81) cells, and CEFs (CRL-12203) were obtained from ATCC and cultured at 37°C in DMEM supplemented with 10% heat inactivated FBS (Hyclone), 100 U/ml penicillin, 100 mg/ml streptomycin, and 10 mM HEPES. Expi293 cells were obtained from Thermo Fisher and cultured shaking (100 RPM) at 37°C and 8% CO2 in Expi293 Expression Medium.
Viruses.
The following alphavirus strains were obtained from the World Reference Center for Emerging Viruses and Arboviruses (University of Texas Medical Branch, Galveston, TX; generous gifts of S. Weaver and K. Plante), propagated in BHK-21 or Vero cells, and titered by focus forming assay (FFA) as described38: BBKV (DAK AR Y 251), CHIKV (181/25), CHIKV Senegal (37997), OCKV (EDS 14), SINV (TR339, Toto, and Girdwood), WHAV (M 78), and WEEV (McMillan). Chimeric viruses including SINV-EEEV (FL93–939)-GFP, SINV-VEEV (TrD-GFP), SINV-WEEV (CBA87)-GFP, SINV-CHIKV (LR-2006)-GFP were constructed by replacing the structural protein genes of SINV TR339 with those from EEEV, VEEV, WEEV, or CHIKV33,39,40. eGFP expressing versions of these viruses and SINV TR339 were generated by inserting an eGFP sequence followed by the Thosea Asigna virus 2A self-cleaving peptide sequence after the capsid gene in the SINV TR339 molecular clone40. The chimeric alphaviruses and SINV TR339 were propagated in Vero cells and titered by focus forming assay (FFA) as described10. Viral RNA was generated by in vitro transcription (mMessage mMachine, Ambion) of linearized cDNA after digestion with XhoI. Viral RNA was introduced to BHK-21 cells by electroporation, supernatants were harvested 24 to 36 hours later, and stock titers were determined by focus-forming assay on Vero CCL81 cells. AURAV (BE AR 10315) was propagated in CEFs and tittered by FFA. AURAV, BBKV, OCKV, SINV, and WHAV antigens were detected using mouse SINV immune ascites fluid or mAb DC2.11237.
Mouse studies.
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01) or University of Pittsburgh (assurance number D16–00118). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering. Wild-type C57BL/6J and BALB/c mice were purchased from the Jackson Laboratory and CD-1 mice were obtained from Charles River Laboratories. Female BALB/c and CD-1 mice and male C57BL/6 mice were used in these studies.
House sparrow sample.
Tissues were collected from a captured adult house sparrow (Passer domesticus) from a ground trap (gift of R, Bowen, Fort Collins, CO). All experiments were performed after approval by the Institutional Animal Care and Use Committee at Colorado State University.
METHOD DETAILS
Recombinant adenoviral vector.
The human Ad5-chicken Mxra8 construct was designed using a codon optimized version of chicken Mxra8 (Gallus gallus; NP_989967) and synthesized commercially (Integrated DNA Technologies). The control Ad5 was reported previously41. Ad5-chicken Mxra8 and Ad5-empty vector control were propagated in 293T cells and purified using cesium chloride density-gradient ultracentrifugation. The virus particle number was determined by spectrophotometry using optical density (260 nm) measurement and plaque assays, as described42. The ability of the Ad5-chicken Mxra8 to induce surface expression of the transgene was established in HEK293 cells 20 h after transduction with Ad5-chicken Mxra8 (MOI, 20). Cells were detached using TrypLE (Thermo Fisher), washed with PBS and stained with a mixture of mouse anti-chicken mAb Mxra8 antibody (6D7, and 11A6 at 2 μg/mL) in PBS, 1% BSA, for 1 h at 4°C followed by APC-conjugated goat anti-mouse secondary antibody (Thermo Fisher, 2 μg/mL) for 30 min at 4°C. After washing and fixation with medium A (Thermo Fisher), data were collected on a MACSQuant X Flow cytometer and analyzed using FlowJo V9.
Mouse experiments.
BALB/c mice were purchased commercially (Jackson Laboratories). Animals were housed in groups and fed standard chow diets. Five-week-old female BALB/c mice were administered 1010 virus particles of hAdV5-chicken Mxra8 or hAdV5-control via intranasal administration. In some experiments, 0.5 mg of anti-IFNAR1 mAb (MAR1–5A3 (18, Leinco) was administered via intraperitoneal route four days after hAdV5 treatment. Five days later mice were inoculated with 104 PFU of SINV. Weights were monitored daily, animals were sacrificed at 40 hpi or 3 dpi, and tissues were harvested.
103 FFU of CHIKV (La Reunion 2006) was mixed with mouse MXRA8-human Fc, Du-D1-Mo-D2-N66R MXRA8-human Fc or LDLRAD3-D1-Fc (50 μg per mouse in PBS) and incubated at 37°C for 30 min before subcutaneous inoculation of 4-week-old male C57BL/6 mice in the footpad. At 72 h post-inoculation, animals were euthanized, and after perfusion with PBS, indicated tissues were collected and processed for viral RNA levels. Joint swelling in the ipsilateral foot was monitored at 72 h post-infection by measuring width × height using digital calipers as previously described43. 103 PFU of WEEV was mixed with 50 μg of Du-D1-Mo-D2-N66R MXRA8 or control (LDLRAD3-DI-human Fc) for 30 min at room temperature before subcutaneous inoculation of 4-week-old CD-1 female mice in the footpad. Mice were monitored daily for survival and clinical signs of disease. Clinical signs were assigned by the following criteria: 0 - healthy; 1 - ruffled fur, mild behavioral changes; 2 - hunched posture, significant behavioral changes; 3 - seizures, ataxia, catatonia; 4 - recumbent moribundity; 5 - death. Mice scoring 3 or higher were immediately euthanized.
Plasmid construction for complementation studies.
MXRA8 cDNA fragments were generated. The MXRA8 signal peptide was replaced with a β2-microglobulin signal peptide (MARSVTLVFLVLVSLTGLYA), FLAG tag (DYKDDDDK) and a short linker (GGS). Nucleotide sequences were codon-optimized, synthesized, and inserted into the lentivirus vector pLV-EF1a using an In-Fusion HD Cloning Kit (Takara) for the following species: mouse Mus musculus (Genbank accession no. NM_024263); turkey, Meleagris gallopavo (XP_010721105.1); duck, Anas platyrhynchos (XM_027443263); chicken, Gallus gallus (NP_989967); house sparrow, Passer domesticus; green sea turtle, Chelonia mydas (XP_007070253.1); and alligator, Alligator mississippiensis (XP_006263071.1). Chimeric Mo-D1-Du-D2 MXRA8, Du-D1-Mo-D2 MXRA8 along with duck C-C’, C”-D, and D-E loop MXRA8 mutants were designed (Fig S10), codon-optimized, synthesized, and inserted into the lentivirus vector pLV-EF1a as described above. Plasmids were transformed into One Shot Stbl3 Chemically Competent E. coli (Thermo Fisher), and bacteria were grown at 30°C on LB Agar plates with carbenicillin (100 μg/ml). Colonies were picked and grown overnight at 30°C in LB supplemented with carbenicillin (100 μg/ml). Plasmids were extracted (Qiagen) and sequenced using the following primers: GCACTTGATGTAATTCTCCTTGGAATTTGC, CTCAAGCCTCAGACAGTGGTTCAAAGT and GGTGGAAAATAACATATAGACAAACGCAC. Primers were used to sequence the following species: Meleagris gallopavo: ATGCTTTTACAGATGGTAACTTCAG and TGAACATCACTGATACTGCCTTTG; Anas platyrhynchos: TCAGGGGAGAATATTTATGCCACAA and TGAATATAACCGATACTGCTTTCGC; Gallus gallus: CAGGGGAGGATACTGATGCC and TGGGCCCCTCTTTATCCGA.
Expression and purification of MXRA8 proteins.
A cDNA fragment encoding residues 22–327 of the chicken MXRA8 extracellular domain (Gallus gallus; NP_989967) was codon-optimized, synthesized, and inserted into the pET21a vector using the NdeI/NotI sites. After sequence confirmation, the plasmid was transformed into BL21(DE3) chemically competent cells (Thermo Fisher). Cells were grown at 37°C to an optimal density (600 nm) of 0.8 and induced with 0.1 mM IPTG for 4 h. Cells were harvested and resuspended in 50 mM Tris-HCl, 1 mM EDTA, 0.01% NaN3, 1 mM DTT, 25% sucrose (TENDS) buffer, and lysed in 50 mM Tris-HCl, 1 mM EDTA, 0.01% NaN3, 1 mM DTT, 200 mM sodium chloride, 1% sodium deoxycholate and 1% Triton X-100. Inclusion bodies were isolated from the cellular lysate after centrifugation at 6,000 × g for 20 min and washed in TENDS buffer supplemented with 100 mM NaCl and 0.5% Triton X-100. A final wash was performed in the same buffer without 0.5% Triton X-100. Inclusion bodies were denatured in in 100 mM Tris-HCl, 6 M guanidinium chloride and 20 mM β-mercaptoethanol for 1 h. Denatured protein was oxidatively refolded overnight at 4°C in 400 mM L-arginine, 100 mM Tris-HCl, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 10 mM EDTA and 200 mM phenylmethylsulphonyl fluoride. Refolded protein was concentrated using a 10,000-molecular weight cut-off stirred cell concentrator (EMD Millipore). Concentrated protein was purified by HiLoad 16/600 Superdex 75 size exclusion chromatography (GE Healthcare) and HiTrap Q HP anion exchange chromatography (GE Healthcare). Purity and oligomeric state were confirmed by SDS-PAGE analysis and size exclusion chromatography.
Chicken MXRA8-Fc, duck MXRA8-Fc, and chimeric Du-D1-Mo-D2 duStalk MXRA8-Fc, Du-D1-Mo-D2 moStalk MXRA8-Fc, Du-D1-Mo-D2-N66R MXRA8-Fc, or Mo-D1-Du-D2 MXRA8-Fc fusion proteins were generated based on a prior protocol used to generate mouse-MXRA8-Fc9. Briefly, cDNA fragments encoding chicken MXRA8 (Gallus gallus; NP_989967; residues 22–328), duck MXRA8 (Anas platyrhynchos; XM_027443263; residues 23–334), Chimeric Du-D1-Mo-D2 duStalk MXRA8 (duck residues 66–196 and 296–334; mouse residues 23–65 and 195–293), chimeric Du-D1-Mo-D2 moStalk MXRA8 (duck residues 66–196; mouse residues 23–65 and 195–334) or chimeric Mo-D1-Du-D2 MXRA8 (mouse residues 65–196; duck residues 29–64 and 199–334) were appended with a GGGGSGGGGS linker and the mouse IgG2b Fc or human Fc region before synthesis (Integrated DNA Technologies) and inserted into the pCDNA3.4 vector. MXRA8-Fc plasmids were diluted in Opti-MEM, incubated with HYPE-5 reagent (OZ Biosciences), and the complex was transfected into 106 cells/ml of Expi-293 cells (Thermo Fisher). Cells were supplemented daily with Expi293 medium and 2% (w/v) Hyclone Cell Boost. Four days post transfection, the supernatant was harvested by centrifuging at 3,000 × g for 15 min and protein was purified using Protein A Sepharose 4B (Thermo Fisher). Chicken MXRA8-Fc fusion protein was eluted using Pierce™ Gentle Ag/Ab Binding and Elution Buffer Kit (Thermo Fisher). After elution, MXRA8-Fc proteins underwent buffer exchange with PBS and stored at 4°C. Purity was confirmed by SDS-PAGE analysis.
A histidine-tagged duck MXRA8 was generated in Expi293 mammalian cells. Briefly, a cDNA fragment encoding duck MXRA8 (Anas platyrhynchos; XM_027443263; Residues 23–328), GGS linker, and C terminal 8X His tag. The duck MXRA8-His tag was synthesized (Integrated DNA Technologies) and inserted into the pCDNA3.4 vector. The duck MXRA8-His plasmid was diluted in Opti-MEM, incubated with HYPE-5 reagent (OZ Biosciences), and the complex was transfected into 106 cells/ml of Expi-293 cells (Thermo Fisher). Cells were supplemented daily with Expi293 medium and 2% (w/v) Hyclone Cell Boost. Four days post-transfection, the supernatant was harvested by centrifuging at 3,000 × g for 15 min, and protein was purified using Cobalt resin (G-Biosciences). After elution with 200 mM imidazole, duck MXRA8-His tag protein underwent buffer exchange with PBS and stored at 4°C. Purity was confirmed by SDS-PAGE analysis.
Sparrow sample collection and sequencing.
Muscle and liver samples were obtained from a captured house sparrow (gift of R, Bowen, Fort Collins, CO). Total RNA was extracted using a MagMax Viral Isolation Kit (Thermo Fisher). First strand cDNA was generated using a SuperScript III First-Strand Synthesis System (Thermo Fisher). House sparrow MXRA8 specific primers were designed from deposited sequences of related species and assembled whole genome sequences. PCR master mixes were prepared in a nucleic acid-free PCR workstation. House sparrow MXRA8 was amplified using PCR with 1X Q5 Reaction Buffer, 200 μM dNTPs, 200 nM Forward primer, 200 nM Reverse primer, 2 μL of cDNA, 1X Q5 High GC Enhancer, and 1 μL of Q5 High-Fidelity DNA Polymerase (NEB). The following amplification protocol was used for both first- and second-round amplifications: (1) 98°C for 30 sec; (2) 98°C for 20 sec; (3) 60–68°C for 30 sec; (4) 72°C for 60 sec; and (5) 72°C for 2 min; with steps 2–4 repeated for 35 cycles. All primer sequences and annealing temperatures are listed in Table S1. PCR products were separated on a 1% agarose gel. PCR products were cloned using Zero Blunt™ TOPO™ PCR Cloning Kit and transformed into One Shot TOP10 Chemically Competent E. coli (Thermo Fisher). Ten selected colonies were sequenced using predefined primers (M13 forward and M13 reverse).
Anti-chicken MXRA8 mAb generation.
Four-week-old BALB/c mice were sequentially immunized and boosted via intravenous route with 20 μg of bacterially-generated, purified chicken MXRA8 at two-week intervals. Four and eight weeks later, mice were boosted with 20 μg of mammalian cell-derived chicken MXRA8-mouse-Fc. After boosting was completed, serum samples were collected and tested for binding to chicken MXRA8 on the surface of complemented ΔMxra8 3T3 cells. Three days after the final boost, the mouse with the highest serum titer (e.g., 1/27,000) for binding chicken MXRA8 underwent a terminal bleed and euthanasia. The spleen of this animal was collected for splenocyte-myeloma fusion and hybridoma production.
Hybridoma supernatants were screened by ELISA for binding to chicken MXRA8. As a second confirmatory assay, we evaluated the binding of hybridoma supernatants to chicken MXRA8 on the surface of complemented ΔMxra8 3T3 cells using flow cytometry. Finally, as a tertiary screen, we tested hybridoma supernatants for blockade of SINV TR339 infection in CEFs. After limiting dilution subcloning, the four clones with the strongest blocking activities (6D7, 9B6, 11A6 and 16D9) were expanded. Antibodies were purified using Protein A Sepharose 4B chromatography (Invitrogen #101042), dialyzed in PBS, concentrated, and sterile-filtered.
Complementation and infection experiments.
Lentiviruses encoding MXRA8 were packaged with psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) vectors in HEK293 cells using FugeneHD (Promega). ΔMxra8 3T3 cells9, ΔMXRA8 CEFs (see below), 293T, HeLa, K562 and Jurkat cells, were transduced with lentiviruses and selected with blasticidin for 7 days. Surface expression of mouse or avian MXRA8 proteins was assessed using rabbit anti-FLAG mAb (Cell Signaling; 1 μL per 500 μL) and Alexa Flour 647 conjugated anti-rabbit IgG (Cell Signaling; 1 μL per 500 μL). Surface expression of mouse MXRA8 also was assessed by flow cytometry after staining with a pool of seven hamster anti-mouse MXRA8 mAbs9 (2 μg/mL), and Alexa Fluor 647 conjugated goat anti-Armenian hamster IgG (2 μg/mL) at 4°C. Surface expression of chicken MXRA8 in some instances was assessed by flow cytometry after staining with a pool of four mouse anti-chicken MXRA8 mAbs (6D7, 9B6, 11A6 and 16D9; 2 μg/mL) and Alexa Flour 647 conjugated goat anti-mouse IgG (2 μg/mL). ΔMxra8 3T3, 293T, HeLa, K562, Jurkat, or CEF cells complemented with avian MXRA8 or murine Mxra8 with surface expression levels of MXRA8 less than 90% after blasticidin selection were enriched further by fluorescence activated cell sorting. Cells (2.5 × 105) were incubated with a rabbit anti-Flag mAb (Cell Signaling) (2 μg/ml) in 1% BSA/PBS for 30 min at 4°C. After 30 min, cells were washed and incubated with Alexa Fluor 647 conjugated goat anti-rabbit IgG (2 μg/mL). After a 30-min incubation, cells were washed, resuspended in PBS supplemented with 2% FBS and 1 mM EDTA, and sorted using a BD FACSAria II. MXRA8+ cells were expanded in culture.
Human LDLRAD3 (NM_174902.4) and human VLDLR (NP_001018066.1) were codon-optimized and synthesized (GeneWiz) and inserted into the lentivirus vector pLV-EF1a-IRES-Hygro (Addgene no. 85134) between the BamHI and MluI restriction enzyme sites using In-Fusion HD Cloning (Takara). The signal peptide of the genes was replaced by human β2-microglobulin signal peptide/FLAG-tag/GGS linker. The plasmids were packaged in HEK-293 cells with psPAX2 (Addgene no. 12260) and pMD2.G (Addgene no. 12259) using Lipofectamine 3000 (Thermo Fisher) and then transduced into Jurkat cells or K562 grown in RPMI-1640 with 10% FBS, 100 U/ml penicillin, 100 U/ml streptomycin and 0.05 mM β-mercaptoethanol. One day later, Jurkat or K562 cells selection with 200 μg/ml of hygromycin (InvivoGen) was initiated for seven days. LDLRAD3 and VLDLR expression was verified with anti-FLAG antibody as described above.
Complemented ΔMxra8 3T3 cells were inoculated with CHIKV 181/25 (MOI 3, 9 h) and SINV-CHIKV-LR (MOI 1, 9.5 h) in DMEM supplemented with 2% FBS or with SINV (TR339, Toto, or Girdwood; MOI 1, 9 h; TR339-GFP MOI 1, 10 h), BBKV (MOI 1, 8 h), OCKV (MOI 1, 8 h), AURAV (MOI 1, 24 h), WHAV (MOI 1, 24 h), SINV-EEEV (MOI 0.1, 9 h), SINV-WEEV (MOI 0.1, 9 h) or SINV-VEEV (TrD) (MOI 0.1, 9 h) in DMEM supplemented with 10% FBS. At indicated time points, cells were harvested, incubated sequentially with Fixation medium A (Thermo Fisher) and Permeabilization medium B (Thermo Fisher), and stained for viral antigen after incubation with the following antibodies: CHIKV (mouse mAb CHK-1144, mouse anti-SINV immune ascites fluid (ATCC) or human anti-E1 DC2.112 and DC2.31537. Cells were washed, incubated with Alexa Fluor 647 conjugated goat anti-mouse IgG (Thermo Fisher) or goat anti-human IgG (Southern Biotech), and analyzed by flow cytometry using a MACSQuant Analyzer 10 (Miltenyi Biotec).
CEFs, ΔMxra8 CEFs, and complemented CEFs cells were inoculated with CHIKV 181/25 (MOI 1, 10 or 24 h), SINV-WEEV (MOI 0.1, 9 h), SINV (MOI 1, 8 h), BBKV (MOI 1, 8 h), OCKV (MOI 1, 22 h), AURAV (MOI 1, 24 h), WHAV (MOI 1, 17 h) and SINV-VEEV (TrD) (MOI 0.1, 9 h) in 10% FBS growth medium. After infection, cells were harvested, fixed, permeabilized, stained with virus-specific antibodies, and analyzed by flow cytometry as described above.
293T cells complemented with chicken, sparrow or murine MXRA8 were inoculated with CHIKV 181/25 (MOI 1, 12 h), CHIKV-37997 (MOI 1, 8 h), SINV (MOI 1, 9 h) and AURAV (MOI 1, 24h) in 10% FBS growth medium. HeLa cells complemented with chicken or mouse MXRA8 were inoculated with CHIKV 181/25 (MOI 1, 14 h), SINV (MOI 1, 12 h), AURAV (MOI 1, 24 h), in 10% FBS growth medium. After infection, all cells were harvested, fixed, permeabilized, stained with virus-specific antibodies, and analyzed by flow cytometry as described above.
Jurkat cells complemented with mouse MXRA8, turkey MXRA8, or human LDLRAD3 were inoculated with CHIKV 181/25 (MOI 10, 16 h), SINV-TR339-GFP (MOI 10, 16 h), or SINV-VEEV-GFP (MOI 10, 16 h) in 2% FBS growth medium. After infection, all cells were harvested, fixed, permeabilized, stained with virus-specific antibodies (if not GFP-tagged), and analyzed by flow cytometry as described above.
K562 cells complemented with mouse MXRA8, turkey MXRA8, or human VLDLR were inoculated with SINV-EEEV-GFP (MOI 10, 16 h) in 2% FBS growth medium. After infection, all cells were harvested, fixed and analyzed by flow cytometry as described above.
For multi-step growth curves, complemented ΔMxra8 3T3 and CEFs were inoculated (MOI 0.01) with SINV TR399 or OCKV for 2 h, washed 3 times with warmed media (37°C), and maintained in 2% FBS growth medium. Viral supernatants were harvested at indicated time points, titered on Vero cells using a focus-forming assay as previously described45. After fixation, cells were permeabilized, and stained with anti-SINV ascites and horseradish peroxidase-conjugated goat anti-mouse IgG. Infected foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 Macroanalyzer (Cellular Technologies).
Virus binding and internalization assays.
For virus binding assays, ΔMxra8 3T3 cells complemented with mouse Mxra8 or chicken MXRA8 or an empty vector were incubated with SINV TR339 (MOI of 1) in DMEM 2% FBS for 1 h on ice. After six rinses with ice cold PBS, cells were lysed in RLT buffer (Qiagen). For attachment inhibition assays, 100 μg/mL of the indicated mAbs were incubated with 3T3 cells for 30 min at 4°C. SINV (MOI, 1) was added to the chilled cells and incubated at 4°C for 1 h. Cells were rinsed six times with ice cold PBS and lysed in RLT buffer (Qiagen). For the internalization assay, virus binding was repeated but after rinsing, DMEM with 2% FBS at 37°C was added for 10, 20, or 30 min. Cells were washed and treated with 100 μg/mL of proteinase K (Invitrogen) for 15 min at 37°C. Proteinase K was rinsed away, and cells were treated with 100 μg/mL of RNAse A for 30 min at 37°C. Cells were rinsed six times with ice cold PBS and lysed in RLT buffer (Qiagen). RNA was isolated using a MagMax Viral Isolation Kit. SINV RNA and GAPDH RNA was quantified using a Taqman RNA-to-Ct 1-step kit with either SINV primers/probe (see above) or murine Gapdh primers/probe: FOR Gapdh: 5′-GTGGAGTCATACTGGAACATGTAG-3′; REV Gapdh: 5′-AATGGTGAAGGTCGGTGTG-3′; and probe: 5′ 6-FAM/TGCAAATGG/ZEN/CAGCCCTGGTG/3′ IABkFQ.
Blocking assays with anti-chicken MXRA8 mAbs.
CEFs (2.5 × 104) were seeded into 96-well plates. Twelve hours later, cells were incubated with mAbs (10 μg/mL) for 1 h at 37 °C in a volume of 50 μL, and then viruses (SINV (MOI 1, 8 h), SINV-WEEV (MOI 0.1, 8 h), AURAV (MOI 1, 24 h), WHAV (MOI 1, 17 h), BBKV (MOI 1, 8 h), OCKV (MOI 1, 22 h) or SINV-VEEV (TrD) (MOI 1, 9 h) in 50 μL were added and incubated for 8 h. Cells were collected, fixed, permeabilized, and E1 protein expression (except for SINV-VEEV-GFP) was monitored by flow cytometry after incubation with anti-E1 DC2.112 and DC2.31537.
Neutralization assays with MXRA8-Fc fusion proteins.
ΔMxra8 3T3 cells (2.5 × 104) complemented with either mouse or duck MXRA8 were seeded into 96-well plates overnight. SINV-CHIKV-LR 2006 virus (MOI of 1), SINV-TR339-GFP (MOI of 1) or SINV-WEEV-GFP (MOI of 0.1) were pre-incubated with increasing doses of LDLR3-D1-Fc10, duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo-D2 MXRA8-Fc, Du-D1-Mo-D2-N66R MXRA8-Fc or Mo-D1-Du-D2 MXRA8-Fc proteins for 1 h at 37°C in a volume of 100 μL. Subsequently, viruses-receptor complexes were added to cells for 9 (SINV-CHIKV) or 10 (SINV-TR339-GFP and SINV-WEEV-GFP) h. Cells then were collected, fixed, and viral antigen expression was measured by flow cytometry.
Surface staining of infected cells with MXRA8-Fc fusion proteins.
Vero cells were inoculated (multiplicity of infection [MOI] of 5 to 10) with SINV-TR339, CHIKV 181/25, MAYV or SINV-VEEV in DMEM supplemented with 2% FBS. After allowing infection to proceed for 14 to 18 h, cells were detached using TrypLE (Thermo Fisher) and washed with PBS. Cells were incubated with LDLRAD3-D1-Fc, Mouse MXRA8-Fc, Duck MXRA8-Fc, Du-D1-Mo-D2-Fc fusion proteins, or human anti-E1 DC2.112 mAb for 30 min at 4°C in PBS, 2% FBS. Cells were washed and incubated with Alexa Fluor 647-conjugated goat anti-human or anti-mouse IgG (1:2000 dilution; Thermo Fisher) for 30 min at 4°C. Cells were washed and resuspended in PBS, 2% FBS, buffer containing 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/mL) to stain dead cells and subjected to flow cytometry analysis using an iQue3 flow cytometer (Sartorius).
BLI-based competition binding assay.
Binding of mouse, duck, or Du-D1-Mo-D2 MXRA8 to captured CHIKV and WEEV VLP was monitored in real-time at 25°C using an GatorPlus device (GatorBio) and analyzed using on-board software (GatorBio) or BiaEvaluation Software (Biacore). All experiments were performed with 1x PBS supplemented with 1% BSA. Anti-mouse IgG Fc biosensors (GatorBio #160004) were incubated with 5 μg/mL of CHK-265 (Fox 2015) or WEEV-209 (unpublished) for 100 sec then, after washing in running buffer for 60 sec, CHIKV VLPs or WEEV VLPs were captured at a nominal concentration of ~20 μg/mL for 300 sec. VLP-coated biosensors were then submerged into the indicated concentrations of monovalent mouse, duck and Du-D1-Mo-D2 MXRA8 proteins cleaved from the Fc-fusions MXRA8 proteins using GlySERIAS (Genovis #A0-GS6).
Gene editing of CEFs.
Chicken Mxra8 was targeted for gene editing in CEFs using sgRNAs to chicken Mxra8 (Chicken Mxra8 sgRNA: ACAGCTCCTACAACCAAGGG). Two sgRNA (CTGAAAAAGGAAGGAGTTGAG and AAGATGAAAGGAAAGGCGTT) that do not target the chicken genome were included as negative controls. The sgRNAs were cloned into lentiCRISPR v.2 (Addgene no. 52961) and packaged in HEK-293 cells with psPAX2 (Addgene no. 12260) and pMD2.G (Addgene no. 12259) using FuGENE® HD (Promega). CEFs were transduced with lentiviruses and selected for 7 days in the presence of 1 μg/mL of puromycin. Clonal cell lines were obtained by limiting dilution. Mxra8 gene editing was confirmed by next generation sequencing on an Illumina HiSeq 2500 platform (Genome Technology Access Center of Washington University) with 300-base-pair paired-end sequencing.
MXRA8 direct binding assays.
Maxisorp ELISA (Thermo Fisher) plates were coated with 4N12 anti-CHIKV mAb (2 μg/mL)46 overnight in sodium bicarbonate buffer, pH 9.3. Plates were washed four times with PBS and 0.05% Tween-20, and blocked with 4% BSA for 1 h at 25°C. CHIKV VLPs (1 μg/mL)14 were added in 2% BSA, and incubated for 2 h at 25°C. Chicken MXRA8-Fc, murine MXRA8-Fc, H77 (anti-HCV) mAb, and CHK-265 mAb were added in serial dilutions in 2% BSA, for 1 h at 25°C. Plates were washed with PBS and 0.05%Tween-20, and incubated with horseradish peroxide conjugated goat anti-mouse IgG (H + L) (1:2,000 dilution, Jackson ImmunoResearch) for 0.5 h at 25°C. After washing, plates were developed with 3,3’−5,5’ tetramethylbenzidine substrate (Thermo Fisher) and stopped with 2 N H2SO4. Plates were read at 450 nM using a TriStar Microplate Reader (Berthold).
Maxisorp ELISA plates were coated directly with WEEV VLPs (10 μg/mL)13 overnight in sodium bicarbonate buffer, pH 9.3. Plates were washed four times with PBS and 0.05% Tween-20, and blocked with 4% BSA for 1 h at 25°C. Chicken MXRA8-Fc, mouse MXRA8-Fc, H77 mAb and CHIK-265 mAb were added in serial dilutions in 2% BSA, for 1 h at 25°C. Plates were washed with PBS and 0.05% Tween-20, and incubated with horseradish peroxide conjugated goat anti-mouse IgG (H + L) (1:2,000 dilution, Jackson ImmunoResearch) for 0.5 h at 25°C. After washing, plates were developed with 3,3’−5,5’ tetramethylbenzidine substrate (Thermo Fisher) and stopped with 2 N H2SO4. Plates were read at 450 nM using a TriStar Microplate Reader (Berthold).
Maxisorp ELISA plates were coated with anti-His tag antibody (2 μg/mL, GenScript)13 overnight in sodium bicarbonate buffer, pH 9.3. Plates were washed four times with PBS and 0.05% Tween-20, and blocked with 4% BSA for 1 h at 25°C. His-tagged mouse MXRA8, duck MXRA8, sparrow MXRA8, and SARS-CoV-2 receptor binding domain (RBD) were added at 10 μg/mL in 2% BSA, for 1 h at 25°C. Plates were washed with PBS and 0.05% Tween-20, and incubated with WEEV VLPs or CHIKV VLPs at 10 μg/mL in 2% BSA, for 1 h at 25°C. Plates were washed with PBS and 0.05% Tween-20, and incubated with anti-WEEV-204 mAb or anti-CHIKV-265 mAb after serial dilutions, in 2% BSA, for 1 h at 25 °C. Plates were washed with PBS and 0.05%Tween-20, and incubated with horseradish peroxide conjugated goat anti-mouse IgG (H + L) (1:2,000 dilution, Jackson ImmunoResearch) for 30 min at 25°C. After washing, plates were developed with 3,3’−5,5’ tetramethylbenzidine substrate (Thermo Fisher) and stopped with 2 N H2SO4. Plates were read at 450 nM using a TriStar Microplate Reader (Berthold).
Viral burden analysis.
RNA was extracted from serum and tissues using the MagMax-96 Viral RNA Isolation Kit (Thermo Fisher). SINV viral RNA levels were quantified by qRT-PCR using a TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher), compared to a SINV and OCKV RNA standard curve, and expressed on a log10 scale as viral focus-forming unit (FFU) equivalents per gram of tissue or milliliter of serum. Primers and probes used are as follows: SINV FOR: 5′ -AAGATCATCGACGCAGTCATC-3′; SINV REV: 5′- GCTGTGGAAGTAACCGAATCT −3′; SINV Probe: 5′-/56 FAM/CCACCTTAC/ZEN/TTCTGCGGCGGATTTA/3IABkFQ/−3’. OCKV FOR: 5′ -AGTTGGCTGTTTGCCCTT-3′; OCKV REV: 5′-CGTGTGCTAGTCAGCATCAT-3′; SINV Probe: 5′-/56-FAM/TAATTAATA/ZEN/GCGACGAGGCGCCGC/3IABkFQ/−3’.
SINV infectious virus burden was evaluated using focus-forming assays as described above. Vero cells were incubated with serial dilutions of homogenized lung tissue samples. After 16 h, cells were fixed, permeabilized and stained as described above. Infected foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 Macroanalyzer (Cellular Technologies).
Cryo-EM sample preparation, data collection, and single particle reconstruction.
WEEV-VLP with and without avian or Du-D1-Mo-D2 MXRA8, and CHIKV with Du-D1-Mo-D2 MXRA8, in molar excess were flash-cooled on holey carbon EM grids in liquid ethane using an FEI Vitrobot (ThermoFisher). Movies of the samples were recorded with EPU software (Thermo Fisher) using a Falcon 4 electron detector (Thermo Fisher) on a Glacios microscope operating at 200 keV. Movies were collected with 48 frames and an electron total dose of 48 e−/Å2 (1 e−/Å2 per frame). The movies were corrected for beam-induced motion using MotionCor247. Contrast transfer function parameters of the electron micrographs were estimated using Gctf 1.1848. Particles were auto-picked from a trained model using crYOLO49. Subsequent 3D-classifications and 3D refinements were performed using RELION-350. Following whole-virus reconstruction, sub-particles representing the icosahedral asymmetric unit were extracted and refined using cryoSPARC51. Following refinements, local resolution was estimated using RELION-3. Additional information regarding the work-flow, number of images, and particles is found in Fig S6. Structures were visualized using ChimeraX52 and PYMOL (https://www.pymol.org/pymol).
Model building and analysis.
The initial models of the WEEV and CHIKV structural proteins (E1, E2, and TM regions) and MXRA8 (duck and Du-D1-Mo-D2) were built as threaded models using SWISS-MODEL53. Threaded models then were simulated using Namdinator using the Molecular Dynamics Flexible Fitting Method to generate a potential to relax the structures into the observed experimental density54. All components were docked into the map of an asymmetric unit as individual rigid bodies. The subunit models then underwent real-space refinement using PHENIX55, first as rigid-body docking, then density morphing, and an iterative procedure of minimization using secondary-structure/torsion-restraints and manual editing using COOT 0.9.656, with additional refinement being performed using Isolde57. A summary of refinement statistics is shown in Table S2. Close-contacts are computed as heavy-atoms within 0.39 nm and were calculated using MDTraj58.
Virus sequence alignments.
All multiple sequence alignments were performed using the MUSCLE algorithm59 and visualized with ALINE60. Sequences were obtained from GenBank as follows: WEEV-E1 (ABD98014.1), SINV-E1 (NP_740677.1), EEEV-E1 (NP_740648.1), VEEV-E1 (NP_741967.1), RRV-E1 (NP_740686.1), MAYV-E1 (NP_740694.1), CHIKV-E1 (NP_690589.2), WEEV-E2 (ABD98014.1), SINV-E2 (NP_740675.1), EEEV-E2 (NP_740646.1), VEEV-E2 (NP_741966.1), RRV-E2 (NP_740684.1), MAYV-E2 (NP_740693.1), CHIKV-E2 (NP_690589.2), duck MXRA8 (NWZ22901.1), sparrow MXRA8 (XP_039583663.1), turkey MXRA8 (XP_042687609.1), chicken MXRA8 (NP_989967.1), mouse MXRA8 (AAH26438.1), and human MXRA8 (NP_115724.1).
Phylogenetic inference.
Structural protein (E1 and E2) sequences were retrieved from the NCBI GenBank for each alphavirus, including CHIKV (QKY67868.1), MAYV (QED21311.1), Una (UNAV, YP_009665989.1), ONNV (AAC97205.1), SFV (NP_463458.1), RRV (AAA47404.1), EEEV (ANB41743.1), Madariaga (MADV, AXV43855.1), VEEV (AGE98294.2), SINV (AAM10630.1), AURAV (NP_632024.1), OCKV (P27285.1), WEEV (QEX51909.1), Buggy Creek virus (BCV, AEJ36227.1), BBKV (AVN98166.1), FMV (YP_003324588.1), HJV (YP_002802300.1), and WHAV (AEJ36239.1). Sequences were aligned via Clustal Omega61, with simple phylogeny inferred via neighbor-joining. Results were visualized in R using the ggtree package55.
Sequences of D1 of MXRA8 were obtained with a BLAST search on the NCBI GenBank. The top 5,000 non-reducent sequences were obtained with an expect threshold of 0.05 and scored with BLOSUM62 for alignments. These sequences were aligned using Biopython, where residues corresponding to the position of duck D1-WEEV E1 contacts were extracted. Extracted sequences were characterized by the fraction of similarity to duck D1 and sorted, which were then inspected and curated manually to include in Fig S11. Sequences shown include Duck (NWZ22901.1), Alligator (XP_006263071.1), Crocidile (XP_019397120.1), Sea Turtle (XP_007070253.1), Turtle (KAG6936352.1), Coelacanth (XP_005986077.1), Lizard (XP_042335573.1), Caecilian (XP_030041825.1), Eel (XP_028663187.1), Komoto Dragon (XP_044301186.1), Snake (XP_034290786.1), Birchir (XP_039612125.1), Carp (XP_042590098.1), Shark (XP_041062630.1), Opossum (XP_007481143.1), Mink (XP_044092840.1), Frog (KAG9482299.1), Toad (XP_040283849.1), Beaver (XP_020011302.1), Possum (XP_036598586.1), Bat (XP_015999954.1), and Mouse (AAH26438.1).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was assigned using Prism Version 8.0 (GraphPad) when P < 0.05. Statistical analysis of viral infection levels was determined by one-way ANOVA with Dunnett’s post-test. Statistical analysis of in vivo experiments was determined by either one-way or two-way ANOVA with a Kruskal-Wallis or Dunnett’s post-test depending on the data distribution and the number of comparison groups. The statistical tests, number of independent experiments, and number of experimental replicates are indicated in the Figure legends.
Supplementary Material
Figure S1. VEEV and EEEV infection are not enhanced by avian or mammalian MXRA8, Related to Figure 1 and Table S1. A. Diagram of MXRA8 cDNA expressed in mouse, human and chicken cells. MXRA8 signal peptide was replaced with a β2-microglobulin signal peptide followed by FLAG tag sequence and a short linker. B. Representative flow cytometry plots showing surface expression of mouse, chicken, turkey and duck MXRA8 with anti-FLAG mAb. C. Sequence relationships between murine and avian MXRA8. Identity (red) and similarity (yellow) matrix indicating the conservation of MXRA8 between species. D-E. ΔMxra8 3T3 cells complemented with chicken or mouse MXRA8 or empty vector were inoculated with MAYV (D) or RRV (E). Infection was assessed using anti-E2 mAbs (see Methods; 3 experiments in triplicate). F. ΔMxra8 3T3 cells complemented with mouse or chicken MXRA8 or empty vector control were inoculated with SINV (Girdwood or Toto) and stained for SINV antigen (3 experiments in triplicate). G. Jurkat cells expressing mouse MXRA8, turkey MXRA8, LDLRAD3, or transduced with an empty vector control were inoculated with CHIKV (strain 181/25), SINV TR339-GFP, or SINV-VEEV-GFP (3 experiments in triplicate). Infection was assessed by anti-E1 antibody staining or GFP expression. H. K562 cells expressing mouse MXRA8, turkey MXRA8, LDLRAD3, VLDLR or transduced with an empty vector control were inoculated with SINV-EEEV-GFP (3 experiments in triplicate). Infection was assessed by GFP expression. I. Representative flow cytometry plot showing surface expression of MXRA8 in CEFs after staining with anti-chicken MXRA8 mAbs (red) or an isotype control (blue). J. CEFs were inoculated with CHIKV 181/25, SINV-WEEV-GFP, or SINV TR339. Infection was assessed by GFP expression, CHIKV E2, or SINV antigen staining and is compared between infected (red) and non-infected cells (blue). D-F: one-way ANOVA with Dunnett’s post-test; mean ± SD: ****, P < 0.0001.
Figure S2. Chicken MXRA8 is required for optimal infection of WHAV, BBKV and OCKV but not EEEV in murine 3T3 cells and CEFs, Related to Figure 1 and Table S1. A. Representative flow cytometry plots showing ΔMxra8 CEFs complemented with empty vector (blue histogram), chicken or mouse MXRA8 (red histograms) and stained with mouse-anti-chicken MXRA8 mAbs or hamster anti-mouse MXRA8 mAbs. Wild type CEFs served as a control (gray histogram). B. ΔMxra8 chicken fibroblasts complemented with chicken or mouse MXRA8 (2 independent clones of each shown) or empty vector control and non-targeted (NT1 and NT2) CEFs were inoculated with SINV-EEEV-GFP. C-F. ΔMxra8 3T3 cells complemented with mouse or chicken MXRA8 or empty vector control were inoculated with WHAV (C), BBKV (D), OCKV (E), or AURAV (F) and stained for viral antigen expression (3 experiments in triplicate). G-J. CEFs were pre-incubated with mouse anti-chicken MXRA8 mAbs for one hour and then inoculated with WHAV (G), BBKV (H), OCKV (I) and AURAV (J) and stained for E1 antigen expression (3 to 4 experiments in triplicate). K. Multi-step growth curves with OCKV in ΔMxra8 CEFs complemented with chicken or mouse MXRA8 or empty vector control. Virus was titrated by focus-forming assay (3 experiments in duplicate). L. ΔMxra8 CEFs complemented with chicken or mouse MXRA8 or empty vector control or non-targeting sgRNA (NT1 and NT2) were inoculated with OCKV. Infection was assessed by viral antigen expression and flow cytometry (3 experiments in triplicate; mean ± SD). M. ΔMxra8 chicken fibroblasts and non-targeted (NT1 and NT2) CEFs were inoculated with AURAV. Infection was assessed with flow cytometry (3 experiments in triplicate; mean ± SD). C-E and G-I and L; one-way ANOVA with Dunnett’s post-test; mean ± SD: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. K: two-way ANOVA with Tukey’s post-test; mean ± SD: ****, P < 0.0001.
Figure S3. Alphavirus infection in human 293T and HeLa cells expressing chicken, sparrow, or mouse MXRA8, Related to Figure 1 and Table S1. A-B. Flow cytometry histograms showing surface expression of chicken and murine MXRA8 in human 293T (A) or HeLa (B) cells after staining with mouse anti-chicken or hamster anti-murine MXRA8 mAbs. Red shading indicates cells complemented with chicken or mouse MXRA8, and blue shading indicates cells complemented with empty vector control. C-J. 293T (C-I) or HeLa cells (D-J) transfected with mouse or chicken MXRA8 or empty vector control were inoculated with SINV (C, D), CHIKV (E, F), AURAV (G, H, 12 hpi; I, J, 24 hpi) and stained for viral antigen (3 experiments in triplicate). K. Flow cytometry histogram showing surface expression of sparrow MXRA8 in human 293T cells using anti-FLAG mAb. Red shading indicates cells expressing sparrow MXRA8, and blue shading indicates cells transfected with empty vector control. L-N. 293T cells were transfected with mouse, chicken, or sparrow MXRA8 or empty vector control and inoculated with SINV (L), CHIKV 181/25 (M), or CHIKV 37997 (N). Cells were stained for viral antigen with specific mAbs (3 experiments in triplicate). C-F, L-N: one-way ANOVA with Dunnett’s post-test; mean ± SD: *, P < 0.05; ****, P < 0.0001.
Figure S4. Chicken MXRA8 promotes SINV infection in mice, Related to Fig 1. A. Five-week-old BALB/c mice were transduced via an intranasal route with 1010 adenoviral particles encoding chicken MXRA8 gene or no transgene. Four days later, mice were inoculated with 0.5 mg of anti-IFNAR1 mAb followed by 104 FFU of SINV one day later via intranasal route. Tissues were harvested at 40 and 72 h post-infection, and infection in the spleen (B) and lung (C-D) were measured by qRT-PCR (B-C; viral RNA) or focus-forming (D; infectious virus) assays (2 experiments, n = 7). B-D: Mann-Whitney test; median values: **, P < 0.01; ***, P < 0.001.
Figure S5. Gold-standard Fourier shell correlation curve resolutions for final maps, Related to Figures 3 and 7. Cutoffs of 0.143 yield a final resolution of: 4.35 Å for WEEV-VLP alone (A), 4.74 Å for WEEV-VLP bound to duck MXRA8 (B), 3.96 Å for CHIKV-VLP bound to Du-D1-Mo-D2 MXRA8 (C), and 3.89 Å for WEEV-VLP bound to Du-D1-Mo-D2 MXRA8 (D).
Figure S6. Cryo-EM processing flow diagram of duck MXRA8 bound to WEEV-VLP, Related to Figure 3. Arrows track the flow of micrographs/particles, and boxes indicate processing steps. Red dots indicate particle centers determined using crYOLO particle picking (top micrograph) or through subparticle extraction using icosahedral symmetry (bottom micrograph).
Figure S7. E1 sequence alignment and contact residues, Related to Figure 4. E1 secondary structure is colored in blue cartoon and domains are colored similarly to Fig 3D. WEEV contact residues (circles) and CHIKV contact residues (diamonds) are colored based on the mode of contact with MXRA8: cyan for wrapped spike heterodimer, red for intraspike heterodimer, and yellow for interspike heterodimer.
Figure S8. E2 sequence alignment and contact residues, Related to Figure 4. E2 secondary structure is colored in dark blue cartoon and domains are colored similarly to Fig 3D. WEEV contact residues (circles) and CHIKV contact residues (diamonds) are colored based on mode of contact with MXRA8: cyan for wrapped spike heterodimer, red for intraspike heterodimer, and yellow for interspike heterodimer.
Figure S9. MXRA8 sequence alignment and contact residues, Related to Figure 4. MXRA8 secondary structure is colored in blue cartoon along with labels for β-strands. Domains 1 and 2 are colored similarly to Fig 3C, with the stalk colored red and transmembrane region colored green. Duck(circles) and mouse (diamonds) contact residues are colored based on the mode of contact with E1-E2 heterodimers: cyan for wrapped spike heterodimer, red for intraspike heterodimer, and yellow for interspike heterodimer.
Figure S10. MXRA8 chimeras and mutants used to assess avian MXRA8 binding mode, Related to Figure 5. MXRA8 secondary structure is colored in the blue cartoon along with labels for β-strands. D1 and D2 are colored similarly to Fig 3C, with the stalk colored red and transmembrane colored green. Sequences include mouse (control), duck, duck D1 with mouse D2, duck D2 with mouse D1, D1 mutations a-d, and D2 mutations a-d, as described in Fig 5D. Double-arginine mutations and loop insertions are depicted with solid red boxes and dashed red boxes, respectively.
Figure S11. Affinity measurements and stalk length effects of chimeric MXRA8 binding to alphaviruses, Related to Figures 6 and 7. A-D. Biolayer interferometry binding analysis of monovalent forms of duck MXRA8 (A), Du-D1-Mo-D2 MXRA8 (B, D), or mouse MXRA8 (C) to immobilized WEEV (A-B) or CHIKV (C-D) VLPs. Representative data are shown in black, with fitted (1:1 Langmuir binding models) in red. Kinetic (KD) affinity constants are displayed (mean ± SD pooled from two to three independent experiments). E-F. Neutralization of SINV-CHIKV (E) or SINV-WEEV (F) infection after pre-treatment with indicated concentrations of duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo D2 duStalk-MXRA8-Fc [duck stalk], or Du-D1-Mo D2 moStalk-MXRA8-Fc [mouse stalk] (two experiments performed in technical duplicates). Infection was analyzed by flow cytometry (GFP expression) and normalized to levels obtained without a MXRA8 inhibitory decoy protein. G-H. Neutralization of SINV-CHIKV (G) or SINV-WEEV (H) infection by duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo D2-MXRA8-Fc, or Du-D1-Mo-D2-N66R-MXRA8-Fc (four experiments, technical duplicates). Infection was analyzed by flow cytometry and normalized to levels obtained without a MXRA8 inhibitory decoy protein.
Figure S12. Expansion of host reservoirs for WEE complex members, Related to Figure 6. A. Sequence alignment of D1 of MXRA8. Sequences are ordered based on similarity to duck MXRA8. Sequence identity of the contacting residues is shown adjacent to the colloquial species name. Duck MXRA8 contacts to WEEV E1 are represented as spheres for wrapped (cyan), intraspike (red), and interspike (yellow) E1-E2 heterodimers. Conserved contacts for each sequence are boxed and colored to represent wrapped (cyan), intraspike (red), and interspike (yellow) contacts. D-E. ΔMxra8 3T3 cells complemented with mouse (pink), chicken (red), duck (blue), alligator (dark gray) or sea turtle (olive green) MXRA8 or empty vector control (gray) were inoculated with SINV-CHIKV-GFP (B) or SINV TR339-GFP (C). Infection was assessed by GFP expression and is compared between infected and non-infected cells (3 experiments, in triplicate). D-E: One-way ANOVA with Dunnett’s post-test; mean ± standard deviation (SD). ****, P < 0.0001.
Video S1. Video cartoon of different engagement strategies of Duck MXRA8 and WEEV versus Mouse MXRA8 and CHIKV, Related to Figures 3 and 4. Each MXRA8 (rainbow) makes unique contacts with the indicated alphavirus E1-E2 heterodimers at the wrapped (light gray), intraspike (gray), or interspike (dark gray) sites. The front view of duck MXRA8 binding to WEEV (left) and mouse MXRA8 binding to CHIKV (right) show distinct orientations and contacting surfaces. The side view shows D1 of duck MXRA8 engaging E1 of the intraspike site, whereas D1 of mouse MXRA8 engages E2 at the wrapped site. The rotated side view shows that each MXRA8 has unique positioning in the binding pocket, with D1 of duck MXRA8 engaging E1 intraspike contacts and D2 of mouse MXRA8 binding E1-E2 interspike contacts. Images for the movie were rendered using ChimeraX52.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mxra8 mAb 1G11.E6 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 1H1.F5 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 3G2.F5 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 4E7.D10 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 7F1.D8 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 8F7.E1 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 9G2.D6 | Zhang et al., 2018 | N/A |
| Anti-chicken MXRA8 mAb 6D7 | This manuscript | N/A |
| Anti-chicken MXRA8 mAb 9B6 | This manuscript | N/A |
| Anti-chicken MXRA8 mAb 11A6 | This manuscript | N/A |
| Anti-chicken MXRA8 mAb 16D9 | This manuscript | N/A |
| Anti-FLAG DYKDDDDK | Cell Signaling | Cat #14793 RRID: AB_2572291 |
| CHK-11 | Pal et al., 2013 | GenBank: AF192908 |
| CHK-48 | Pal et al., 2013 | N/A |
| CHK-265 mAb | Pal et al., 2013 | N/A |
| DC2.112 | Quiroz et al., 2019 | N/A |
| DC2.315 | Quiroz et al., 2019 | N/A |
| Sindbis virus immune ascitic fluid | ATCC | Cat #V-560–701–562 |
| H77 (anti-HCV E2) | Sabo et al., 2011 | N/A |
| ZIKV NS1 | Native Antigen | ZIKVSU-NS1–100 |
| Alexa Fluor 647 conjugated goat anti-mouse IgG | Thermo Fisher | A21235; RRID: AB_2535804 |
| Alexa Fluor 647 conjugated goat anti-human IgG | Thermo Fisher | A21445; RRID: AB_2535862 |
| Peroxidase conjugated goat anti-mouse IgG (H + L) | Jackson ImmunoResearch | 115–035–062; RRID: AB_2338504 |
| Anti-Mouse IgG (whole molecule)–Peroxidase antibody | Sigma-Aldrich | Cat #A5278 AB_258232 |
| Bacterial and virus strains | ||
| Western equine encephalitis virus (strain McMillan) | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| CHIKV 181/25 | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| CHIKV 37997 | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| SINV-CHIKV (chimera: La Reunion 2006) | This study | N/A |
| Mayaro virus (strain BeH407) | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| Ross River virus (strain T48) | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| SINV TR339 | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| SINV TR339-GFP | This study | N/A |
| SINV Toto | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| SINV Girdwood | Gift from Mark T. Heise at UNC-Chapel Hill | N/A |
| Babanki | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| Ockelbo | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| Whataroa | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| Aura | World Reference Center for Emerging Viruses and Arboviruses | N/A |
| SINV-WEEV (chimera CBA87)-GFP | Sun et al., 2020 | N/A |
| SINV-EEEV (chimera FL93–939)-GFP | Sun et al., 2020 | N/A |
| SINV-VEEV (Trinidad Donkey)-GFP | Sun et al., 2020 | N/A |
| Biological samples | ||
| House sparrow (Passer domesticus) muscle tissue | Colorado State University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Chikungunya virus-like particles (strain 37997) | Akahata et al., 2010 | N/A |
| WEEV virus-like practices | Ko et al., 2019 | N/A |
| Mouse Mxra8 (residues 22–336) fused to mouse IgG2b Fc region | Zhang et al., 2018 | N/A |
| Chicken MXRA8 (residues 23–334) fused to mouse IgG2b Fc region | This study | N/A |
| Histidine-tagged duck (residues 23–334) | This study | N/A |
| Histidine-tagged mouse (residues 23–331) | This study | N/A |
| Histidine-tagged sparrow (residues 23–334) | This study | N/A |
| Cobalt resin | G-Biosciences | 786–932 |
| Mouse Mxra8 (residues 22–336) fused to human IgG1 Fc region | This study | N/A |
| Duck MXRA8 (residues 23–334) fused to human IgG1 Fc region | This study | N.A |
| Chimeric Du-D1-Mo-D2 duck Stalk MXRA8 (Duck residues 66–196 and 296–334; Mouse: residues 23–65 and 195–293) fused to human IgG1 | This study | N/A |
| Chimeric Du-D1-Mo-D2 mouse Stalk MXRA8 (Duck residues 66–196; Mouse: residues 23–65 and 195–334) fused to human IgG1 | This study | N/A |
| Chimeric Du-D1-Mo-D2 N66R MXRA8 fused to human IgG1 | This study | N/A |
| Chimeric Mo-D1-Du-D2 MXRA8 (Mouse residues 65–196; Duck: residues 29–64 and 199–334) fused to human IgG1 | This study | N/A |
| Human LDLRAD3-D1 fused to human IgG1 Fc region | Ma et al., 2020 | N/A |
| GlySERIAS Immobilized Microspin | Genovis | N/A |
| Deposited data | ||
| WEEV VLP structure | This study | PDB: 8DAQ |
| WEEV VLP + Duck MXRA8 structure | This study | PDB: 8DAN |
| WEEV VLP + Du-D1-Mo-D2 structure | This study | PDB: 8SQN |
| CHIKV VLP + Du-D1-Mo-D2 structure | This study | PDB: 8EWF |
| Experimental models: Cell lines | ||
| NIH/3T3 | ATCC | CRL-1658 |
| ΔMxra8 3T3 cells | Zhang et al., 2018 | B/A |
| HEK-293 | ATCC | CRL-1573 |
| BHK-21 | ATCC | CCL-10 |
| Vero | ATCC | CCL-81 |
| HeLa | ATCC | CCL-1 |
| K562 | ATCC | CRL-3344 |
| Jurkat | ATCC | TIB-152 |
| Chicken embryonic fibroblasts (CEF) | ATCC | CRL-12203 |
| Expi293F | Thermo Fisher | A14527 |
| Experimental models: Organisms/strains | ||
| BALB/c | Jackson Laboratory | # 000651 |
| C57BL/6J | Jackson Laboratory | # 000664 |
| CD-1 mice | Charles River | # 022 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | V9 |
| FlowJo | FlowJo, LLC | Versions 9 and 10 |
| ggtree | Guangchuang et al., 2017 | V3.4.4 |
| Relion | Zivanov et al, 2018 | Version 3 |
| cryoSPARC | Punjani et al., 2017 | Version 3.3.2 |
| MotionCor2 | Zheng et al, 2017 | Version 1.4.0 |
| GCTF | Zhang et al., 2016 | Version 1.18 |
| crYOLO | Wagner et al., 2019 | Version 1.8.4 |
| ChimeraX | Goddard et al., 2018 | Version 1.2.5 |
| PYMOL | https://www.pymol.org/pymol | Version 2.2.3 |
| SWISS-MODEL | Waterhouse et al., 2018 | N/A |
| Namdinator | Kidmose et al., 2019 | N/A |
| PHENIX | Adams et al., 2010 | 1.20.1 |
| COOT | Emsley et al., 2010 | Version 0.9.6 |
| Isolde | Croll, 2018 | Version 1.1.0 |
| MDTraj | McGibbon et al., 2015 | Version 1.9 |
| MUSCLE | Edgar, 2004 | Version 3.8.31 |
| ALINE | Bond and Schüttelkopf, 2009 | 1.0.025 |
| Biopython | Cock et al, 2009 | Version 1.79 |
HIGHLIGHTS.
Avian but not mammalian MXRA8 is an entry receptor for WEE complex alphaviruses
Mammalian but not avian MXRA8 binds to Semliki Forest complex alphaviruses
Avian and mammalian MXRA8 bind different alphaviruses in flipped orientations
Chimeric avian-mouse MXRA8 inhibits alphaviruses from multiple antigenic groups
ACKNOWLEDGEMENTS
This study was supported by NIH grants R01AI114816, R01AI123348, R01AI095436, R01 AI067380, NIAID contract HHSN272201700060C, DTRA contract MCDC2103-011 and a Doris Duke Physician Scientist fellowship award. Doris Duke foundation funds were not used for any of the experiments with animals or animal-derived cell lines. We thank Richard Bowen for providing sparrow tissue samples and Kevin Carlton and John Mascola from the Vaccine Research Center of the National Institutes of Allergy and Infectious Diseases (NIH) for a gift of the WEEV VLPs.
DECLARATION OF INTERESTS
M.S.D. is a consultant or member of a Scientific Advisory Board for Inbios, Ocugen, Vir Biotechnology, Topspin Therapeutics, and Moderna. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Emergent BioSolutions, Moderna, Generate Biomedicines, Vir Biotechnology, and Immunome. D.H.F. is a founder of Courier Therapeutics and has received unrelated funding support from Emergent BioSolutions and Mallinckrodt Pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Weaver SCS, D.W. (2011). Alphavirus Infections. In Tropical Infectious Diseases: Principles, Pathogens and Practice, D.H.W. Guerrant Richard L., Weller Peter F., ed. (Saunders WB), pp. 519–524. 10.1016/B978-0-7020-3935-5.00078-1. [DOI] [Google Scholar]
- 2.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, and Frolov I (2007). The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81, 2472–2484. JVI.02073–06 [pii] 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Kang W, Shirako Y, Rumenapf T, Strauss EG, and Strauss JH (1997). Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J Virol 71, 613–623. 10.1128/JVI.71.1.613-623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn CS, Lustig S, Strauss EG, and Strauss JH (1988). Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci U S A 85, 5997–6001. 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strauss JHSEG (1997). Recombination in Alphaviruses. Seminars in VIROLOGY 8,, 85–94, VI9701151044. [Google Scholar]
- 6.Kim AS, Zimmerman O, Fox JM, Nelson CA, Basore K, Zhang R, Durnell L, Desai C, Bullock C, Deem SL, et al. (2020). An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis. Cell Host Microbe 27, 428–440.e429. 10.1016/j.chom.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basore K, Kim AS, Nelson CA, Zhang R, Smith BK, Uranga C, Vang L, Cheng M, Gross ML, Smith J, et al. (2019). Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor. Cell 177, 1725–1737 e1716. 10.1016/j.cell.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Zhao Z, Chai Y, Jin X, Li C, Yuan F, Liu S, Gao Z, Wang H, Song J, et al. (2019). Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein. Cell 177, 1714–1724 e1712. 10.1016/j.cell.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Zhang R, Kim AS, Fox JM, Nair S, Basore K, Klimstra WB, Rimkunas R, Fong RH, Lin H, Poddar S, et al. (2018). Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574. 10.1038/s41586-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma H, Kim AS, Kafai NM, Earnest JT, Shah AP, Case JB, Basore K, Gilliland TC, Sun C, Nelson CA, et al. (2020). LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature. 10.1038/s41586-020-2915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark LE, Clark SA, Lin C, Liu J, Coscia A, Nabel KG, Yang P, Neel DV, Lee H, Brusic V, et al. (2022). VLDLR and ApoER2 are receptors for multiple alphaviruses. Nature 602, 475–480. 10.1038/s41586-021-04326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allison AB, Stallknecht DE, and Holmes EC (2015). Evolutionary genetics and vector adaptation of recombinant viruses of the western equine encephalitis antigenic complex provides new insights into alphavirus diversity and host switching. Virology 474, 154–162. 10.1016/j.virol.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko SY, Akahata W, Yang ES, Kong WP, Burke CW, Honnold SP, Nichols DK, Huang YS, Schieber GL, Carlton K, et al. (2019). A virus-like particle vaccine prevents equine encephalitis virus infection in nonhuman primates. Sci Transl Med 11. 10.1126/scitranslmed.aav3113. [DOI] [PubMed] [Google Scholar]
- 14.Chang LJ, Dowd KA, Mendoza FH, Saunders JG, Sitar S, Plummer SH, Yamshchikov G, Sarwar UN, Hu Z, Enama ME, et al. (2014). Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet 384, 2046–2052. 10.1016/s0140-6736(14)61185-5. [DOI] [PubMed] [Google Scholar]
- 15.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, and Nabel GJ (2010). A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med 16, 334–338. nm.2105 [pii] 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fassbinder-Orth CA, Barak VA, Rainwater EL, and Altrichter AM (2014). Buggy Creek virus (Togaviridae: Alphavirus) upregulates expression of pattern recognition receptors and interferons in House Sparrows (Passer domesticus). Vector Borne Zoonotic Dis 14, 439–446. 10.1089/vbz.2013.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosimann ALP, de Siqueira MK, Ceole LF, and Nunes Duarte Dos Santos C (2018). A new Aura virus isolate in Brazil shows segment duplication in the variable region of the nsP3 gene. Parasit Vectors 11, 321. 10.1186/s13071-018-2907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, and Schreiber RD (2006). Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res 26, 804–819. [DOI] [PubMed] [Google Scholar]
- 19.Strauss JH, Strauss EG, and Kuhn RJ (1995). Budding of alphaviruses. Trends Microbiol 3, 346–350. [DOI] [PubMed] [Google Scholar]
- 20.Jose J, Snyder JE, and Kuhn RJ (2009). A structural and functional perspective of alphavirus replication and assembly. Future Microbiol 4, 837–856. 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chmielewski D, Schmid MF, Simmons G, Jin J, and Chiu W (2022). Chikungunya virus assembly and budding visualized in situ using cryogenic electron tomography. Nature microbiology 7, 1270–1279. 10.1038/s41564-022-01164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin J, Galaz-Montoya JG, Sherman MB, Sun SY, Goldsmith CS, O’Toole ET, Ackerman L, Carlson LA, Weaver SC, Chiu W, and Simmons G (2018). Neutralizing Antibodies Inhibit Chikungunya Virus Budding at the Plasma Membrane. Cell Host Microbe 24, 417–428.e415. 10.1016/j.chom.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H, Zhao Z, Chai Y, Jin X, Li C, Yuan F, Liu S, Gao Z, Wang H, Song J, et al. (2019). Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein. Cell 177, 1714–1724.e1712. 10.1016/j.cell.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Nir Y, Lasowski Y, Avivi A, and Cgoldwasser R (1969). Survey for antibodies to arboviruses in the serum of various animals in Israel during 1965–1966. Am J Trop Med Hyg 18, 416–422. 10.4269/ajtmh.1969.18.416. [DOI] [PubMed] [Google Scholar]
- 25.Craighead JE, Shelokov A, and Peralta PH (1962). The lizard: a possible host for eastern equine encephalitis virus in Panama. American journal of hygiene 76, 82–87. 10.1093/oxfordjournals.aje.a120266. [DOI] [PubMed] [Google Scholar]
- 26.Thomas LA, Eklund CM, and Rush WA (1958). Susceptibility of garter snakes (Thamnophis spp.) to western equine encephalomyelitis virus. Proc Soc Exp Biol Med 99, 698–700. 10.3181/00379727-99-24468. [DOI] [PubMed] [Google Scholar]
- 27.Bowen GS (1977). Prolonged western equine encephalitis viremia in the Texas tortoise (Gopherus berlandieri). Am J Trop Med Hyg 26, 171–175. 10.4269/ajtmh.1977.26.171. [DOI] [PubMed] [Google Scholar]
- 28.Cao D, Ma B, Cao Z, Zhang X, and Xiang Y (2023). Structure of Semliki Forest virus in complex with its receptor VLDLR. Cell 186, 2208–2218 e2215. 10.1016/j.cell.2023.03.032. [DOI] [PubMed] [Google Scholar]
- 29.Wang KS, Schmaljohn AL, Kuhn RJ, and Strauss JH (1991). Antiidiotypic antibodies as probes for the Sindbis virus receptor. Virology 181, 694–702. 10.1016/0042-6822(91)90903-o. [DOI] [PubMed] [Google Scholar]
- 30.Wang KS, Kuhn RJ, Strauss EG, Ou S, and Strauss JH (1992). High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol 66, 4992–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klimstra WB, Nangle EM, Smith MS, Yurochko AD, and Ryman KD (2003). DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol 77, 12022–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, and Cherry S (2011). Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104. 10.1016/j.chom.2011.06.009 S1931–3128(11)00218–6 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimstra WB, Ryman KD, and Johnston RE (1998). Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72, 7357–7366. 10.1128/JVI.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heil ML, Albee A, Strauss JH, and Kuhn RJ (2001). An amino acid substitution in the coding region of the E2 glycoprotein adapts Ross River virus to utilize heparan sulfate as an attachment moiety. J Virol 75, 6303–6309. 10.1128/JVI.75.14.6303-6309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernard KA, Klimstra WB, and Johnston RE (2000). Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276, 93–103. [DOI] [PubMed] [Google Scholar]
- 36.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, and Rey FA (2010). Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712. nature09555 [pii] 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 37.Kim AS, Kafai NM, Winkler ES, Gilliland TC Jr., Cottle EL, Earnest JT, Jethva PN, Kaplonek P, Shah AP, Fong RH, et al. (2021). Pan-protective anti-alphavirus human antibodies target a conserved E1 protein epitope. Cell 184, 4414–4429.e4419. 10.1016/j.cell.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fox JM, Long F, Edeling MA, Lin H, van Duijl-Richter MKS, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, et al. (2015). Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 163, 1095–1107. 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim AS, Austin SK, Gardner CL, Zuiani A, Reed DS, Trobaugh DW, Sun C, Basore K, Williamson LE, Crowe JE Jr., et al. (2019). Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nature microbiology 4, 187–197. 10.1038/s41564-018-0286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun C, Gardner CL, Watson AM, Ryman KD, and Klimstra WB (2014). Stable, high-level expression of reporter proteins from improved alphavirus expression vectors to track replication and dissemination during encephalitic and arthritogenic disease. J Virol 88, 2035–2046. 10.1128/jvi.02990-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, and McCray PB Jr. (2005). ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 79, 14614–14621. 10.1128/jvi.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittereder N, March KL, and Trapnell BC (1996). Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol 70, 7498–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawman DW, Stoermer KA, Montgomery SA, Pal P, Oko L, Diamond MS, and Morrison TE (2013). Chronic joint disease caused by persistent chikungunya virus infection is controlled by the adaptive immune response. J Virol 87, 13878–13888. 10.1128/JVI.02666-13 JVI.02666–13 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, et al. (2013). Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog 9, e1003312. 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brien JD, Lazear HM, and Diamond MS (2013). Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol 31, 15D 13 11–15D 13 18. 10.1002/9780471729259.mc15d03s31. [DOI] [PubMed] [Google Scholar]
- 46.Smith SA, Silva LA, Fox JM, Flyak AI, Kose N, Sapparapu G, Khomandiak S, Ashbrook AW, Kahle KM, Fong RH, et al. (2015). Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell Host Microbe 18, 86–95. 10.1016/j.chom.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, et al. (2019). SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications biology 2, 218. 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 52.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–w303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kidmose RT, Juhl J, Nissen P, Boesen T, Karlsen JL, and Pedersen BP (2019). Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531. 10.1107/s2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta crystallographica. Section D, Structural biology 74, 519–530. 10.1107/s2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CX, Schwantes CR, Wang LP, Lane TJ, and Pande VS (2015). MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophysical journal 109, 1528–1532. 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bond CS, and Schüttelkopf AW (2009). ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr 65, 510–512. 10.1107/s0907444909007835. [DOI] [PubMed] [Google Scholar]
- 61.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. VEEV and EEEV infection are not enhanced by avian or mammalian MXRA8, Related to Figure 1 and Table S1. A. Diagram of MXRA8 cDNA expressed in mouse, human and chicken cells. MXRA8 signal peptide was replaced with a β2-microglobulin signal peptide followed by FLAG tag sequence and a short linker. B. Representative flow cytometry plots showing surface expression of mouse, chicken, turkey and duck MXRA8 with anti-FLAG mAb. C. Sequence relationships between murine and avian MXRA8. Identity (red) and similarity (yellow) matrix indicating the conservation of MXRA8 between species. D-E. ΔMxra8 3T3 cells complemented with chicken or mouse MXRA8 or empty vector were inoculated with MAYV (D) or RRV (E). Infection was assessed using anti-E2 mAbs (see Methods; 3 experiments in triplicate). F. ΔMxra8 3T3 cells complemented with mouse or chicken MXRA8 or empty vector control were inoculated with SINV (Girdwood or Toto) and stained for SINV antigen (3 experiments in triplicate). G. Jurkat cells expressing mouse MXRA8, turkey MXRA8, LDLRAD3, or transduced with an empty vector control were inoculated with CHIKV (strain 181/25), SINV TR339-GFP, or SINV-VEEV-GFP (3 experiments in triplicate). Infection was assessed by anti-E1 antibody staining or GFP expression. H. K562 cells expressing mouse MXRA8, turkey MXRA8, LDLRAD3, VLDLR or transduced with an empty vector control were inoculated with SINV-EEEV-GFP (3 experiments in triplicate). Infection was assessed by GFP expression. I. Representative flow cytometry plot showing surface expression of MXRA8 in CEFs after staining with anti-chicken MXRA8 mAbs (red) or an isotype control (blue). J. CEFs were inoculated with CHIKV 181/25, SINV-WEEV-GFP, or SINV TR339. Infection was assessed by GFP expression, CHIKV E2, or SINV antigen staining and is compared between infected (red) and non-infected cells (blue). D-F: one-way ANOVA with Dunnett’s post-test; mean ± SD: ****, P < 0.0001.
Figure S2. Chicken MXRA8 is required for optimal infection of WHAV, BBKV and OCKV but not EEEV in murine 3T3 cells and CEFs, Related to Figure 1 and Table S1. A. Representative flow cytometry plots showing ΔMxra8 CEFs complemented with empty vector (blue histogram), chicken or mouse MXRA8 (red histograms) and stained with mouse-anti-chicken MXRA8 mAbs or hamster anti-mouse MXRA8 mAbs. Wild type CEFs served as a control (gray histogram). B. ΔMxra8 chicken fibroblasts complemented with chicken or mouse MXRA8 (2 independent clones of each shown) or empty vector control and non-targeted (NT1 and NT2) CEFs were inoculated with SINV-EEEV-GFP. C-F. ΔMxra8 3T3 cells complemented with mouse or chicken MXRA8 or empty vector control were inoculated with WHAV (C), BBKV (D), OCKV (E), or AURAV (F) and stained for viral antigen expression (3 experiments in triplicate). G-J. CEFs were pre-incubated with mouse anti-chicken MXRA8 mAbs for one hour and then inoculated with WHAV (G), BBKV (H), OCKV (I) and AURAV (J) and stained for E1 antigen expression (3 to 4 experiments in triplicate). K. Multi-step growth curves with OCKV in ΔMxra8 CEFs complemented with chicken or mouse MXRA8 or empty vector control. Virus was titrated by focus-forming assay (3 experiments in duplicate). L. ΔMxra8 CEFs complemented with chicken or mouse MXRA8 or empty vector control or non-targeting sgRNA (NT1 and NT2) were inoculated with OCKV. Infection was assessed by viral antigen expression and flow cytometry (3 experiments in triplicate; mean ± SD). M. ΔMxra8 chicken fibroblasts and non-targeted (NT1 and NT2) CEFs were inoculated with AURAV. Infection was assessed with flow cytometry (3 experiments in triplicate; mean ± SD). C-E and G-I and L; one-way ANOVA with Dunnett’s post-test; mean ± SD: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. K: two-way ANOVA with Tukey’s post-test; mean ± SD: ****, P < 0.0001.
Figure S3. Alphavirus infection in human 293T and HeLa cells expressing chicken, sparrow, or mouse MXRA8, Related to Figure 1 and Table S1. A-B. Flow cytometry histograms showing surface expression of chicken and murine MXRA8 in human 293T (A) or HeLa (B) cells after staining with mouse anti-chicken or hamster anti-murine MXRA8 mAbs. Red shading indicates cells complemented with chicken or mouse MXRA8, and blue shading indicates cells complemented with empty vector control. C-J. 293T (C-I) or HeLa cells (D-J) transfected with mouse or chicken MXRA8 or empty vector control were inoculated with SINV (C, D), CHIKV (E, F), AURAV (G, H, 12 hpi; I, J, 24 hpi) and stained for viral antigen (3 experiments in triplicate). K. Flow cytometry histogram showing surface expression of sparrow MXRA8 in human 293T cells using anti-FLAG mAb. Red shading indicates cells expressing sparrow MXRA8, and blue shading indicates cells transfected with empty vector control. L-N. 293T cells were transfected with mouse, chicken, or sparrow MXRA8 or empty vector control and inoculated with SINV (L), CHIKV 181/25 (M), or CHIKV 37997 (N). Cells were stained for viral antigen with specific mAbs (3 experiments in triplicate). C-F, L-N: one-way ANOVA with Dunnett’s post-test; mean ± SD: *, P < 0.05; ****, P < 0.0001.
Figure S4. Chicken MXRA8 promotes SINV infection in mice, Related to Fig 1. A. Five-week-old BALB/c mice were transduced via an intranasal route with 1010 adenoviral particles encoding chicken MXRA8 gene or no transgene. Four days later, mice were inoculated with 0.5 mg of anti-IFNAR1 mAb followed by 104 FFU of SINV one day later via intranasal route. Tissues were harvested at 40 and 72 h post-infection, and infection in the spleen (B) and lung (C-D) were measured by qRT-PCR (B-C; viral RNA) or focus-forming (D; infectious virus) assays (2 experiments, n = 7). B-D: Mann-Whitney test; median values: **, P < 0.01; ***, P < 0.001.
Figure S5. Gold-standard Fourier shell correlation curve resolutions for final maps, Related to Figures 3 and 7. Cutoffs of 0.143 yield a final resolution of: 4.35 Å for WEEV-VLP alone (A), 4.74 Å for WEEV-VLP bound to duck MXRA8 (B), 3.96 Å for CHIKV-VLP bound to Du-D1-Mo-D2 MXRA8 (C), and 3.89 Å for WEEV-VLP bound to Du-D1-Mo-D2 MXRA8 (D).
Figure S6. Cryo-EM processing flow diagram of duck MXRA8 bound to WEEV-VLP, Related to Figure 3. Arrows track the flow of micrographs/particles, and boxes indicate processing steps. Red dots indicate particle centers determined using crYOLO particle picking (top micrograph) or through subparticle extraction using icosahedral symmetry (bottom micrograph).
Figure S7. E1 sequence alignment and contact residues, Related to Figure 4. E1 secondary structure is colored in blue cartoon and domains are colored similarly to Fig 3D. WEEV contact residues (circles) and CHIKV contact residues (diamonds) are colored based on the mode of contact with MXRA8: cyan for wrapped spike heterodimer, red for intraspike heterodimer, and yellow for interspike heterodimer.
Figure S8. E2 sequence alignment and contact residues, Related to Figure 4. E2 secondary structure is colored in dark blue cartoon and domains are colored similarly to Fig 3D. WEEV contact residues (circles) and CHIKV contact residues (diamonds) are colored based on mode of contact with MXRA8: cyan for wrapped spike heterodimer, red for intraspike heterodimer, and yellow for interspike heterodimer.
Figure S9. MXRA8 sequence alignment and contact residues, Related to Figure 4. MXRA8 secondary structure is colored in blue cartoon along with labels for β-strands. Domains 1 and 2 are colored similarly to Fig 3C, with the stalk colored red and transmembrane region colored green. Duck(circles) and mouse (diamonds) contact residues are colored based on the mode of contact with E1-E2 heterodimers: cyan for wrapped spike heterodimer, red for intraspike heterodimer, and yellow for interspike heterodimer.
Figure S10. MXRA8 chimeras and mutants used to assess avian MXRA8 binding mode, Related to Figure 5. MXRA8 secondary structure is colored in the blue cartoon along with labels for β-strands. D1 and D2 are colored similarly to Fig 3C, with the stalk colored red and transmembrane colored green. Sequences include mouse (control), duck, duck D1 with mouse D2, duck D2 with mouse D1, D1 mutations a-d, and D2 mutations a-d, as described in Fig 5D. Double-arginine mutations and loop insertions are depicted with solid red boxes and dashed red boxes, respectively.
Figure S11. Affinity measurements and stalk length effects of chimeric MXRA8 binding to alphaviruses, Related to Figures 6 and 7. A-D. Biolayer interferometry binding analysis of monovalent forms of duck MXRA8 (A), Du-D1-Mo-D2 MXRA8 (B, D), or mouse MXRA8 (C) to immobilized WEEV (A-B) or CHIKV (C-D) VLPs. Representative data are shown in black, with fitted (1:1 Langmuir binding models) in red. Kinetic (KD) affinity constants are displayed (mean ± SD pooled from two to three independent experiments). E-F. Neutralization of SINV-CHIKV (E) or SINV-WEEV (F) infection after pre-treatment with indicated concentrations of duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo D2 duStalk-MXRA8-Fc [duck stalk], or Du-D1-Mo D2 moStalk-MXRA8-Fc [mouse stalk] (two experiments performed in technical duplicates). Infection was analyzed by flow cytometry (GFP expression) and normalized to levels obtained without a MXRA8 inhibitory decoy protein. G-H. Neutralization of SINV-CHIKV (G) or SINV-WEEV (H) infection by duck MXRA8-Fc, mouse MXRA8-Fc, Du-D1-Mo D2-MXRA8-Fc, or Du-D1-Mo-D2-N66R-MXRA8-Fc (four experiments, technical duplicates). Infection was analyzed by flow cytometry and normalized to levels obtained without a MXRA8 inhibitory decoy protein.
Figure S12. Expansion of host reservoirs for WEE complex members, Related to Figure 6. A. Sequence alignment of D1 of MXRA8. Sequences are ordered based on similarity to duck MXRA8. Sequence identity of the contacting residues is shown adjacent to the colloquial species name. Duck MXRA8 contacts to WEEV E1 are represented as spheres for wrapped (cyan), intraspike (red), and interspike (yellow) E1-E2 heterodimers. Conserved contacts for each sequence are boxed and colored to represent wrapped (cyan), intraspike (red), and interspike (yellow) contacts. D-E. ΔMxra8 3T3 cells complemented with mouse (pink), chicken (red), duck (blue), alligator (dark gray) or sea turtle (olive green) MXRA8 or empty vector control (gray) were inoculated with SINV-CHIKV-GFP (B) or SINV TR339-GFP (C). Infection was assessed by GFP expression and is compared between infected and non-infected cells (3 experiments, in triplicate). D-E: One-way ANOVA with Dunnett’s post-test; mean ± standard deviation (SD). ****, P < 0.0001.
Video S1. Video cartoon of different engagement strategies of Duck MXRA8 and WEEV versus Mouse MXRA8 and CHIKV, Related to Figures 3 and 4. Each MXRA8 (rainbow) makes unique contacts with the indicated alphavirus E1-E2 heterodimers at the wrapped (light gray), intraspike (gray), or interspike (dark gray) sites. The front view of duck MXRA8 binding to WEEV (left) and mouse MXRA8 binding to CHIKV (right) show distinct orientations and contacting surfaces. The side view shows D1 of duck MXRA8 engaging E1 of the intraspike site, whereas D1 of mouse MXRA8 engages E2 at the wrapped site. The rotated side view shows that each MXRA8 has unique positioning in the binding pocket, with D1 of duck MXRA8 engaging E1 intraspike contacts and D2 of mouse MXRA8 binding E1-E2 interspike contacts. Images for the movie were rendered using ChimeraX52.
Data Availability Statement
All data supporting the findings of this study are available within the paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not include original code. Cryo-EM data has been deposited in public databases (PDB: 8DAQ [WEEV VLP], 8DAN [WEEV VLP + Duck MXRA8], 8SQN [WEEV VLP + Du-D1-Mo-D2], and 8EWF [CHIKV VLP + Du-D1-Mo-D2]).


