SUMMARY
Pollen-pistil interactions establish interspecific/intergeneric prezygotic hybridization barriers in plants. Rejection of undesired pollen at the stigma is crucial to avoid outcrossing but can be overcome with support of mentor pollen. The mechanisms underlying this hybridization barrier are largely unknown. Here in Arabidopsis, we demonstrate that receptor-like kinases FERONIA/CURVY1/ANJEA/HERK1 and cell wall proteins LRX3/4/5 interact on papilla cell surfaces with autocrine stigmatic RALF1/22/23/33 peptide ligands (sRALFs) to establish a lock that blocks the penetration of undesired pollen tubes. Compatible pollen-derived RALF10/11/12/13/25/26/30 peptides (pRALFs) act as a key, outcompeting sRALFs and enabling pollen tube penetration. By treating Arabidopsis stigmas with synthetic pRALFs, we unlock the barrier, facilitating pollen tube penetration from distantly-related Brassicaceae species and resulting in interspecific/intergeneric hybrid embryo formation. Therefore, we uncover a ‘lock-and-key’ system governing the hybridization breadth of interspecific/intergeneric crosses in Brassicaceae. Manipulating this system holds promise for facilitating broad hybridization in crops.
Keywords: Pollen-pistil interaction, interspecific/intergeneric hybridization barrier, RALF, CrRLK1L receptors, pollen mentor effect, reproductive isolation, Brassicaceae
Graphical Abstract
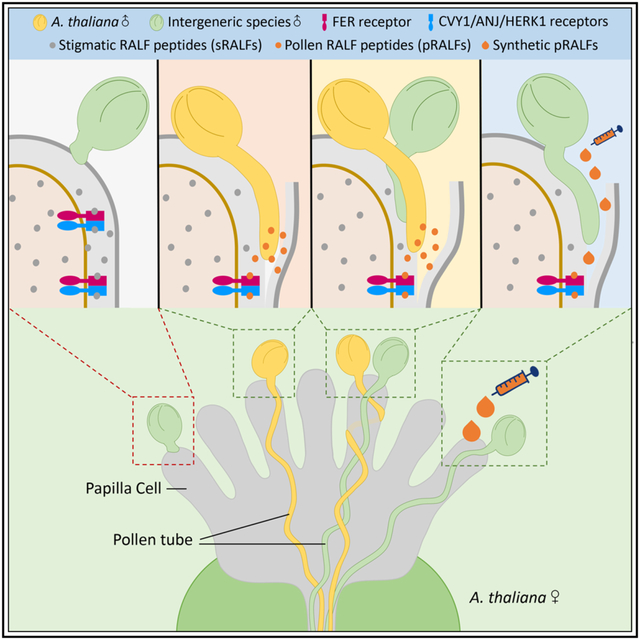
In Brief
A stigmatic intergeneric hybridization barrier is established via sRALF peptides/CrRLK1L receptors in concert with LRX proteins to block the penetration of incompatible pollen tubes, which can be overcome by pRALF peptides derived from compatible pollen.
INTRODUCTION
As per zoologist Ernst Mayr, the ‘classical’ concept of biological species centers on interbreeding and is defined thus: ‘a species consists of groups or populations that are capable to breed and to produce fertile offspring, but are reproductively isolated from other such groups or populations.1 Nevertheless, it is now widely accepted that natural hybridization can occur between different species, occasionally leading to the emergence of new species. This phenomenon, known as hybrid speciation, is particularly prevalent in flowering plants (angiosperms).2,3 Many important cultivated plants, such as wheat (Triticum aestivum), upland and sea-island cotton (Gossypium hirsutum and G. barbadense), oilseed rape (Brassica napus), leaf mustard (B. juncea) and others, originated from natural hybrids.4 The establishment of a new species necessitates genetic isolation, which is achieved through pre- and/or post-zygotic reproductive hybridization barriers.5,6,7,8,9,10
In the context of pre-zygotic reproductive isolation, pollen-pistil interactions impose a series of interspecific/intergeneric hybridization barriers.11 The surface of stigma cells, which serves as the receptive site for pollen grains, is the initial point of compatibility determination. Therefore, pollen-stigma interactions, encompassing pollen adhesion, pollen hydration, pollen germination, pollen tube penetration into the stigma, and pollen tube growth into the style, constitute significant interspecific/intergenric hybridization barriers in plants. These interactions are crucial since pollen from distantly related plant species remains unrecognized by stigma cells and fails to germinate tubes or penetrate the stigma/style. Our current understanding of how pollen-stigma interactions function as interspecific/intergeneric hybridization barriers is limited.
Recent research in Brassica rapa revealed that the stigmatic S-locus receptor kinase (SRK) receptor, known for regulating self-incompatibility (SI) in Brassicaceae, contributes to the rejection of intra- and inter-specific pollen via FERONIA (FER)-ROS signaling.12 This finding showed that this SI system could serve as an interspecific hybridization barriers in crops containing SRKs.13,14 Nonetheless, there is an increasing need to identify other pollen-stigma interactions that act as interspecific/intergeneric hybridization barriers in the majority of flowering plants that lack stigmatic SI systems. In Arabidopsis thaliana, STIGMATIC PRIVACY 1 (SPRI1), a stigmatic protein, was implicated in establishing interspecies incompatibility in the Brassicaceae.15 The interactors and the underlying molecular mechanism of SPRI1 remained elusive. Recent findings also showed that POLLEN COAT PROTEIN B-class (PCP-B) peptides carried by compatible pollen grains are recognized by the stigmatic receptor FER in a species-specific manner, promoting rapid hydration of conspecific pollen.12,16 However, brief delayed hydration still permits interspecific pollen germination and tube penetration/growth,12 hence not constituting a strict interspecific/intergeneric hybridization barrier.
To overcome the stringent pollen-stigma hybridization barriers, plant breeders discovered that applying conspecific pollen alongside heterospecific pollen onto the stigma can break the recognition barrier, enabling foreign pollen tubes to penetrate the stigma. This phenomenon, termed the “pollen mentor effect”17, has applications in agriculture to breach interspecific/intergeneric hybridization barriers and create hybrids that were previously unattainable. However, the underlying molecular mechanism and the nature of the pollen factor(s) are unknown. When using viable conspecific pollen, a major challenge is that it predominantly yields conspecific offspring rather than hybrids due to the competitive advantage of conspecific pollen.18,19 As a result, gamma-irradiation, X-rays or chemical treatment are commonly adopted to deactivate mentor pollen prior to application.17 Alternatively, protein extracts from conspecific pollen can induce the mentoring effect. The successful deployment of these strategies implies that breaking the stigmatic interspecific/intergeneric hybridization barrier via utilizing the pollen mentor effect might rely on certain pollen factors that are not disabled by radiation and/or chemical treatment.20 It was speculated that the mentor pollen provides either (i) some recognition proteins enabling incompatible pollen to germinate, (ii) a P-factor (from pollen) that interacts with an S-factor (from stigma) to render the accessibility of incompatible pollen, and/or (iii) certain pollen tube growth supporting substances.17 We suspect that such factor(s) may represent polymorphic protein-based signaling molecules derived from pollen grains.
In recent years, evidence has emerged demonstrating that peptide signals and their corresponding receptors govern cell-to-cell communication, playing important roles in plant growth, reproduction, and immunity regulation.21,22,23,24 During fertilization, female-male interactions, including recognition between pollen/pollen tubes and the stigma, the style, the transmitting tract, the septum and the ovule, heavily depend on peptide/receptor-mediated signaling pathways.22,25 The first step in female-male interactions is recognition between pollen and the stigma, which controls SI responses in Brassica species12,26,27,28 and pollen hydration in A. thaliana16 via pollen-secreted peptides and stigmatic receptors. While these peptide/receptor-mediated signaling pathways are known to influence pollen-stigma recognition, the use of peptides/receptors beyond SRK-mediated SI systems to prevent interspecific/intergeneric hybridization in plants lacking SI systems and/or even with SI systems has not been explored. The identity of pollen-derived “mentor factors” also remains unknown.
In this study, we identified a group of seven pollen-secreted RAPID ALKALINIZATION FACTOR peptides (pRALFs) and demonstrated their role in opening the stigmatic hybridization barrier established by interactions among stigmatic CrRLK1L receptors, stigma-secreted RALF peptides (sRALFs) and stigmatic LRX cell wall proteins. This signaling pathway diverges from previously described pollen-pistil communication systems. Disrupting the stigmatic hybridization barrier via genetic approaches or exogenous application of synthetic pRALF peptides allowed broader interspecific crosses, producing intergenric hybrid embryos. This study uncovers a comprehensive molecular mechanism that establishes an intergeneric hybridization barrier during pollen tube penetration into stigmas in plants. It also demonstrate that this stigmatic hybridization barrier is a significant hurdle to intergeneric crosses. Altogether, we can now explain the genetic basis of the “pollen mentor effect” and demonstrate an easy and practical strategy to overcome the stigmatic intergeneric hybridization barrier, which can be used in the future to achieve wider hybridizations in crop plants.
RESULTS
Seven pollen-specific RALF peptides (pRALFs) promote pollen tube penetration on A. thaliana stigmas
To identify possible pollen mentor effectors, we first conducted RNA sequencing (RNA-seq) analysis on mature pollen grains to identify putative secreted and pollen-specific polymorphic proteins that could play a role in pollen-stigma communication. Multiple RALF peptide genes were found to be highly and specifically expressed (Figure S1A), and they cluster into five clades in the phylogenetic tree (Figure S1B). Three clades contain previously characterized RALF members (RALF4/19, RALF6/7/16, RALF36/37) that function in maintenance of pollen tube integrity, establishment of the polytubey block and pollen tube reception, respectively.29,30,31 Therefore, we focused on the other two RALF clades. After confirming their high expression levels in pollen/pollen tubes by assaying promoter activity and fluorescent fusion proteins (Figures 1A, 1B, S1C and S1D), we systematically knocked out these RALF genes via CRISPR/Cas9 technology. Based on phylogenic clustering, we created, for example, a triple mutant r8 r9 r15 (knocking out RALF8, 9, and 15, simultaneously, Figures S2A and S2B) of one clade and mutants of different orders of the other clade. The latter mutants include a quadruple mutant r10 r11 r12 r13 (Figure S2C and S2D), a triple mutant r25 r26 r30 (Figures S2E and S2F), and a septuple mutant r10 r11 r12 r13 r25 r26 r30 (abbreviated as ralf sept, Figures 1C and S2G). We did not find an obvious phenotype when either r8 r9 r15 triple or r10 r11 r12 r13 quadruple mutant pollen was crossed to wild-type (WT) pistils (Figures 1D, 1E and S2H). In contrast, r25 r26 r30 triple and ralf sept mutant pollen that can hydrate and germinate normally on WT stigmas (Figures 1F, 1G and S2I) had difficulties in penetrating papilla cells at 3 hours after pollination (HAP), whilst the ralf sept mutant pollen completely failed to penetrate (Figures 1D and 1E). Cryo-scanning electron microscopy (CSEM) results confirmed the penetration failure of ralf sept mutant pollen into stigmas (Figure 1H). It should be noted that the penetration defect of ralf sept mutant pollen was significantly more severe than that of r25 r26 r30 mutant pollen, indicating a genetic redundancy among the seven pollen-expressed pRALFs. Penetration failure of ralf sept pollen tubes into stigmas likely resulted from abnormal recognition/communication with the wild-type stigmas, which was further supported by in vitro assays, in which germination of ralf sept pollen grains and tube growth were indistinguishable from those of wild type plants (Figures 1I and 1J). Pollen penetration defects can be partially or largely complemented by expressing pRALF11 or pRALF26 in ralf sept mutant (Figures 1D and 1E), supporting the hypothesis of a genetic redundancy of the seven pRALFs. Notably, exogenous application of 10 μM synthetic pRALF11 or pRALF26 peptide onto stigmas could partially or largely rescue the penetration defects of ralf sept mutant pollen, whereas synthetic RALF4 or RALF7, two unrelated pollen-expressed RALF peptides, could not (Figures 1D, 1E and S2J). This further indicates a highly critical role of pRALF peptides in pollen-stigma communication.
Figure 1. Seven pollen-specific RALF peptides (pRALFs) promote pollen-tube penetration in Arabidopsis.
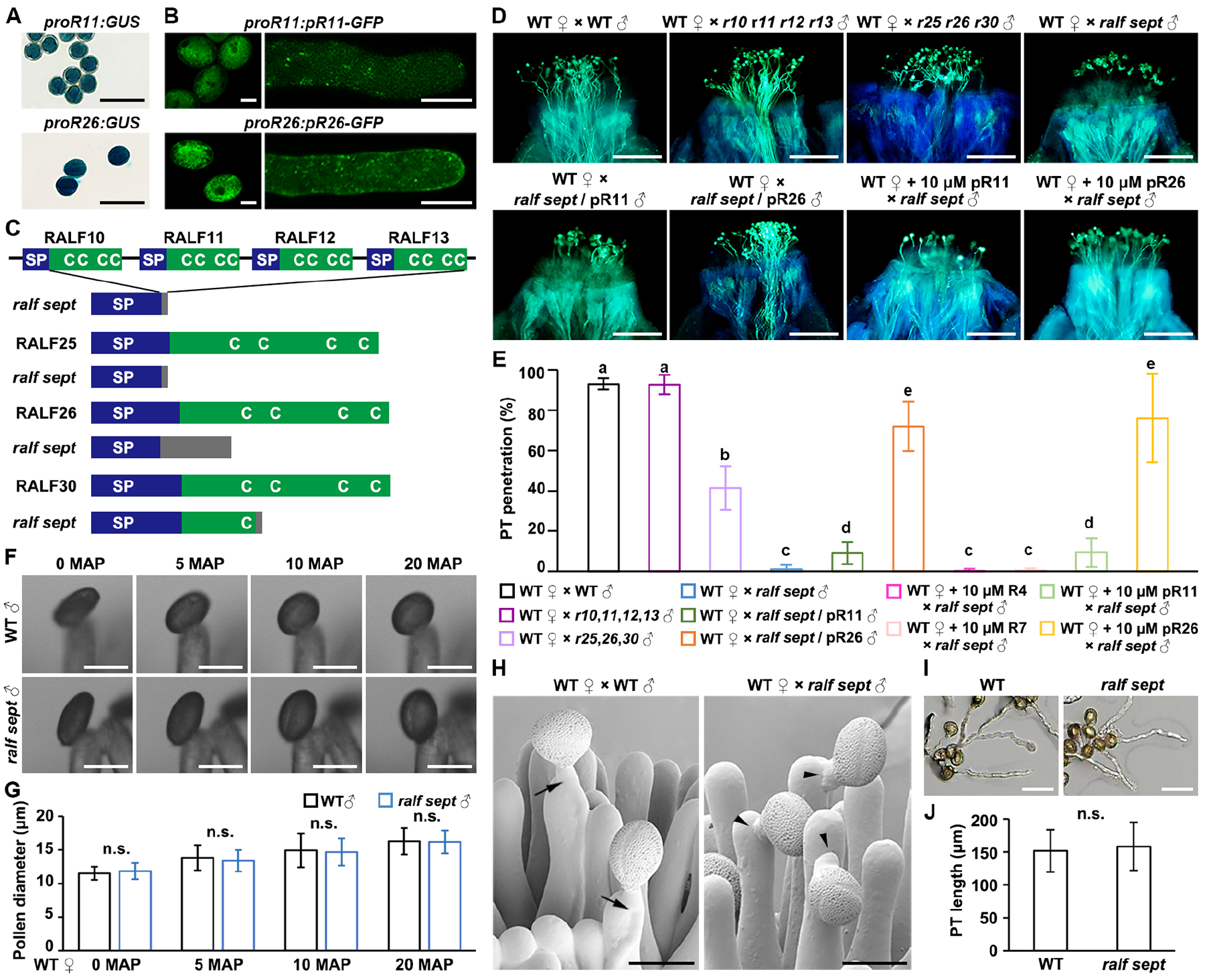
(A) GUS staining images show promoter activities of pRALF11 and pRALF26 (abbreviated as pR11 and pR26, respectively) in pollen. Scale bars, 50 μm.
(B) Mature pollen grains and pollen tubes expressing protein fusions of pR11/pR26-GFP driven by their respective native promoters. Scale bars, 10 μm.
(C) Schematic diagrams describe specific mutations created by CRISPR/Cas9-edited mutagenesis in the ralf10 ralf11 ralf12 ralf13 ralf25 ralf26 ralf30 septuple mutant (abbreviated as ralf sept). Native peptide structures were aligned above respective mutations. C, conserved cysteine residues; SP, signal peptide. Gray boxes indicate missense sequences due to frame shift or fragment deletion.
(D) Aniline blue staining shows status of pollen tube penetration in the wild-type (WT) stigmas at 3 hours after pollination (HAP). Pollen applied include WT, r10 r11 r12 r13, r25 r26 r30, ralf sept, pR11-complemented ralf sept, or pR26-complemented ralf sept. +10 μM pR11/26 indicate WT stigmas that were pre-treated with synthetic pR11 or pR26 peptides, respectively. Scale bars, 200 μm.
(E) Statistical analysis of pollen tube penetration assayed in (D) with negative controls shown in Figure S2J. Experiments were repeated at least three times. Different letters represent significant differences between groups (P<0.001).
(F) Representative bright field images show pollination-induced pollen hydration of WT or ralf sept pollen on WT stigmas. MAP, minutes after pollination. Scale bars, 20 μm.
(G) Measurement and statistical analysis of pollen diameter during pollen hydration in (F). Equatorial diameters of pollen grains were measured.
(H) Cryo-Scanning Electron Microscopic (CSEM) images of WT stigmas pollinated with WT or ralf sept pollen at 3 HAP. Black arrows point to WT pollen tubes penetrating the papilla cells and black arrow heads point to ralf sept pollen tubes failing to penetrate the papilla cells. Scale bars, 25 μm.
(I) In vitro growth of WT and ralf sept pollen tubes. Images were collected after incubation for 6 hours. Scale bars, 50 μm.
(J) Statistical analysis of pollen tube length in (I). Experiments were repeated at least three times. N>100 for each group. Data in (E, G and J) are mean values ± SD. n.s., not significant, P>0.1 (Student’s t test).
FER/CURVY (CVY1)/ANJ/HERCULES RECEPTOR KINASE 1 (HERK1) receptors control pollen tube penetration at the stigma
Then, how can pollen-derived RALF peptides regulate pollen tube penetration into stigmas? RALF peptides have been previously reported to interact with CrRLK1L family receptors.29,30,31,32,33,34 To identify the stigmatic receptors that perceive the seven pRALFs, RNA-seq analysis was conducted with stigma tissues. We found that four CrRLK1L receptors including FER from one clade and CVY1, ANJ, as well as HERK1 from another clade are expressed in stigma papilla cells (Figures 2A, 2B, 2C and S3A). We next examined single fer-435, single cvy136 and double anj herk137 mutants, respectively. Genetic analysis showed that WT pollen could penetrate stigmas of all mutants (Figures 2D, 2E, S3B and S3C). However, in striking contrast to their failures in penetrating WT and cvy1 stigmas (Figures S3D and S3E), ralf sept mutant pollen tubes could fully penetrate papilla cells of the fer-4 mutant, and partially penetrate papilla cells of the anj herk1 mutant (Figures 2F and 2G). By further knocking out cvy1 in the anj herk1 mutant background (Figure S3F), ralf sept mutant pollen tubes could now fully penetrate papilla cells of the cvy1 anj herk1 triple mutant (Figures 2F and 2G) comparable to WT levels (Figures 2D and 2E). For comparison, when another stigma-expressed CrRLK1L receptor THESEUS1 (THE1) was investigated, ralf sept mutant pollen tubes could not penetrate papilla cells of the the1-4 mutant38 (Figures S3D and S3E). These findings revealed a critical role of CrRLK1L receptors in specifically sensing paracrine pRALF signals. The fact that the penetration defect of ralf sept pollen tubes was completely rescued by a fer-4 single mutation or by the combined mutation of cvy1, anj and herk1 indicates that the CVY1, ANJ, and HERK1 receptors function redundantly to control pollen tube penetration, and that FER functions differently from CVY1/ANJ/HERK1, supporting the hypothesis that FER serves as a scaffold for other receptors in the assembly of receptor complexes.34 In summary, the role of CrRLK1L receptors in sensing paracrine pRALF signals is indispensable only in the absence of pRALFs, hinting at the existence of an inhibitory system functioning through these receptors to regulate pollen tube penetration.
Figure 2. FER/CVY1/ANJ/HERK1 receptors perceive pollen derived pRALFs.
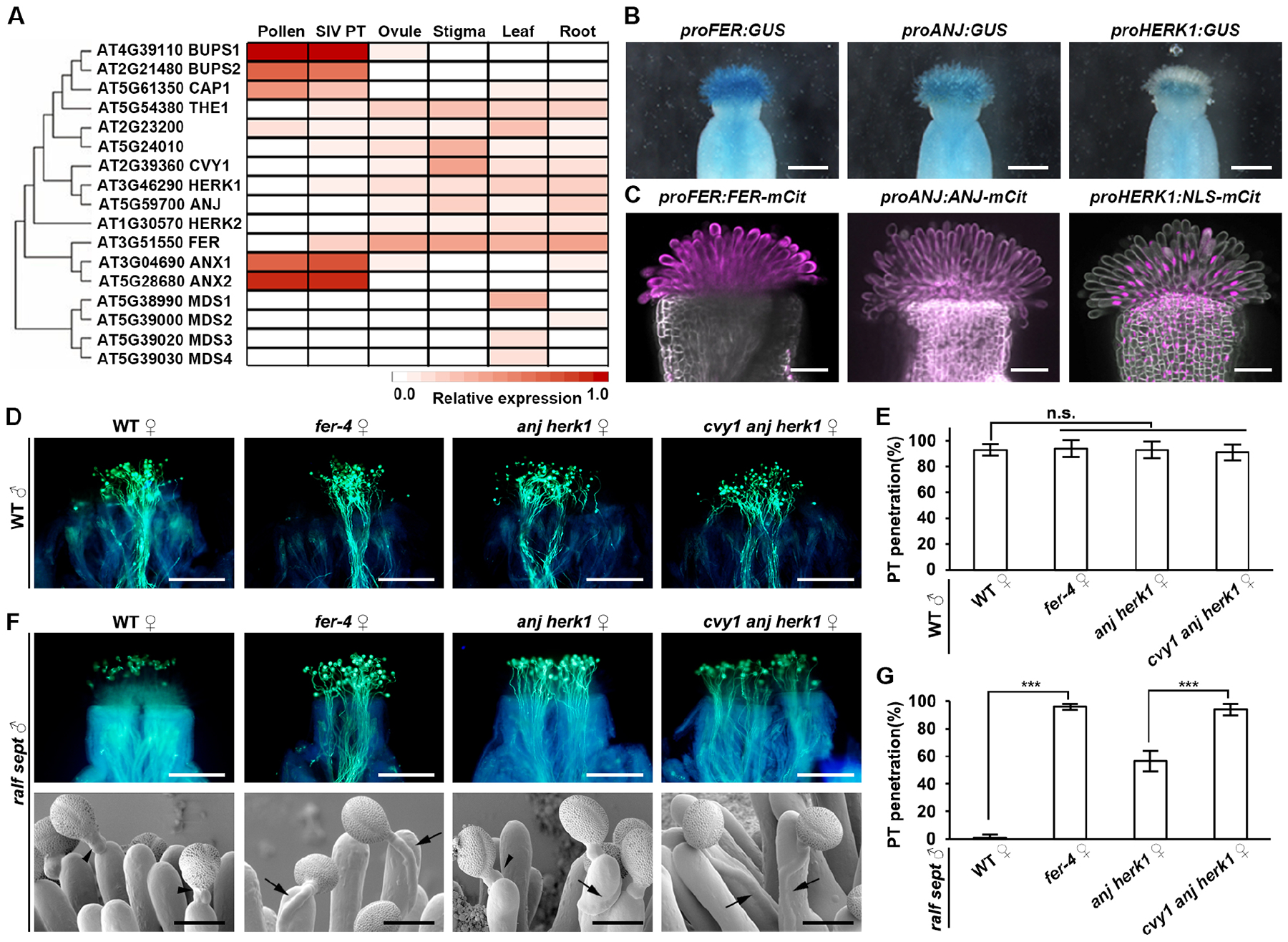
(A) Phylogenetic tree of the Arabidopsis CrRLK1L family and expression heatmap of each member in different tissues.
(B) Results of GUS staining show promoter activity of FER, ANJ and HERK1 in the stigmas. Scale bars, 0.25 mm.
(C) Fluorescence signals show fusion reporter protein of indicated transgenes in the stigmas. mCit, mCitrine; NLS-mCit, mCitrine with nuclear localization sequence. Scale bars, 0.1 mm.
(D) Aniline blue staining shows penetration of WT pollen tubes at 3 HAP in the stigmas of WT, fer-4, anj herk1, and cvy1 anj herk1, respectively. Scale bars, 200 μm.
(E) Statistical analysis of pollen tube penetration shown in (D).
(F) Aniline blue staining and CSEM images showing the penetration status of ralf sept pollen tubes at 3 HAP in the stigmas of WT, fer-4, anj herk1, and cvy1 anj herk1, respectively. Black arrow heads point to ralf sept pollen tubes failing to penetrate and black arrows point to ralf sept pollen tubes penetrating the stigma. Scale bars for aniline blue staining, 200 μm. Scale bars for CSEM, 25 μm.
(G) Statistical analysis of pollen tube penetration shown in (F). Data in (E and G) are mean values ± SD; *** shows P<0.001 (Student’s t test). n.s., not significant, P>0.1 (Student’s t test). Each of the above assays was repeated at least three times.
See also Figure S3.
Autocrine stigmatic RALF (sRALF) signaling controls pollen tube penetration at the stigma
We further hypothesized that the inhibitory role of these receptors would require the involvement of other peptide ligands, likely derived from the pistil side as autocrine signals. Indeed, we identified four stigma-expressing RALF peptides (sRALFs), sRALF1, 22, 23, and 3316 (Figures S1B, S4A and S4B). RALF peptides were previously classified into two major distinct groups39, one of which contain pRALFs that we characterized above and which do not show the typical seedling or root growth inhibitory property of the family.40 However, the other group includes sRALFs which possess the growth inhibitory property. Protein sequence alignment showed that sRALFs are divergent from pRALFs by their conserved S1P cleavage site as well as YISY and YY motifs (Figure 3A). To perform genetic tests, we obtained the r33–3 single mutant16 and by CRISPR/Cas9 technology generated r22 r33 double, r1 r22 r33 triple, and r1 r22 r23 r33 quadruple mutants (abbreviated as ralf quad, Figures 3B and S4C). Intriguingly, the phenotypes of these sRALF mutants highly resembled those of the fer-4 and cvy1 anj herk1 receptor mutants, respectively, described above. More specifically, ralf sept mutant pollen tubes could partially penetrate the papilla cells of r33–3 single, r22 r33 double or r1 r22 r33 triple mutants, but fully penetrated the papilla cells of the ralf quad mutant (Figures 3C and 3D), similar to WT pollen tubes that fully penetrated ralf quad stigmas (Figures S4D and S4E). Therefore, the more sRALF genes were mutated, the higher rates for the ralf sept mutant pollen were observed to penetrate into sRALF mutant stigmas, supporting the hypothesis that all four sRALFs act redundantly and contribute to the establishment of the stigmatic gate.
Figure 3. Autocrine RALFs function through FER/CVY1/ANJ/HERK1 receptors to form an inhibitory system at the stigma.
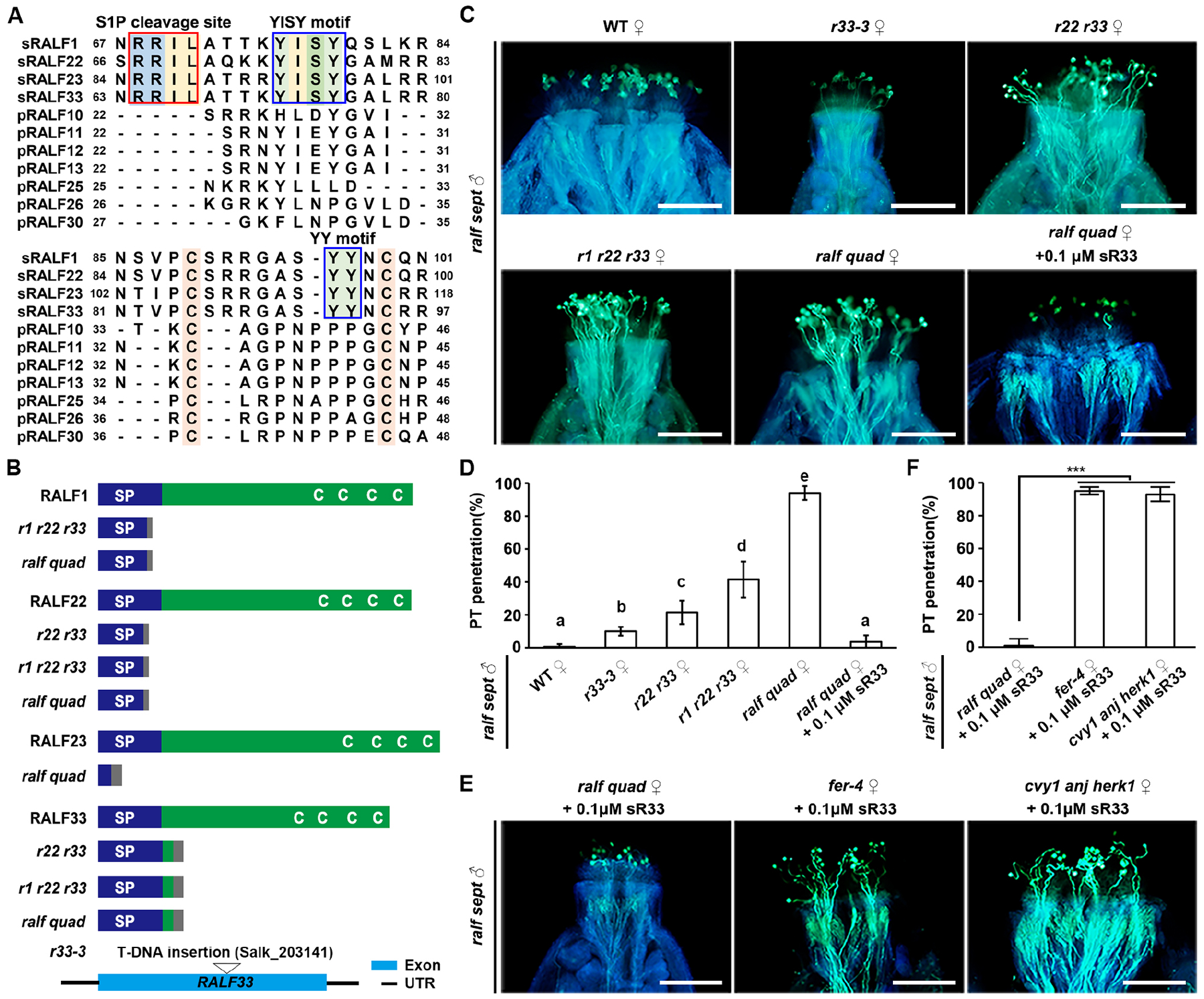
(A) Amino-acid alignment of pRALFs and sRALFs. Conserved residues are highlighted with colors. Boxes show conserved motifs as indicated.
(B) Schematic diagrams describing wild-type and CRISPR/Cas9-generated mutated peptide structures of sRALF1, 22, 23 and 33 in the double, triple and quadruple sRALF mutants. (Bottom) T-DNA insertional position in RALF33. C, conserved cysteine residues (Cs); SP, signal peptides. Gray boxes indicate missense sequences due to frame shift mutations.
(C) Aniline blue staining showing penetration status of ralf sept pollen tubes at 3 HAP in the stigmas of WT and sRALF mutants as indicated (ralf quad = r1 r21 r22 r33), as well as penetration of ralf quad stigmas pretreated with 0.1 μM synthetic sR33. Scale bars, 200 μm.
(D) Statistical analysis of pollen tube penetration shown in (C). Different letters represent significant differences between groups (P<0.001).
(E) Aniline blue staining showing penetration status of ralf sept pollen tubes at 3 HAP into the stigmas of ralf quad, fer-4 or cvy1 anj herk1, all of which were pretreated with 0.1 μM synthetic sR33. Scale bars, 200 μm.
(F) Statistical analysis of pollen tube penetration assayed in (F). Data in (D and F) are mean values ± SD; *** shows P<0.001 (Student’s t test). Each of the above assays was repeated at least three times.
See also Figure S4.
Importantly, exogenous application of 0.1 μM synthetic sRALF33 peptide could completely restore the penetration blockage of ralf sept mutant pollen tubes in ralf quad mutant stigmas (Figures 3C and 3D). This directly supports the biological importance of sRALFs in an autocrine signaling pathway with CrRLK1L receptors. Moreover, exogenous application of synthetic sRALF33 could not block penetration of ralf sept pollen tubes into stigmas of either fer-4 single or cvy1 anj herk1 triple mutants (Figures 3E and 3F). This finding indicates that sRALF33 signaling requires the presence of FER and CVY1/ANJ/HERK1 receptors in the same genetic pathway.
pRALFs compete for receptor binding to open the stigmatic lock
Interestingly, we further found that this sRALF33-mediated blockage could be gradually lifted by the addition of an increasing concentration of synthetic pRALF26 peptide (Figures 4A and 4B), indicating that these two groups of RALF peptides act antagonistically. In vitro Co-IP assays showed that both autocrine sRALF22/33 and paracrine pRALF11/26 peptides interact with the same receptors (Figures 4C and 4D). Microscale thermophoresis (MST) analysis further revealed that selected sRALF33, pRALF11 or pRALF26 could interact with FER, CVY1, ANJ, and HERK1, respectively, with high affinity as indicated by low equilibrium dissociation constants (Kd) (Figures 4E–4H). Moreover, pRALF26 was able to compete with sRALF33 for receptor binding (Figures 4I and 4J), which clearly demonstrates the combating feature of these peptides in signaling. Thus, we propose that on the stigma, sRALFs bind to the receptor complex FER-CVY1/ANJ/HERK1 to establish a ‘lock’ that prevents pollen tubes from penetrating stigmatic papilla cells. On the other side, pRALFs secreted from compatible pollen compete with autocrine sRALFs and serve as a ‘key’ to open the stigmatic ‘lock’, thereby enabling the penetration of pollen tubes. As a result, pollen grains lacking compatible paracrine pRALFs or similar peptides would not be able to unlock the autocrine ligand-receptor signaling gate that prevents pollen tube penetration.
Figure 4. pRALFs compete with sRALFs for FER-CVY1/ANJ/HERK1-receptor binding.
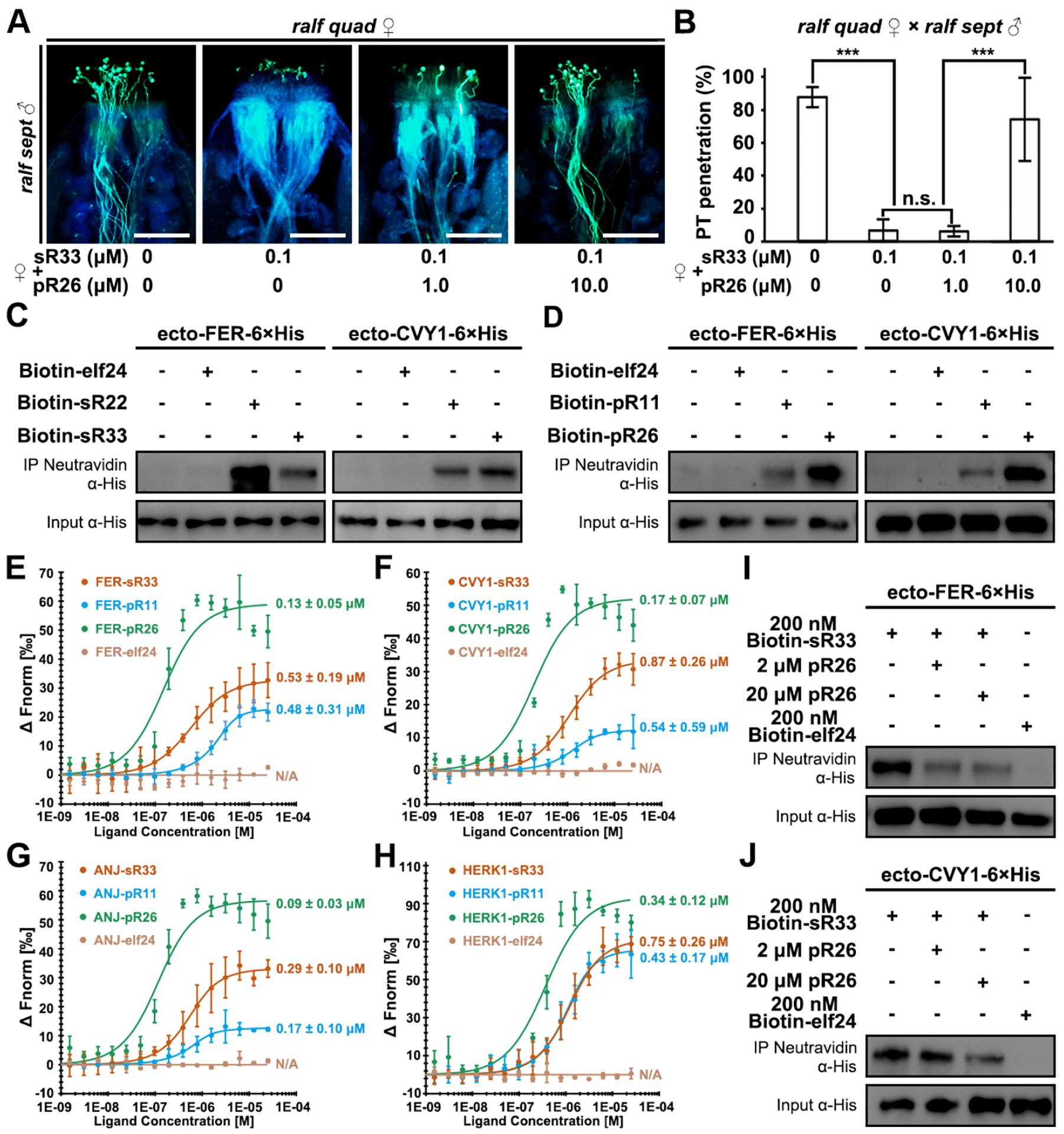
(A) Aniline blue staining showing competition results at 3 HAP. The ralf quad mutant stigmas were pre-treated with combinations of differentially concentrated sR33 and/or pR26 synthetic peptides (concentrations specified below). Scale bars, 200 μm.
(B) Statistical analysis of pollen tube penetration shown in (A).
(C) Pull-down assays showing binding activities between 6×His-tagged ectodomains of FER or CVY1 with biotinylated peptides, elf24 (negative control) and sR22/33, respectively.
(D) Pull-down assays showing binding activities between 6×His-tagged ectodomains of FER or CVY1 with biotinylated peptides, elf24 (negative control) and pR11/26, respectively.
(E-H) MST analyses show high binding affinities of sR33, pR11, and pR26, but not of elf24 (negative control) with the ectodomains of FER (E), CVY1 (F), ANJ (G), and HERK1 (H), respectively.
(I) Competitive pull-down assays showing changes in binding activities between 6×His-tagged ectodomains of FER and sR33 in the presence of an increasing amount of pR26.
(J) Competitive pull-down assays showing changes in binding activities between 6×His-tagged ectodomains of CVY and sR33 in the presence of an increasing amount of pR26.
Data in (B) are mean values ± SD; *** shows P<0.001 (Student’s t test). n.s., not significant, P>0.1 (Student’s t test). Each of the above assays was repeated at least three times.
LRX cell wall proteins are essential for the stigmatic lock
Next, we investigated the mechanism how antagonistic RALF peptides control pollen tube penetration into the stigma. Because the two stigmatic peptides sRALF23/33 and the two receptors FER/ANJ characterized in this study were also previously reported to regulate the level of reactive oxygen species (ROS) to control the timing of pollen hydration at the stigma16, we next investigated whether ROS levels also regulate the pollen tube penetration process. After treating stigmas with either ROS scavengers or the NADPH oxidase inhibitor DPI, both of which lower ROS levels in vivo, we found that the penetration defects of ralf sept pollen tubes into WT stigmas were not altered (Figures S5A–S5C). Our RNA-seq analysis identified two NADPH oxidases of the RBOH family, RBOHD and RBOHF, that showed expression in the stigma (Figure S5D). The mutation of RBOHD was previously reported to promote pollen hydration.16 We therefore characterized rbohd single41 and rbohd rbohf double mutants42, and found that, although stigmatic ROS levels were significantly reduced in both mutants compared to that in WT stigmas (Figures 5A and 5B), the penetration defects of ralf sept pollen tubes were not altered (Figures 5C and 5D). These results collectively indicate that ROS does not directly contribute to the pRALF-sRALF-mediated control of pollen tube penetration.
Figure 5. Cell-wall LRX proteins participate in the establishment of the stigmatic lock.
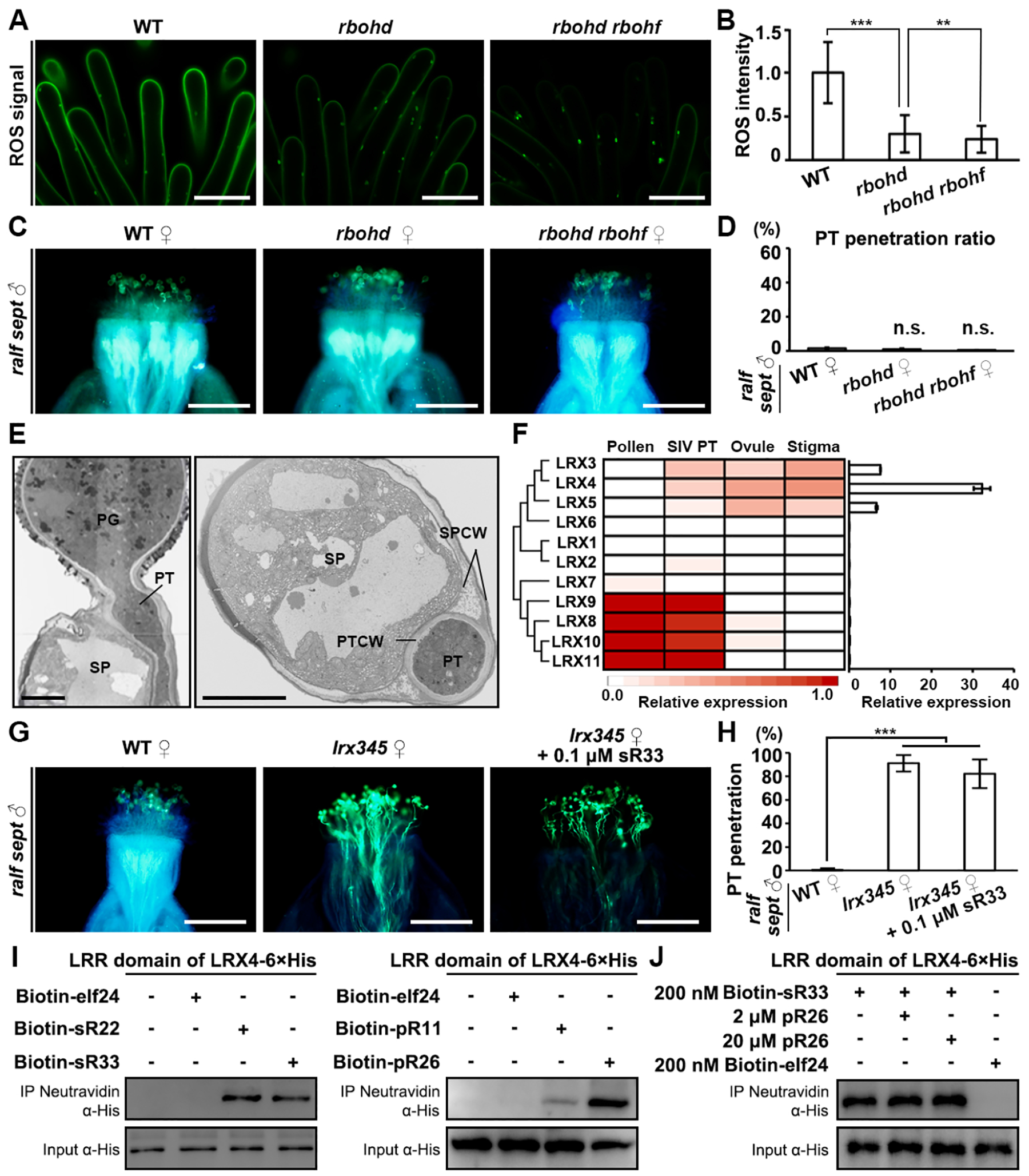
(A) H2DCF-DA staining shows ROS levels in unpollinated papilla cells of WT, rbohd and rbohd rbohf, respectively. Scale bars, 10 μm.
(B) Quantification of stigmatic ROS level shown in (A). Data are mean values ± SD; *** shows P<0.001, ** shows P<0.01 (Student’s t test).
(C) Aniline blue staining showing penetration status of ralf sept pollen tubes at 3 HAP in the stigmas of WT, rbohd and rbohd rbohf, respectively. Scale bars, 200 μm.
(D) Statistical analysis of the pollen tube penetration phenotype shown in (A). n.s., not significant, P>0.1 (Student’s t test).
(E) TEM images of a wild-type pollen tube penetrating a wild-type papilla cell (penetration occurs between the cell wall and the plasma membrane). Left, longitudinal section; Right, cross section. PG, pollen grain; PT, pollen tube; SP, stigma papilla; SPCW, stigma papilla cell wall; PTCW, pollen tube cell wall. Scale bar, 5 μm.
(F) Relative expression levels of each LRX gene in designated tissues. Left, heatmap; Right, real-time PCR results.
(G) Aniline blue staining showing penetration status of ralf sept pollen tubes at 3 HAP in the stigmas of WT, lrx345, and lrx345 pretreated with 0.1 μM synthetic sR33, respectively. Scale bars, 200 μm.
(H) Statistical analysis of the pollen tube penetration phenotype shown in (G). Data are mean values ± SD; *** shows P<0.001 (Student’s t test). All the assays were performed at least three times.
(I) Pull-down assays showing binding activities between 6×His-tagged LRR domain of LRX4 and biotinylated elf24 (negative control), sRALFs (22/33, left panel) and pRALFs (11/26, right panel), respectively.
(J) Competitive pull-down assays showing lack of changes in binding activities between sR33 with 6×His-tagged LRR domain of LRX4 in the presence of an increasing amount of pR26.
See also Figure S5.
Considering the way how pollen tubes penetrate the stigmatic cell wall and grow in the loosened space between the wall and the plasma membrane43,44 (Figure 5E), it is plausible to hypothesize that stigmatic cell-wall proteins may participate in the regulation of penetration of compatible pollen tubes. It was previously reported that cell wall-associated LEUCINE-RICH REPEAT-EXTENSIN (LRX) proteins were able to interact with RALF peptides.30,45,46,47,48,49,50 We therefore investigated three LRX protein genes, LRX3, 4 and 5, which are expressed in the stigma (Figure 5F). Notably, genetic analysis showed that ralf sept mutant pollen tubes could penetrate stigmas of lrx3 lrx4 lrx5 (abbreviated as lrx345) triple mutants50 (Figures 5G and 5H), thus highly resembling their behavior on the pistils of the stigmatic receptor and sRALF mutants, respectively. Moreover, exogenous application of synthetic sRALF33, which blocked ralf sept mutant pollen tubes to penetrate the ralf quad mutant pistils, could not block penetration of the same ralf sept pollen tubes into lrx345 stigmas (Figures 5G and 5H). These results indicate that LRX3/4/5 proteins are reciprocally required for the sRALF-FER-CVY1/ANJ/HERK1-mediated signaling pathway to establish the stigmatic lock. We further tested protein-protein interactions by in vitro pull-down assays and found that both autocrine sRALF22/33 and paracrine pRALF11/26 peptides interacted with the LRR domain of LRX4 (Figure 5I), but the pRALFs could not compete with sRALFs for LRX binding (Figure 5J). This differs from their competitiveness for receptor binding (Figures 4I and 4J). Considering the amino acid sequence variations between pRALFs and sRALFs, it is well possible that pRALFs and sRALFs bind with different affinities or to different domains of LRX proteins, leading to differential regulation on the wall properties of papilla cells.48,51 It was previously reported that both CrRLK1L receptors and LRX proteins were involved in RALF-mediated regulation of cell wall integrity29,30, but the exact molecular link between RALF-binding LRXs and CrRLK1Ls was not clear.48,52 Elucidation of the structure of the sRALF/pRALF-FER-CVY1/ANJ/HERK1 receptor complexes in association with LRX proteins will be challenging, but could ultimately help to understand the antagonistic RALF peptide-mediated unlocking mechanism for pollen tube penetration.
Pollen tube penetration and fertilization from distantly-related species are enabled by the pRALF-mediated mentor effect
According to this ‘lock-and-key’ model, the antagonism of pRALF- and sRALF-mediated CrRLK1L signaling would allow compatible pollen that produces similar pRALF peptides to penetrate the stigma but prevent alien/foreign pollen lacking similar peptides from penetrating, so that an interspecific/intergeneric hybridization barrier can be established. To test this hypothesis, we collected pollen from different Brassicaceae species including Arabidopsis lyrata, Capsella rubella, Erysium cheiranthoides, Cardamine flexuosa, Rorippa indica, and Descurainia sophia to pollinate A. thaliana stigmas. Notably, pollen from A. lyrata and C. rubella could penetrate, whereas pollen from E. cheiranthoides, C. flexuosa, R. indica and D. sophia could not penetrate (Figure 6A), which is consistent with the evolutionary divergence time between these species.53 The two Arabidopsis species separated approximately 5 million years ago (mya). A. thaliana and C. rubella diverged 14.6 mya, while divergence of the latter four species occurred already about 16.6 to 27.4 mya.53 This indicated that closely related species may produce similar paracrine pRALFs as effective keys. However, when the female stigmatic lock is disrupted by fer-4, cvy1 anj herk1, ralf quad or lrx345 mutations in A. thaliana, pollen from distant species, such as C. flexuosa, E. cheiranthoides, R. indica and D. sophia, was able to penetrate (Figures 6B–6F). Thus, the proposed ‘lock-and-key’ system indeed functions as an interspecific/intergeneric prezygotic barrier in A. thaliana that prevents pollen tube penetration from distantly related species in the Brassicaceae family.
Figure 6. Stigmatic penetration of pollen tubes from distantly-related Brassicaceae species is controlled by a FER-CVY1/ANJ/HERK1-LRX-composed stigmatic “lock”.
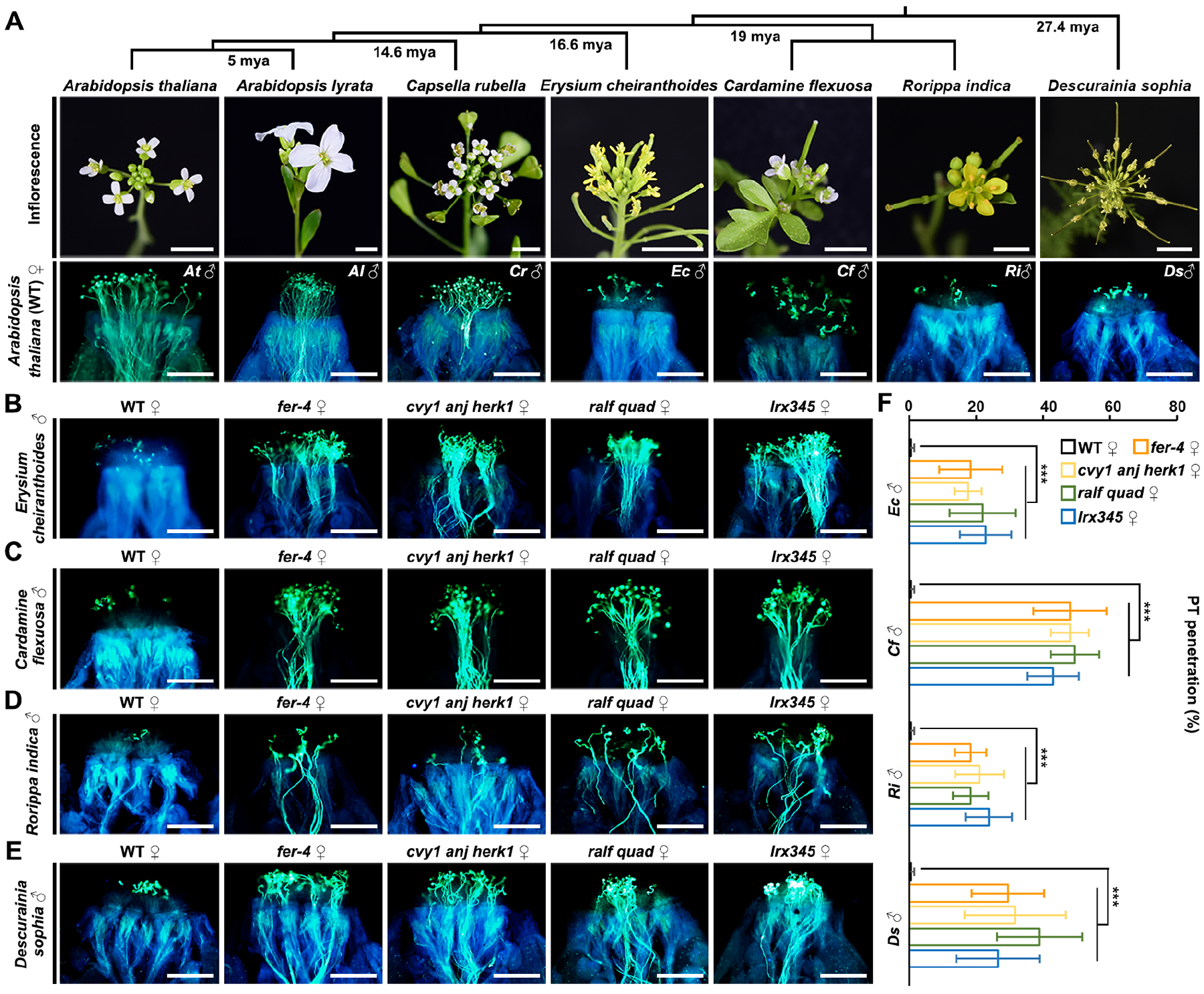
(A) Pollen-tube penetration assay (aniline blue staining) on A. thaliana pistils applied with pollen from multiple Brassicaceae species as indicated. Phylogenetic relations and inflorescence images of selected Brassicaceae species are shown above. Ri and Ds, 6 HAP, others, 3 HAP. Mya, million years ago.
(B-E) Aniline blue staining shows the pollen tube penetration status using indicated female and male genotypes. Pollen tubes of Erysium cheiranthoides (B), Cardamine flexuosa (C), Rorippa indica (D) and Descurainia sophia (E) in the stigmas of WT, fer-4, cvy1 anj herk1, ralf quad, or lrx345 mutants (from left to right), respectively. (B and C), 3HAP; (D and E), 6HAP. All assays were repeated at least three times. Scale bars in (A-E), 200 μm.
(F) Statistical analysis of the pollen tube penetration ratio shown in (B-E). *** shows P<0.001 (Student’s t test).
According to this ‘lock-and-key’ working model, paracrine pRALFs are apparently the most critical factors to unlock such an interspecific/intergeneric prezygotic hybridization barrier. To further verify this hypothesis, we treated A. thaliana stigmas with the ‘key’ from the pollen side, synthetic pRALF26 peptide as an example that is capable of unlocking the autocrine signaling-mediated barrier on A. thaliana stigmas. Strikingly, pollen from distant species E. cheiranthoides, C. flexuosa, R. indica and D. sophia could now efficiently penetrate the pRALF26-treated stigmatic papilla cells of A. thaliana (Figure 7A). More strikingly, we successfully obtained hybrid embryos after pollen from E. cheiranthoides or D. sophia was deposited to A. thaliana pistils either pre-treated with the pollen “key” pRALF26 or after disruption of the stigmatic “lock” (Figure 7B–7E). We noted that hybrid embryos were not obtained in the other two wide crosses due to reproductive barriers at the pollen tube guidance and reception stage, respectively (Figure S6). This result demonstrates that in the plants where the major reproductive barrier occurs at the stigma, removal of this stigmatic barrier could lead to fertilization and production of hybrid embryos and, thus, the opportunity for successful interspecific/intergeneric hybridization. In summary, synthetic pRALF26 peptide triggered wider outcrossing successfully and thus recapitulated a typical pollen mentor effect. Altogether these findings elucidated that the FER-CVY1/ANJ/HERK1-RALF complex in concert with LRX proteins are the major molecular components of the pollen mentor effect in the Brassicaceae (Figure 7F and 7G).
Figure 7. Pollen tube penetration and fertilization from distantly-related species are enabled by the pRALF-mediated pollen mentor effect.
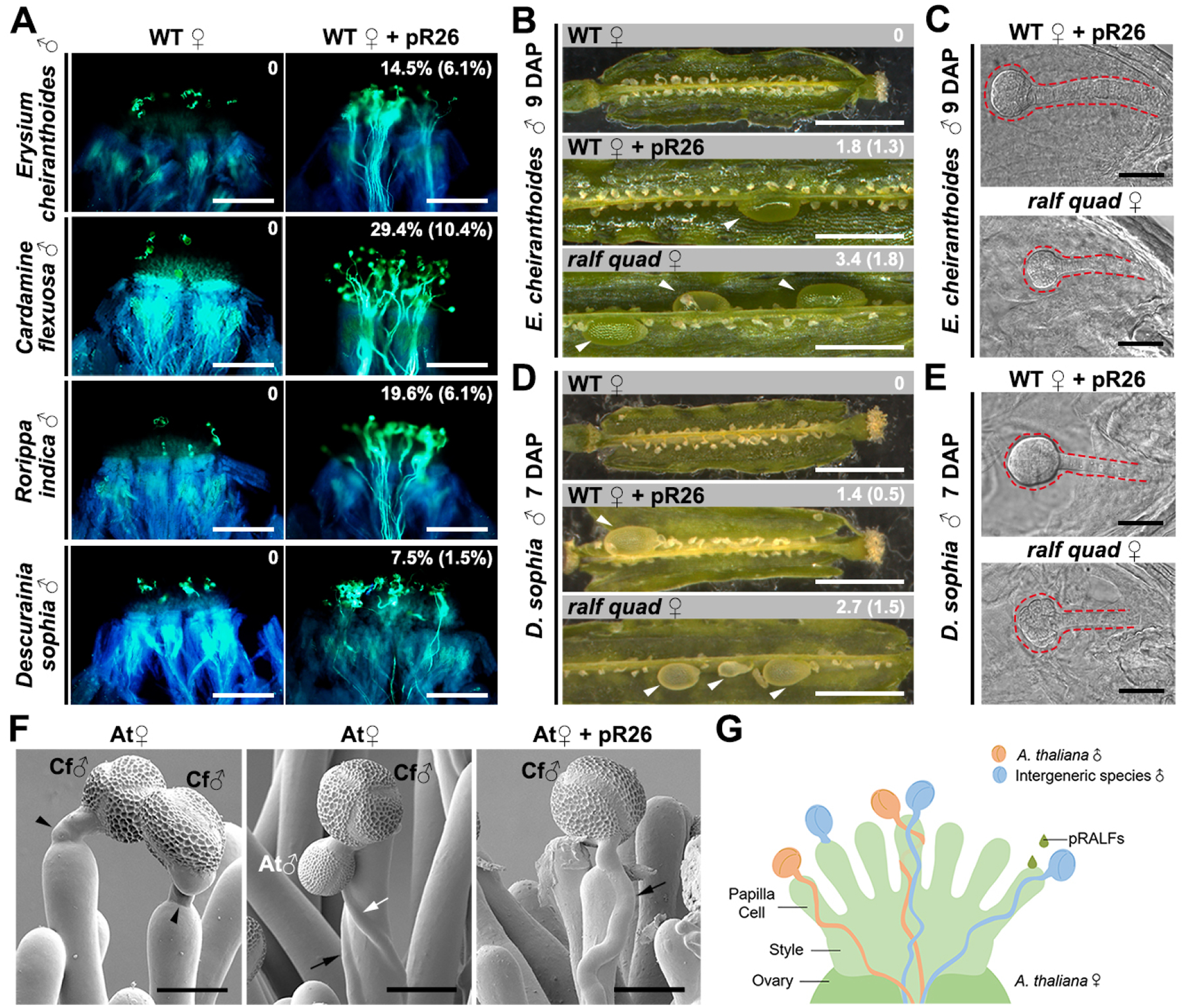
(A) Erysium cheiranthoides (3 HAP), Cardamine flexuosa (3 HAP), Rorippa indica (6 HAP) or Descurainia sophia (6 HAP) pollen tubes on WT (A. thaliana) stigmas pre-treated with/without 10 μM of synthetic pR26 peptide. The percentage values, shown as average percentage (s.d.), indicate average pollen tube penetration ratio of the corresponding pollen in A. thaliana pistils. Scale bars, 200 μm.
(B-E) Treatment of stigmas with the “key” pRALF26 and opening the stigmatic “lock” by knocking out sRALFs, respectively, broke the intergeneric reproductive barrier. In (B) and (D), arrowheads indicate enlarged ovules. Values in (B) and (D), shown as average (s.d.), indicate the number of enlarged ovules in the pod. (C) and (E) show DIC images of hybrid embryos. Dashed lines highlight the hybrid embryos formed 9 and 7 days after pollination (DAP), respectively. Scale bars, 1 mm (siliques) and 50 μm (embryos). Each experiment was repeated at least three times with consistent results.
(F) CSEM images of A. thaliana stigma (At) pollinated with Cardamine flexuosa (left), C. flexuosa and A. thaliana pollen (middle), as well as C. flexuosa pollen after pretreatment with synthetic pRALF26 peptide at 3 HAP (right). Black arrows point to C. flexuosa pollen tubes and white arrows point to A. thaliana pollen tubes penetrating papilla cells, whereas black arrow heads point to C. flexuosa pollen tubes failing to penetrate papilla cells. Scale bars, 25 μm.
(G) Graphic diagram describing the pollen mentor effect (allowing incompatible pollen tubes (blue) to penetrate and grow towards the transmitting tract. This can be induced by paracrine pRALF peptides, which are either derived from compatible pollen tubes (orange) or after treatment of synthetic peptides. External application of pRALFs enhances the efficiency of the pollen mentor effect, allowing distantly related intergeneric species to hybridize with A. thaliana and generate conventionally unattainable hybrids. See also Figure S6.
DISCUSSION
Pollen-stigma recognition represents a crucial interspecific/intergeneric reproductive barrier in distant hybridization of flowering plants. In this study, we demonstrated that by manipulating the corresponding RALF peptide signals as well as components in the CrRLK1L receptor complex and LRX cell wall proteins, we successfully broke through this stigmatic barrier. This manipulation resulted in successful fertilization between distantly-related species and the production of hybrid embryos. CrRLK1L family receptors, known for their roles developmental processes such as root hair growth35 and immunity responses34, have also been implicated in multiple aspects of reproduction, including pollen hydration16, maintenance of pollen tube integrity29,30,54, polytubey block31,55, and pollen tube reception.31,37,56,57 Among the CrRLK1L family receptors, FER appears to play pleiotropic roles, participating in nearly all of the aforementioned processes, while other FER-interacting CrRLK1L receptors contribute to specific roles in distinct biological contexts. FER is thought to act as a pivotal scaffold that perceives RALF signals and recruits other receptors and ligands to achieve specificity.34 This specificity has been demonstrated in the functions of ANJ for pollen hydration16 and ANJ/HERK1 for the polytubey block and pollen tube reception.31,37 In this study, we identified specific functions of the CVY1, ANJ and HERK1 receptors in papilla cells, and further supported FER’s role as a scaffold in receptor complexes.
Significantly, it has recently been reported that the stigma-expressed FER receptor also plays a role in SRK-dependent SI responses and interspecific isolation in Chinese cabbage by regulating stigmatic ROS production.12,58 This finding seems to underscore a general trend in CrRLK1L signaling, as ROS levels—acting as downstream effectors—have been reported to undergo changes in several CrRLK1L receptor-mediated processes, including immunity40, pollen hydration16, pollen tube integrity59,60 and pollen tube reception.61 For example, RALF33-FER/ANJ-mediated signaling maintains elevated stigmatic ROS levels, while decreasing ROS levels promotes pollen hydration in A. thaliana16 and self/interspecific pollination in Chinese cabbage.12,58 Intriguingly, in contrast to these cases, our study demonstrates that reducing ROS levels does not promote pollen penetration into the stigma. This suggests that in A. thaliana and potentially other species lacking stigmatic SI systems, ROS might not be a component of the downstream signaling pathway modulated by the counteractive interactions of pRALFs/sRALFs and LRX proteins in controlling pollen tube penetration. The investigation now shifts towards comprehending the variations in FER receptor complexes across different processes and identifying the downstream signaling constituents. This is pivotal for better understanding how the aforementioned ‘lock-and-key’ mechanism rculminates in a discernible outcome that ultimately leads to the alteration of stigmatic cell walls, thus influencing pollen tube penetration.
It is plausible that RALFs indeed represent the specific components steering the assembly of distinct FER-complexes and outcomes. By analyzing the sequence properties of mature peptides from 51 plant species, RALF peptides can be broadly classified into two major groups. One group displays ubiquitous expression across various tissues (Clade I, II, and III), while the other group exhibits specific expression in reproductive tissues (Clade IV).39 However, RALF members within Clade IV have been less extensively studied. An recent investigation revealed that RALF6/7/16/36/37 from this clade are involved in regulating the polytubey block and pollen tube reception.31 Interestingly, the four sRALF peptides reported in this study belong to Clade I/II RALF members, all of which encompass the S1P protease cleavage sites and YISY/YY motifs crucial for interacting with LLG co-receptors or LRX cell wall proteins.39,48 Conversely, pRALFs belong to Clade IV and lack these motifs. The antagonism and functional divergence elucidated in our study between pRALFs and sRALFs can, to some extent, be attributed to their distinct protein domains and biophysical properties. These factors might contribute to the modification and composition of receptor complexes, ultimately leading to diverse and specific downstream signaling pathways. In the future, advancements in resolving protein structures with greater resolution for the divergent ligand-receptor binding hold the promise to gain deeper insights into the functional antagonism of these peptides, thereby unveiling the mechanisms governing the regulation of pollen tube penetration in the papilla cells of the stigma.
In this study, we have further identified LRX proteins as another group of factors involved in the antagonistic signaling between sRALFs and pRALFs. LRX proteins have been previously implicated in multiple CrRLK1L receptor-mediated processes, including root hair growth47, vacuole expansion45, maintenance of pollen tube integrity30 and immunity.52 Accumulating evidence suggests that LRX proteins play a conserved role in regulating cell wall properties through RALF-binding interactions. For example, RALF4 and RALF19 bind to LRX8 to regulate pollen tube integrity30; RALF22 and RALF23 bind to LRX3 and LRX4 to modulate salt tolerance.50 In our study, LRX3, LRX4, and LRX5 proteins were found to bind to sRALF peptides, establishing the stigmatic reproductive barrier. Previous structural analyses have highlighted the importance of the loop region in RALF4 for LRX binding, with the conserved YY motif (Tyr83 and Tyr84) being critical for strong binding affinity.48 Notably, the loop regions of the four sRALFs, but not those of the pRALFs, exhibit high similarity to RALF4 and contain the conserved YY motif. Although lacking this motif, our pull-down assays revealed that pRALFs could still bind to LRX proteins, albeit potentially with a lower binding affinity and possibly at a different or overlapping position. This interaction might ultimately lead to the destabilization of LRX dimers48 and/or modified LRX function. We anticipate that further investigation into the structures of pRALF-LRX and/or FER/CVY1/ANJ/HERK1-sRALF/pRALF complexes will ultimately provide a mechanistic understanding of how LRX proteins are regulated by different RALF ligands.
As the stigma serves as the primary site for pollen interaction that establishes an interspecific/intergeneric barrier, breaking this stigmatic barrier is a critical step in plant breeding and agriculture to achieve successful interspecific/intergeneric hybridization. In the past, the pollen mentor effect was adopted after its discovery17, yet with limited success. This study has finally unraveled the molecular mechanism underlying a more general interspecific/intergeneric reproductive barrier, shedding light on the pollen mentor effect. The demonstration that treating stigmas with paracrine pRALFs can recapitulate the pollen mentor effect will undoubtedly drive future applications and refinements of this technology. It may also be extended to other plant species, facilitating efficient interspecific/intergeneric cross-breeding. Notably, the finding that pollen tubes from distantly-related species could only partially penetrate stigmas with a defective lock (as observed in fer, cvy1 anj herk1, ralf quad and lrx345 mutants) or following pRALF26 treatment implies that additional components and/or mechanisms could also contribute to the stigmatic reproductive barrier. We have indeed identified more peptides with unknown functions that exhibit high expression in pollen and pollen tubes (Figure S1A). Investigating whether these peptides also contribute to the stigmatic reproductive barrier is a promising avenue for future research. Lastly, it is crucial to note that while uncovering the molecular ‘lock-and-key’ mechanisms at the pollen-stigma interface, we have also discovered a valuable tool for future crop breeding. However, due to the existence of multiple reproductive barriers prior to and after double fertilization, hybrid speciation necessitates overcoming all pre-zygotic and post-zygotic barriers. Our observation that hybrid embryos were obtained only in two out of four intergeneric crosses, with relatively low efficiency, emphasizes the need for additional efforts to remove all barriers, enhancing the generation of new hybrid species, especially beyond the Brassicaceae family. In conclusion, alongsaide unraveling the molecular intricacies of the pollen-stigma interface, we have uncovered a significant and promising tool for future crop breeding, with potential to contribute to global food security.
Limitations of the Study
Our study has unveiled a “lock and key” recognition mechanism on the stigma, involving pollen-derived “key” pRALF ligands and a stigmatic “lock” established through sRALF-FER/ANJ/HERK1 signaling, which includes the participation of LRX cell wall proteins. This mechanism acts as an interspecific/intergeneric reproductive barrier between pollen and the stigma during the penetration of pollen tubes into the stigma in Brassicaceae. However, whether other pollen-stigma recognition mechanisms contribute to the establishment of this interspecific/intergeneric reproductive barrier and the extent to which these identified mechanisms are conserved in plant species from different families, including SI species, remain to be clarified. Our observations indicate that pollen from distantly-related species could only partially penetrate stigmas with a compromised lock or upon treatment with pRALF26, suggesting the presence of additional mechanisms. It is now crucial to unravel the structural basis of the antagonistic interaction between ‘key’ pRALFs and ‘lock’ sRALFs and to identify downstream signaling pathways. These tasks demand substantial research efforts beyong the scope of this study. Furthermore, the precise role of LRX proteins in the ‘lock’ mechanism remains to be fully elucidated. Although investigating modifications of cell wall proteins and their response to membrane signaling poses significant challenges, clarifying these modifications will ultimately provide a comprehensive understanding of the mechanism underlying the opening of the “lock”.
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
• RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Li-Jia Qu (qulj@pku.edu.cn).
Materials Availability
Constructs and reagents in this study will be made available upon request, but a completed Materials Transfer Agreement may be required if there is potential for commercial application.
Data and Code Availability
The raw sequence data generated during this study are available at the Genome Sequence Archive62 in National Genomics Data Center63, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA010659, https://ngdc.cncb.ac.cn/gsa).
• EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Bacterial strains
E. coli DH5α was cultured at 37°C in LB medium for plasmid DNA extraction. E. coli BL21 (DE3) was cultured at 18°C in LB medium for protein expression. A. tumefaciens GV3101 was cultured at 28°C in LB medium for plant transformation.
Plant materials
Arabidopsis thaliana (Arabidopsis, Columbia-0 ecotype) was used as wild-type (WT) plant. Single mutant fer-4, double mutant anj herk1 and triple mutant lrx3/4/5 were described previously. Mutants of pRALFs and sRALFs were obtained by CRISPR/Cas9 technology in this study. Arabidopsis lyrata and Capsella rubella were provided by Ya-Long Guo (Institute of Botany, Chinese Academy of Sciences). Cardamine flexuosa and Rorippa indica plants were collected on the Peking University campus and Erysium cheiranthoides and Descurainia sophia plants were collected in the suburb of Beijing. For expression pattern observation and complementary experiments, transgenic plants were obtained through Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method.64
• METHOD DETAILS
Plant growth conditions
Plants were grown in a growth chamber at 22 ± 2°C with LED lights (GPL production modules DR/W and DR/B/FR, Philips) under long-day conditions (16 hr light/ 8 hr dark).
Phylogenic analysis, sequence alignment and heatmap plotting
Protein sequences of the RALF family and CrRLK1L family of A. thaliana (Table S2) were downloaded from the Arabidopsis Information Resource (http://www.arabidopsis.org/). Phylogenetic analysis was performed by using MEGA X (http://www.megasoftware.net/). Full-length amino acid sequences were used for ClustalW sequence alignment. Based on conserved residues, Neighbor-Joining trees were constructed with 500 replicates of bootstrap. Phylogenetic relationship of Cruciferous species used in Fig. 4 is based on a previous report.53
Gene expression pattern in several designated tissues and the whole small peptide list were obtained from Huang et al..65 Read counts were converted to reads per kilobase per million mapped reads (RPKMs) and log-transformed. We selected the top 30 highly-expressed peptide genes of dry pollen to perform the heatmap. Heatmap exhibits row related values of the genes.
Plasmid construction and plant transformation
To generate GFP reporters, genomic sequences of each RALF gene containing the promoter region and coding sequence was cloned into pDONR221 and ultimately cloned into the vector pK7FWG0, which was developed from pK7FWG266 by using BP and LR reaction following the manufacturer’s protocol (Invitrogen). To generate the construct proRALF:RALF-LAT52:GFP for the complementation experiment (used in Fig. 1D), the same genomic sequence of RALF11 or RALF26 was cloned into the destination vector pB7M34GW66 at a position close to the LAT52:GFP sequence using BP and LR clonases (Invitrogen). The promoter region of each gene is indicated by the primers listed in Table S1. Obtained constructs were transformed into ralf sept plants.
For promoter-GUS reporter assays, promoters of each RALF or receptor gene were cloned into pDONR221, and then pB7GUSWG067 by using BP and subsequent LR reactions. Obtained constructs were transformed into WT plants. A. thaliana plants were transformed through the floral dip method with the Agrobacterium GV3101 strain64.
Receptor reporter lines (pFER:FER-mCit, pANJ:ANJ-mCit, pHERK1:NLS-mCit) used in this study are the same as previously reported31.
Plasmid construction and transgenic screening for CRISPR/Cas9-mediated mutagenesis
All ralf mutants were obtained by using the previously reported egg cell-specific promoter-controlled CRISPR/Cas9 system.68,69 For the ralf1 ralf22 ralf23 ralf33 quadruple mutant, four fragments containing eight sgRNAs were amplified from pCBC-DT1T2 by using the primers RALF1-BsF/F0-s1 and RALF1-BsR/R0-s4, RALF22-BsF/F0-s1 and RALF22-BsR/R0-s4, RALF23-BsF/F0-s1 and RALF23-BsR/R0-s4, RALF33-BsF/F0-s1 and RALF23-BsR/R0-s4, and then cloned into pENTR-MSR68 to produce pENTR-RALF1, pENTR-RALF22, pENTR-RALF23 and pENTR- RALF33, respectively. The two dual-spacers were finally constructed into the vector pENTR-RALF1-22-23-33 by digestion and ligation using Spe I/Hind III and Xba I/Hind III restriction enzymes, respectively. The fragment containing eight sgRNAs was cloned into pHEE401E by digestion and ligation using Spe I/Hind III,69 producing the destination vector pHEE401E-RALF1-22-23-33. This construct was transformed into WT plants by using the Agrobacterium-mediated floral dip method.64 To obtain the ralf10 ralf11 ralf12 ralf13 ralf25 ralf26 ralf30 septuple mutants, sgRNAs targeting RALF25, RALF26 and RALF30 were cloned into PHEE401E as described above. The vector was transformed into WT plants to generate an r25 r26 r30 triple mutant. Then RALF10_sgRNA1_U6_26t-U6_29p-RALF13_sgRNA2 was amplified from pCBC-DT1T2 by using the primers R10–13-BsF/F0 and R10–13-BsR/R0. The dual-spacers were cloned into pHEE401E binary vector69 to produce the plasmid pHEE401E-RALF10–13, which was transformed either into WT or r25 r26 r30 triple mutant, and screened for r10 r11 r12 r13 quadruple mutants and ralf sept mutants.
To obtain cvy1 anj herk1 mutants, the fragment containing two sgRNAs amplified from pCBC-DT1T2 with primers of CVY1-BsF/F0-s3 and CVY1-BsR/R0-s4 was cloned into pHEE401E by Golden-Gate Cloning,69 producing the destination vector pHEE401E-CVY1, which was transformed into the anj herk1 double mutant37 and screened for triple mutants.
To efficiently identify mutated transgenic plants, T0 generation seeds were screened on plates supplemented with 37.5 μg/mL hygromycin. In the T1 generation, plants with effective mutations were identified by direct sequencing of PCR products by using identifier-primers (CVY1-CRI-F/R, RALF1-CRI-F/R, RALF22-CRI-F/R, RALF23-CRI-F/R, RALF33-CRI-F/R, RALF25-CRI-F/R, RALF26-CRI-F/R, RALF30-CRI-F/R and RALF10–13-CRI-F/R). The mutation pattern was reconfirmed in the T2 generation. Cas9-free plants were identified by Hyg-IDF/ Hyg-IDR. Cas9-free seeds were confirmed by growing on antibiotics-resistant plates.
RNA isolation and qRT-PCR
To quantify the expression levels of RALF family genes and LRX family genes, Col-0 flowers were emasculated at stage 12. Stigmatic papilla cells from three emasculated pistils were excised 24 hr later and subjected to RNA extraction with a Dynabeads™ mRNA DIRECTTM Micro Purification Kit (Thermo Scientific). Pollen RNA was extracted by the same method using mature pollen of Col-0. Eluted RNA was reverse-transcribed using Superscript™ II reverse transcriptase Kit (Thermo Scientific). Each cDNA was amplified by KAPA HiFi HotStart ReadyMix (Roche) for 20 cycles and purified by Agencourt AMPure XP beads (Beckman Coulter). Relative abundance of RALF and LRX transcripts, respectively, was detected using qRT-PCR with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) with the primer pairs listed in Table S1. The following program was used: 95°C for 5 min, followed by 40 cycles at 95°C for 10 sec, and 55°C for 30 sec. ACTIN8 (At1g49240) was used as internal control to calculate relative expression levels of each gene. Numbers were presented as means ± SD from three replicates.
Data Processing of RNA-seq Analysis
Data of the pollen and ovule that were collected from a previously published paper.65 For other RNA sequencing data used in this study, mRNA profiles of semi-in vivo pollen tubes (SIV PT), stigma, 12 hr-imbibed seeds, 14 day-old seedlings, and 14 day-old roots and leaves were performed using wild-type Arabidopsis thaliana ecotype Col-0 as material. SIV PTs were collected following semi-in vivo pollen germination.18 Flowers at stage 12 were emasculated and stigmas collected 24 hr after emasculation. Total RNA was isolated from each sample by TRIzol reagent (Invitrogen, San Diego, CA, USA). Libraries for RNA-seq were prepared following Illumina RNA kit standard procedure (Illumina, San Diego, CA, USA) and sequenced using Illumina HiSeq 2000 (Illumina). After the low-quality reads were discarded, 12–36 million paired reads per sample were collected and mapped against the A. thaliana reference genome (TAIR10) using TopHat (v2.0.14). Counting of genes and differential gene analyses were performed using Cufflinks (v2.2.1) as described.70 0.2 for FPKM values was set as a threshold to determine whether genes are expressed.
GUS staining and microscopic observation
Histochemical GUS staining was performed as previously reported.18 Briefly, inflorescences or pistils were incubated in 90% acetone on ice for 30 minutes. Then samples were rinsed by phosphate buffer [50 mM Na2HPO4/NaH2PO4 (pH7.0), 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6] twice for 15 minutes each time, transferred into a staining solution containing 2 mM X-gluc (Sigma), infiltrated under the vacuum for 2 hours and incubated in 37°C. 70% ethanol was used to terminate staining. Samples were rehydrated by 50%, 30%, 10% of ethanol and finally ddH2O. Images were captured using an M205FA microscope (Leica).
For protein expression observation of male expressed RALF11 and RALF26, pollen grains were germinated on pollen germination medium containing 1.4% agar in Petri dishes under 28°C for 6 hours. Fluorescence of pollen grains and pollen tubes was imaged using an A1R confocal laser scanning microscopy (Nikon) with a 40× objective (WI/1.15). GFP was excited at 488 nm with emitted light measured at 500–550 nm.
Confocal microscopy of receptor reporter lines was performed as previously reported.31 Briefly, mCitrine (displayed in magenta) was excited using the 488 nm diode laser and emission was captured from 499–552 nm. mScarlet (displayed in gray) was excited using a 561 nm diode laser and emission was captured from 570–632 nm in a two-track process. Images of the stigma were acquired with a VisiScope Spinning Disc system from Visitron using a 40× objective, and processed using Zeiss ZEN 3.1, ZEN blue 3.4.91 and Fiji (Image J).
Pollination assay, Cryo-Scanning Electron Microscopy and aniline blue staining
For observation and analysis of pollen tube penetration, pollen grains were collected from freshly opened flowers, and 20 or 50 pollen were placed with the help of an eyelash pen to emasculated pistils at stage 13. For inter-specific/inter-generic pollination, emasculated Arabidopsis pistils were detached from the flower at stage 13 and placed on pollen germination medium containing 1.5% agar in Petri dishes. 20 or 50 fresh pollen grains from other species were applied on stigma evenly with the help of an eyelash pen, and pistils were collected at 3 hours after pollination (HAP) (for Arabidopsis lyrata, Capsella rubella, Erysium cheiranthoides and Cardamine flexuosa) and 6 HAP (for Rorippa indica and Descurainia sophia) for observation.
For aniline blue staining, siliques were harvested at corresponding hours after pollination and fixed in acetic acid/ethanol 1:3 for more than two hours in vacuum, then rehydrated through a series of ethanol (70%, 50%, 30%) and ddH2O for 15 min each time. After treatment with 8 M NaOH overnight, samples were rinsed with ddH2O twice and stained with aniline blue solution (0.3% decolorized aniline blue, in 108 mM K3PO4) for more than 2 hours in the dark. Stained samples were observed under a fluorescence microscope (Olympus BX51) equipped with an ultraviolet filter set. Pollen tubes that entered the style were considered to successfully penetrate the stigma. The analysis was repeated for at least three times.
For treatment with pRALF peptides, flowers were emasculated at stage 12. After 24 hours, 1 μL 10 μM pRALF11 and pRALF26 peptide, respectively, was applied to WT stigmas (at stage 13). 10–15 minutes later, when the applied peptide solution was air-dried on the stigma, the treatment was repeated once. Synthetic pollen-expressed RALF4 and RALF7 peptides were used as negative controls. Pollination was performed when the stigma was air-dried again. The same treatment was used for sRALF33 treatment which was applied to emasculated stigmas of WT, ralf quad, fer-4, cvy1 anj herk1 and lrx345 mutants, respectively. For sRALF33 and pRALF26 co-treatment, 1 μL pre-mixed peptides of indicated concentrations was applied twice before pollination.
For Cryo-Scanning Electron Microscopy (Cryo-SEM), a system containing the FEI Helios NanoLab G3 UC scanning electron microscope (ThermoFisher Scientific) and the Quorum PP3010T workstation (Quorum Technologies), which has a cryo preparation chamber connected directly to the microscope, was used. Pollinated pistils were collected at 1 HAP and frozen in subcooled liquid nitrogen (-210°C). Frozen pistils were transferred in vacuum to the cold stage of the chamber, where sublimation (-90°C, 5 min) and sputter coating (10 mA, 60 sec) with platinum were conducted. Finally, pistils were transferred to another cold stage in the scanning electron microscope for image collection. Images of pollinated stigmas were recorded using the electron beam at 5 KV and 0.2 nA with a working distance of 10 mm. Resolution of final data was 3072 × 2048. The analysis was repeated for at least three times.
To obtain hybrid embryos by interspecific pollination, stage-12 A. thaliana flowers were emasculated. Twenty-four hours after emasculation, pollen of Erysium cheiranthoides or Descurainia sophia was applied to mature A. thaliana stigmas. The number of enlarged ovules with developing embryos was counted at 9 and 7 days after pollination (DAP), respectively.
ROS staining and quantification in stigmatic papilla cells
ROS staining of stigmatic papilla cells was performed according to a previously described protocol.16 Arabidopsis flowers were emasculated at stage 12. After 24 hours, pistils were dissected from flowers, incubated in 500 nM H2DCF-DA (2’,7’-dichlorofluorescein diacetate, Invitrogen) for 2 min, and washed three times with ddH2O. Imaging was carried out using an A1R confocal laser scanning microscopy (Nikon) with a 40× objective (WI/1.15). Fluorescence was excited at 488 nm with emitted light measured at 500–550 nm. Imaging was carried out with the same laser power for fluorescence-intensity measurement and quantification. Fluorescence intensities were obtained along the cell edge of papilla cells using ImageJ software. Mean values were calculated from at least five papilla cells in about 8–10 independent pistils. Experiments were repeated at least three times with different batches of plants.
Recombinant protein expression, purification and peptide synthesis
C-terminal His-tagged ectoFER, ectoCVY1, ectoANJ and ectoHERK1 were expressed and purified according to a previously described protocol.31 N-terminal His-tagged LRR domain of LRX4 (LRR4) was expressed in the BL21 E. coli strain and extracted through a similar procedure as described above.31,71 Protein extract was purified using Ni-NTA (Novagen) from the supernatant. Purified proteins were eluted by elution buffer containing 25 mM Tris-HCl pH8.0, 150 mM NaCl, and 250 mM imidazole.
Biotinylated RALF and elf24 peptides used in this study (Table S3) were synthesized by Scilight Biotechnology LLC with a purity higher than 95%. All peptides were diluted in sterile ddH2O.
Pull-down assays between RALF peptides, RLK ectodomains and LRR domain of LRX4
Purified His-tagged RLK ectodomains and LRX4 LRR domain (LRR4), respectively, were mixed with biotinylated RALF peptides in 500 μL pull-down buffer (20 mM Tris-HCl, pH7.5, 500 mM NaCl, 1% IGEPAL) and incubated at 4°C for 0.5 hrs. RLK ectodomain samples (at a final concentration of 50 nM) and RALF peptides (sRALFs at a final concentration of 250 nM, pRALFs at a final concentration of 1 μM) were used in one tube. For pull-down assays of LRR4 with sRALFs, 50 nM His-tagged LRR4 and 250 nM sRALF22 or sRALF33 were mixed in one tube. Then 30 μL Streptavidin Magnetic Beads (New England Biolabs) were added in each sample for 1 h incubation at 4°C and washed with 1.0 mL pull-down buffer for 5–6 times. Bound proteins were eluted with 40 μL SDS loading buffer from beads at 100 °C for 10 min. SDS-PAGE and western blot analysis were performed to exam interactions. Mouse anti-His antibody (TransGen Biotech, 1:5,000 for detection) and goat anti-mouse IgG HRP conjugated secondary antibody (Cwbio, CW0102S, 1:3,000 for detection) were used in western blots. All experiments were repeated for at least three times.
Microscale thermophoresis (MST) assays
MST assays were applied to determine binding affinities. 200 nM purified His-tagged ectoFER, ectoANJ, ectoHERK1 or ectoCVY1 was labeled with 100 nM His-labeling dye solution as described in the kit manual (Monolith™ His-Tag Labeling Kit RED-tris-NTA, MO-L008) for 30 min at room temperature in HEPES buffer (10 mM HEPES, 150 mM NaCl, 0.05% P20). Labeled proteins were mixed with prepared gradient-diluted RALF peptide at a concentration ranging from 0.0062 to 200 μM in HEPES buffer (final concentration of protein samples was 50 nM). Samples were centrifuged at 13,000 rpm and 4°C for 10 min and loaded into capillaries (Monolith™ NT.115 Standard Treated Capillaries, MO-K022). All measurements were performed using a Monolish NT.115 device (NanoTemper Technologies) at medium MST power and 80% LED power. Raw data were analyzed by MO Affinity Analysis software (V2.2.4). All experiments were repeated for at least three times.
Pull-down assays to examine peptide competition
For the interaction competition assay, 50 nM His-tagged ectoFER/ectoCVY1 and LRX4 LRR domain, respectively, were incubated with 200 nM sRALF33 or 200 nM sRALF33 with pRALF26 of indicated concentrations in pull-down buffer (50 mM Tris-HCl, pH7.5, 500 mM NaCl, 0.1% NP-40) at 4°C for 0.5 hrs. Then, 30 μL Streptavidin Magnetic Beads (New England Biolabs) were added to incubate at 4°C for 1 h. All samples were washed 5–6 times with pull-down buffer. After elution, protein samples were subjected to SDS-PAGE and western blot analysis to exam interactions.
• QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments involving measurements/quantifications, imaging and quantifications from images were repeated at least three times with similar results. Data plotting and statistical tests were performed in Excel. In all graphs, asterisks indicate statistical significance (n.s., not significant, *P<0.05; **P<0.01; ***P<0.001) tested by Student’s t test (two groups). In box plots, data are mean values ± SD.
Supplementary Material
(A) Heatmap of the top 30 small peptide genes highly and specifically expressed in pollen and pollen tubes in a descending order.
(B) Phylogenetic tree and related heatmap of all RALF family members in the different tissues indicated. Blue boxes contain RALF genes highly expressed in the male gametophyte (pollen grains and semi in vitro grown pollen tubes, SIV PT).
(C) GUS staining results showing promoter activities of RALF genes indicated. Scale bars, 5 mm.
(D) GUS signals resulting from promoter activities of RALF12, RALF13, RALF25 or RALF30 in mature pollen grains. Scale bars, 50 μm.
(A-G) Mutation forms in different RALF mutants. (A), (C), (E), and (G), sequencing results showing specific mutations identified in r8 r9 r15 triple, r10 r11 r12 r13 quadruple, r25 r26 r30 triple, and ralf sept mutants, respectively. Red boxes indicate insertions and red arrows indicate deletions. (B), (D) and (F), schematics showing CRISPR/Cas9-edited mutations resulting in disrupted RALF peptides in r8 r9 r15 triple mutant, r10 r11 r12 r13 quadruple mutant, and r25 r26 r30 triple mutant, respectively. SP, signal peptides; gray boxes indicate missense sequences due to frameshift mutations.
(H) Aniline blue staining showing the status of WT and r8 r9 r15 pollen tube growth at 3 HAP in a wild-type stigma. Scale bars, 200 μm.
(I) TEM image showing germinated ralf sept pollen tubes that fail to penetrate wild-type papilla cells. Note that due to the loose contact, the contact area was dissected during tissue preparation for TEM. PG, pollen grain; PT, pollen tube; SP, stigma papilla; PTCW, pollen tube cell wall; SPCW, stigma papilla cell wall. Scale bar, 5 μm.
(J) Aniline blue staining of ralf sept pollen tube penetration at 3HAP on stigmas pre-treated with synthetic RALF4 or RALF7 peptides (negative controls). Scale bars, 200 μm.
(A) GUS staining shows promoter activity of CVY1 in stigmas. Scale bar, 0.5 mm.
(B) Aniline blue staining showing WT pollen tube penetration at 3 HAP in WT, cvy1 and the1-4 stigmas, respectively.
(C) Statistical analysis of pollen tube penetration in (A). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test).
(D) Aniline blue staining showing ralf sept mutant pollen-tube penetration at 3 HAP in WT, cvy1 and the1-4 stigmas, respectively.
(E) Statistical analysis of pollen tube penetration in (C). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test). In (B)-(E), all assays were repeated at least three times. Scale bars, 200 μm.
(F) Schematic diagrams describing CRISPR/Cas9-mediated mutations of CVY1 in the anj herk1 mutant background at the nucleotide (top) and protein (bottom) levels, respectively. Early sc, early stop codon; SP, signal peptide; TM, transmembrane domain.
(A) Real-time PCR analysis to investigate transcript levels of pRALFs (RALF11, 13, 25, 26, 30) and sRALFs (RALF1, 22, 23, 33) in the stigma.
(B) Real-time PCR analysis to show transcript levels of above pRALF and sRALF in pollen.
(C) Sequencing results show specific mutations of sRALFs in a ralf quad mutants. Red arrows indicate deletions, the red box indicates insertion and the blue box indicates inversion.
(D) Aniline blue staining showing WT pollen tube penetration at 3 HAP in ralf quad stigmas. Scale bars, 200 μm.
(E) Statistical analysis of pollen tube penetration in stigmas shown in (D). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test).
Assays in (A, B, D) were repeated with three biological replicates.
(A) Aniline blue staining showing pollen tube penetration in WT stigma, WT stigmas pretreated with ROS scavengers (KI and CuCl2) or NADPH oxidase inhibitor DPI at 3 HAP pollinated with WT and ralf sept pollen, respectively. Each assay was repeated at least three times. Scale bars, 200 μm.
(B and C) Statistical analysis of pollen tube penetration in the stigmas shown in (A). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test).
(D) Phylogenetic tree of the RBOH protein family in Arabidopsis (left) and heatmap showing relative expression levels of each RBOH member in designated tissues.
Aniline blue staining showing pollen tubes of E. cheiranthoides, C. flexuosa, R. indica and D. sophia in A. thaliana ralf quad pistils at 48 HAP. Red asterisks indicate enlarged ovules after successful fertilization. White arrows point to pollen tubes that have targeted ovules. Red arrowheads indicate ovules that failed to attract pollen tubes. Yellow arrows indicate ovules with defective pollen tube reception that showed a pollen tube overgrowth phenotype. Scale bars, 200 μm.
Table S1: Primers used in the research, related to STAR Methods
Table S3: Sequences used for protein expression and synthesis, related to STAR Metheods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-His | Transgen Biotech | HT501–01 |
| Goat anti-mouse antibody | CWBIO | CW0102 |
| Bacterial and virus strains | ||
| Escherichia coli (E. coli) DH5α competent cell | Lab stock | N/A |
| E. coli BL21 (DE3) competent cell | Transgen Biotech | CD601–02 |
|
Rhizobium radiobacter (Agrobacterium tumefaciens) GV3101 pMP90 competent cell |
Lab stock | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Biotin-pRALF11/26, Biotin-sRALF22/33, RALF4, RALF7 | Scilight Biotechnology LLC (Beijing, China) | N/A |
| Biotin-elf24 | Scilight Biotechnology LLC (Beijing, China) | N/A |
| pRALF11/26, sRALF33 | Scilight Biotechnology LLC (Beijing, China) | N/A |
| KI | Sangon Biotech | A100512–0050 |
| CuCl2 | Sangon Biotech | A603090–0250 |
| DPI | Meilunbio | MB1817–1 |
| H2DCF-DA | Invitrogen | C6827 |
| FER-6×His | This paper | N/A |
| CVY1–6×His | This paper | N/A |
| ANJ-6×His | This paper | N/A |
| HERK1–6×His | This paper | N/A |
| LRR4–6×His | This paper | N/A |
| Critical commercial assays | ||
| Gateway Cloning Kit | Invitrogen | |
| Deposited data | ||
| N/A | N/A | N/A |
| Experimental models: Cell lines | ||
| N/A | N/A | N/A |
| Experimental models: Organisms/strains | ||
| Arabidopsis: Col-0 | Lab stock | N/A |
| Arabidopsis: r8 r9 r15 | This paper | N/A |
| Arabidopsis: r10 r11 r12 r13 | This paper | N/A |
| Arabidopsis: r25 r26 r30 | This paper | N/A |
| Arabidopsis: ralf sept | This paper | N/A |
| Arabidopsis: r33–3 | Liu et al.16 | N/A |
| Arabidopsis: r22 r33 | This paper | N/A |
| Arabidopsis :r1 r22 r33 | This paper | N/A |
| Arabidopsis: ralf quad | This paper | N/A |
| Arabidopsis: fer-4 | Duan et al.35 | N/A |
| Arabidopsis: anj herk1 | Galindo-Trigo et al.37 | N/A |
| Arabidopsis: cvy1 | Lab stock | N/A |
| Arabidopsis: the1-4 | Lab stock | N/A |
| Arabidopsis: cvy1 anj herk1 | This paper | N/A |
| Arabidopsis: lrx3/4/5 | Zhao et al.50 | N/A |
| Arabidopsis lyrata | From Ya-Long Guo | N/A |
| Capsella rubella | From Ya-Long Guo | N/A |
| Cardamine flexuosa | Lab stock | N/A |
| Rorippa indica | Lab stock | N/A |
| Erysium cheiranthoides | Lab stock | N/A |
| Descurainia sophia | Lab stock | N/A |
| Oligonucleotides | ||
| Primers used in this study are listed in Table S1 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pR11/pR26pro:GUS | This paper | N/A |
| Plasmid: pR11/pR26pro: pR11/pR26-GFP | This paper | N/A |
| Plasmid: pR11/pR26pro: pR11/pR26-LAT52:GFP | This paper | N/A |
| Plasmid: FER/ANJ/HERK1/CVY 1pro:GUS | This paper | N/A |
| Plasmid: pHEE401E-RALF1-22-23-33 | This paper | N/A |
| Plasmid: pHEE401E-RALF10–13 | This paper | N/A |
| Plasmid: pHEE401E-RALF25-26-30 | This paper | N/A |
| Plasmid: pHEE401E-CVY1 | This paper | N/A |
| Plasmid: pET28GW-LRR4 | This paper | N/A |
| Software and algorithms | ||
| ImageJ | N/A | https://imagei.nih.gov/ii/ |
| cellSens | N/A | https://lifescience.evidentscientific.com.cn/en/software/cellsens/ |
| Bioedit | N/A | https://bioedit.software.informer.com/7.2/ |
| R version 4.1.3 | N/A | https://www.r-proiect.org/ |
| MEGA X | N/A | https://megasoftware.net/ |
Highlights.
Stigmatic sRALFs establish an intergeneric barrier ‘lock’ to prevent hybridization
sRALFs act through CrRLK1L receptor complexes and LRXs cell wall proteins
As a ‘key’, compatible pollen-derived pRALFs outcompete sRALFs to unlatch the ‘lock’
Application of synthetic pRALFs breaks the stigmatic barrier, enabling plant outcrosses
ACKNOWLEGMENTS
We thank Chunzhao Zhao from Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences and Jiangkang Zhu from Southern University of Science and Technology for sharing the lrx3 lrx4 lrx5 mutant, Chao Li from Eastern China Normal University for sharing the ralf33-3 mutant, Shangwei Zhong from Peking University for sharing the rbohd rbohf mutant, Yalong Guo from Institute of Botany (CAS) for providing Capsella rubella plants, Bin Xia from Peking University for suggestions on activity of synthetic peptides, Zongcheng Lin from Huazhong Agricultural University for discussion on SI-related questions, Zengxiang Ge, Qingpei Huang and Xin Wang from Peking University for help in CRISPR/Cas9 vector construction, RNA sequencing, and heatmap plotting, respectively. This work was supported by the National Natural Science Foundation of China (Grant No. 31991202 and 31830004 to L.-J.Q., and 32122014 and 32070854 to S.Z.), the Young Elite Scientists Sponsorship Program by China Association of Science & Technology (2019QNRC001 to S.Z.), and the National Institute of Health (R35 GM131827 to J.D.). L.-J.Q. was supported by the New Cornerstone Science Foundation, and T.D. was supported by the German Research Foundation DFG via SFB924.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
A patent application (No. 202311098597.3) for the pRALFs and their potential applications in achieving distant hybridization in plants has been submitted to the State Intellectual Property Office of China.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at:
REFERENCES
- 1.Mayr E (1942). Systematics and the origin of species: from the viewpoint of a zoologist. Nature 151, 347–348. DOI: 10.1038/151347A0 [DOI] [Google Scholar]
- 2.Arnold ML (1997). Natural Hybridization and Evolution. Oxford University Press, New York, USA. DOI: 10.5860/choice.35-0883 [DOI] [Google Scholar]
- 3.Hegarty MJ, and Hiscock SJ (2005). Hybrid speciation in plants: new insights from molecular studies. New Phytol. 165, 411–423. DOI: 10.1111/j.1469-8137.2004.01253.x [DOI] [PubMed] [Google Scholar]
- 4.Dar JA, Beigh ZA, and Wani AA (2017) Polyploidy: Evolution and Crop Improvement. In: Bhat T, and Wani A (eds) Chromosome Structure and Aberrations. Springer, New Delhi. DOI: 10.1007/978-81-322-3673-3_10 [DOI] [Google Scholar]
- 5.Coyne JA, and Orr HA (2004). Speciation. Sinauer, Sunderland, MA, USA. pp 125–178. [Google Scholar]
- 6.Jacobsen MW, Smedegaard L, Sorensen SR, Pujolar JM, Munk P, Jonsson B, Magnussen E, and Hansen MM (2017). Assessing pre- and post-zygotic barriers between North Atlantic eels (Anguilla anguilla and A. rostrata). Heredity 118, 266–275. DOI: 10.1038/hdy.2016.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowry DB, Modliszewski JL, Wright KM, Wu CA, and Willis JH (2008). The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. T. R. Soc. B 363, 3009–3021. DOI: 10.1098/rstb.2008.0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nosil P, Vines TH, and Funk DJ (2005). Perspective: Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59, 705–719. DOI: 10.1554/04-428 [DOI] [PubMed] [Google Scholar]
- 9.Ramsey J, Bradshaw HD, and Schemske DW (2003). Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57, 1520–1534. DOI: 10.1111/j.0014-3820.2003.tb00360.x [DOI] [PubMed] [Google Scholar]
- 10.Rieseberg LH, and Willis JH (2007). Plant speciation. Science 317, 910–914. DOI: 10.1126/science.1137729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedinger PA, Broz AK, Tovar-Mendez A, and McClure B (2017). Pollen-pistil interactions and their role in mate selection. Plant Physiol. 173, 79–90. DOI: 10.1104/pp.16.01286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Yang L, Yang L, Wu X, Cui X, Zhang L, Hui J, Zhao Y, Yang H, Liu S, et al. (2023). Stigma receptors control intraspecies and interspecies barriers in Brassicaceae. Nature 614, 303–308. DOI: 10.1038/s41586-022-05640-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan ZJ, Zhong S and Qu L-J (2023) Insights into pollen–stigma recognition: Self-incompatibility mechanisms serve as interspecies barriers in Brassicaceae? aBiotech. 4, 176–179. DOI: 10.1007/s42994-023-00105-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song ZH, Zhong S, and Qu L-J (2023). FERONIA and reactive oxygen species: regulators in the self-incompatibility response and in interspecific pollination. Mol. Hort 3, 10. DOI: 10.1186/s43897-023-00058-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii S, Tsuchimatsu T, Kimura Y, Ishida S, Tangpranomkorn S, Shimosato-Asano H, Iwano M, Furukawa S, Itoyama W, Wada Y, et al. (2019). A stigmatic gene confers interspecies incompatibility in the Brassicaceae. Nat. Plants 5, 731–741. DOI: 10.1038/s41477-019-0444-6 [DOI] [PubMed] [Google Scholar]
- 16.Liu C, Shen LP, Xiao Y, Vyshedsky D, Peng C, Sun X, Liu ZW, Cheng LJ, Zhang H, Han ZF, et al. (2021). Pollen PCP-B peptides unlock a stigma peptide-receptor kinase gating mechanism for pollination. Science 372, 171–175. DOI: 10.1126/SCIENCE.ABC6107 [DOI] [PubMed] [Google Scholar]
- 17.Knox RB, Gaget M, and Dumas C (1987). Mentor pollen techniques. Int. Rev. Cytol 107, 315–332. DOI: 10.1016/S0074-7696(08)61080-3 [DOI] [Google Scholar]
- 18.Zhong S, Liu ML, Wang ZJ, Huang QP, Hou SY, Xu YC, Ge ZX, Song ZH, Huang JY, Qiu XY, et al. (2019). Cysteine-rich peptides promote interspecific genetic isolation in Arabidopsis. Science 364, eaau9564. DOI: 10.1126/science.aau9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong S, and Qu L-J (2019). Cysteine-rich peptides: signals for pollen tube guidance, species isolation and beyond. Science China Life Sciences 62, 1243–1245. DOI: 10.1007/s11427-019-9819-3 [DOI] [PubMed] [Google Scholar]
- 20.Rejón J, Delalande F, Schaeffer-Reiss C, Alché J, Rodríguez-García M, Van Dorsselaer A, and Castro A (2016). The pollen coat proteome: At the cutting edge of plant reproduction. Proteomes 4, 5. DOI: 10.3390/proteomes4010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu L-J, Li L, Lan Z, and Dresselhaus T (2015). Peptide signalling during the pollen tube journey and double fertilization. J. Exp. Bot 66, 5139–5150. DOI: 10.1093/jxb/erv275 [DOI] [PubMed] [Google Scholar]
- 22.Zhong S, and Qu L-J (2019). Peptide/receptor-like kinase-mediated signaling involved in male–female interactions. Curr. Opin. Plant Biol 51, 7–14. DOI: 10.1016/j.pbi.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 23.Hater F, Nakel T, and Gross-Hardt R (2020). Reproductive Multitasking: The Female Gametophyte. Annual Review of Plant Biology, Vol 71, 2020 71, 517–546. DOI: 10.1146/annurev-arplant-081519-035943 [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Zhao Z, Zheng X, Shan W, and Fan J (2022). How a single receptor-like kinase exerts diverse roles: lessons from FERONIA. Mol. Horticulture 2, 25. 10.1186/s43897-022-00046-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dresselhaus T, and Franklin-Tong N (2013). Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036. DOI: 10.1093/mp/sst061 [DOI] [PubMed] [Google Scholar]
- 26.Schopfer CR, Nasrallah ME, and Nasrallah JB (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. DOI: 10.1126/science.286.5445.1697 [DOI] [PubMed] [Google Scholar]
- 27.Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, and Hinata K (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916. DOI: 10.1038/35002628 [DOI] [PubMed] [Google Scholar]
- 28.Franklin-Tong N (2002). Receptor-ligand interaction demonstrated in Brassica self-incompatibility. Trend. Genet 18, 113–115. DOI: 10.1016/S0168-9525(01)02613-0 [DOI] [PubMed] [Google Scholar]
- 29.Ge ZX, Bergonci T, Zhao YL, Zou YJ, Du S, Liu MC, Luo XJ, Ruan H, Garcia-Valencia LE, Zhong S, et al. (2017). Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1599. DOI: 10.1126/science.aao3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martinez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, et al. (2017). RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358, 1600–1603. DOI: 10.1126/science.aao5467 [DOI] [PubMed] [Google Scholar]
- 31.Zhong S, Li L, Wang ZJ, Ge ZX, Li QY, Bleckmann A, Wang JZ, Song ZH, Shi YH, Liu TX, et al. (2022). RALF peptide signaling controls the polytubey block in Arabidopsis. Science 375, 290–296. DOI: 10.1126/science.abl4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge ZX, Dresselhaus T, and Qu L-J (2019). How CrRLK1L receptor complexes perceive RALF signals. Tred. Plant Sci 24, 978–980. DOI: 10.1016/j.tplants.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 33.Haruta M, Sabat G, Stecker K, Minkoff BB, and Sussman MR (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. DOI: 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, and Zipfel C (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. DOI: 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- 35.Duan QH, Kita D, Li C, Cheung AY, and Wu HM (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 107, 17821–17826. DOI: 10.1073/pnas.1005366107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gachomo EW, Baptiste LJ, Kefela T, Saidel WM, and Kotchoni SO (2014). The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biol. 14. 221. DOI: 10.1186/s12870-014-0221-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galindo-Trigo S, Blanco-Tourinan N, DeFalco TA, Wells ES, Gray JE, Zipfel C, and Smith LM (2020). CrRLK1L receptor-like kinases HERK1 and ANJEA are female determinants of pollen tube reception. EMBO Rep. 21, e48466. DOI: 10.15252/embr.201948466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merz D, Richter J, Gonneau M, Sanchez-Rodriguez C, Eder T, Sormani R, Martin M, Hematy K, Hofte H, and Hauser MT (2017). T-DNA alleles of the receptor kinase THESEUS1 with opposing effects on cell wall integrity signaling. J. Exp. Bot 68, 4583–4593. DOI: 10.1093/jxb/erx263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell L, and Turner SR (2017). A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci 8, 37. DOI: 10.3389/fpls.2017.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abarca A, Franck CM, and Zipfel C (2021). Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 187, 996–1010. DOI: 10.1093/plphys/kiab308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma LY, Zhang H, Sun LR, Jiao YH, Zhang GZ, Miao C, and Hao FS (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot 63, 305–317. DOI: 10.1093/jxb/err280 [DOI] [PubMed] [Google Scholar]
- 42.Torres MA, Dangl JL, and Jones JD (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522. DOI: 10.1073/pnas.012452499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kandasamy MK, Nasrallah JB, and Nasrallah ME (1994). Pollen-pistil interactions and developmental regulation of pollen-tube growth in Arabidopsis. Development 120, 3405–3418. DOI: 10.1242/dev.120.12.3405 [DOI] [Google Scholar]
- 44.Riglet L, Rozier F, Kodera C, Bovio S, Sechet J, Fobis-Loisy I, and Gaude T (2020). KATANIN-dependent mechanical properties of the stigmatic cell wall mediate the pollen tube path in Arabidopsis. eLife 9, e57282. DOI: 10.7554/eLife.57282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dünser K, Gupta S, Herger A, Feraru MI, Ringli C, and Kleine-Vehn J (2019). Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J. 38, e100353. DOI: 10.15252/embj.2018100353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabrice TN, Vogler H, Draeger C, Munglani G, Gupta S, Herger AG, Knox P, Grossniklaus U, and Ringli C (2018). LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol. 176, 1981–1992. DOI: 10.1104/pp.17.01374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herger A, Gupta S, Kadler G, Franck CM, Boisson-Dernier A, and Ringli C (2020). Overlapping functions and protein-protein interactions of LRR-extensins in Arabidopsis. PLOS Genet. 16, e1008847. DOI: 10.1371/journal.pgen.1008847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moussu S, Broyart C, Santos-Fernandez G, Augustin S, Wehrle S, Grossniklaus U, and Santiago J (2020). Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc. Natl. Acad. Sci. USA 117, 7494–7503. DOI: 10.1073/pnas.2000100117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sede AR, Borassi C, Wengier DL, Mecchia MA, Estevez JM, and Muschietti JP (2018). Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett. 592, 233–243. DOI: 10.1002/1873-3468.12947 [DOI] [PubMed] [Google Scholar]
- 50.Zhao CZ, Zayed O, Yu ZP, Jiang W, Zhu PP, Hsu CC, Zhang LR, Tao WA, Lozano-Duran R, and Zhu JK (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 115, 13123–13128. DOI: 10.1073/pnas.1816991115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, and Chai J (2019). Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274. DOI: 10.1038/s41586-019-1409-7 [DOI] [PubMed] [Google Scholar]
- 52.Gronnier J, Frank CM, Stegmann M, DeFalco TA, Abarca A, von Arx M, Dünser K, Lin W, Yang Z, Cleine-Vehn J, et al. (2022). Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. eLife 11, e74162. DOI: 10.7554/eLife.74162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang CH, Sun RR, Hu Y, Zeng LP, Zhang N, Cai LM, Zhang Q, Koch MA, Al-Shehbaz I, Edger PP, et al. (2016). Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol 33, 394–412. DOI: 10.1093/molbev/msv226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, Lu J, Zhang Y, Guo J, Lin W, Norman JMN, Qin Y, Zhu X, and Yang ZB (2021). Membrane receptor-mediated mechano-transduction maintains cell integrity during pollen tube growth within the pistil. Dev. Cell 56, 1030–1042. DOI: 10.1016/j.devcel.2021.02.030 [DOI] [PubMed] [Google Scholar]
- 55.Duan Q, Liu MJ, Kita D, Jordan SS, Yeh FJ, Yvon R, Carpenter H, Federico AN, Garcia-Valencia LE, Eyles SJ, et al. (2020). FERONIA controls pectin- and nitric oxide-mediated male-female interaction. Nature 579, 561–566. 10.1038/s41586-020-2106-2 [DOI] [PubMed] [Google Scholar]
- 56.Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, and Grossniklaus U (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660. DOI: 10.1126/science.1143562 [DOI] [PubMed] [Google Scholar]
- 57.Gao QF, Wang C, Xi YS, Shao QL, Li LG, and Luan S (2022). A receptor-channel trio conducts Ca2+ signalling for pollen tube reception. Nature 607, 534–539. DOI: 10.1038/s41586-022-04923-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang LL, Huang JB, Su SQ, Wei XC, Yang L, Zhao HH, Yu JQ, Wang J, Hui JY, Hao SY, et al. (2021). FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr. Biol 31, 3004–3072. DOI: 10.1016/j.cub.2021.04.060 [DOI] [PubMed] [Google Scholar]
- 59.Lassig R, Gutermuth T, Bey TD, Konrad KR, and Romeis T (2014). Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J. 78, 94–106. DOI: 10.1111/tpj.12452 [DOI] [PubMed] [Google Scholar]
- 60.Ge ZX, Cheung AY, and Qu L-J (2019). Pollen tube integrity regulation in flowering plants: insights from molecular assemblies on the pollen tube surface. New Phytol. 222, 687–693. DOI: 10.1111/nph.15645 [DOI] [PubMed] [Google Scholar]
- 61.Duan QH, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, and Cheung AY (2014). Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun 5, 3129. DOI: 10.1038/ncomms4129 [DOI] [PubMed] [Google Scholar]
- 62.Chen TT, Chen X, Zhang SS, Zhu JW, Tang BX, Wang AK, Dong LL, Zhang ZW, Yu CX, Sun YL, et al. (2021). The genome sequence archive family: Toward explosive data growth and diverse data types. Genom Proteom Bioinf 19, 578–583. DOI: 10.1016/j.gpb.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue YB, Bao YM, Zhang Z, Zhao WM, Xiao JF, He SM, Zhang GQ, Li YX, Zhao GP, Chen RS, et al. (2022). Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucl Acids Res 50, D27–D38. DOI: 10.1093/nar/gkab951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clough SJ, and Bent AF (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. DOI: 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 65.Huang QP, Dresselhaus T, Gu HY, and Qu L-J (2015). Active role of small peptides in Arabidopsis reproduction: Expression evidence. J. Integr. Plant Biol 57, 518–521. DOI: 10.1111/jipb.12356 [DOI] [PubMed] [Google Scholar]
- 66.Karimi M, Inze D, and Depicker A (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trend. Plant Sci 7, 193–195. DOI: 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- 67.Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, and Forner J (2013). GreenGate - A novel, versatile, and efficient cloning system for plant transgenesis. PLOS One 8, e83043. DOI: 10.1371/journal.pone.0083043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engler C, Kandzia R, and Marillonnet S (2008). A one pot, one step, precision cloning method with high throughput capability. PLOS One 3, e3647. DOI: 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, and Chen QJ (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 961–974. DOI: 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. DOI: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ge ZX, Zhao YL, Liu MC, Zhou LZ, Wang LL, Zhong S, Hou SY, Jiang JH, Liu TX, Huang QP, et al. (2019). LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr Biol 29, 3256–3265.e5. DOI: 10.1016/j.cub.2019.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Heatmap of the top 30 small peptide genes highly and specifically expressed in pollen and pollen tubes in a descending order.
(B) Phylogenetic tree and related heatmap of all RALF family members in the different tissues indicated. Blue boxes contain RALF genes highly expressed in the male gametophyte (pollen grains and semi in vitro grown pollen tubes, SIV PT).
(C) GUS staining results showing promoter activities of RALF genes indicated. Scale bars, 5 mm.
(D) GUS signals resulting from promoter activities of RALF12, RALF13, RALF25 or RALF30 in mature pollen grains. Scale bars, 50 μm.
(A-G) Mutation forms in different RALF mutants. (A), (C), (E), and (G), sequencing results showing specific mutations identified in r8 r9 r15 triple, r10 r11 r12 r13 quadruple, r25 r26 r30 triple, and ralf sept mutants, respectively. Red boxes indicate insertions and red arrows indicate deletions. (B), (D) and (F), schematics showing CRISPR/Cas9-edited mutations resulting in disrupted RALF peptides in r8 r9 r15 triple mutant, r10 r11 r12 r13 quadruple mutant, and r25 r26 r30 triple mutant, respectively. SP, signal peptides; gray boxes indicate missense sequences due to frameshift mutations.
(H) Aniline blue staining showing the status of WT and r8 r9 r15 pollen tube growth at 3 HAP in a wild-type stigma. Scale bars, 200 μm.
(I) TEM image showing germinated ralf sept pollen tubes that fail to penetrate wild-type papilla cells. Note that due to the loose contact, the contact area was dissected during tissue preparation for TEM. PG, pollen grain; PT, pollen tube; SP, stigma papilla; PTCW, pollen tube cell wall; SPCW, stigma papilla cell wall. Scale bar, 5 μm.
(J) Aniline blue staining of ralf sept pollen tube penetration at 3HAP on stigmas pre-treated with synthetic RALF4 or RALF7 peptides (negative controls). Scale bars, 200 μm.
(A) GUS staining shows promoter activity of CVY1 in stigmas. Scale bar, 0.5 mm.
(B) Aniline blue staining showing WT pollen tube penetration at 3 HAP in WT, cvy1 and the1-4 stigmas, respectively.
(C) Statistical analysis of pollen tube penetration in (A). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test).
(D) Aniline blue staining showing ralf sept mutant pollen-tube penetration at 3 HAP in WT, cvy1 and the1-4 stigmas, respectively.
(E) Statistical analysis of pollen tube penetration in (C). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test). In (B)-(E), all assays were repeated at least three times. Scale bars, 200 μm.
(F) Schematic diagrams describing CRISPR/Cas9-mediated mutations of CVY1 in the anj herk1 mutant background at the nucleotide (top) and protein (bottom) levels, respectively. Early sc, early stop codon; SP, signal peptide; TM, transmembrane domain.
(A) Real-time PCR analysis to investigate transcript levels of pRALFs (RALF11, 13, 25, 26, 30) and sRALFs (RALF1, 22, 23, 33) in the stigma.
(B) Real-time PCR analysis to show transcript levels of above pRALF and sRALF in pollen.
(C) Sequencing results show specific mutations of sRALFs in a ralf quad mutants. Red arrows indicate deletions, the red box indicates insertion and the blue box indicates inversion.
(D) Aniline blue staining showing WT pollen tube penetration at 3 HAP in ralf quad stigmas. Scale bars, 200 μm.
(E) Statistical analysis of pollen tube penetration in stigmas shown in (D). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test).
Assays in (A, B, D) were repeated with three biological replicates.
(A) Aniline blue staining showing pollen tube penetration in WT stigma, WT stigmas pretreated with ROS scavengers (KI and CuCl2) or NADPH oxidase inhibitor DPI at 3 HAP pollinated with WT and ralf sept pollen, respectively. Each assay was repeated at least three times. Scale bars, 200 μm.
(B and C) Statistical analysis of pollen tube penetration in the stigmas shown in (A). Data are mean values ± SD; n.s. shows P>0.1 (Student’s t test).
(D) Phylogenetic tree of the RBOH protein family in Arabidopsis (left) and heatmap showing relative expression levels of each RBOH member in designated tissues.
Aniline blue staining showing pollen tubes of E. cheiranthoides, C. flexuosa, R. indica and D. sophia in A. thaliana ralf quad pistils at 48 HAP. Red asterisks indicate enlarged ovules after successful fertilization. White arrows point to pollen tubes that have targeted ovules. Red arrowheads indicate ovules that failed to attract pollen tubes. Yellow arrows indicate ovules with defective pollen tube reception that showed a pollen tube overgrowth phenotype. Scale bars, 200 μm.
Table S1: Primers used in the research, related to STAR Methods
Table S3: Sequences used for protein expression and synthesis, related to STAR Metheods
Data Availability Statement
The raw sequence data generated during this study are available at the Genome Sequence Archive62 in National Genomics Data Center63, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA010659, https://ngdc.cncb.ac.cn/gsa).


